Summary
Impairments in the differentiation process can lead to skin diseases that can afflict ∼20% of the population. Thus, it is of utmost importance to understand the factors that promote the differentiation process. Here we identify the transcription factor KLF3 as a regulator of epidermal differentiation. Knockdown of KLF3 results in reduced differentiation gene expression and increased cell cycle gene expression. Over half of KLF3's genomic binding sites occur at active enhancers. KLF3 binds to active enhancers proximal to differentiation genes that are dependent upon KLF3 for expression. KLF3's genomic binding sites also highly overlaps with CBP, a histone acetyltransferase necessary for activating enhancers. Depletion of KLF3 causes reduced CBP localization at enhancers proximal to differentiation gene clusters, which leads to loss of enhancer activation but not priming. Our results suggest that KLF3 is necessary to recruit CBP to activate enhancers and drive epidermal differentiation gene expression.
Subject Areas: Molecular Genetics, Molecular Mechanism of Gene Regulation, Transcriptomics
Graphical Abstract
Highlights
-
•
KLF3 is necessary for epidermal differentiation and barrier formation of the skin
-
•
KLF3 binds to enhancers and is necessary for their activation
-
•
The histone acetyltransferase CBP is necessary for epidermal differentiation
-
•
KLF3 recruits CBP to enhancers to induce differentiation gene expression
Molecular Genetics; Molecular Mechanism of Gene Regulation; Transcriptomics
Introduction
The human epidermis is a stratified epithelial tissue that serves as a protective interface between the body and the surrounding environment (Gonzales and Fuchs, 2017). Not only does it act as a barrier against detrimental factors such as pathogen exposure and physical harm but also allows the body to retain water and prevent dehydration. These protective attributes rely on the proper homeostasis and differentiation of the tissue. The epidermis is maintained by stem and progenitor cells that reside in the basal layer (Ge and Fuchs, 2018). These cells are capable of generating new cells destined for differentiation, while also maintaining the proliferative population. Cells that leave the proliferative state and enter the differentiation program undergo various stages of differentiation as they migrate outward and ultimately form the cornified envelope (stratum corneum). The cornified envelope is composed of enucleated cells containing bundled keratin filaments, crosslinked proteins, and lipids extruded into the extracellular matrix that together create the skin barrier. Defects in differentiation and thus formation of this barrier can lead to a variety of skin diseases such as ichthyosis, atopic dermatitis, and psoriasis, which can impact up to 20% of the population (Lopez-Pajares et al., 2013).
Proper keratinocyte differentiation relies on an effective exit out of the cell cycle and the induction of the differentiation gene expression program. Work from our lab as well as others have shown that differentiation gene expression is reliant on a variety of regulatory factors, including lineage determining transcription factors (LDTFs) such as KLF4, MAF/MAFB, CEBPA/B, ZNF750, and GRHL3 (Boxer et al., 2014; Chen et al., 2014; Hopkin et al., 2012; Lopez et al., 2009; Lopez-Pajares et al., 2015; Mistry et al., 2012; Segre et al., 1999; Sen et al., 2012; Ting et al., 2005). Although many of these important factors have been identified, the search continues for transcription factors that promote differentiation.
Krüppel-like factors are transcription factors characterized by three C2H2 (two cysteine, two histidine) zinc finger domains that facilitate their binding to DNA (Kaczynski et al., 2003). These domains are highly conserved among the various KLF proteins. The KLF family of transcription factors can be distinguished by variations in their N-terminal domains, which allow them to have unique protein-protein interactions and activities.
Krüppel-like factor 3 (KLF3), also known as basic Krüppel-like factor (BKLF), has been implicated in the regulation of differentiation in multiple cellular contexts (Pearson et al., 2011). For example, murine embryonic fibroblasts knocked out for KLF3 show a greater ability to differentiate into adipocytes upon stimulation with adipogenic factors, suggesting that KLF3 inhibits adipogenesis (Sue et al., 2008). In contrast, KLF3 is necessary for B cell development and the induction of skeletal muscle differentiation genes (Himeda et al., 2010; Vu et al., 2011). These findings suggest that KLF3 can regulate various differentiation processes (Pearson et al., 2011). However, whether it promotes or inhibits differentiation may rely on the cellular context, its genomic binding sites, and/or interacting proteins. It is currently unclear what role KLF3 has in the differentiation and homeostasis of human epithelial tissue.
LDTFs can recruit epigenetic writers and readers to remodel chromatin for gene activation or repression. For example, ZNF750 was found to promote differentiation and suppress proliferation gene expression through interactions with epigenetic factors such as CTBP1/2, RCOR1, and KDM1A (Boxer et al., 2014). Similarly, GRHL3 is able to recruit the Trithorax complex (including MLL2, a histone-lysine methyltransferase) through WDR5 to a variety of epidermal differentiation genes and activate their expression through H3K4 methylation (Hopkin et al., 2012). Although these studies have elucidated how some LDTFs recruit epigenetic factors to regulate epidermal differentiation, it is not known whether epigenomic writers of active enhancers have any role in this process.
Enhancers are essential for cell-type specific, developmental, and differentiation gene expression (Smallwood and Ren, 2013). Two types of enhancers have been described, namely primed and active, which can be identified by associated histone modifications. Primed enhancers are marked by mono-methylation of H3K4 (H3K4me1), whereas active enhancers contain both H3K4me1 and acetylated H3K27 (H3K27ac) (Creyghton et al., 2010; Heintzman et al., 2007; Rada-Iglesias et al., 2011). P300 and CBP are highly homologous histone acetyl transferases enriched at active enhancers and promote their activation through acetylation of H3K27 (Visel et al., 2009; Wang et al., 2009). Inhibition of these epigenomic writers lead to inactivation of enhancers and downregulation of target genes. CBP and P300 have been shown to be functionally redundant in many studies; however, there are studies where they have been found to have distinct and opposite roles (Holmqvist and Mannervik, 2013; Ramos et al., 2010; Roth et al., 2001). CBP has been shown in hematopoietic stem cells to promote stem cell self-renewal, whereas P300 is necessary for their differentiation (Rebel et al., 2002). Whether P300 or CBP plays any role in the skin and how they localize to active enhancers is a question of active investigation.
In this study we identify KLF3 as an LDTF that promotes epidermal differentiation. We show that KLF3 is expressed throughout the human epidermis but is upregulated in the differentiated layers. Functionally, KLF3 promotes differentiation gene expression and suppresses cell-cycle associated genes. KLF3 does this independently and is not redundant with closely related transcription factor KLF4. Chromatin immunoprecipitation followed by high throughput sequencing (ChIP-Seq) of KLF3 revealed that KLF3 binding occurred at active enhancers. Based on this enhancer association, we sought to compare the transcriptional programs of KLF3 to P300 and CBP by RNA-Seq. We found that KLF3's transcriptional regulation correlates closely with that of CBP but not P300. ChIP-Seq of CBP-binding sites shows substantial overlap with KLF3 at active enhancers. Loss of KLF3 prevented CBP binding to KLF3-bound active enhancers. Our results demonstrate that KLF3 is necessary for CBP's localization to active enhancers to promote epidermal differentiation.
Results
KLF3 Expression Is Upregulated during Epidermal Differentiation
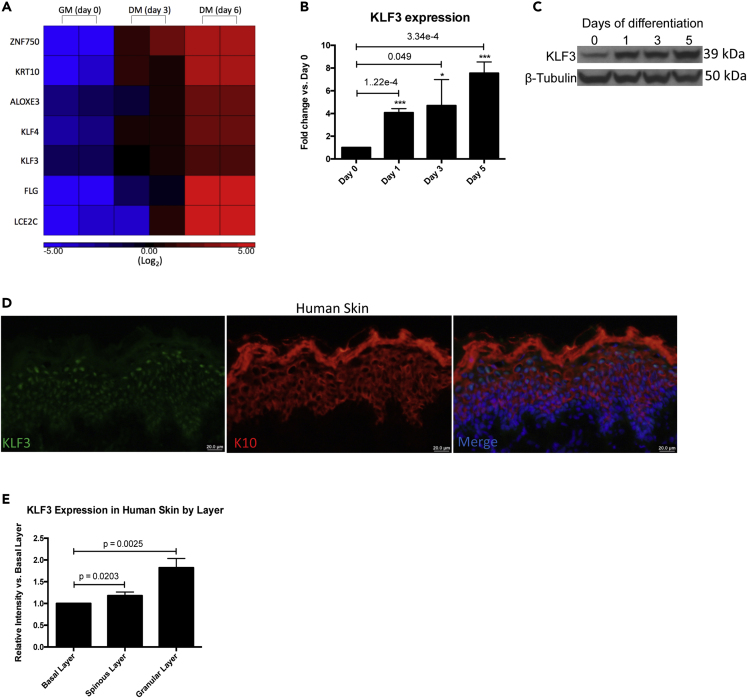
To identify potential regulators of epidermal differentiation, we searched for transcription factors whose mRNA levels were upregulated during this process. Analysis of a previously generated RNA sequencing (RNA-Seq) dataset showed that KLF3 transcripts were upregulated during differentiation (Figure 1A and S1A) (Kretz et al., 2012). This increase in expression is similar to other known transcription factors involved in promoting epidermal differentiation including KLF4 and ZNF750 as well as differentiation structural genes FLG, KRT10, and LCE2C (Figure 1A). To validate KLF3 expression levels, primary human keratinocytes were induced to differentiate by seeding in full confluence and high calcium. Keratinocytes were differentiated for 1, 3, or 5 days, and KLF3 expression levels were analyzed. Compared with the undifferentiated control (Day 0), KLF3 expression was elevated on both the mRNA and protein levels after just one day of differentiation and progressively increased throughout the time course (Figures 1B and 1C). In human skin, KLF3 protein localized to the nucleus and was found throughout the epidermis. However, KLF3 expression was highest in the differentiated layers (such as the granular layer) of the epidermis, which co-expressed the differentiation protein keratin 10 (K10) (Figures 1D and 1E). To confirm the specificity of the KLF3 antibody, primary human keratinocytes were transfected with control siRNA (CTLi) or KLF3 siRNA (KLF3i) and induced to differentiate for 3 days. Efficient KLF3 knockdown and the specificity of the antibody on the protein level was confirmed by western blot (Figure S1B). Control and knockdown cells were also used to regenerate human skin by seeding the cells on devitalized human dermis. This allows the cells to establish cell-cell and cell-basement membrane contact all in three dimensions, which allows the stratification of the tissue and the accurate representation of the gene expression program of human epidermis (Li et al., 2019; Li and Sen, 2015; Noutsou et al., 2017). KLF3 protein expression was increased in the upper, more differentiated layers of the epidermis, which was in contrast to P63 staining, which was primarily found in the basal layer of the epidermis (Figure S1C). KLF3 knockdown samples showed complete loss of KLF3 staining, demonstrating the specificity of the KLF3 antibody (Figure S1C). Together, these data demonstrate that KLF3 is expressed throughout the human epidermis but is elevated in the differentiated layers.
