Abstract
Background
Most acute respiratory infection (ARI) research focuses on severe disease and overlooks the burden of community-managed illness. For community-based studies, home-based specimen collection by parents could be a resource-saving alternative to collection by healthcare workers (HCWs). In this study, we compared parent and HCW groups for their likelihood to collect specimens and the timeliness and quality of such collection.
Methods
In this unblinded randomized controlled trial, parents from Brisbane, Australia, were taught to identify new ARI episodes in their children aged <2 years. When their child had a new ARI, parents either collected a nasal swab from the child (P group) or contacted an HCW who visited to obtain a nasopharyngeal swab (HCW group). We compared the likelihood and timeliness of specimen collection and respiratory pathogen detection. A nested diagnostic study compared paired specimen collections from children in the HCW group.
Results
Included were 76 incident ARI episodes from 31 children and 102 episodes from 33 children in the P and HCW groups, respectively. The proportions of ARIs for which a specimen was collected were similar (P group, 69.7%; HCW group, 72.5%; P = .77), and pathogens were detected in 93.8% and 77.5% of the specimens, respectively (P = .03). The period between ARI onset and specimen collection was shorter in the P group than in the HCW group (mean difference, 1.9 days [95% confidence interval, 0.7–3.0 days]; P < .001). For the 69 paired specimens, viral loads were lower in the parent-collected swabs (mean cycle threshold difference, 4.5 [95% confidence interval, 3.1–5.9]; P < .001).
Conclusions
Parents and HCWs obtained samples in similar proportions of ARI episodes, but the parents collected the samples fewer days after ARI onset and with a resulting higher likelihood of pathogen identification. This method can be used in population-based epidemiological studies of ARI as a resource-saving alternative.
Trial Registration
ClinicalTrials.gov identifier NCT00966069.
Keywords: acute respiratory infections, children, nasal swabs, parent, specimen collection
Parents and HCWs obtained samples in similar proportions of ARI episodes, but the parents collected the samples fewer days after ARI onset and with a resulting higher likelihood of pathogen identification.
Acute respiratory infections (ARIs), the most frequent illnesses in childhood, caused a loss of >84.9 million disability-adjusted life-years globally in children aged <5 years in 2015 [1]. Viral ARIs in early childhood are also the major cause of acute asthma exacerbations and might play a role in asthma inception in susceptible individuals at high risk for it [2–4]. Most ARI research focuses on severe disease, particularly hospitalizations, and often overlooks the burden of community-managed disease and associated economic costs, which are heavily influenced by work days lost by the parent or caregiver [5, 6]. Community-based research of ARIs in children is needed to capture the full spectrum of ARI severity, to comprehensively assess the cost-effectiveness of preventive and therapeutic options, and to improve our understanding of the developing immune system [7].
Respiratory specimen collection in the home by a household member might facilitate community-based ARI research; yet, research on this topic has been limited. Results of a pilot randomized controlled trial (RCT) in the Netherlands in 2006 suggested that swabs were almost twice as likely to be collected during an ARI by parents than by healthcare workers (HCWs) (43% vs 24%, respectively) [8]. In addition, parent-collected swabs were 1.2 times more likely to test positive for any virus (80% vs 67%, respectively). However, because of the small number of participants, neither of these differences was statistically significant [8]. The quality and acceptability of parent-collected nasal specimens for virus detection in young children were subsequently demonstrated [9–13], and several studies have used parental sampling, although they were without validation [14–16]. Studies with a small number of participants have involved swabs collected simultaneously by a parent and an HCW from the same child [13] or within 24 hours of one another [17].
To prepare for a larger, community-based study [18–22], we sought to compare the likelihood and timeliness of swab obtainment and the quality of specimens collected by parents and HCWs. Our primary hypothesis was that allocation to a parent-collected nasal swab specimen group would increase the proportion of identified ARIs for which a specimen was collected over that of an HCW group collecting nasopharyngeal specimens.
METHODS
Study Design and Subjects
We conducted a parallel-group RCT to compare the relative proportions, timeliness, and quality of parent-collected nasal swabs versus those of HCW-collected nasopharyngeal swabs (NPSs) during an ARI episode. Within the HCW arm of the study, we also conducted a nested diagnostic study to compare the quality and diagnostic performance of the swabs collected by parents and HCWs at the same time from the same child. Children were eligible for the study if they were healthy, living in Brisbane (a subtropical capital city in Australia) without chronic disease, born at ≥36 weeks’ gestation, and aged <2 years between September 1, 2009, and February 26, 2010.
The Queensland Children’s Health Services Ethics Committee approved this study (HREC/09/QRCH/42).
RCT Component
At the enrollment (initial) visit, after we obtained written informed consent from the parent or guardian, each child was randomly assigned (1:1 ratio) into a parent-collected-swab group (P group) or an HCW-collected-swab group (HCW group).
During the initial visit, all parents were taught by a study nurse how to (1) recognize symptoms of an ARI, (2) complete a daily symptom diary, and (3) collect an anterior nasal specimen [15, 16, 18].
