Abstract
BACKGROUND:
For adults with chest pain, the electrocardiogram (ECG) and measures of serum biomarkers are used to screen and diagnose myocardial necrosis. These measurements require time that can delay therapy and affect prognosis. Our objective was to investigate the feasibility and utility of saliva as an alternative diagnostic fluid for identifying biomarkers of acute myocardial infarction (AMI).
METHODS:
We used Luminex and lab-on-a-chip methods to assay 21 proteins in serum and unstimulated whole saliva procured from 41 AMI patients within 48 h of chest pain onset and from 43 apparently healthy controls. Data were analyzed by use of logistic regression and area under curve (AUC) for ROC analysis to evaluate the diagnostic utility of each biomarker, or combinations of biomarkers, in screening for AMI.
RESULTS:
Both established and novel cardiac biomarkers demonstrated significant differences in concentrations between patients with AMI and controls without AMI. The saliva-based biomarker panel of C-reactive protein, myoglobin, and myeloperoxidase exhibited significant diagnostic capability (AUC = 0.85, P < 0.0001) and in conjunction with ECG yielded strong screening capacity for AMI (AUC = 0.96) comparable to that of the panel (brain natriuretic peptide, troponin-I, creatine kinase-MB, myoglobin; AUC = 0.98) and far exceeded the screening capacity of ECG alone (AUC approximately 0.6). En route to translating these findings to clinical practice, we adapted these unstimulated whole saliva tests to a novel lab-on-a-chip platform for proof-of-principle screens for AMI.
CONCLUSIONS:
Complementary to ECG, saliva-based tests within lab-on-a-chip systems may provide a convenient and rapid screening method for cardiac events in prehospital stages for AMI patients.
About 13.2 million individuals in the US have coronary artery disease, 7.8 million have suffered an acute myocardial infarction (AMI),10 and 6.8 million have symptoms of angina pectoris (1). Patients with acute chest pain in the emergency present important diagnostic, economic, and operational challenges. Electrocardiogram (ECG) is the mainstay initial screening test that identifies chest pain–related ST elevation myocardial infarction (STEMI), which typically arises from blocked arteries. For this reason ECG is usually performed by paramedics before the patient’s hospital arrival (2). However, fewer than one-third of patients admitted to emergency departments exhibit definitive ECG evidence of myocardial injury, and the remainder must be observed to exclude the possibility of a non-ST-segment elevation myocardial infarction (NSTEMI). To detect or exclude myocardial necrosis in such patients, serial measurements of biochemical markers in serum are required, which often include myoglobin (MYO), creatine kinase-MB (CK-MB), total CK and cardiac troponins T and I (cTnT, cTnI) (3). Rates of release of biomarkers differ depending on intracellular location, protein size, and local blood and lymphatic flow characteristics (4).
Currently, AMI is defined by detection of rise and/or fall of cardiac biomarkers (preferably cTnI) with at least 1 value above the 99th percentile of the upper reference limit together with evidence of myocardial ischemia (based on ECG or imaging) (5, 6). Despite tremendous progress in developing new diagnostic and screening methods, a substantial number of AMI cases today are missed or diagnosed too late to offer effective therapies. According to a recent report, in many emergency rooms it takes approximately 60 min for 25% of AMI patients to be examined (7). Likewise, there is a compelling need to improve triage and minimize delays associated with seeking care, transportation to the emergency department, and performance of reperfusion by either pharmacological or catheter-based approaches.
Despite demonstrated improvements in turnaround time through use of point-of-care (POC) approaches (8–11), implementation of these approaches is limited to only a fraction of emergency rooms worldwide (12 ). Frequently cited reasons for the slow rate of adoption of these new promising POC cardiac diagnostic options include perceived lack ofprecision, concerns with methods of standardization, and difficulties with clinical data management of such devices (12). These reasons and recent advances with ultrasensitive laboratory-based assays have secured troponin as the preferred marker in serum in preference to MYO and CK-MB (13).