Figure 1.
KLF3 Expression Is Increased during Epidermal Differentiation
(A) Heatmap of replicate RPKM normalized, Log2 transformed RNA-sequencing data showing the relative expression of KLF3 and a panel of differentiation genes in keratinocytes cultured in growth media (GM) versus differentiation media (DM, day 3 and day 6 differentiation). Red color denotes increased expression and blue color denotes decreased expression.
(B) RT-qPCR analysis of KLF3 mRNA levels in a time course of keratinocyte differentiation done in two-dimensions, with day 0 representing undifferentiated keratinocytes and days 1–5 representing progressively differentiated keratinocytes. n = 3. Data are graphed as the mean ± SD. Statistics: t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) Western blot of KLF3 protein levels in a time course of keratinocyte differentiation with B-Tubulin as a loading control. Representative image is shown, n = 3.
(D) Immunofluorescent staining of KLF3 (green) and KRT10 (red, a marker of the differentiated layers) in human skin biopsy. Merged image includes Hoechst staining of nuclei. n = 3. Scale bar: 20 μm.
(E) Quantitation of the signal intensity of KLF3 staining in the basal, spinous, and granular layer from the images acquired in (D). Data are graphed as the mean ± SD. Statistics: t test.
KLF3 Is Necessary for Epidermal Differentiation
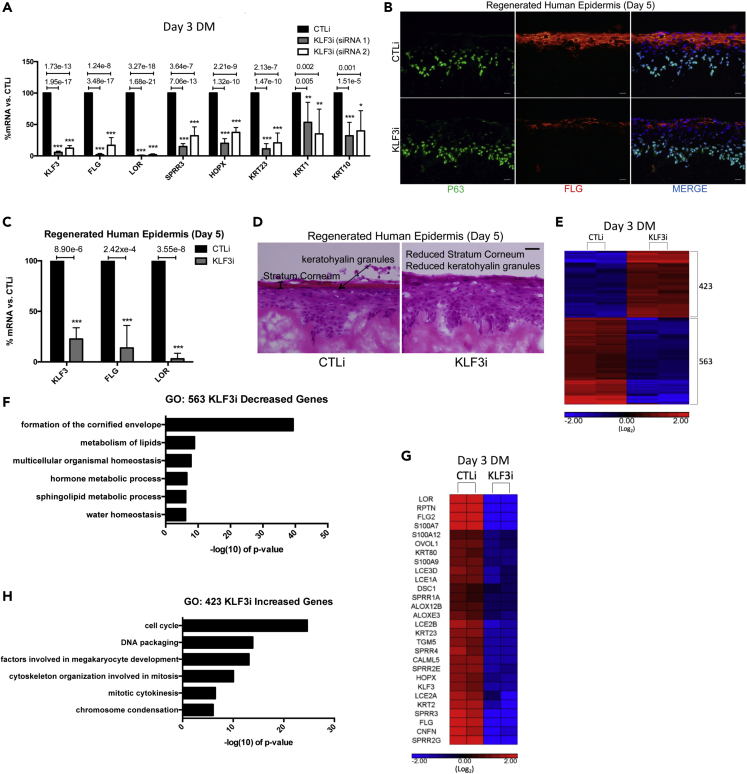
Because KLF3 expression is increased during differentiation, we sought to determine if KLF3 plays a functional role in epidermal differentiation. Knockdown of KLF3 using 2 distinct siRNAs in primary human keratinocytes blocked differentiation gene expression (Figure 2A). These include genes such as filaggrin (FLG), loricrin (LOR), SPRR3, HOPX, KRT1, KRT10, and KRT23, many of which have been implicated in diseases of the skin (Figure 2A) (Koseki et al., 2019; Mariotto et al., 2016). Knockdown of KLF3 also slightly increased apoptosis from ∼2.8% to ∼4.5% (Figure S1D). To determine if KLF3 is necessary for differentiation in a tissue context, control and KLF3-depleted cells were seeded on devitalized human dermis to regenerate skin. Immunofluorescent staining and RT-QPCR from these samples show a dramatic reduction in the protein and mRNA levels for the late differentiation markers FLG and LOR (Figures 2B, 2C, and S1E). Importantly, KLF3 depletion did not impact the expression of the basal layer transcription factor P63 (Figures 2B and S1E). This loss of late differentiation proteins resulted in a diminished cornified envelope and thus likely barrier breach in KLF3 knockdown tissue (Figure 2D).
Figure 2.
KLF3 Is Necessary for Human Epidermal Differentiation
(A) RT-qPCR quantifying the relative mRNA expression levels of a panel of epidermal differentiation genes in CTLi and KLF3i keratinocytes after 3 days of differentiation. Two separate siRNAs (siRNA 1 and siRNA 2) targeting different regions of KLF3 were used (n = 6). Data are graphed as the mean ± SD. Statistics: t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(B) Immunofluorescent staining of late differentiation marker FLG (red) and basal layer marker P63 (green) in day 5 regenerated human epidermis treated with control (CTLi) or KLF3 targeting (KLF3i) siRNA. Merged image includes Hoechst staining of nuclei. n = 3. Scale bar: 20 μm.
(C) RT-qPCR quantifying the relative mRNA expression levels of LOR and FLG in CTLi and KLF3i day 5 regenerated human epidermis. n = 4. Data are graphed as the mean ± SD. Statistics: t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Hematoxylin and eosin staining of CTLi and KLF3i day 5 regenerated human epidermis. n = 3, scale bar: 20 μm.
(E) Heatmap generated for replicate (n = 2) RPKM normalized RNA-Sequencing data from CTLi and KLF3i keratinocytes differentiated for 3 days. The expression of genes significantly (FDR ≤0.05 and fold change ≥2 versus CTLi) increased (red) or decreased (blue) are displayed (log2 scale).
(F) Gene ontology (GO) term enrichment for the 563 genes significantly decreased upon KLF3 knockdown.
(G) Heatmap showing the relative expression levels of a panel of differentiation-associated genes from the CTLi and KLF3i RNA sequencing datasets (log2 scale). n = 2.
(H) Gene ontology term enrichment for the 423 genes significantly increased in expression upon KLF3 knockdown.
To understand the genome wide gene expression changes associated with KLF3 depletion during differentiation, we performed RNA-Seq in replicates for control or KLF3 knockdown cells placed in differentiation conditions for 3 days. Five hundred sixty-three genes were found to be decreased (≥2 fold change and FDR ≤ 0.05, ANOVA) in KLF3-depleted samples (Figures 2E and Table S1). Gene ontology (GO) analysis of these genes shows enrichment for terms related to epidermal differentiation, such as formation of the cornified envelope, metabolism of lipids, and water homeostasis (Figure 2F). This list of genes includes structural differentiation genes such as KRT, LCE, S100, and SPRR family proteins, as well as proteins associated with important differentiation processes such as lipid metabolism (ALOX12B, ALOXE3) and desquamation (KLKs) (Figure 2G). Four hundred twenty-three genes were increased in KLF3 knockdown cells and were enriched for GO terms primarily associated with cell division (Figures 2E and 2H, Table S1). Although there was an increase in cell-cycle associated GO terms, the proliferative capacity of the basal layer did not significantly change upon KLF3 depletion (Figures S1F–S1G). Validation of the RNA-Seq results by RT-qPCR also showed decreased expression of differentiation genes and increased expression of proliferation genes such as CDKL2 and CCNB1 (Figure S2A). To determine which stage of the differentiation process KLF3 was regulating, we compared the KLF3 gene expression signature with a previously published time course of epidermal differentiation, which defined progenitor, early, and late differentiation gene signatures (Lopez-Pajares et al., 2015). Of the 563 genes decreased upon KLF3 knockdown, 23.6% (133/563: p < 2.156 × 10−122) overlapped with the late differentiation gene signature, whereas only 0.53% (3/563) overlapped with the early differentiation signature, which was not significant (Figure S2B). Of the 423 genes increased upon KLF3 knockdown, 17% (72/423: p < 2.699e-40) overlapped with the progenitor gene expression signature, whereas only 1.65% (7/423) and 1.89% (8/423) overlapped with the early and late differentiation signatures, respectively (Figure S2B). These results suggest that KLF3 is necessary to induce late differentiation gene expression while suppressing progenitor genes.
