Abstract
Genome engineering using programmable nucleases such as transcription activator-like effector nuclease (TALEN), and clustered regularly interspaced short palindromic repeat-associated protein nine facilitated the introduction of genetic alterations at specific genomic sites in various cell types. These tools have been applied to cancer modeling to understand the pathogenic effects of the growing catalog of mutations found in human cancers. Pertaining to brain tumors, neural progenitor cells derived from human induced pluripotent stem cells (iPSCs) engineered with different combinations of genetic driver mutations observed in distinct molecular subtypes of glioblastomas, the most common form of primary brain cancer in adults, give rise to brain tumors when engrafted orthotopically in mice. These glioblastoma models recapitulate the transcriptomic signature of each molecular subtype and authentically resemble pathobiology of glioblastoma, including inter- and intra-tumor heterogeneity, chromosomal aberrations, and extrachromosomal DNA amplifications. Similar engineering with genetic mutations found in medulloblastoma and atypical teratoid rhabdoid tumors in iPSCs have led to genetically trackable models that bear clinical relevance to these pediatric brain tumors. These models have contributed to improved comprehension of the genetic causation of tumorigenesis and offered a novel platform for therapeutic discovery. Studied in the context of three-dimensional cerebral organoids, these models have aided in the study of tumor invasion as well as therapeutic responses. In summary, modeling brain tumors through genome engineering enables not only the establishment of authentic tumor avatars driven by bona fide genetic mutations observed in patient samples but also facilitates functional investigations of particular genetic alterations in an otherwise isogenic background.
Keywords: brain tumor, CRISPR/Cas9, genome engineering, glioma, modeling
Introduction
The development of genome engineering technologies using programmable nucleases, including zinc finger nucleases (ZFN),1) transcription activator-like effector nuclease (TALEN),2-4) and cluster regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein nine (Cas9) system5,6) have aided in the generation of genetically engineered murine models previously unattainable through conventional homologous recombination-mediated gene-targeting.7) When applied to induced pluripotent stem cells (iPSCs),8) these technologies have transformed the field of disease modeling.9) For example, Reinhardt et al.10) generated iPSCs from patients with Parkinson’s disease harboring LRRK2 mutations as well as normal controls, and then either corrected or introduced the mutations using ZFN genome editing. The mutated or mutation-corrected iPSCs were then differentiated to dopaminergic neurons, revealing that mutant LRRK2 induced ERK activation leading to dopaminergic neurodegeneration.
Such approaches through combinations of genome engineering and stem cell technologies have paved the way for sophisticated cancer models driven by pertinent mutations uncovered in human clinical tumor specimens. As an example, Heckl et al.11) modified Nf1, Ezh2, Dnmt3a, Tet2, and Runx1 in mouse hematopoietic stem cells to model acute myeloid leukemia. In other studies, colon organoids7) from human intestinal crypt stem cells introduced with different combinations of genetic alterations in APC, SMAD4, TP53, KRAS, and PIK3CA genes, which are commonly affected in colorectal cancers, were generated.12,13) These organoid models harbored features of colorectal cancers such as aneuploidy, formed tumors in vivo upon xenotransplantation, revealed genetic alterations underlying invasion, and accurately predicted drug responses.14)
Applications of genome editing techniques have led to paradigm shifts in the modeling of adult and pediatric brain cancers. Prior to these efforts, disease modeling relied heavily on genetically engineered mouse (GEM) models, human astrocyte-derived models, and patient-derived xenografts (PDX). While the utility of these models is undisputed, each harbor intrinsic limitations that restrict the interpretation of human relevance or generalizability. In this article, we review advances in brain tumor modeling through genome engineering (Table 1) and discuss the relative merits of this approach to previously available models.
Table 1.