Parents were taught to keep a daily symptom diary for the study child and to identify when a new (at least 3 symptom-free days after the previous episode) ARI occurred. An ARI was defined as the presence of at least 1 (fever, wheezing, shortness of breath, pulmonary congestion, moist cough, pneumonia, or ear infection) or 2 (nasal discharge or congestion, sore throat, cough, muscle aches, chills, headache, irritability, decreased activity, or vomiting) specific symptoms [15, 16].
When an ARI occurred, those in the P group were asked to obtain an anterior nasal swab and mail it back to the research laboratory. Those in the HCW group were asked to notify research staff to make an appointment for a home visit by an HCW as soon as possible for collection of an NPS. These NPS specimens were returned by the HCW to the research laboratory immediately after the home visit.
The primary outcome for the RCT was the proportions of identified ARIs for which a specimen was collected in the P and HCW groups. Secondary outcomes were the timeliness and quality of swab obtainment. The timeliness of swab return was measured as the number of days between the onset of an ARI and when the specimen was collected. We measured specimen quality in 2 ways, by comparing the likelihoods of pathogen identification and by comparing the endogenous retrovirus 3 (ERV3, a marker of human DNA) loads, determined by semi-quantitative estimates of viral load determined by real-time polymerase chain reaction (PCR) assay cycle threshold (Ct) values, in these two groups.
Nested Diagnostic Study Component
When a child in the HCW group received a home visit after a reported ARI, the child underwent specimen collection from both nostrils; an NPS specimen was collected from 1 nostril by the HCW (described as part of the RCT study component), and an anterior nasal swab was collected from the other nostril by a parent. The anterior nasal swab was mailed back to the research laboratory.
The primary outcome measure for the nested diagnostic study was the proportion of specific agreement of pathogen detection in paired swabs. The secondary outcome was the agreement in ERV3 loads between parent-collected and HCW-collected swabs [18].
Laboratory Testing
All study swabs received in the laboratory were catalogued and stored at −80°C until they underwent analysis. As described elsewhere [9, 18, 23], stored specimens were thawed and tested by PCR assay for sample quality using ERV3, 17 respiratory viruses, and 3 bacteria. Ct values from positive real-time PCR assays are inversely proportional to the amplified ERV3 nucleic acid in the NPS sample and provide a semi-quantitative estimate of viral load [23].
Sample Size
The sample-size calculation was based on the primary outcome of interest for the RCT component of the study, which was the difference between the proportions of identified ARIs for which a specimen was collected in the 2 groups. To show a difference of 25 percentage points, we estimated that 60 subjects (30 per group) were required (power, 80%; α, .05). This estimate was based on the following assumptions: an average of 4 ARIs per subject over the course of the study, a within-individual intraclass correlation coefficient of 0.15 [16, 24], specimen collection in 50% of the ARI episodes in the HCW-collected group [8], an attrition rate of 25% in each group, and 80% usable symptom-diary data.
Data Analyses
Descriptive analyses of demographic and clinical data are presented (according to group) as frequencies with proportions, means with standard deviation (SD), or medians with interquartile range (IQR) and compared by the χ2 test, t test, or Poisson regression, depending on distribution of the data. From the symptom-diary data we calculated a crude prevalence of symptoms as the number of days with symptoms in relation to the total number of days provided. We determined the number of incident ARIs (ARI events present at the initial visit were excluded), the duration of ARIs (total and average per participant), and ARI rates per child-year. The mean durations of ARI episodes in the P and HCW groups were compared using a generalized linear model with Gaussian family and identity link. Robust variance estimates were calculated with sandwich estimators used to account for repeated episodes within children. Effect estimates are presented as the mean between-group difference with its 95% confidence interval (CI). ARI rates in the P and HCW groups were compared using Poisson regression, and effect estimates are presented as incidence rate ratios with the 95% CI.
For the RCT component of the analysis, we excluded swabs that could not be linked to an ARI episode on the basis of the symptom-diary data (because no data were available or the swab was obtained >7 days after the first day of the ARI episode). We analyzed the first swab if more than 1 swab was obtained during the same ARI episode. The associations between group and the proportion of identified ARIs with a swab collected and between group and the proportion of swabs with a pathogen-, virus-, or bacterium-positive finding were estimated using generalized linear models with binomial family and identity link with robust variance estimates. The associations between group and timeliness and between group and ERV3 Ct values of swabs were estimated using a generalized linear model with Gaussian family and identity link. Effect estimates are presented as absolute between-group mean differences and their 95% CI.