Because KLF4 is critical for epidermal differentiation and related to KLF3, we wanted to determine the relationship between the two transcription factors and determine if they regulate each other's expression levels. Knockdown of KLF4 had no impact on the protein expression of KLF3 in regenerated skin (Figures S1C and S1H). Similarly, knockdown of KLF3 had no impact on KLF4 expression (Figures S1C and S1H). KLF3 also did not co-precipitate with KLF4, suggesting that they do not associate with each other or impact each other's expression (Figure S2C). To determine if these factors regulated a common gene expression program, we used our previously published RNA-Seq data on KLF4 knockdown cells and compared it with the KLF3 gene expression profile (Jones et al., 2020). KLF4 knockdown resulted in 772 downregulated genes and 1,265 upregulated genes (Figure S3A, Table S2). The RNA-Seq results were also validated by RT-qPCR (Figure S3B). Gene ontology for KLF4i-decreased genes showed enrichment for genes related to epidermis development, whereas cell-cycle genes are enriched in the upregulated gene set (Figures S3C and S3D). Greater than half of KLF3 differentially regulated genes were also found in the KLF4 knockdown data with 298 co-downregulated (p < 1.375 × 10−313) and 226 co-upregulated genes (p < 1.243e-180) (Figures S3E and S3F). The KLF3i and KLF4i RNA-Seq datasets are also highly correlated with a Pearson coefficient of ∼0.86 (Figure S3G). Because of their similar gene expression signatures, we wanted to determine if there are redundant genes that they co-regulate. RNA-Seq was performed on KLF3/KLF4 double-knockdown cells with 741 genes downregulated, whereas 958 genes were upregulated with results validated by RT-qPCR (Figures S3H–S3I, Table S3). Interestingly, only 95 downregulated (enriched for drug catabolic process) and 151 upregulated (enriched for interferon signaling) genes were uniquely identified in the double-knockdown samples (Figures S3J–S3M). Notably, there was no enrichment for differentiation or cell-cycle GO terms in those genes (Figure S3L–S3M). Thus, although KLF3 and KLF4 regulate many of the same differentiation and cell cycle genes, they cannot compensate for the loss of one another. Furthermore, KLF3 and KLF4 redundantly regulate a small subset of genes (246 unique genes that are differentially regulated only upon KLF3/KLF4 double-knockdown) together, which are not differentiation or cell-cycle related. Together, these data suggest that KLF3 is essential for promoting epidermal differentiation and is non-redundant with KLF4 in regulating this process.
KLF3 Binds to Active Enhancers to Drive Epidermal Differentiation Gene Expression
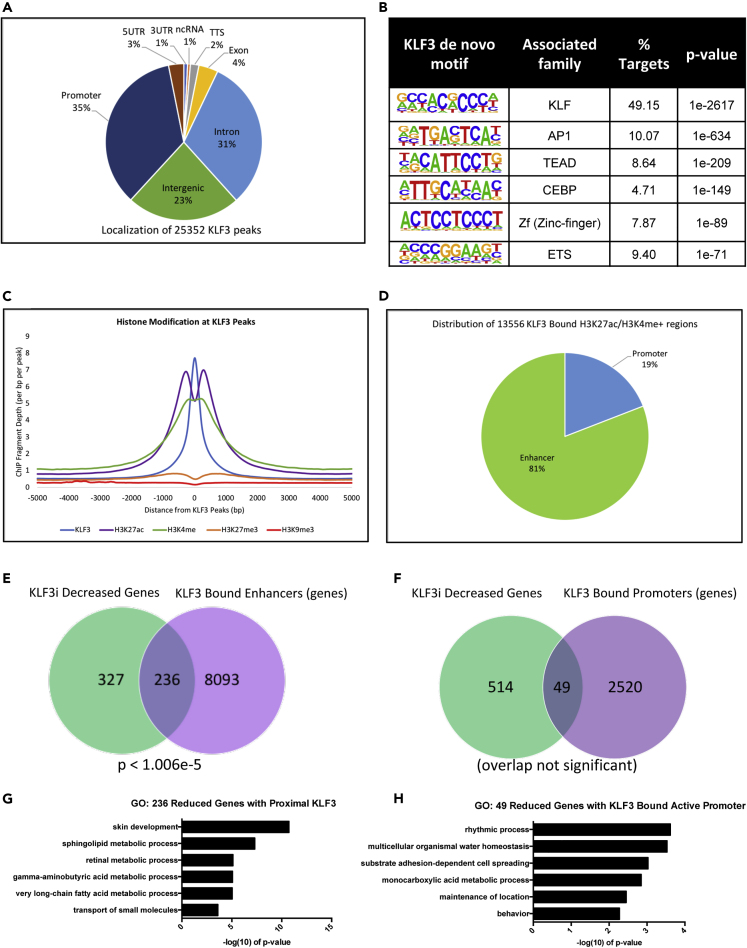
To investigate how KLF3 may be regulating gene expression during differentiation, we determined the genomic binding sites for KLF3 by performing chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq). ChIP-Seq was performed on replicate KLF3 pulldowns from primary human keratinocytes differentiated for 3 days. We identified 25,352 KLF3 peaks found in both replicate datasets (p value < 0.0001, FDR <0.001, 4x enrichment versus input), which were primarily located at promoter (35%), intergenic (23%), and intronic (31%) regions (Figure 3A, Table S4). A search for consensus binding motifs within KLF3 peak locations showed a significant enrichment for KLF family motifs, further confirming the specificity of the pull-down (Figure 3B). Interestingly, KLF3 peak locations were also enriched for AP1, CEBP, and ETS factor family motifs, which have all been implicated in epidermal differentiation (Figure 3B) (Ezhkova et al., 2009; Lopez et al., 2009; Rubin et al., 2017). This suggests that lineage determining transcription factors bind to similar sites in the genome.
Figure 3.
KLF3 Binds to Active Enhancers Proximal to Differentiation Genes
(A) Genomic localization of the 25,352 KLF3 bound peaks identified by HOMER. KLF3 ChIP Seq was performed in replicates (n = 2) from day 3 differentiated keratinocytes.
(B) de novo motif enrichments and their associated family of factors identified within the 25,352 KLF3 peaks.
(C) Mean density profile displaying histone marks (H3K27ac:purple, H3K4me:green, H3K27me3:orange, and H3K9me3:red) centered around KLF3 peaks (blue). The 25,352 KLF3 peaks were used as the reference, and the profiles are displayed +/− 5kb from the KLF3 peak centers.
(D) Percent distribution of the 13,556 KLF3-bound peaks that overlapped with H3K27ac/H3K4me positive regions. These 13,556 regions with KLF3/H3K27ac/H3K4me binding were annotated using HOMER and all regions not mapped to promoters were considered enhancers.
(E) Venn diagram showing the number of genes decreased in KLF3i that have a proximal KLF3-bound H3K27ac/H3K4me positive enhancer(s). Overlap significance in the Venn diagram was calculated using hypergeometric distribution p values.
(F) Venn diagram showing the number of genes decreased in KLF3i that have a KLF3-bound H3K27ac/H3K4me positive promoter.
(G) Gene ontology (GO) term enrichment for the 236 genes decreased in KLF3i that have a KLF3-bound H3K27ac/H3K4me positive, proximal enhancer(s).
(H) Gene ontology term enrichment for the 49 genes decreased in KLF3i that have a KLF3-bound H3K27ac/H3K4me positive promoter.
Based on KLF3's enrichment for motifs related to other LDTFs, we compared the KLF3 ChIP-Seq data with previously published datasets for these factors in differentiation conditions. This included ChIP-Seq data for KLF4, ZNF750, GRHL3, MAF, and MAFB (Boxer et al., 2014; Hopkin et al., 2012; Lopez-Pajares et al., 2015). Pearson correlation coefficients were generated comparing each of these datasets, revealing that the KLF3 sequencing data correlates well with these other transcription factors, with the most significant correlation being with KLF4 (Pearson correlation score of 0.77 and 0.80) (Figure S4A). Based on the positive correlation with these LDTFs, we sought to quantify the shared binding between KLF3 and the various factors. KLF3-bound peaks were overlapped with each individual LDTF-bound peaks (Figure S4B). The highest overlap was with MAFB where 45.4% of its peaks overlapped those of KLF3. It should be noted that there were very few MAFB peaks to begin with (6,489), which could account partially for the high overlap. Thirty-five percent of KLF4 peaks directly overlapped with KLF3 and they co-occupy 15,203 common binding sites. Both factors have similar binding profiles near genes found on the epidermal differentiation complex (EDC), such as the S100 family (Figure S4C). These results suggest that KLF3 binds to shared regions with KLF4 and other LDTFs, further demonstrating the importance of KLF3 to the differentiation process.
To further investigate how KLF3 regulates gene expression, we wanted to characterize the type of regions that KLF3 binds in the genome. Histone marks for open chromatin regions (H3K27ac and H3K4me) and closed chromatin regions (H3K27me3 and H3K9me3) obtained from the ENCODE data for primary human keratinocytes were plotted around KLF3 peak regions, which revealed an association of KLF3 with open but not closed chromatin (Figure 3C) (Gerstein et al., 2012). Because H3K27ac and H3K4me co-localization identifies active enhancers or promoters, we sought to investigate how many of these regions KLF3 binds (Calo and Wysocka, 2013). We compared H3K4me data (representing primed regions) and H3K27ac data (representing active regions) from differentiated keratinocytes (Klein et al., 2017; Rubin et al., 2017). Overlap of these datasets with KLF3 showed that over 53% (13,556/25,352) of KLF3-bound regions contained both H3K4me and H3K27ac peaks (Figure 3D). To determine the distribution of KLF3 at active enhancer versus promoter regions, KLF3-bound H3K4me/H3K27ac positive regions were mapped back to annotated promoters (HG19:REFSEQ transcription start sites). Promoter mapped regions were separated from the 13,556 active regions, with the remaining regions being KLF3-bound active enhancers. KLF3-bound H3K4me/H3K27ac regions were found to be highly enriched for enhancers at 81%, whereas only 19% of these active regions mapped to promoters (Figure 3D). These KLF3-bound active enhancer and promoter regions were then mapped back to their nearest genes, which identified 2,569 genes with KLF3-bound promoters and 8,329 genes with proximal KLF3-bound enhancers (with some genes having both). These gene lists were overlapped with the 563 genes significantly decreased in expression by KLF3 knockdown (Figures 3E and 3F). Forty-two percent (236/563, p < 1.006 × 10−5) of the genes decreased upon KLF3 knockdown had proximal KLF3 bound enhancers. Importantly, the most significant GO term for the 236 genes decreased by KLF3 knockdown that have a proximal KLF3 enhancer binding is skin development (Figure 3G). In contrast only 8.7% (49/563, not significant) of the genes decreased upon KLF3 depletion had KLF3 bound at its promoter (Figure 3H). Those 49 genes do not have enrichment for epidermal-differentiation-associated GO terms but instead were enriched for rhythmic process (Figure 3H). This suggests that KLF3 regulation of differentiation genes is primarily enhancer dependent. To validate the binding of KLF3 to enhancer regions, KLF3 ChIP-qPCR was performed on control and KLF3 knockdown cells (Figure S4D). Control samples show KLF3 enrichment at differentiation relevant sites, such as enhancers proximal to IVL, HOPX, ALOX12B, CALML5, LOR, KRT78, OVOL1, ELF3, S100A7, KLK9, and KRT80 (Figure S4D). KLF3 knockdown ablated KLF3 binding to those genomic regions, which demonstrates the robustness and specificity of the KLF3 bound regions (Figure S4D).