Brain tumor models generated through genome engineering
| Authors (year) | Modified genes | Modalities | Species | Materials | Tumors modeled |
|---|---|---|---|---|---|
| Duan et al.41) (2015) | PTEN | TALEN | Human | ESCs | GBM |
| Zuckermann et al.61) (2015) | Ptch1, Trp53, Pten, Nf1 | CRISPR/Cas9 | Mouse | Embryonic brains | Medulloblastoma, GBM |
| Bian et al.56) (2018) | MYC, CDKN2A, CDKN2B, EGFR, NF1, PTEN, TP53 | CRISPR/Cas9, SB-transposon | Human | Cerebral organoids | GBM, CNS-PNET |
| Ogawa et al.57) (2018) | TP53, HRAS | CRISPR/Cas9 | Human | Cerebral organoids | GBM (mesenchymal subtype) |
| Huang et al.43) (2019) | GSE1, KDM3B | CRISPR/Cas9 | Human | NESCs (Gorlin syndrome) | Medulloblastoma (SHH subtype) |
| Terada et al.45) (2019) | TP53, SMARCB1 | CRISPR/Cas9 | Human | iPSCs | AT/RT |
| Koga et al.47) (2020) | PTEN, NF1, TP53, PDGFRA | CRISPR/Cas9 | Human | iPSCs | GBM (mesenchymal, proneural subtypes) |
| Yu et al.63) (2020) | Trp53, Nf1, Pten, Pik3ca | CRISPR/Cas9 PB-transposon | Mouse | Embryonic brains | GBM |
AT/RT: atypical teratoid rhabdoid tumor, CNS-PNET: central nervous system primitive neuroectodermal tumor, CRISPR: clusters of regularly interspaced short palindromic repeats, ESC: embryonic stem cell, GBM: glioblastoma, iPSC: induced pluripotent stem cell, NESC: neuroepithelial stem cell, PB: PiggyBac, SB: Sleeping Beauty, SHH: Sonic Hedgehog, TALEN: transcription activator-like effector nuclease.
Non-genome Engineering-derived Brain Tumor Models
The earlier models of brain cancer include tumors that formed in rat or murine brains treated with DNA damaging mutagens. Rat C6 glioma cell line, which was induced by exposure to methylnitrosourea,15,16) produces glioma-like tumors when injected in rat brains17) and has been frequently utilized as a syngeneic model in glioma research.18) Similar efforts using murine models exposed to methylcholanthrene have led to the generation of the GL261, and CT-2A glioblastoma cell lines.19-21) While these syngeneic models aid in the investigations of tumor immune response, they harbor increased mutational burden and exaggerated immune response relative to those observed in human disease.22,23) Moreover, these tumors tend to form a well-defined mass rather than the invasive histology seen in human gliomas.24)
The second class of brain cancer models involves tumors that arose consequent to the introduction of transgenes. Danks et al. introduced SV40 T antigen under control of glial fibrillary acidic protein promoter and succeeded in transforming mouse astrocytes in 1995.25) Later, Holland et al.26,27) introduced oncogenes such as EGFR and CDK4 by somatic cell gene delivery using replication-competent avian leukosis virus splice acceptor (RCAS) viral vectors and their receptor, Tva, to generate glioma models. Importantly, the RCAS-Tva system has also been utilized to model pediatric brain tumors such as medulloblastoma.28) Different models of glioblastomas, the most aggressive form of gliomas, have been generated through the introduction of oncogenes such as Src, K-ras, H-ras, PDGFB, and EGFRvIII.29,30) These GEM models have been fundamental in dissecting molecular mechanisms underlying genetic carcinogenesis. However, whether insights derived from these studies are pertinent to human disease remains an open question. Pertinent to this concern, there are significant discrepancies in experiments where drugs were simultaneously tested against human and murine models of glioblastoma.31) These therapeutic differences can extend orders of magnitude.