For the diagnostic test component of the analysis, we analyzed swab pairs obtained from the same child during the same visit in the HCW group. We performed descriptive analyses of the detection of no, any, and the same pathogens in swab pairs. The positive agreement was calculated using the formula 2a/(2a + b + c) and the negative agreement as 2d/(2d + b + c), in which a, b, c, and d are the standard cell labels for a 2 × 2 table [25]. Agreement was classified according to the scale suggested by Landis and Koch (1977) for Cohen’s κ value [26]. Asymptotic 95% CIs were calculated on the basis of standard errors that were calculated using the formulae given by Mackinnon [27] (see supplementary material). We further compared the ERV3 Ct values of paired swabs using a generalized linear model with Gaussian family and identity link. In addition, we calculated the mean difference of the ERV3 Ct values between paired swabs and used the limits-of-agreement method for assessing the agreement between them [28]. Analyses were performed by using Stata 12 for Windows (Stata Corp, College Station, Texas) and Excel 2010 for Windows (Microsoft, Redmond, Washington). Fuller descriptions of recruitment, randomization, study procedures, laboratory testing, and data analyses are provided in the supplementary material.
This trial was registered under ClinicalTrials.gov identifier NCT00966069.
RESULTS
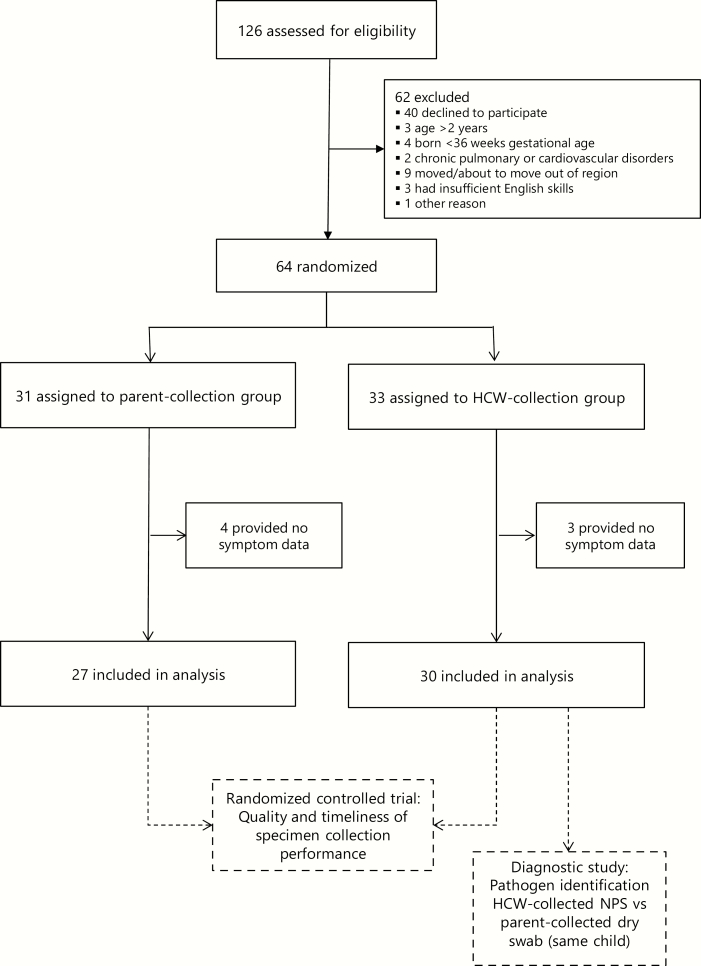
Overall, 126 children were assessed for eligibility, 64 of whom were randomly assigned (P group, 31; HCW group, 33) (Figure 1). Overall, the sociodemographic characteristics in the 2 groups were similar (Table 1).
Figure 1.
Trial profile. Abbreviations: HCW, healthcare worker; NPS, nasopharyngeal swab.
Table 1.
Sociodemographic Characteristics of the Study Subjects and Number and Duration of Incident ARI Episodes According to Group
| Characteristic | P Group (N = 31) | HCW Group (N = 33) | P |
|---|---|---|---|
| Male sex (n [%]) | 15 (48.4) | 18 (54.5) | .62 |
| Age at study entry (first visit) (mean [SD]) (mo) | 15.6 (6.3) | 15.2 (5.0) | .78 |
| Ever breastfed (n [%]) | 27 (87.1) | 31 (93.9) | .26 |
| Child ever received influenza vaccine (P group, n = 30; HCW group, n = 31) (n [%]) | 5 (16.1) | 5 (15.2) | .86 |
| No childcare (n [%])a | 14 (45.2) | 12 (36.4) | .56 |
| Adults (≥16 years old) in the household (median [IQR]) (n) | 2 (2-2) | 2 (2-2) | .57 |
| Other children (aged <16 years) belonging to the household (median [IQR]) (n) | 1 (0–1) | 1 (0–2) | .05 |
| No smoking adults in household (n [%]) | 24 (77.4) | 24 (72.7) | .62 |
| Primary carer employed (n [%]) | 18 (58.1) | 19 (57.6) | .97 |
| Income (n [%])b | .71 | ||
| <26 000 AUD | 4 (12.9) | 2 (6.1) | |
| 26 000 to <52 000 AUD | 7 (22.6) | 6 (18.2) | |
| 52 000 to <94 000 AUD | 10 (32.3) | 11 (33.3) | |
| ≥94 000 AUD | 10 (32.3) | 14 (42.4) | |
| Incident ARI episodes | |||
| Person-days contributed (n) | 4835 | 6109 | .27 |
| Incident ARI episodes (n) | 76 | 102 | |
| Duration of ARI episodes (mean [SD]) (days) | 9.9 (12.6) | 11.1 (17.0) | .61 |
| ARI incidence rate per child-year (95% CI) | 6.9 (5.5–8.7) | 7.4 (6.1–9.0) | .33 |
Abbreviations: ARI, acute respiratory infection; AUD, Australian dollars; CI, confidence interval; HCW group, healthcare worker collection group; IQR, interquartile range; P group, parent collection group; SD, standard deviation.