CBP and KLF3 Control a Gene Expression Program that Is Necessary for Epidermal Differentiation
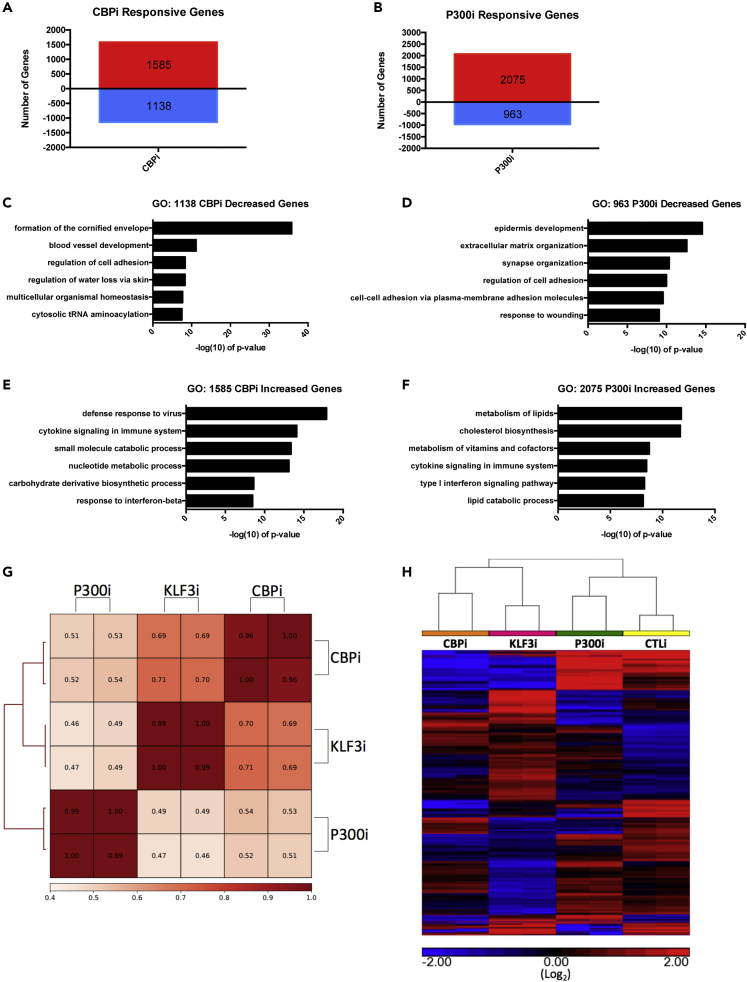
Because a majority of KLF3-bound H3K27ac/H3K4me regions are active enhancers, we wanted to determine if epigenetic factors associated with enhancers are critical for epidermal differentiation. Enhancer regions are often bound by CBP/P300, which promotes the acetylation of H3K27, which activates enhancers primed with H3K4me (Visel et al., 2009; Wang et al., 2009). To determine whether these epigenetic factors have a role in epidermal differentiation and identify possible functional correlations with KLF3, RNA-Seq was performed on CBP and P300 knockdown cells.
CBP and P300 each regulated several thousand genes. One thousand five hundred eighty-five genes were upregulated and 1,138 genes were diminished in expression upon CBP knockdown (Figure 4A, Table S5). Surprisingly, P300 knockdown resulted in twice as many upregulated (2,075) genes as downregulated (963) ones, suggesting it may also function as a repressor in keratinocytes (Figure 4B, Table S6). Both the CBP and P300 knockdown RNA-Seq datasets were validated by RT-qPCR (Figures S5A and S5B). GO term enrichments generated for downregulated genes revealed both factors regulate the epidermal differentiation gene expression program, with significant terms including formation of the cornified envelope and epidermis development for CBP and P300, respectively (Figures 4C and 4D). However, the significance of the CBP-related term is much higher than that of P300 (Figures 4C and 4D). Although genes upregulated by CBP knockdown did not enrich for terms related to differentiation (Figure 4E), genes increased by P300 knockdown enriched for the terms metabolism of lipids and cholesterol biosynthesis, which are both involved in the formation of the cornified envelope and critical for the differentiation process (Figures 4E and 4F). This suggests that P300 is involved in both the suppression and promotion of different parts of the differentiation program. This also suggests that P300 and CBP control non-redundant transcriptional programs in the skin, and CBP is more essential for promoting the differentiation program.
Figure 4.
CBP and KLF3 Regulate a Similar Gene Expression Program
(A) Histogram summarizing the gene expression changes in CBP knockdown (CBPi) keratinocytes when compared with controls (CTLi) after three days of differentiation. RNA Seq was performed in replicates. Number of increased genes is displayed in red, whereas decreased genes are shown in blue. Differentially expressed genes were selected with FDR ≤0.05 and fold change ≥2 versus CTLi.
(B) Histogram summarizing the gene expression changes identified by RNA sequencing in P300 knockdown (P300i) keratinocytes when compared with controls (CTLi) after 3 days of differentiation. RNA Seq was performed in replicates.
(C) Gene ontology (GO) term enrichment for the 1,138 genes significantly decreased in expression upon CBP knockdown.
(D) Gene ontology term enrichment for the 963 genes significantly decreased in expression upon P300 knockdown.
(E) Gene ontology term enrichment for the 1,585 genes significantly increased in expression upon CBP depletion.
(F) Gene ontology term enrichment for the 2,075 genes significantly increased in expression upon P300 depletion.
(G) Heatmap plot of Pearson correlation coefficients comparing replicate KLF3i, CBPi, and P300i RNA sequencing datasets (RPKM normalized).
(H) Unsupervised hierarchical clustering was performed on the RPKM normalized RNA-Sequencing datasets for CTLi, KLF3i, CBPi, and P300i keratinocytes differentiated for 3 days (log2 scale). Heatmap is focused on the KLF3-regulated genes. Increased genes are displayed in red, whereas decreased genes are shown in blue.
To determine whether KLF3 and the epigenetic factors regulated differentiation through the same genes, the downregulated genes were overlapped with each other. Over 50% (282/563) of the KLF3-knockdown-reduced genes were also downregulated upon CBP knockdown (p < 2.722 × 10−231), whereas only 133 were downregulated in the P300 knockdown (p < 5.875 × 10−67), showing a more significant transcriptional overlap with CBP (Figures S5C and S5D). Pearson correlations of the RPKM-normalized RNA-Seq datasets also demonstrated that CBP (avg. = 0.70) correlated more with KLF3 than P300 (avg. = 0.48) (Figure 4G). Interestingly, CBP and P300 correlated with each other (avg. = 0.52) less significantly than KLF3 with CBP (avg. = 0.70), suggesting that CBP and P300 play non-redundant roles in the skin (Figure 4G). As an orthogonal confirmation of these relationships, a gene expression heatmap based on unsupervised hierarchical clustering was generated using KLF3 knockdown responsive genes as the reference (Figure 4H). KLF3 clustered with CBP, whereas P300 clustered with the control samples. RT-qPCR also demonstrated that CBP knockdown using two unique siRNAs blocked the expression of the same critical differentiation genes as KLF3, whereas P300 depletion had much less impact (Figures S5E, S5F, and 2A). In regenerated human skin, CBP knockdown phenocopied KLF3i with loss of expression of FLG and LOR on both the protein and mRNA levels (Figures S5G and S5H). Together, these data show that KLF3 shares a similar knockdown phenotype and transcriptional network with CBP.