Human cell models are essential to address such biological differences presented by murine platforms. Rich et al.32) and Sonoda et al.33) engineered human astrocytes with combinations of TERT and HRAS expression and inhibition of the TP53 pathway by simian virus 40 (SV40) T antigen or by human papillomavirus (HPV) E6 and E7 and succeeded in establishing high-grade glioma models. These models enable investigations focused on gliomagenic mechanisms in the context of human cells. To the extent that viral expression of SV40 and HPV is not found in most human glioblastomas,34) it remains unclear whether the physiology of these tumors is clinically relevant. Additionally, there are many features of clinical glioblastoma which have not been carefully scrutinized in these models, including inter- and intra-tumoral heterogeneity. As other examples, various monolayer cell lines derived from human gliomas were established in serum-containing media.35) These models are easy to expand for experimental use, but again lack typical histological features such as heterogeneity and in vivo invasive potential and are not ideal as some studies suggest genomic deviation from the original patient sample.24,36)
PDX models overcome disadvantages of established monolayer cell lines by maintaining original phenotypes observed in clinical samples upon orthotopic engraftment, thus enabling studies on inter- and intra-tumoral heterogeneity,37,38) and effects of targeted therapies.39) However, the heterogeneity of the PDX models serves as a double-edged sword, making experimental standardization difficult due to vast variability in background mutations present in each clinical sample.
In summary, valuable insights have been gained through various brain tumor models. Undoubtedly, they will continue to be utilized for multiple purposes. It is crucial, however, to keep in mind the various caveats associated with these different models. As genome engineering technologies emerge, these new tools may offer answers in addressing these limitations.
Brain Tumor Models Derived from Genome-Engineered Human Stem Cells
Using TALEN-mediated homologous recombination to delete PTEN, a tumor suppressor gene affected in 36% of glioblastoma patients,40) Duan et al. generated glioma models from human embryonic stem cells (ESCs) differentiated to neural stem cells (NSCs). When engrafted in immunocompromised mice, these PTEN-null NSCs formed neoplastic lesions and presented sensitivity to mitomycin C. Transcriptomically, the PTEN-null NSCs showed differential expression of PAX7 compared with wild-type control, which was validated in the Cancer Genome Atlas (TCGA) dataset.41) This was the first model to show that disruption of a glioblastoma- associated tumor suppressor leads to the reprogramming of human NSCs toward a cancer stem cell-like phenotype. However, PTEN alterations are seldomly observed solely by themselves in human glioblastomas and are almost always accompanied by other oncogenic events.42)
Later, Huang et al.43) generated neuroepithelial cells (NESCs) from iPSCs derived from patients with Gorlin syndrome, a tumor predisposition syndrome caused by mutations in PTCH1, which is associated with an increased risk of medulloblastoma. In their study, CRISPR/Cas9 disruption of GSE1, which is commonly co-mutated in adult medulloblastoma, resulted in accelerated tumorigenesis. Interestingly, the tumors obtained by engraftment of GSE1 knockout NESCs into the cerebellum of mice clustered closer to the Sonic Hedgehog (SHH) subtype of medulloblastoma driven by SHH pathway activation that occurs due to disruption of PTCH1. As an example of another brain tumor model, Terada et al. disrupted SMARCB1, which is recurrently affected in atypical teratoid rhabdoid tumors (AT/RT),44) to model this malignant pediatric brain cancer.45) This study presented a potential use of brain tumor cells derived from genetically engineered human iPSCs for drug screening.