aNo childcare if neither formal (regulated care outside the child’s home) nor informal (nonregulated care provided by family or friends) care was used.
bIncome categories were based on 2009 Australian Bureau of Statistics income quartiles [38].
The participants provided 10 944 days of daily symptom data (P group, 4835 days; HCW group, 6109 days; average days per child, 156.0 [P group] and 185.1 [HCW group] [P = .27, Student t test]). Seven children provided no data (P group, 4; HCW group, 3), so specimens collected from these children were excluded from further analyses (Figure 1). At least 1 solicited symptom was reported for children on 27% of the total study days (2916 child-days). The most common symptom was nasal discharge (19% of all study days), followed by cough (11%).
Study children experienced 178 incident ARI episodes (P group, 76; HCW group, 102). The incidence and duration of ARIs in both groups were similar (Table 1).
Randomized Controlled Trial
There were 53 (P group) and 74 (HCW group) swabs available during 76 and 102 incident ARI episodes, respectively, resulting in similar proportions of incident ARIs for which a specimen was collected (69.7% vs 72.5%, respectively; mean difference, 2.8% [95% CI, [minus]6.2% to 21.8%]; P = .77) (Table 2). No safety issues in relation to swab collection were reported. We found that the average period between ARI onset and specimen collection was significantly shorter in the P group (mean, 3.0 days; SD, 2.7 days) than in the HCW group (mean, 4.9 days; SD, 2.8 days; mean difference, 1.9 days [95% CI, 0.7–3.0 days]; P < .001). Swabs collected in the P group had higher ERV3 Ct values (ie, lower ERV3 loads) than those collected in the HCW group (Table 2). The proportion of swabs from which any pathogen was detected during incident ARIs and the proportion of swabs from which any bacterium was detected during incident ARIs was higher in the P group than in the HCW group (any pathogen, 93.8% vs 77.5%, and any bacterium, 91.7% vs 73.2%, respectively; Table 2).
Table 2.
Numbers and Proportions of Specimens Returned at Incident ARI Episodes, ERV3 Ct Values, and Pathogen Detection According to Group
| Comparsion between parent and HCW collection groups | P Group (n = 27) |
HCW Groupa (n = 30) |
Mean Difference (95% CI) | P |
|---|---|---|---|---|
| Specimen collected during incident ARI episode | ||||
| Specimens collected/incident ARI episodes (n/N) | 53/76 | 74/102 | ||
| Incident ARIs for which a specimen was collected (%) | 69.7 | 72.5 | 2.8 (−6.2 to 21.8) | .77 |
| Timeliness of specimen collection | ||||
| Time between ARI symptom onset and specimen collection (mean [SD]) (days) | 3.0 (2.7) | 4.9 (2.8) | 1.9 (0.7 to 3.0) | .001 |
| Quality of specimen collection | ||||
| ERV3 Ct value (mean [SD]) | 31.1 (3.6) | 27.1 (3.0) | −4.0 (−5.6 to −2.4) | <.001 |
| Incident ARIs for which a pathogen-positive swab was available (%) | 59.2b | 53.9c | −5.3 (−25.5 to 14.9) | .61 |
| Swabs with any pathogen detected during incident ARIs (%) | 93.8 | 77.5 | −16.3 (−31.1 to −1.5) | .03 |
| Swabs from which any virus was detected during incident ARIs (%) | 41.7 | 29.6 | −12.1 (−26.9 to 2.7) | .11 |
| Swabs from which any bacterium was detected during incident ARIs (%) | 91.7 | 73.2 | −18.4 (−33.1 to −3.8) | <.001 |
Abbreviations: ARI, acute respiratory infection; CI, confidence interval; Ct, cycle threshold; ERV3, endogenous retrovirus 3; HCW group, healthcare worker collection group; P group, parent collection group; SD, standard deviation.
aHCW-collected nasopharyngeal specimens only.
bIncluded were 76 incident ARIs; 53 swabs were collected, 48 swabs were analyzed, and 45 swabs tested positive for a pathogen.
cIncluded were 102 incident ARIs; 74 swabs were collected, 71 swabs were analyzed, and 55 swabs tested positive for a pathogen.