With recent release of the saliva proteome, there is increased interest in the use oforal fluid samples for the diagnosis of a variety of oral and systemic diseases (14–18). Indeed, saliva as a noninvasive diagnostic medium has a number of advantages relative to ease of collection, storage, and overall fluid management that make it well-suited for POC applications. Saliva has been used for measuring electrolytes, drugs, cytokines, hormones, enzymes, antibodies, microbes, and RNAs (19–21). However, oral fluids have not yet been reported for the diagnosis of AMI. Furthermore, little is known about the salivary biomarker expression levels for the various cardiac indications. One challenge associated with oral fluids is that biomarker concentrations are often significantly lower than in serum counterparts, making measurement more challenging with traditional analytical approaches.
Through the last decade, our team has made sustained efforts to combine and adapt lab-on-a-chip (LOC), microfluidic, microelectromechanical systems and nano-biochip tools for practical implementation of highly sensitive and accurate miniaturized sensors that are suitable for variety of important applications, including multiplex analysis of minute amounts of bioanalytes in serum and saliva (22–32). These integrated test systems can complete all aspects of sample processing and separation and analyte detection and are amenable to POC applications.
Chemically sensitized bead “microreactors” within the LOC system were recently applied for measurement of C-reactive protein (CRP) and other biomarkers of inflammation in saliva, demonstrating significantly lower detection level (by >3 decade orders of magnitude) for CRP than high-sensitivity CRP ELISA methods, allowing for measurement of inflammatory biomarkers related to select disease states (27).
We report here for the first time the measurement of salivary biomarkers associated with AMI and explore the possibility of using these new salivary biomarkers in novel nano-biochip ensembles for screening chest pain patients for AMI. Our initial objective was to determine if serum biomarkers commonly associated with AMI diagnosis can be detected reliably using unstimulated whole saliva (UWS). Persons who did not have chest pain were recruited as controls for the first part of this study to demonstrate feasibility of measuring protein concentrations of both standard and novel biomarkers in saliva and serum. Choice of this control group was expected to amplify potential differences between salivary samples of AMI and non-AMI patients and thus allow for more efficient identification of potential biomarkers for use in subsequent larger studies.
Materials and Methods
STUDY DESIGN, PATIENT RECRUITMENT AND SAMPLE COLLECTION
A cross-sectional clinical case-control study was implemented, and 56 patients were recruited within 48 h of onset of symptoms of AMI, along with 59 age- and sex-matched non-AMI controls at hospitals of the University of Kentucky and University of Louisville. STEMI were based on ECG elevation of ST-segments by >0.1 mV in contiguous leads in patients with ischemic symptoms and increased cardiac biomarkers (>99th percentile of the upper reference limit for TnI, cutoff 0.04 μ-g/L). Diagnoses NSTEMI were made for patients with inconclusive ECG, but ischemic symptoms such as ECG changes consistent with ischemia (depression of the ST segments or new left bundle branch block), new pathologic Q-waves, or evidence of perfusion defects on stress test, followed by confirmatory positive TnI test.
Recruitment was coordinated with the cardiac care team and balancing needs of patients, including pain management, reperfusion and family support. All study participants were at least 18 years old. Exclusion criteria were fever, stroke, immune disorders, use of steroidal medications, organ complications/failure, and inability to provide saliva. Rights of all study participants were protected by institutional review boards at the participating sites. All participants gave informed consent before sample collection, and samples tested were deidentified to ensure privacy.
Demographic information was obtained, medical records reviewed, oral evaluation performed, and biological fluids obtained (blood and UWS) from each study participant. Samples were transported on ice to a local laboratory, centrifuged, divided into aliquots, and stored at −80 °C until analyzed. Samples from the University of Louisville were shipped on dry ice to University of Kentucky laboratories on a bimonthly basis. Samples were analyzed in duplicate for cardiac enzymes and a panel of 21 biomarkers. Analyses were performed by using Luminex, ELISA, or Beckman Access in the CLIA-certified University of Kentucky hospital clinical chemistry laboratory within 3 months of storage.
Oral health was assessed visually at the bedside or in the dental operatory for controls. Oral health was scored as poor, fair, or good based on presence or absence of dental complaints, degree of mucosal inflammation, and extent of decay and periodontal disease.