CBP Genome Occupancy Correlates with KLF3
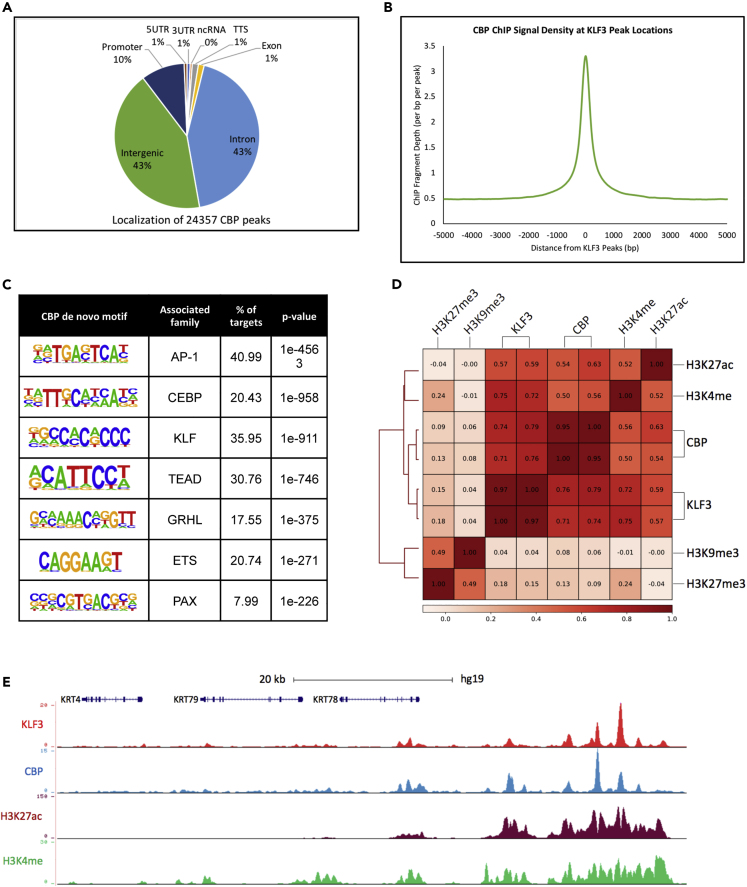
Because of the similarities between KLF3 and CBP transcriptional regulation during differentiation, we wanted to investigate how extensively these factors share genomic binding sites. ChIP-Seq was performed on replicate CBP pull-downs from day 3 differentiated keratinocytes. Twenty-three thousand three hundred fifty-seven replicate CBP peaks were identified, which primarily localized to intergenic (43%) and intronic (43%) genomic sites (Figure 5A, Table S7). This is consistent with reports that CBP is primarily localized to enhancers (Wang et al., 2009). To determine if CBP and KLF3 peaks can co-localize, signal from the CBP ChIP Seq dataset was plotted using KLF3 peak locations as a reference (Figure 5B). The summit of the CBP signal is found at the peak of KLF3 bound regions, suggesting that they bind to the same locations in the genome. To confirm this co-localization, CBP- and KLF3-bound peak regions were overlapped with each other (Figure S6A). This revealed 8,461 shared peak locations between KLF3 and CBP. Thus ∼35% (8,461/24,357) of the CBP bound peaks and ∼33% (8,461/25,352) of the KLF3 bound peaks are shared with each other (Figure S6A). de novo motif analysis of CBP-bound regions also showed significant enrichment for KLF family binding motifs (Figure 5C). Motifs for AP1, CEBP, and ETS factors were also enriched in these datasets, as they were for KLF3 binding, suggesting that these important factors may co-localize and regulate similar regions (Figures 5C and 3B). Pearson correlations comparing these ChIP-Seq datasets also demonstrated strong positive correlation between KLF3 and CBP (avg. = 0.75) (Figure 5D). Importantly, there was also strong positive correlation between KLF3 and CBP with the chromatin marks of active enhancers, H3K4me and H3K27ac (Figure 5D). In contrast, there was no correlation with the marks H3K27me3 or H3K9me3, which represent inactive regions (Figure 5D). Visualization of the ChIP-seq profiles for KLF3 and CBP show co-localization at active enhancer regions (marked by H3K27ac and H3K4me) proximal to critical epidermal differentiation genes such as the keratin (KRT) family, SPRR4, and IVL (Figures 5E and S6B). This suggests that KLF3 and CBP co-localize at genomic regions that may regulate epidermal differentiation.
Figure 5.
CBP and KLF3 Binds to Similar Sites in the Genome
(A) Genomic localization of the 24,357 CBP-bound peaks identified by HOMER. CBP ChIP Seq was performed in replicates from day 3 differentiated keratinocytes.
(B) Mean density profile displaying CBP genomic binding centered around KFL3 peaks. The 25,352 KLF3 peaks were used as the reference, and the profile of CBP binding is displayed +/− 5kb from the KLF3 peak centers.
(C) de novo motif enrichments and their associated family of transcription factors identified within the 24,357 CBP peaks.
(D) Heatmap plot of Pearson correlation coefficients between replicate KLF3 and CBP ChIP sequencing datasets and histone marks data (H3K27ac, H3K4me, H3K27me3, and H3K9me3) (RPKM normalized).
(E) UCSC genome browser tracks displaying KLF3 (red) and CBP (blue) ChIP sequencing profiles near a cluster of keratin genes. H3K27ac (maroon) and H3K4me (green) are included to represent open and active chromatin.
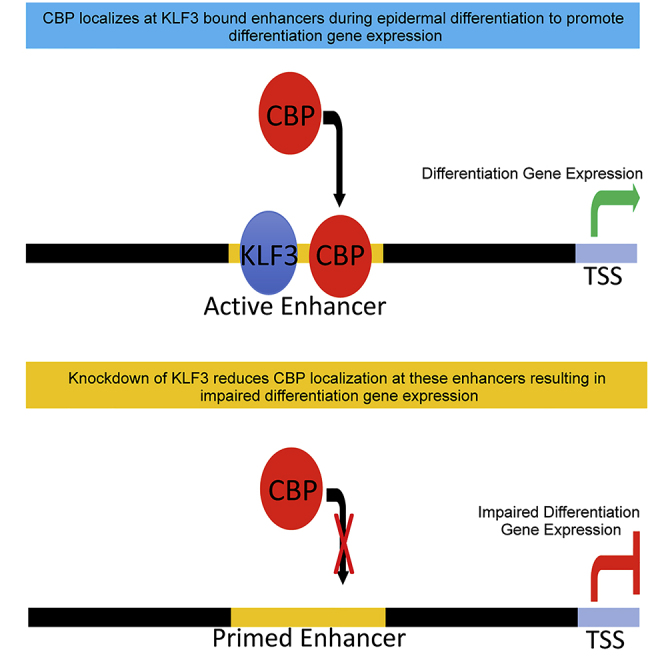
KLF3 Promotes CBP Localization at Active Enhancers Proximal to Differentiation Genes
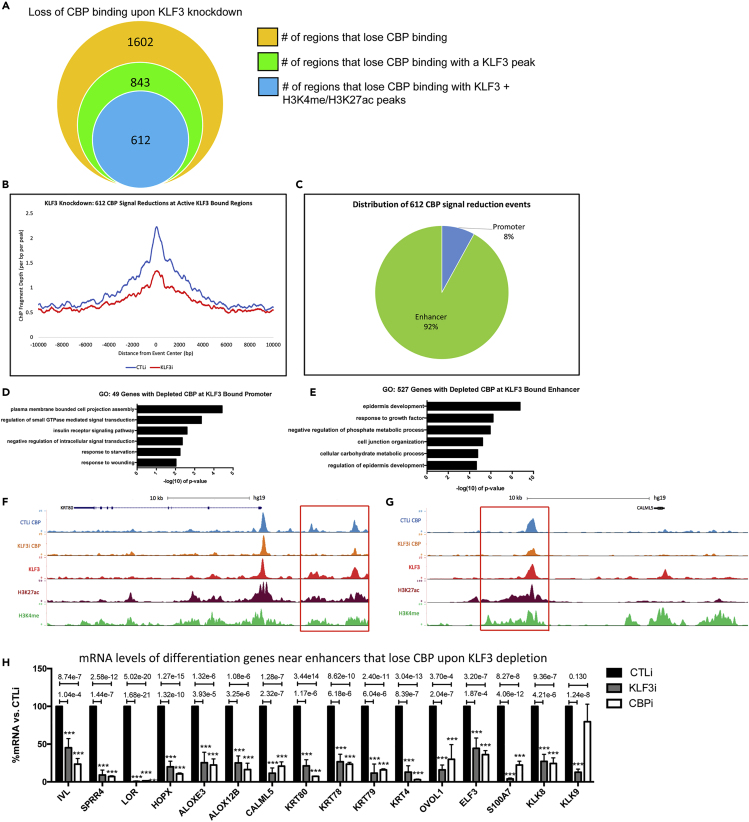
Because LDTFs can recruit epigenetic factors to promote gene expression changes, we sought to investigate whether KLF3 can promote the localization of CBP at active enhancer regions in differentiated keratinocytes. To do this, we knocked down KLF3 in differentiated keratinocytes and performed ChIP-Seq on CBP to determine if its localization to KLF3-bound active enhancers is affected. DiffReps was used to detect the loss or gain of CBP binding upon KLF3 depletion (Shen et al., 2013). KLF3 knockdown resulted in 1,602 regions of significantly reduced CBP binding, whereas 2,186 regions gained CBP binding (Figure 6A and S6C, Table S8). Next, we compared whether the losses in CBP binding occurred within KLF3-bound regions. About 52.6% (843/1,602) of the CBP-reduced binding occurred within KLF3 peaks regions (Figure 6A). In addition, the vast majority of the reductions in CBP (72.6%, 612/843) signal at KLF3 peaks occur in active regions of chromatin (H3K4me/H3K27ac positive, promoters included) (Figure 6A). Plotting of the CBP signal density at the 612 sites of KLF3-bound active regions (H3K4me/H3K27ac positive) showed a dramatic reduction in CBP binding upon KLF3 knockdown (Figure 6B). The loss of CBP binding was also not due to global loss of CBP protein levels, as KFL3 depletion did not significantly impact its expression (Figure S6D). We also tested whether KLF3 knockdown led to gains in CBP binding at KLF3-bound regions. Only 15% (328/2,186) of the KLF3-bound regions gained CBP binding upon KLF3 knockdown (Figure S6C). These results suggest that KLF3 is necessary for the localization of CBP to its binding sites.
Figure 6.
KLF3 Is Necessary for CBP Localization at Enhancers Proximal to Differentiation Genes
(A) Summary of significant CBP ChIP signal reduction events in day 3 differentiated keratinocytes knocked down for KLF3. The orange circle represents the total number of regions in the genome that lose CBP binding upon KLF3 knockdown. The green circle represents the number of regions that lose CBP binding that occur at a KLF3 peak upon KLF3 depletion. The blue circle represents the regions that lose CBP binding, which also contain KLF3, H3K4me, and H3K27ac binding upon KLF3 knockdown. CBP ChIP Seq was performed in replicates in CTLi and KLF3i cells. Significant signal changes were identified by Diffreps.
(B) Mean density profile displaying CBP binding centered around KLF3/H3K4me/H3K27ac peaks. The CBP mean density is plotted around the 612 regions identified in (6A:blue circle). The profile of CBP binding is shown +/− 10kb from the centers of the 612 regions in CTLi (blue) and KLF3i (red) cells.