Nested Diagnostic Study
Of the 74 paired swabs obtained in the HCW group, 69 pairs were analyzed for viruses and bacteria. Of these swabs, no pathogen was identified in 4 (6%) pairs and at least 1 pathogen was identified in both swabs in 47 (68%) pairs.
The positive and negative agreements for at least 1 pathogen, at least 1 virus, and at least 1 bacterium detected were 0.84 (95% CI, 0.77–0.91) and 0.31 (95% CI 0.08–0.54), 0.63 (95% CI, 0.46–0.81) and 0.85 (95% CI, 0.77–0.92), and 0.82 (95% CI, 0.74–0.90) and 0.39 (95% CI, 0.17–0.60), respectively. Additional single-pathogen results are shown in Table 3.
Table 3.
Agreement in Pathogen Detection in 69 Paired Nasal Swabs Obtained Within the HCW Groupa
| Parameter | Parent-Collected Swab Positive, HCW-Collected Swab Negative (n) | Parent-Collected Swab Negative, HCW-Collected Swab Positive (n) | Parent- and HCW-Collected Swabs Positive (n) | Parent- and HCW-Collected Swabs Negative (n) | Positive Agreement (95% CI) | Negative Agreement (95% CI) |
|---|---|---|---|---|---|---|
| At least 1 pathogen identified | 12 | 6 | 47 | 4 | 0.84 (0.77–0.91) | 0.31 (0.08–0.54) |
| At least 1 virus identified | 9 | 6 | 13 | 41 | 0.63 (0.46–0.81) | 0.85 (0.77–0.92) |
| At least 1 bacterium identified | 13 | 6 | 44 | 6 | 0.82 (0.74–0.90) | 0.39 (0.17–0.60) |
| Viruses detected | ||||||
| Rhinovirus | 10 | 1 | 1 | 57 | 0.15 (0.00–0.42) | 0.91 (0.86–0.96) |
| Parainfluenza virus III | 1 | 1 | 1 | 66 | 0.5 (0–1.00) | 0.99 (0.96–1.01) |
| Respiratory syncytial virus A | 1 | 0 | 1 | 67 | 0.67 (0.05–1.00) | 0.99 (0.98–1.00) |
| Respiratory syncytial virus B | 0 | 0 | 2 | 67 | 1 (1.00–1.00) | 1 (1.00–1.00) |
| Human coronavirus NL63 | 0 | 0 | 1 | 68 | 1 (1.00–1.00) | 1 (1.00– 1.00) |
| Human coronavirus HKU1 | 0 | 1 | 0 | 68 | 0 (0.00–0.00) | 0.99 (0.98–1.01) |
| Human metapneumovirus | 0 | 1 | 0 | 68 | 0 (0.00– 0.00) | 0.99 (0.98–1.00) |
| Adenoviruses | 1 | 3 | 0 | 65 | 0 (0.00–0.00) | 0.97 (0.94–1.00) |
| Human polyomavirus WUV | 0 | 3 | 1 | 65 | 0.40 (0.00–0.94) | 0.98 (0.95–1.00) |
| Human polyomavirus KIV | 3 | 1 | 2 | 63 | 0.5 (0.08–0.92) | 0.97 (0.94–1.00) |
| Human bocavirus 1 | 0 | 0 | 1 | 68 | 1 (1.00–1.00) | 1 (1.00–1.00) |
| Bacteria | ||||||
| Haemophilus influenzae | 10 | 10 | 18 | 31 | 0.64 (0.50– 0.79) | 0.76 (0.65–0.86) |
| Streptococcus pneumoniae | 13 | 6 | 32 | 18 | 0.77 (0.67–0.87) | 0.65 (0.51–0.80) |
| Moraxella catarrhalis | 16 | 11 | 27 | 15 | 0.67 (0.55–0.79) | 0.53 (0.37–0.68) |
Abbreviations: CI, confidence interval; HCW group, healthcare worker collection group.
aInfluenza virus A, influenza virus B, parainfluenza virus I, parainfluenza virus II, human coronavirus OC43, and human coronavirus 229E were not found in any swab.
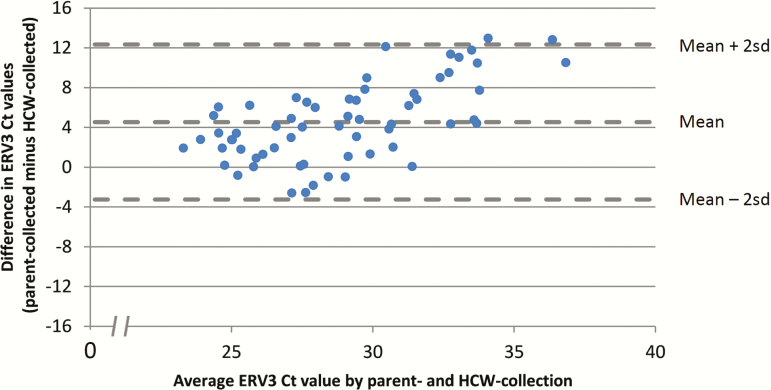
The mean ERV3 Ct value was higher (ie, lower load) in parent-collected nasal swabs (31.2; SD, 4.8) than in the HCW-collected NPS specimens (26.7; SD, 2.6) (mean difference, −4.5 [95% CI, −5.9 to −3.1]; P < .001). Figure 2 is a Bland-Altman plot that shows the difference in viral loads between the paired observations. The 95% limits of agreement were −12.3 and 3.3.