More recently, our group established glioblastoma models by introducing different combinations of genetic alterations observed in different molecular subtypes of glioblastoma42,46) into human iPSCs.47) In this study, neural progenitor cells (NPCs) were differentiated from iPSCs harboring CRISPR/Cas9-induced combinatory alterations of PTEN/NF1 and TP53/PDGFRA, which are commonly observed in mesenchymal and proneural glioblastoma molecular subtypes, respectively. Here, gene-edited NPCs gave rise to GBM-like tumors upon orthotopic engraftments in immunocompromised animals. The tumors were confirmed to have histological features of glioblastoma by meticulous pathological assessment, and presented the transcriptomic signatures of mesenchymal and proneural subtypes, respectively. Our study proved that introducing different combinations of driver genetic alterations in cells with isogenic backgrounds results in tumor models presenting distinct phenotypes. Furthermore, the single-cell RNA sequencing analyses revealed that these genetically engineered human iPSC-derived models presented inter- and intra-tumor heterogeneity as observed in patient samples.48) Importantly, these models showed prominent chromosomal abnormality accompanied with extrachromosomal DNA amplifications, which are commonly seen in glioblastoma samples.49-52) As the tumor cells derived from these in vivo models grew in sphere condition in vitro and formed secondary tumors upon re-engraftments, these models were suitable for testing of drug sensitivity and assessments of longitudinal tumor evolution.47)
Such varieties of brain tumor models show significant potential of modeling numerous types of brain tumors driven by different genetic drivers through the introduction of defined alterations in isogenic human backgrounds, facilitated by the power of genome engineering. Limitations include a lack of immune components due to engraftment of these models in immunocompromised animals (Table 2).
Table 2.
Advantages and limitations of genome-engineered brain tumor models from different platforms
| Platforms | Authors | Advantages | Limitations |
|---|---|---|---|
| Human stem cells | Duan et al.41) Huang et al.43) Terada et al.45) Koga et al.47) |
Feasibility in experimental
standardization in isogenic background Enabling human tumor biology investigation |
Restrictions in the assessment of immune environment |
| Human cerebral organoids | Ogawa et al.57) Huang et al.43) |
Enabling limited use of
animals. Feasibility in the assessment of tumor–tumor microenvironment interactions in three-dimensional context in vitro |
Restrictions in the assessment of immune
environment Lack of physiological backgrounds e.g., blood vessels |
| Mouse embryonic brains | Zuckermann et
al.61) Yu et al.63) |
Spontaneous tumor formation in syngeneic backgrounds enabling assessment of immune interactions | Restrictions in the interpretation of
human relevance. Technical hurdles of in utero electroporation. |
Brain Tumor Models in Genome- Engineered Human Cerebral Organoids
As shown in the studies of colorectal cancer models in colon organoids, tissue organoids are potential tools for modeling and investigating cancers in three-dimensional contexts.12,13) In the field of neuroscience, Lancaster et al. established the methods of generating cerebral organoids from human pluripotent stem cells.53,54) Lincous et al. generated glioblastoma organoid models by combining patient-derived glioma stem cells and cerebral organoids derived from human ESCs and proved that such models serve as a robust tool to investigate biological behaviors of glioblastoma invasion.55)
Bian et al.56) and Ogawa et al.57) introduced genome engineering into cerebral organoids to model brain tumors in vitro. Bian et al. introduced genetic edits at the early stages of the cerebral organoid formation using combinations of the Sleeping Beauty (SB) transposon system58-60) to insert multiple copies of oncogenes, thus mimicking their overexpression, and CRISPR/Cas9 for disruption of tumor suppressor genes. Organoids electroporated with the combinations of constructs of MYCOE (OE indicates overexpression), CDKN2A–/–/CDKN2B–/–/EGFROE/EGFRvIIIOE, NF1–/–/PTEN–/–/TP53–/– , and EGFRvIIIOE/CDKN2A–/–/PTEN–/– each resulted in overgrowth of electroporated cells indicating neoplastic transformation. Organoids with MYCOE presented transcriptomic signatures of central nervous system primitive neuroectodermal tumors. The other combinations were associated with transcriptome signatures seen in human glioblastomas and exhibited distinct drug sensitivity in vivo, suggesting the potential use of these models for future drug screening.56) In another model, Ogawa et al.57) introduced a cassette of HRASG12V and a fluorescent protein, tdTomato, at the TP53 locus in cerebral organoids using CRISPR/Cas9 to overexpress a mutant HRAS while disrupting TP53. Overgrowth of transformed tdTomato-positive cells suggested neoplastic transformation. The transformed cells in these organoid models were transplantable to the brains of immunocompromised mice and cerebral organoids as well.