(C) Percent distribution of the 612 H3K27ac/H3K4me positive regions containing KLF3 peaks with significant CBP depletion upon KLF3 knockdown. These 612 regions were annotated using HOMER and all regions not mapped to promoters were considered enhancers.
(D) Gene ontology (GO) term enrichment for the 49 genes that lose CBP binding at their active promoter (H3K4me/H3K27ac positive) containing a KLF3 peak.
(E) Gene ontology term enrichment for the 527 genes that lose CBP binding at a proximal enhancer (H3K27ac/H3K4me positive) containing a KLF3 peak. (F) UCSC genome browser tracks displaying CBP binding (CBP ChIP Seq profiles) from CTLi (blue) and KLF3i (orange) cells, near differentiation gene KRT80. KLF3 (red), H3K27ac (maroon), and H3K4me (green) binding are also shown.
(G) UCSC genome browser tracks displaying CBP binding (CBP ChIP Seq profiles) from CTLi (blue) and KLF3i (orange) cells, near differentiation gene CALML5. KLF3 (red), H3K27ac (maroon), and H3K4me (green) binding are also shown.
(H) RT-qPCR quantifying the relative mRNA expression levels of epidermal differentiation genes near enhancers that lose CBP genomic binding upon KLF3 knockdown in CTLi, KLF3i, and CBPi keratinocytes after 3 days of differentiation. N = 4 except for LOR and HOPX in KLF3i samples where N = 6. Statistics: t test, ∗∗∗p < 0.001. Mean values are shown with error bars = SD.
Based on this loss of CBP at KLF3-bound active chromatin regions, we wanted to explore whether these reductions are primarily occurring at enhancer or promoter regions. The 612 KLF3/H3K27ac/H3K4me-bound regions that lost CBP binding upon KLF3 knockdown was mapped back to annotated promoters. This revealed that a small portion (8%) of these events occurred at promoters, whereas 92% occurred at enhancers (Figure 6C). This suggests that the KLF3-dependent localization of CBP occurs primarily at active enhancer regions. Mapping these regions to proximal genes revealed only 49 genes associated with promoter reductions, whereas 527 genes have proximal enhancer reductions. Gene ontology analysis was performed for both sets of genes (Figures 6D and 6E). Although CBP reductions at the 49 active promoters did not enrich for differentiation genes, the 527 enhancer-associated genes had epidermis development as the most significant term (Figures 6D and 6E). Validation of these results by ChIP-qPCR demonstrate that KLF3 is necessary for CBP to localize to enhancers proximal to differentiation genes such as IVL, HOPX, ALOX12B, CALML5, LOR, KRT78, OVOL1, ELF3, S100A7, KLK9, and KRT80 (Figure S6E). Visualization of these enhancer sites that are proximal to important epidermal differentiation genes confirms the reduction of CBP signal (Figures 6F, 6G, S7A, and S7B). Importantly, depletion of KLF3 or CBP resulted in loss of mRNA levels of differentiation genes near enhancers that lose CBP binding upon KLF3 knockdown (Figures 6F–6H, S6E, S7A, and S7B). This suggests that KLF3 dependent localization of CBP to enhancers proximal to differentiation genes is essential for their expression. Because depletion of KLF3 reduced CBP binding at enhancer regions proximal to epidermal differentiation genes, we wanted to determine whether this would impact either priming (H3K4me) or activation (H3K27ac) of enhancers. Loss of KLF3 caused a significant reduction in H3K27ac but not H3K4me at the same enhancers as CBP reduction (IVL, HOPX, ALOX12B, CALML5, LOR, KRT78, OVOL1, ELF3, S100A7, KLK9, and KRT80) (Figures S7C and S7D). This suggests that KLF3 is necessary for enhancer activation but not priming. To determine if KLF3 directly recruited CBP to enhancers for activation, co-immunoprecipitation (co-IP) experiments were performed. KLF3 did not co-IP out CBP, suggesting that KLF3 may not directly recruit CBP to enhancer regions and may instead be necessary to set up the local region to allow for CBP to localize to KLF3 bound enhancers (Figure S7E).
Discussion
Here, we sought to identify factors that are involved in regulating epidermal differentiation. Through the analysis of a previously generated RNA-seq dataset comparing undifferentiated with differentiated keratinocytes, we identified the transcription factor KLF3 as being upregulated during epidermal differentiation. KLF3 expression was expressed throughout the epidermis but increased in the upper differentiated layers, suggesting a possible role in regulating differentiation.
Next, we characterized the KLF3 knockdown phenotype and found that depletion of KLF3 blocked differentiation gene expression in both two-dimensional cultures and three-dimensional regenerated human skin. Global gene expression profiling of KLF3 knockdown cells showed 563 genes downregulated that were important for terminal differentiation including formation of the cornified envelope. In addition, 423 genes were upregulated upon KLF3 depletion, which were genes involved in proliferation. This suggests that KLF3's normal function is to activate differentiation gene expression while suppressing proliferation. Because KLF3 is a related transcription factor to KLF4, which is a well-characterized transcription factor necessary for epidermal differentiation, we decided to test functional redundancies between them (Mistry et al., 2014; Segre et al., 1999). Comparison of KLF3 knockdown with that of KLF4 knockdown revealed that over half of the KLF3 gene signature overlapped with that of KLF4 with significant shared regulation of differentiation genes. However, the double-knockdown of these factors revealed non-redundant functions in this regulation, as knockdown of either factor alone can impact the expression of differentiation genes, but genes only impacted by the double-knockdown do not enrich for functions related to differentiation. Instead there were only 95 and 151 genes uniquely downregulated and upregulated respectively in the KLF3/KLF4 double-knockdown cells. These differentially regulated genes were not enriched for differentiation genes but rather genes involved in drug catabolic process or interferon response. This suggests that these factors cannot compensate for one another to promote differentiation gene expression as loss of either factor results in a blockade of differentiation. It has been previously shown that KLF factors can utilize unique binding sites to regulate the same genes, as well as compete for common binding sites to balance transcriptional activity (Ilsley et al., 2017). However, this competitive activity was shown in murine erythroid cells, where KLF3 acts primarily as a repressor competing for binding sites with the activator KLF1 to limit KLF1-induced gene expression. It is unclear if KLF3 and KLF4 are competing for common sites in human keratinocytes, as they do not have opposing roles but instead similar roles in promoting differentiation and suppressing cell-cycle genes. It is possible that they simultaneously occupy binding sites to promote similar gene expression programs but provide unique functional activities at these genomic loci. Thus, although KLF3 and KLF4 share many genomic binding sites and regulation of transcription, their functions at these sites may not be redundant.
Although KLF3 genome occupancy has been studied, the previous methods used for ChIP-Seq analysis primarily involved pulling down overexpressed exogenous KLF3, and these experiments were predominantly performed using murine cell lines (Burdach et al., 2014; Ilsley et al., 2017). Endogenous KLF3 has been pulled down in human primary erythroblasts; however, sequencing was not performed on those samples (Funnell et al., 2014). This study presents the ChIP-seq of endogenous KLF3 in primary human cells. Genome-wide profiling of KLF3-binding sites through ChIP-seq revealed that KLF3 shares many binding sites with other epidermal LDTFs including KLF4, ZNF750, MAF, and MAFB. For example, 35% of KLF4 and 45.4% of MAFB peaks directly overlapped with KLF3 peaks, suggesting that epidermal LDTFs regulate similar regions of the genome to control differentiation. Surprisingly, the majority of KLF3 bound peaks are enriched for H3K4me and H3K27ac signal (histone marks of open and active chromatin regions). This suggests that in differentiating keratinocytes, KLF3 may be performing activities that contribute to the promotion of differentiation gene activation and transcription. This is in contrast to previous studies classifying KLF3 as a transcriptional repressor (predominantly in murine cells), which showed KLF3's binding and repressive activity primarily through EMSA and luciferase reporter assays focused on single-gene targets in addition to the regulation of gene expression identified through RT-qPCR or microarrays (Burdach et al., 2014; Crossley et al., 1996; Eaton et al., 2008; Funnell et al., 2012, 2014; Mak et al., 2014). KLF3 has also been proposed to be a repressor based on interactions with CTBP proteins, which can promote downstream chromatin-modifying activity to repress gene expression (Dewi et al., 2015; Turner et al., 2003). However, it has recently been demonstrated that CTBP proteins can act as activators when forming complexes with epidermal transcription factors such as KLF4 and ZNF750 to promote differentiation gene expression (Boxer et al., 2014). Thus, although it is possible that KLF3 has this repressive activity at certain genes (i.e. cell cycle genes) in keratinocytes, we sought to further investigate its association with open and active chromatin regions throughout the genome because over 53% (13,556/25,352) of KLF3 binding occurs there. We found that 81% of KLF3-bound H3K4me/H3K27ac positive regions were enhancers, whereas only 19% were promoters. When these regions were mapped to proximal genes and compared with the KLF3 knockdown, we found that there were only 49 genes with KLF3-bound, active promoters that have decreased expression upon KLF3 knockdown. These 49 genes were also not enriched for GO terms such as epidermal differentiation but instead were enriched for rhythmic process and multicellular organismal water homeostasis. Strikingly, 42% (236/563) of the genes that decreased in expression upon KLF3 knockdown had proximal KLF3-bound enhancers. Furthermore, these genes were highly enriched for GO terms related to skin development. This suggests that KLF3-dependent differentiation gene expression may be due to KLF3 activity at nearby enhancers and not by KLF3 activity at their promoters.