Figure 2.
Bland-Altman plot displaying the difference in ERV3 Ct values between paired nasal swabs (parent-collected nasal swab and HCW-collected nasopharyngeal specimen) obtained within the HCW group (n = 62). Abbreviations: Ct, cycle threshold; ERV3, endogenous retrovirus 3; HCW, healthcare worker; SD, standard deviation.
DISCUSSION
Studying ARIs is important, not only for their direct effect on health and economic burden but also because of their influence on chronic lung disease and the increasing recognition of their role in the ontogeny of the developing human immune system. Respiratory specimen collection in the home conducted by a household member might facilitate studies in this by providing a resource-saving and potentially bias-reducing alternative to collection by HCWs. This alternative offers the possibility of performing richer studies to assess pathogen acquisition at various life stages and the role that viruses and bacteria play in respiratory health.
In our study, sampling by parents led to a decrease in the time between ARI onset and swab collection, which is believed to improve virus detection [11]. Although our overall virus-detection rate was similar to that reported by a study of rhinitis episodes in children who attended childcare [29], it was lower than that reported by other similar studies [11, 12, 24]. The bacterium-detection rate was similar to that reported from other upper-airway studies in young children [30] and higher than that reported for adults [31].
Our results did not confirm our primary hypothesis, which was based on the RCT findings in Dutch infants [8], because specimen collection by the parents did not increase the proportion of swabs obtained compared to that obtained by HCWs. Possible reasons for this result could be cultural and/or methodologic differences: in 2 other Australian studies, the sample collection rates were similarly high [11] or even higher [24]. van der Zalm et al [8] suggested that the lower proportions of swab obtainment in the HCW group might have been caused by parents being too busy to call or simply forgetting to call the study coordinator. However, we did not find these lower proportions in the HCW group in our study. Differences between these studies included the age of the children and the sampling method. Children in our study were slightly older (up to the age of 2 years) than those in the Dutch RCT, in which infants were followed during their first year of life. In addition, the Australian parents were asked to collect an anterior nasal swab, whereas the Dutch parents were expected to obtain a more invasive nasopharyngeal mucus sample, which might be expected to lead to a lower proportion of sampling initially and during subsequent ARIs. Parents in our study were also aware of the research question, which might not have been the case in the Dutch study.
Although self-collection or parent collection of specimens is now common in research, few data about how specimens are returned to the laboratory after collection have been published. Virus- and bacterium-detection rates were demonstrably better in the parent-collection arm of this study, during which specimens were mailed to the laboratory using the standard postal service. In a previous study, we compared paired specimens collected in central Australia at the same time from the same individual that were either frozen immediately for the journey or returned by air and surface mail at an ambient temperature to our laboratory. We found no effect on overall virus detection despite the difference in maximum temperature experienced in transit for each set of swabs (frozen, −5°C; surface mail, 30°C) [32]. However, we did find that bacterial detection seemed to be reduced in the mailed specimens [33], which is at odds with a recent study from Western Australia, which found that exposure to ambient conditions (maximum temperature during the study, 33°C) for up to 14 days and parent collection did not result in reduced bacterial detection in specimens collected from 20 in-hospital very premature infants [34]. Additional work is required to assess the effect of specimen transport from the home to the laboratory on specimen quality and pathogen detection.
Although the parents were trained to identify the start of an ARI episode, it is not clear if they identified every ARI episode correctly. Some episodes included multiple swabs collected even though the parents were asked to collect only 1 swab per episode as early as possible after the start. As a consequence, in cases in which no samples were received for an ARI episode, we were unable to determine whether this result was from missed sampling or from not having identified the start of an ARI episode. However, because it occurred in both groups, it should not have interfered with our overall conclusions.
Our results provide support for the notion that after simple training, parents can collect specimens suitable for pathogen analysis in community-based studies. A key advantage of parental sampling, compared with collection by HCWs or trained research staff, is lower cost from a reduced need for HCW home visits. In addition, compliance should improve, because a parent collecting and mailing swabs is easier and less time-consuming than arranging a timely home visit convenient for both parents and staff. Our findings of substantial positive and almost perfect negative agreement for the detection of any virus between paired swabs obtained from the same child at the same visit by a parent (anterior nasal swab) and an HCW (NPS specimen) were slightly lower for the positive agreement (0.85, calculated from the numbers provided) but better in terms of negative agreement (0.39) than that reported from a Hong Kong study that compared the finding of 5 viruses in nasal swab and nasopharyngeal aspirate specimens [35].