In sum, cerebral organoid brain tumor models may offer opportunities for multiplex genome engineering and provide a novel platform for drug screening in vitro. These models afford opportunities for in vitro investigations on interactions between tumor cells and brain microenvironment, which cannot be done using conventional in vitro models, although cerebral organoids still lack some physiological components such as an immune microenvironment and blood vessels (Table 2).
Spontaneous Mouse Brain Tumor Models Using Genome Engineering
To overcome the laborious and time-consuming processes of generating GEM models, Zuckermann et al.,61) using an in utero electroporation technique of mouse embryonic brains, developed a spontaneous mouse brain tumor model by introducing Cas9 and small guide RNAs expressing plasmids to target various genes. In utero electroporation of CRISPR constructs targeting Ptch1 resulted in high tumor formation efficiency. The transcriptome of these tumor models clustered together with a previously published medulloblastoma GEM model with Ptch1 alterations. They further tested different combinations of target genes to model glioblastoma and confirmed the combination of CRISPR constructs targeting Trp53, Nf1, and Pten generate glioblastoma-like tumors in eight out of eight animals. Similarly, Yu et al. induced mouse in vivo brain tumors by in utero electroporation of CRISPR/Cas9 constructs targeting Trp53 and Nf1 together with PiggyBac62) transposable vectors harboring different variants of Pik3ca mutations.63)
These models generated through in utero genome engineering proved that this approach is an efficient way to establish in vivo syngeneic tumor models with potentially numerous combinations of genetic alterations, although limitations of this approach include technical challenges in manipulating embryos in utero (Table 2).
Functional Analyses of Genetic Alterations in Isogenic Backgrounds
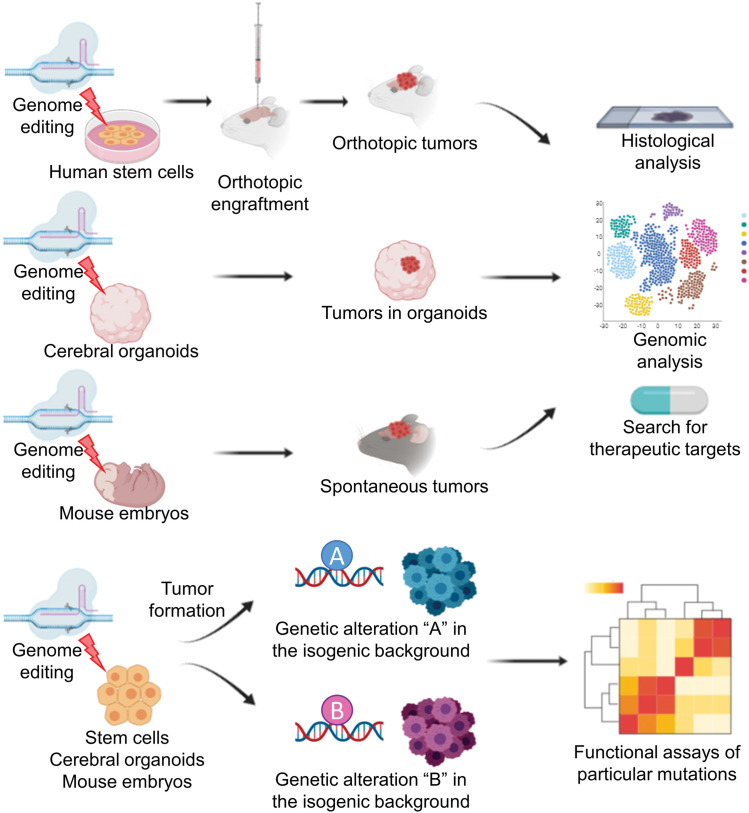
One of the benefits of utilizing genome engineering for tumor modeling is the feasibility of introducing designed genetic alterations into any materials such as human stem cells, cerebral organoids, and mouse embryonic brains for downstream applications (Fig. 1). Such efficient genomic modifications enable functional testing of specific genetic alterations in isogenic backgrounds (Fig. 1). As proved in our models and others, different combinations of genetic alterations introduced in these isogenic platforms result in distinct phenotypes of brain tumors.47,61) Huang et al. analyzed tumorigenic functions of co-occurring mutations in conjunction with PTCH1 alterations found in adult medulloblastoma patients. Among those co-mutated genes, GSE1 and KDM3B were disrupted using CRISPR/Cas9 in an isogenic background of NESCs derived from Gorlin syndrome patients, which showed that alterations in GSE1, but not KDM3B, accelerate tumorigenesis.43) This study effectively utilized genome engineering tools to validate the tumorigenic function of potential driver mutations whose roles in particular tumor formation were previously unknown. Yu et al. efficiently screened 27 variants of Pik3ca mutations in the background of Trp53 and Nf1 knockout and showed that C420R and H1047R mutations of this gene result in hyperexcitability of the surrounding brain.63)
Fig. 1.