Because of this enhancer association, we sought to investigate the functional relationship between KLF3 and the enhancer-associated epigenomic writers CBP and P300. CBP and P300 are well-known chromatin-modifying proteins that have been shown to be necessary for enhancer activity as well as acting as transcriptional co-activators to promote gene expression (Raisner et al., 2018). However, their roles in epidermal differentiation remain unclear. These factors are often considered redundant in their functions due to their largely overlapping genomic binding sites as well as redundant functions in promoting stem cell self-renewal such as in embryonic stem cells (Fang et al., 2014; Ramos et al., 2010). However, there is also evidence that these epigenetic factors are non-redundant. Knockout of either gene leads to embryonic lethality in mice, which precluded further analysis in adults (Tanaka et al., 2000; Yao et al., 1998). Conditional knockout in murine hematopoietic stem cells (HSC) have shown that CBP is necessary for HSC self-renewal, whereas p300 is important for HSC differentiation (Rebel et al., 2002). Very little is known about P300 and CBP function in the skin. However, P300 and CBP are frequently mutated (loss of function) in human cutaneous squamous cell carcinomas (Attar and Kurdistani, 2017; Li et al., 2015). Heterozygous reduction of Cbp can cooperate with oncogenic Ras to drive epidermal thickening, suggesting a tumor suppressor role for Cbp in the murine skin (Ichise et al., 2019). We show here that CBP and P300 do not play redundant roles in epidermal differentiation. Although knockdown of P300 blocked the expression of a subset of differentiation genes, it also induced the expression of terminal differentiation genes involved in lipid metabolism and cholesterol biosynthesis, which are critical to the formation of the epidermal barrier. This suggests that P300 can repress and activate different aspects of the epidermal differentiation program. In contrast, knockdown of CBP blocked epidermal differentiation in 2D and regenerated human skin and shared a similar transcriptional program as KLF3. Based on this, we sought to explore the relationship between CBP and KLF3.
We found that CBP bound to 24,357 sites in the genome with 86% of those localized to intergenic or intronic regions that are indicative of enhancers. In contrast, only a small minority (10%) mapped back to promoter regions. CBP bound sites were enriched for transcription factor motifs such as the CEBP, KLF, GRHL, and ETS family. Importantly, ∼35% (8,461/24,357) of CBP-bound sites directly overlapped with KLF3-bound regions. Because of this, we investigated whether KLF3 is necessary for CBP localization at KLF3-bound enhancer regions. Remarkably, 53% (843/1,602) of the regions where CBP lost binding upon KLF3 knockdown occurred at KLF3-bound sites. Of these 843 regions, 73% (612/843) of the regions were also bound with H3K4me/H3K27ac (with 92% being in active enhancers). Notably, knockdown of KLF3 led to a loss of H3K27ac at enhancers proximal to epidermal differentiation genes without any impacts on H3K4me. This suggests that KLF3 is not necessary for enhancer priming but important for enhancer activation likely through the acetyltransferase activity of CBP. Because KLF3 is necessary for CBP localization to KLF3-bound enhancers, we asked whether KLF3 directly recruited CBP to enhancers. Interestingly, KLF3 and CBP do not directly associate with each other through endogenous co-IP experiments. It is possible that the KLF3 and CBP interaction is extremely transient or weak. Another possibility is that KLF3 is needed to set up the chromatin structure with associated proteins at the KLF3-bound enhancers, which then allows CBP to localize. Together these data suggest that KLF3 is necessary for epidermal differentiation by promoting the proper localization of CBP to enhancers.
Primer and siRNA sequences for this work can be found in Table S9.
Limitations of the Study
One of the limitations of the study is that the RNA-Seq data for KLF3i, KLF4i, KLF3i/KLF4i, CBPi, and P300i were done as replicates. However, the RNA-Seq datasets were extensively validated by RT-qPCR in this study.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, George Sen (gsen@health.ucsd.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The datasets generated from this study such as RNA-Seq and ChIP-Seq have been deposited with GEO accession number: GSE142225.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH R01AR072590 and R01AR066530) to G.L. Sen.
Author Contributions
J.J and G.L.S conceived the project, designed the experiments, and wrote the paper. J.J, Y.C., M.T., J.T.L, and J. L. performed the experiments. All authors contributed to the reading and editing of the manuscript.
Declaration of Interests
The authors declare no conflicts of interest.
Published: July 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101320.
Supplemental Information
RNA-Seq comparing control (CTLi) and KLF3 knockdown (KLF3i) cells. Samples were sequenced in replicate (n = 2). Significant changes between CTLi and KLF3i cells were identified by ANOVA. Differentially expressed genes were filtered for a p value with FDR <0.05 and an average fold change >2 (+/−).
RNA-Seq comparing control (CTLi) and KLF4 knockdown (KLF4i) cells. Samples were sequenced in replicate (n = 2). Significant changes between CTLi and KLF4i cells were identified by ANOVA. Differentially expressed genes were filtered for a p value with FDR <0.05 and an average fold change >2 (+/−).
RNA-Seq comparing control (CTLi) and KLF4/KLF3 double knockdown (KLF3i/KLF4i) cells. Samples were sequenced in replicate (n = 2). Significant changes between CTLi and KLF3i/KLF4i cells were identified by ANOVA. Differentially expressed genes were filtered for a p value with FDR <0.05 and an average fold change >2 (+/−).
KLF3 ChIP-Seq performed in duplicates on differentiated primary human keratinocytes. Significant peaks were mapped back to nearest genes.
RNA-Seq comparing control (CTLi) and CBPi knockdown (CBPi) cells. Samples were sequenced in replicate (n = 2). Significant changes between CTLi and CBPi cells were identified by ANOVA. Differential genes were filtered for a p value with FDR <0.05 and an average fold change >2 (+/−).
RNA-Seq comparing control (CTLi) and P300i knockdown (P300i) cells. Samples were sequenced in replicate (n = 2). Significant changes between CTLi and P300i cells were identified by ANOVA. Differential genes were filtered for a p value with FDR <0.05 and an average fold change >2 (+/−).
CBP ChIP-Seq performed in duplicates on differentiated primary human keratinocytes. Significant peaks were mapped back to nearest genes.
CBP ChIP-Seq was performed in CTLi and KLF3i cells placed in differentiation medium for 3 days. The differentially CBP bound regions between CTLi and KLF3i samples were identified using Diffreps. N = 2.
All the RT-QCPR primers, ChIP-QPCR primers, and siRNA sequences used in the manuscript are listed in this table.
References
- Attar N., Kurdistani S.K. Exploitation of EP300 and CREBBP lysine acetyltransferases by cancer. Cold Spring Harb. Perspect. Med. 2017;7:1–15. doi: 10.1101/cshperspect.a026534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer L.D., Barajas B., Tao S., Zhang J., Khavari P.A. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 2014;28:2013–2026. doi: 10.1101/gad.246579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdach J., Funnell A.P., Mak K.S., Artuz C.M., Wienert B., Lim W.F., Tan L.Y., Pearson R.C., Crossley M. Regions outside the DNA-binding domain are critical for proper in vivo specificity of an archetypal zinc finger transcription factor. Nucleic Acids Res. 2014;42:276–289. doi: 10.1093/nar/gkt895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E., Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol. Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Mistry D.S., Sen G.L. Highly rapid and efficient conversion of human fibroblasts to keratinocyte-like cells. J. Invest. Dermatol. 2014;134:335–344. doi: 10.1038/jid.2013.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley M., Whitelaw E., Perkins A., Williams G., Fujiwara Y., Orkin S.H. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 1996;16:1695–1705. doi: 10.1128/mcb.16.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewi V., Kwok A., Lee S., Lee M.M., Tan Y.M., Nicholas H.R., Isono K., Wienert B., Mak K.S., Knights A.J. Phosphorylation of Kruppel-like factor 3 (KLF3/BKLF) and C-terminal binding protein 2 (CtBP2) by homeodomain-interacting protein kinase 2 (HIPK2) modulates KLF3 DNA binding and activity. J. Biol. Chem. 2015;290:8591–8605. doi: 10.1074/jbc.M115.638338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S.A., Funnell A.P., Sue N., Nicholas H., Pearson R.C., Crossley M. A network of Kruppel-like Factors (Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J. Biol. Chem. 2008;283:26937–26947. doi: 10.1074/jbc.M804831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E., Pasolli H.A., Parker J.S., Stokes N., Su I.H., Hannon G., Tarakhovsky A., Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F., Xu Y., Chew K.K., Chen X., Ng H.H., Matsudaira P. Coactivators p300 and CBP maintain the identity of mouse embryonic stem cells by mediating long-range chromatin structure. Stem Cells. 2014;32:1805–1816. doi: 10.1002/stem.1705. [DOI] [PubMed] [Google Scholar]
- Funnell A.P., Norton L.J., Mak K.S., Burdach J., Artuz C.M., Twine N.A., Wilkins M.R., Power C.A., Hung T.T., Perdomo J. The CACCC-binding protein KLF3/BKLF represses a subset of KLF1/EKLF target genes and is required for proper erythroid maturation in vivo. Mol. Cell. Biol. 2012;32:3281–3292. doi: 10.1128/MCB.00173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell A.P., Vernimmen D., Lim W.F., Mak K.S., Wienert B., Martyn G.E., Artuz C.M., Burdach J., Quinlan K.G., Higgs D.R. Differential regulation of the alpha-globin locus by Kruppel-like Factor 3 in erythroid and non-erythroid cells. BMC Mol. Biol. 2014;15:8. doi: 10.1186/1471-2199-15-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Fuchs E. Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat. Rev. Genet. 2018;19:311–325. doi: 10.1038/nrg.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales K.A.U., Fuchs E. Skin and its regenerative powers: an alliance between stem cells and their niche. Dev. Cell. 2017;43:387–401. doi: 10.1016/j.devcel.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Himeda C.L., Ranish J.A., Pearson R.C., Crossley M., Hauschka S.D. KLF3 regulates muscle-specific gene expression and synergizes with serum response factor on KLF binding sites. Mol. Cell. Biol. 2010;30:3430–3443. doi: 10.1128/MCB.00302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist P.H., Mannervik M. Genomic occupancy of the transcriptional co-activators p300 and CBP. Transcription. 2013;4:18–23. doi: 10.4161/trns.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkin A.S., Gordon W., Klein R.H., Espitia F., Daily K., Zeller M., Baldi P., Andersen B. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 2012;8:e1002829. doi: 10.1371/journal.pgen.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichise T., Yoshida N., Ichise H. CBP/p300 antagonises EGFR-Ras-Erk signalling and suppresses increased Ras-Erk signalling-induced tumour formation in mice. J. Pathol. 2019;249:39–51. doi: 10.1002/path.5279. [DOI] [PubMed] [Google Scholar]