For rhinoviruses, we observed a slight positive and an almost perfect negative agreement between the parent- and HCW-collected swabs, meaning that rhinoviruses were detected more often in parent-collected swabs, which might have been a result of the different sampling sites. However, point inoculation of rhinovirus suspension in adult volunteers led to higher virus-recovery rates for samples from the nasopharynx than in those from the turbinates [36]. The almost perfect positive agreement and fair negative agreement for any bacterium detected are supported by findings from other studies [37].
Interesting to note is that Ct values for ERV3 in the parent-collected swabs were higher (ie, lower load) than those in the HCW-collected swabs. However, the Ct values in the parent-collected specimens remained adequate for pathogen identification [9]. Additional studies of this nature are required to better understand the role of sample quality and collection site in pathogen detection.
In summary, our results did not show sampling by parents to be superior to sampling by HCWs in terms of proportions of incident ARIs for which a specimen was collected. However, we found that parents collected specimens earlier in the course of the ARI episodes and that these samples had higher pathogen yields than those from the HCWs. As a consequence, having parents collect nasal swab specimens from young children is a potential resource-saving strategy to use in population-based studies on viral infections.
Supplementary Material
Notes
Acknowledgment. We thank the nursing, laboratory, and other research staff who assisted with this study.
Financial support. This study was supported by a grant from the Children’s Hospital Foundation Queensland (grant 10290) and a doctoral fellowship of the German Academic Exchange Service (DAAD).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Global health estimates (GHE) 2015: DALYs by age, sex and cause. Available at: http://www.who.int/healthinfo/global_burden_disease/GHE2015_DALY_Global_2000_2015.xls Accessed December 26, 2018.
- 2. Raedler D, Schaub B. Immune mechanisms and development of childhood asthma. Lancet Respir Med 2014; 2:647–56. [DOI] [PubMed] [Google Scholar]
- 3. Rubner FJ, Jackson DJ, Evans MD, et al. Early life rhinovirus wheezing, allergic sensitization, and asthma risk at adolescence. J Allergy Clin Immunol 2017; 139:501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sigurs N, Aljassim F, Kjellman B, et al. Asthma and allergy patterns over 18 years after severe RSV bronchiolitis in the first year of life. Thorax 2010; 65:1045–52. [DOI] [PubMed] [Google Scholar]
- 5. Lambert SB, Allen KM, Carter RC, Nolan TM. The cost of community-managed viral respiratory illnesses in a cohort of healthy preschool-aged children. Respir Res 2008; 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yin JK, Salkeld G, Lambert SB, et al. Estimates and determinants of economic impacts from influenza-like illnesses caused by respiratory viruses in Australian children attending childcare: a cohort study. Influenza Other Respir Viruses 2013; 7:1103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wolsk HM, Følsgaard NV, Birch S, et al. Picornavirus-induced airway mucosa immune profile in asymptomatic neonates. J Infect Dis 2016; 213:1262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van der Zalm MM, Uiterwaal CS, de Jong BM, et al. Viral specimen collection by parents increases response rate in population-based virus studies. J Allergy Clin Immunol 2006; 117:955–6. [DOI] [PubMed] [Google Scholar]
- 9. Alsaleh AN, Whiley DM, Bialasiewicz S, et al. Nasal swab samples and real-time polymerase chain reaction assays in community-based, longitudinal studies of respiratory viruses: the importance of sample integrity and quality control. BMC Infect Dis 2014; 14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van der Gugten AC, van der Zalm MM, Uiterwaal CS, et al. Human rhinovirus and wheezing: short and long-term associations in children. Pediatr Infect Dis J 2013; 32:827–33. [DOI] [PubMed] [Google Scholar]
- 11. Lambert SB, Allen KM, Nolan TM. Parent-collected respiratory specimens—a novel method for respiratory virus and vaccine efficacy research. Vaccine 2008; 26:1826–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zoch B, Karch A, Dreesman J, et al. Feasibility of a birth cohort study dedicated to assessing acute infections using symptom diaries and parental collection of biomaterials. BMC Infect Dis 2015; 15:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Esposito S, Molteni CG, Daleno C, et al. Collection by trained pediatricians or parents of mid-turbinate nasal flocked swabs for the detection of influenza viruses in childhood. Virol J 2010; 7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Byington CL, Ampofo K, Stockmann C, et al. Community surveillance of respiratory viruses among families in the Utah Better Identification of Germs-Longitudinal Viral Epidemiology (BIG-LoVE) study. Clin Infect Dis 2015; 61:1217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lambert SB, Allen KM, Druce JD, et al. Community epidemiology of human metapneumovirus, human coronavirus NL63, and other respiratory viruses in healthy preschool-aged children using parent-collected specimens. Pediatrics 2007; 120:e929–37. [DOI] [PubMed] [Google Scholar]