Overview of brain tumor modeling using genome engineering. Introducing brain-tumor-associated mutations into human stem cells, cerebral organoids, or mouse embryonic brains give rise to various brain tumor models that can be utilized for histological, genomic analyses, and search for novel therapeutic targets. These models allow comparative functional assessment of particular genetic alterations in otherwise isogenic backgrounds.
As shown, introducing genome engineering into brain tumor modeling enables efficient investigations of genetic functions of mutations associated with tumorigenesis and tumor progression. When applied in the context of synthetic lethality, these models have the potential to accelerate the development of precision medicine as it pertains to brain tumor treatment.
Conclusion
The available literature suggests the feasibility and utility of genome engineering as a tool to model the mutations uncovered through the interrogation of human brain tumor specimens. The approach is flexible and can be applied to stem cells, organoids, and through in utero electroporation. In these contexts, genome engineering has enabled next-generation brain tumor models that should contribute to the accelerated discovery of effective therapeutics for brain tumor patients.
Footnotes
Conflicts of Interest Disclosure
The authors declare no conflict of interest.
References
- 1).Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD: Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11: 636–646, 2010 [DOI] [PubMed] [Google Scholar]
- 2).Miller JC, Tan S, Qiao G, et al. : A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148, 2011 [DOI] [PubMed] [Google Scholar]
- 3).Hockemeyer D, Wang H, Kiani S, et al. : Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29: 731–734, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P: Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 29: 149–153, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Cong L, Ran FA, Cox D, et al. : Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J: RNA-programmed genome editing in human cells. Elife 2: e00471, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS: Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature 317: 230–234, 1985 [DOI] [PubMed] [Google Scholar]
- 8).Takahashi K, Yamanaka S: Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676, 2006 [DOI] [PubMed] [Google Scholar]
- 9).Hockemeyer D, Jaenisch R: Induced pluripotent stem cells meet genome editing. Cell Stem Cell 18: 573–586, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Reinhardt P, Schmid B, Burbulla LF, et al. : Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell 12: 354–367, 2013 [DOI] [PubMed] [Google Scholar]
- 11).Heckl D, Kowalczyk MS, Yudovich D, et al. : Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol 32: 941–946, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Drost J, van Jaarsveld RH, Ponsioen B, et al. : Sequential cancer mutations in cultured human intestinal stem cells. Nature 521: 43–47, 2015 [DOI] [PubMed] [Google Scholar]
- 13).Matano M, Date S, Shimokawa M, et al. : Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 21: 256–262, 2015 [DOI] [PubMed] [Google Scholar]
- 14).Tuveson D, Clevers H: Cancer modeling meets human organoid technology. Science 364: 952–955, 2019 [DOI] [PubMed] [Google Scholar]
- 15).Benda P, Lightbody J, Sato G, Levine L, Sweet W: Differentiated rat glial cell strain in tissue culture. Science 161: 370–371, 1968 [DOI] [PubMed] [Google Scholar]
- 16).Schmidek HH, Nielsen SL, Schiller AL, Messer J: Morphological studies of rat brain tumors induced by N-nitrosomethylurea. J Neurosurg 34: 335–340, 1971 [DOI] [PubMed] [Google Scholar]
- 17).Auer RN, Del Maestro RF, Anderson R: A simple and reproducible experimental in vivo glioma model. Can J Neurol Sci 8: 325–331, 1981 [DOI] [PubMed] [Google Scholar]
- 18).Grobben B, De Deyn PP, Slegers H: Rat C6 glioma as experimental model system for the study of glioblastoma growth and invasion. Cell Tissue Res 310: 257–270, 2002 [DOI] [PubMed] [Google Scholar]