MEASUREMENT OF BIOMARKERS BY LUMINEX AND BECKMAN ACCESS
Standard cardiac biomarkers brain natriuretic peptide (BNP), MYO, CK-MB, and cTnI were measured with a Beckman Access (imprecision <10% CV in plasma or serum). A Luminex IS-100 instrument was used for multiplexed detection relevant to cardiovascular disease and was used with kits available from Bead- lyte Technology (Millipore) for the following tests: CRP; interleukin-6 (IL-6); monocyte chemoattractant protein-1 (MCP-1); IL-1β; myeloperoxidase (MPO); soluble cluster of differentiation ligand (sCD40L); tumor necrosis factor-α (TNF-α); regulated on activation, normal T expressed and secreted (RANTES); fractalkine; soluble vascularization cellular adhesion molecule-1 (sVCAM-1); epithelial cell–derived neutrophil-activating peptide 78 (ENA-78); IL-18; E-selectin; growth related protein-α (Gro-α); adiponectin; soluble intercellular adhesion molecule-1 (sICAM-1); and matrix metalloproteinase-9 (MMP-9) (imprecision 3.7% to 17.2% in plasma or serum for all biomarkers).
DATA ANALYSIS
Data mining steps were completed by consolidating information into homogenous datasets to maximize the number of patients for which a complete biomarker panel was available. The process resulted in an 88-patient dataset for serum, composed of 42 controls, 23 NSTEMI, and 23 STEMI patients, along with an 84-patient dataset for saliva (43 controls and 16 NSTEMI and 25 STEMI patients) from the initial 115 patients. Inability to collect sufficient sample volume was the primary reason for loss of some of patients to the study. Also, some samples were required for development, validation, and testing of the LOC system.
Nonparametric Wilcoxon–Mann–Whitney tests were used to evaluate differences between median biomarker concentrations detected in saliva of non-AMI controls and AMI patients. Medcalc V. 9.5.2.0 software was used for logistic regression and ROC analysis. The ROC curves were constructed and area under curve (AUC) computed, either from single biomarker concentrations or for multimarker panels. SE and 2-tailed P values at 95% confidence levels were determined.
LOC MULTIPLEXED TEST
Design, fabrication, and testing methods for LOC structures have been described in detail in previous reports (23, 24, 26, 27, 30, 31). New modifications to procedures and methods for multiplexed AMI diagnosis are described in the Supplemental Data section (see Lab-on-a-Chip (LOC) Multiplexed Test in Supplemental Materials in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol55/issue8]).
Results
Demographics of the control group, (59 persons, mean age 49.3 years, range 37–79 years; 34 females and 25 males; 48 white, 8 African American, and 3 Hispanics) were similar to the AMI group (56 persons, mean age 54.8 years, range 29–84 years; 36 females and 20 males; 47 white and 9 African American). The AMI patients had 8 fewer teeth and slightly poorer oral health (data not shown), and mean body mass indices were identical (i.e., 28.5).
A total of 21 protein biomarkers with relevance to cardiac disease patient classification that have a strong precedent in reported literature (33, 34) were studied (see online Supplemental Fig. 1, biomarker cascade) from samples obtained between 12 and 48 h after onset of AMI-related chest pain [mean (SD) time: 30.3 (14.1) h]. The greatest ratio in serum protein expression for AMI is seen for cTnI (115), followed by CK-MB (6.5), BNP (5.4), CRP (4.3), MPO (2.5), and MYO (1.8), all exhibiting P < 0.0001 [Fig. 1 (red bars)]. Biomarkers MMP-9 (1.6, P = 0.004) and sCD40L (1.4, P = 0.37), though providing some discriminatory potential, yielded more modest ratios. The most downregulated proteins in serum were fractalkine (0.51, P = 0.49), IL-6 (0.54, P = 0.030), and Gro-α (0.60, P = 0.060). Serum-based analysis of established biomarkers yielded strong diagnostic capabilities, as demonstrated by AUC values (see Supplemental Table 1): cTnI (0.99), CK-MB (0.93), BNP (0.90), and MYO (0.77). These values are consistent with previously reported values (8) for these biomarkers (0.94, 0.91, 0.85, 0.78, respectively) in a study involving more than 2000 patients. Combinations of these biomarkers gave AUCs between 0.99 and 1.00 (see online Supplemental Table 1).
Fig. 1. Biomarker expression and diagnostic accuracy values.