- Ilsley M.D., Gillinder K.R., Magor G.W., Huang S., Bailey T.L., Crossley M., Perkins A.C. Kruppel-like factors compete for promoters and enhancers to fine-tune transcription. Nucleic Acids Res. 2017;45:6572–6588. doi: 10.1093/nar/gkx441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J., Chen Y., Tiwari M., Li J., Ling J., Sen G.L. BRD4 is necessary for differentiation downstream of epidermal lineage-determining transcription factors. J. Invest. Dermatol. 2020 doi: 10.1016/j.jid.2020.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski J., Cook T., Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R.H., Lin Z., Hopkin A.S., Gordon W., Tsoi L.C., Liang Y., Gudjonsson J.E., Andersen B. GRHL3 binding and enhancers rearrange as epidermal keratinocytes transition between functional states. PLoS Genet. 2017;13:e1006745. doi: 10.1371/journal.pgen.1006745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseki R., Morii W., Noguchi E., Ishikawa M., Yang L., Yamamoto-Hanada K., Narita M., Saito H., Ohya Y. Effect of filaggrin loss-of-function mutations on atopic dermatitis in young age: a longitudinal birth cohort study. J. Hum. Genet. 2019;64:911–917. doi: 10.1038/s10038-019-0628-y. [DOI] [PubMed] [Google Scholar]
- Kretz M., Webster D.E., Flockhart R.J., Lee C.S., Zehnder A., Lopez-Pajares V., Qu K., Zheng G.X., Chow J., Kim G.E. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen Y., Xu X., Jones J., Tiwari M., Ling J., Wang Y., Harismendy O., Sen G.L. HNRNPK maintains epidermal progenitor function through transcription of proliferation genes and degrading differentiation promoting mRNAs. Nat. Commun. 2019;10:4198. doi: 10.1038/s41467-019-12238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sen G.L. Generation of genetically modified organotypic skin cultures using devitalized human dermis. J. Vis. Exp. 2015:e53280. doi: 10.3791/53280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.Y., Hanna G.J., Laga A.C., Haddad R.I., Lorch J.H., Hammerman P.S. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin. Cancer Res. 2015;21:1447–1456. doi: 10.1158/1078-0432.CCR-14-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez R.G., Garcia-Silva S., Moore S.J., Bereshchenko O., Martinez-Cruz A.B., Ermakova O., Kurz E., Paramio J.M., Nerlov C. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat. Cell Biol. 2009;11:1181–1190. doi: 10.1038/ncb1960. [DOI] [PubMed] [Google Scholar]
- Lopez-Pajares V., Qu K., Zhang J., Webster D.E., Barajas B.C., Siprashvili Z., Zarnegar B.J., Boxer L.D., Rios E.J., Tao S. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev. Cell. 2015;32:693–706. doi: 10.1016/j.devcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pajares V., Yan K., Zarnegar B.J., Jameson K.L., Khavari P.A. Genetic pathways in disorders of epidermal differentiation. Trends Genet. 2013;29:31–40. doi: 10.1016/j.tig.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak K.S., Burdach J., Norton L.J., Pearson R.C., Crossley M., Funnell A.P. Repression of chimeric transcripts emanating from endogenous retrotransposons by a sequence-specific transcription factor. Genome Biol. 2014;15:R58. doi: 10.1186/gb-2014-15-4-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotto A., Pavlova O., Park H.S., Huber M., Hohl D. HOPX: the unusual homeodomain-containing protein. J. Invest. Dermatol. 2016;136:905–911. doi: 10.1016/j.jid.2016.01.032. [DOI] [PubMed] [Google Scholar]
- Mistry D.S., Chen Y., Sen G.L. Progenitor function in self-renewing human epidermis is maintained by the exosome. Cell Stem Cell. 2012;11:127–135. doi: 10.1016/j.stem.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry D.S., Chen Y., Wang Y., Zhang K., Sen G.L. SNAI2 controls the undifferentiated state of human epidermal progenitor cells. Stem Cells. 2014;32:3209–3218. doi: 10.1002/stem.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutsou M., Li J., Ling J., Jones J., Wang Y., Chen Y., Sen G.L. The cohesin complex is necessary for epidermal progenitor cell function through maintenance of self-renewal genes. Cell Rep. 2017;20:3005–3013. doi: 10.1016/j.celrep.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R.C., Funnell A.P., Crossley M. The mammalian zinc finger transcription factor Kruppel-like factor 3 (KLF3/BKLF) IUBMB Life. 2011;63:86–93. doi: 10.1002/iub.422. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner R., Kharbanda S., Jin L., Jeng E., Chan E., Merchant M., Haverty P.M., Bainer R., Cheung T., Arnott D. Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep. 2018;24:1722–1729. doi: 10.1016/j.celrep.2018.07.041. [DOI] [PubMed] [Google Scholar]
- Ramos Y.F., Hestand M.S., Verlaan M., Krabbendam E., Ariyurek Y., van Galen M., van Dam H., van Ommen G.J., den Dunnen J.T., Zantema A. Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res. 2010;38:5396–5408. doi: 10.1093/nar/gkq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebel V.I., Kung A.L., Tanner E.A., Yang H., Bronson R.T., Livingston D.M. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl. Acad. Sci. U S A. 2002;99:14789–14794. doi: 10.1073/pnas.232568499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S.Y., Denu J.M., Allis C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Rubin A.J., Barajas B.C., Furlan-Magaril M., Lopez-Pajares V., Mumbach M.R., Howard I., Kim D.S., Boxer L.D., Cairns J., Spivakov M. Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat. Genet. 2017;49:1522–1528. doi: 10.1038/ng.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre J.A., Bauer C., Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Sen G.L., Boxer L.D., Webster D.E., Bussat R.T., Qu K., Zarnegar B.J., Johnston D., Siprashvili Z., Khavari P.A. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev. Cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Shao N.Y., Liu X., Maze I., Feng J., Nestler E.J. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013;8:e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood A., Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr. Opin. Cell Biol. 2013;25:387–394. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sue N., Jack B.H., Eaton S.A., Pearson R.C., Funnell A.P., Turner J., Czolij R., Denyer G., Bao S., Molero-Navajas J.C. Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol. Cell. Biol. 2008;28:3967–3978. doi: 10.1128/MCB.01942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Naruse I., Hongo T., Xu M., Nakahata T., Maekawa T., Ishii S. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech. Dev. 2000;95:133–145. doi: 10.1016/s0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- Ting S.B., Caddy J., Hislop N., Wilanowski T., Auden A., Zhao L.L., Ellis S., Kaur P., Uchida Y., Holleran W.M. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308:411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- Turner J., Nicholas H., Bishop D., Matthews J.M., Crossley M. The LIM protein FHL3 binds basic Kruppel-like factor/Kruppel-like factor 3 and its co-repressor C-terminal-binding protein 2. J. Biol. Chem. 2003;278:12786–12795. doi: 10.1074/jbc.M300587200. [DOI] [PubMed] [Google Scholar]
- Visel A., Blow M.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T.T., Gatto D., Turner V., Funnell A.P., Mak K.S., Norton L.J., Kaplan W., Cowley M.J., Agenes F., Kirberg J. Impaired B cell development in the absence of Kruppel-like factor 3. J. Immunol. 2011;187:5032–5042. doi: 10.4049/jimmunol.1101450. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zang C., Cui K., Schones D.E., Barski A., Peng W., Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T.P., Oh S.P., Fuchs M., Zhou N.D., Ch'ng L.E., Newsome D., Bronson R.T., Li E., Livingston D.M., Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA-Seq comparing control (CTLi) and KLF3 knockdown (KLF3i) cells. Samples were sequenced in replicate (n = 2). Significant changes between CTLi and KLF3i cells were identified by ANOVA. Differentially expressed genes were filtered for a p value with FDR <0.05 and an average fold change >2 (+/−).
RNA-Seq comparing control (CTLi) and KLF4 knockdown (KLF4i) cells. Samples were sequenced in replicate (n = 2). Significant changes between CTLi and KLF4i cells were identified by ANOVA. Differentially expressed genes were filtered for a p value with FDR <0.05 and an average fold change >2 (+/−).
RNA-Seq comparing control (CTLi) and KLF4/KLF3 double knockdown (KLF3i/KLF4i) cells. Samples were sequenced in replicate (n = 2). Significant changes between CTLi and KLF3i/KLF4i cells were identified by ANOVA. Differentially expressed genes were filtered for a p value with FDR <0.05 and an average fold change >2 (+/−).
KLF3 ChIP-Seq performed in duplicates on differentiated primary human keratinocytes. Significant peaks were mapped back to nearest genes.
RNA-Seq comparing control (CTLi) and CBPi knockdown (CBPi) cells. Samples were sequenced in replicate (n = 2). Significant changes between CTLi and CBPi cells were identified by ANOVA. Differential genes were filtered for a p value with FDR <0.05 and an average fold change >2 (+/−).
RNA-Seq comparing control (CTLi) and P300i knockdown (P300i) cells. Samples were sequenced in replicate (n = 2). Significant changes between CTLi and P300i cells were identified by ANOVA. Differential genes were filtered for a p value with FDR <0.05 and an average fold change >2 (+/−).
CBP ChIP-Seq performed in duplicates on differentiated primary human keratinocytes. Significant peaks were mapped back to nearest genes.
CBP ChIP-Seq was performed in CTLi and KLF3i cells placed in differentiation medium for 3 days. The differentially CBP bound regions between CTLi and KLF3i samples were identified using Diffreps. N = 2.
All the RT-QCPR primers, ChIP-QPCR primers, and siRNA sequences used in the manuscript are listed in this table.
Data Availability Statement
The datasets generated from this study such as RNA-Seq and ChIP-Seq have been deposited with GEO accession number: GSE142225.