- 19).Ausman JI, Shapiro WR, Rall DP: Studies on the chemotherapy of experimental brain tumors: development of an experimental model. Cancer Res 30: 2394–2400, 1970 [PubMed] [Google Scholar]
- 20).Szatmári T, Lumniczky K, Désaknai S, et al. : Detailed characterization of the mouse glioma 261 tumor model for experimental glioblastoma therapy. Cancer Sci 97: 546–553, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Zimmerman HM, Arnold H: Experimental brain tumors. I. Tumors produced with methylcholanthrene. Cancer Res 1: 919–938, 1941 [Google Scholar]
- 22).Szulzewsky F, Arora S, de Witte L, et al. : Human glioblastoma-associated microglia/monocytes express a distinct RNA profile compared to human control and murine samples. Glia 64: 1416–1436, 2016 [DOI] [PubMed] [Google Scholar]
- 23).Genoud V, Marinari E, Nikolaev SI, et al. : Responsiveness to anti-PD-1 and anti-CTLA-4 immune checkpoint blockade in SB28 and GL261 mouse glioma models. Oncoimmunology 7: e1501137, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Huszthy PC, Daphu I, Niclou SP, et al. : In vivo models of primary brain tumors: pitfalls and perspectives. Neuro-oncology 14: 979–993, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Danks RA, Orian JM, Gonzales MF, et al. : Transformation of astrocytes in transgenic mice expressing SV40 T antigen under the transcriptional control of the glial fibrillary acidic protein promoter. Cancer Res 55: 4302–4310, 1995 [PubMed] [Google Scholar]
- 26).Holland EC, Hively WP, DePinho RA, Varmus HE: A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev 12: 3675–3685, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Holland EC, Hively WP, Gallo V, Varmus HE: Modeling mutations in the G1 arrest pathway in human gliomas: overexpression of CDK4 but not loss of INK4a-ARF induces hyperploidy in cultured mouse astrocytes. Genes Dev 12: 3644–3649, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Schüller U, Heine VM, Mao J, et al. : Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14: 123–134, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Stylli SS, Luwor RB, Ware TM, Tan F, Kaye AH: Mouse models of glioma. J Clin Neurosci 22: 619–626, 2015 [DOI] [PubMed] [Google Scholar]
- 30).Chow LM, Endersby R, Zhu X, et al. : Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell 19: 305–316, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Shi Y, Lim SK, Liang Q, et al. : Gboxin is an oxidative phosphorylation inhibitor that targets glioblastoma. Nature 567: 341–346, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Rich JN, Guo C, McLendon RE, Bigner DD, Wang XF, Counter CM: A genetically tractable model of human glioma formation. Cancer Res 61: 3556–3560, 2001 [PubMed] [Google Scholar]
- 33).Sonoda Y, Ozawa T, Hirose Y, et al. : Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res 61: 4956–4960, 2001 [PubMed] [Google Scholar]
- 34).Wang Z, Hao Y, Zhang C, et al. : The landscape of viral expression reveals clinically relevant viruses with potential capability of promoting malignancy in lower- grade glioma. Clinical Cancer Res 23: 2177–2185, 2017 [DOI] [PubMed] [Google Scholar]
- 35).Westphal M, Meissner H: Establishing human glioma-derived cell lines. Methods Cell Biol 57: 147–165, 1998 [DOI] [PubMed] [Google Scholar]
- 36).Li A, Walling J, Kotliarov Y, et al. : Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res 6: 21–30, 2008 [DOI] [PubMed] [Google Scholar]
- 37).Neftel C, Laffy J, Filbin MG, et al. : An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178: 835–849.e21, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Wang Q, Hu B, Hu X, et al. : Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32: 42–56.e6, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Patrizii M, Bartucci M, Pine SR, Sabaawy HE: Utility of glioblastoma patient-derived orthotopic xenografts in drug discovery and personalized therapy. Front Oncol 8: 23, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Cancer Genome Atlas Research Network: Comprehensive genomic characterization defines human glioblastoma genes and core pathways Nature 455: 1061–1068, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Duan