- 16. Lambert SB, O’Grady KF, Gabriel SH, Nolan TM. Respiratory illness during winter: a cohort study of urban children from temperate Australia. J Paediatr Child Health 2005; 41:125–9. [DOI] [PubMed] [Google Scholar]
- 17. Vargas CY, Wang L, Castellanos de Belliard Y, et al. Pilot study of participant-collected nasal swabs for acute respiratory infections in a low-income, urban population. Clin Epidemiol 2016; 8:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lambert SB, Ware RS, Cook AL, et al. Observational Research in Childhood Infectious Diseases (ORChID): a dynamic birth cohort study. BMJ Open 2012; 2:e002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sarna M, Alsaleh A, Lambert SB, et al. Respiratory viruses in neonates: a prospective, community-based birth cohort study. Pediatr Infect Dis J 2016; 35:1355–7. [DOI] [PubMed] [Google Scholar]
- 20. Sarna M, Ware RS, Sloots TP, et al. The burden of community-managed acute respiratory infections in the first 2-years of life. Pediatr Pulmonol 2016; 51:1336–46. [DOI] [PubMed] [Google Scholar]
- 21. Sarna M, Lambert SB, Sloots TP, et al. Viruses causing lower respiratory symptoms in young children: findings from the ORChID birth cohort. Thorax 2018; 73:969–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sarna M, Ware RS, Lambert SB, et al. Timing of first respiratory virus detections in infants: a community-based birth cohort study. J Infect Dis 2018; 217:418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yuan CC, Miley W, Waters D. A quantification of human cells using an ERV-3 real time PCR assay. J Virol Methods 2001; 91:109–17. [DOI] [PubMed] [Google Scholar]
- 24. Kusel MM, de Klerk NH, Holt PG, et al. Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life: a birth cohort study. Pediatr Infect Dis J 2006; 25:680–6. [DOI] [PubMed] [Google Scholar]
- 25. de Vet HCW, Dikmans RE, Eekhout I. Specific agreement on dichotomous outcomes can be calculated for more than two raters. J Clin Epidemiol 2017; 83:85–9. [DOI] [PubMed] [Google Scholar]
- 26. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977; 33:159–74. [PubMed] [Google Scholar]
- 27. Mackinnon A. A spreadsheet for the calculation of comprehensive statistics for the assessment of diagnostic tests and inter-rater agreement. Comput Biol Med 2000; 30:127–34. [DOI] [PubMed] [Google Scholar]
- 28. Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986; 1:307–10. [PubMed] [Google Scholar]
- 29. Rodrigues F, Foster D, Nicoli E, et al. Relationships between rhinitis symptoms, respiratory viral infections and nasopharyngeal colonization with Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in children attending daycare. Pediatr Infect Dis J 2013; 32:227–32. [DOI] [PubMed] [Google Scholar]
- 30. Hendley JO, Hayden FG, Winther B. Weekly point prevalence of Streptococcus pneumoniae, Hemophilus influenzae and Moraxella catarrhalis in the upper airways of normal young children: effect of respiratory illness and season. APMIS 2005; 113:213–20. [DOI] [PubMed] [Google Scholar]
- 31. Obasi CN, Barrett B, Brown R, et al. Detection of viral and bacterial pathogens in acute respiratory infections. J Infect 2014; 68:125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O’Grady KA, Torzillo PJ, Rockett RJ, et al. Successful application of a simple specimen transport method for the conduct of respiratory virus surveillance in remote Indigenous communities in Australia. Trop Med Int Health 2011; 16:766–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O’Grady KA, Whiley DM, Torzillo PJ, et al. Mailed versus frozen transport of nasal swabs for surveillance of respiratory bacteria in remote Indigenous communities in Australia. BMC Infect Dis 2013; 13:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Townsi N, Simpson SJ, Sikazwe CT, et al. Environmental exposure and parental collection does not affect detection or semi-quantitative load assessment of bacteria in nasal swab specimens from children. Infect Dis (Lond) 2018; 50:468–71. [DOI] [PubMed] [Google Scholar]
- 35. Sung RY, Chan PK, Choi KC, et al. Comparative study of nasopharyngeal aspirate and nasal swab specimens for diagnosis of acute viral respiratory infection. J Clin Microbiol 2008; 46:3073–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winther B, Gwaltney JM Jr, Mygind N, et al. Sites of rhinovirus recovery after point inoculation of the upper airway. JAMA 1986; 256:1763–7. [PubMed] [Google Scholar]
- 37. Luna PN, Hasegawa K, Ajami NJ, et al. The association between anterior nares and nasopharyngeal microbiota in infants hospitalized for bronchiolitis. Microbiome 2018; 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Australian Bureau of Statistics. Household income and income distribution. 6523.0. 2009–10. Available at: http://www.abs.gov.au/ausstats/subscriber.nsf/0/DBE855896D8CA36DCA2578FB0018533C/$File/65230_2009-10.pdf Accessed December 26, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.