S, Yuan G, Liu X, et al. : PTEN deficiency reprogrammes human neural stem cells towards a glioblastoma stem cell-like phenotype. Nat Commun 6: 10068, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Brennan CW, Verhaak RG, McKenna A, et al. : TCGA Research Network: The somatic genomic landscape of glioblastoma. Cell 155: 462–477, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Huang M, Tailor J, Zhen Q, et al. : Engineering genetic predisposition in human neuroepithelial stem cells recapitulates medulloblastoma tumorigenesis. Cell Stem Cell 5: 433–446, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Versteege I, Sévenet N, Lange J, et al. : Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394: 203–206, 1998 [DOI] [PubMed] [Google Scholar]
- 45).Terada Y, Jo N, Arakawa Y, et al. : Human pluripotent stem cell-derived tumor model uncovers the embryonic stem cell signature as a key driver in atypical teratoid/rhabdoid tumor. Cell Rep 26: 2608–2621.e6, 2019 [DOI] [PubMed] [Google Scholar]
- 46).Verhaak RG, Hoadley KA, Purdom E, et al. : Cancer Genome Atlas Research Network: Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Koga T, Chaim IA, Benitez JA, et al. : Longitudinal assessment of tumor development using cancer avatars derived from genetically engineered pluripotent stem cells. Nat Commun 11: 550, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Patel AP, Tirosh I, Trombetta JJ, et al. : Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344: 1396–1401, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Nathanson DA, Gini B, Mottahedeh J, et al. : Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 343: 72–76, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Turner KM, Deshpande V, Beyter D, et al. : Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543: 122–125, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Wu S, Turner KM, Nguyen N, et al. : Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575: 699–703, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).deCarvalho AC, Kim H, Poisson LM, et al. : Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet 50: 708–717, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Lancaster MA, Knoblich JA: Generation of cerebral organoids from human pluripotent stem cells. Nature Protocols 9: 2329, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Lancaster MA, Renner M, Martin CA, et al. : Cerebral organoids model human brain development and microcephaly. Nature 501: 373–379, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Linkous A, Balamatsias D, Snuderl M, et al. : Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep 26: 3203–3211.e5, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Bian S, Repic M, Guo Z, et al. : Genetically engineered cerebral organoids model brain tumor formation. Nat Methods 15: 631–639, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Ogawa J, Pao GM, Shokhirev MN, Verma IM: Glioblastoma model using human cerebral organoids. Cell Rep 23: 1220–1229, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA: Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436: 272–276, 2005 [DOI] [PubMed] [Google Scholar]
- 59).Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA: Mammalian mutagenesis using a highly mobile somatic sleeping beauty transposon system. Nature 436: 221–226, 2005 [DOI] [PubMed] [Google Scholar]
- 60).Ivics Z, Hackett PB, Plasterk RH: Izsvák Z: Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91: 501–510, 1997 [DOI] [PubMed] [Google Scholar]
- 61).Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, et al. : Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nature Communications 6: 7391, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Yusa K, Zhou L, Li MA, Bradley A, Craig NL: A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci USA 108: 1531–1536, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Yu K, Lin CJ, Hatcher A, et al. : PIK3CA variants selectively initiate brain hyperactivity during gliomagenesis. Nature 578: 166–171, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]