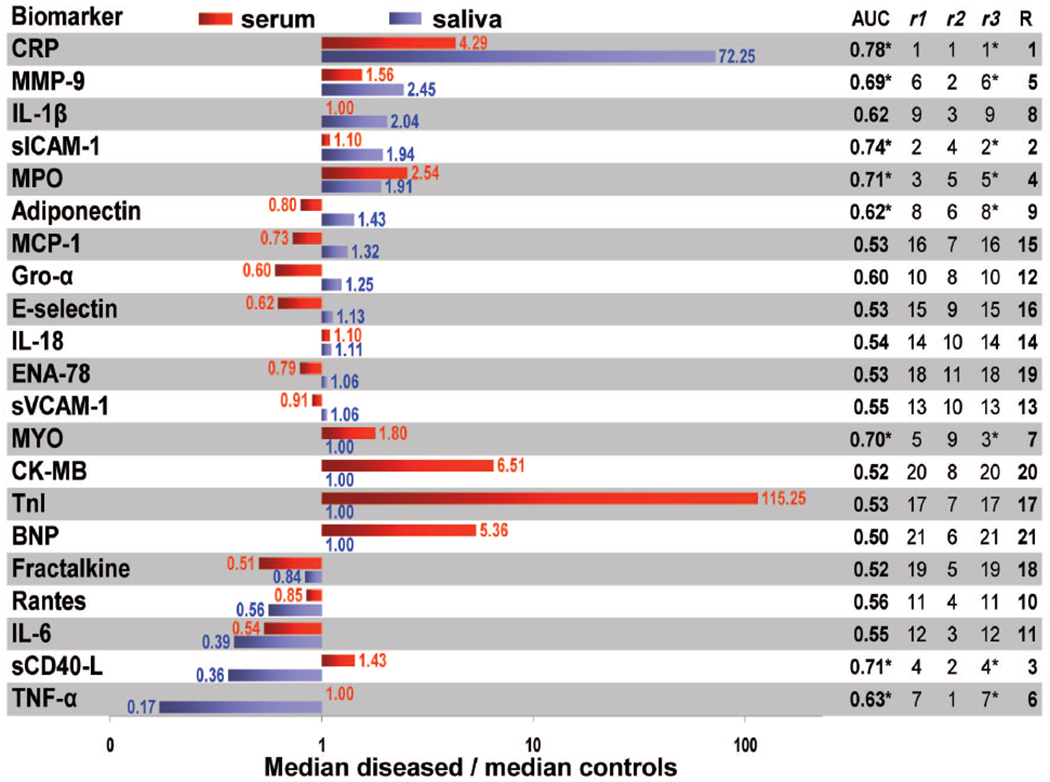
Bar graphs on the left show ratios of median concentrations of biomarkers for the diseased patients over those of non-AMI controls for serum (red) and saliva (blue); ratio values are indicated next to each bar. The AUC values for saliva are provided on the right (*P < 0.05). Here r1 is the rank obtained by each biomarker according to its AUC, r2 is the ranking obtained from the ratio of median diseased over median non-AMI controls (up- and downregulated biomarkers are ranked equally), and r3 ranking results from the P value related to the statistical significance of the difference between the medians of diseased and control populations. An aggregate ranking (R) is also provided based on the averages of r1, r2, and r3.
The concentrations of novel biomarkers in saliva were evaluated for their preliminary capacity to serve as alternative biomarkers for AMI screening accuracy. In UWS (Fig. 1, blue bars), CRP showed the highest ratio in median concentrations of AMI/control (72, P < 0.0001), followed by MMP-9 (2.5, P = 0.0029), IL-1β (2.0, P = 0.0659), sICAM-1 (1.9, P = 0.0001), MPO (1.9, P = 0.0008), adiponectin (1.4, P = 0.052), monocyte chemoattractant protein 1 (1.3, P = 0.66), and Gro-α (1.2, P = 0.16). The most downregulated proteins in whole saliva were TNF-α (0.17, P = 0.038), sCD40L (0.36, P = 0.0005), and IL-6 (0.39, P = 0.40).
Identifying a series of biomarkers that can be measured reliably in saliva enabled the use of logistic regression as a screening tool for this initial determination of the most useful multimarker panels for AMI assessment. A series of 4 ROC curves for AMI classification using various logistic regression algorithms, are presented in Fig. 2. The first model studied included all salivary biomarker inputs regardless of statistical or biological significance. This “enter” model appears to yield the most accurate diagnostic capabilities of studied cases (AUC = 0.97; 95% CI, 0.89–1.00; P = 0.0001), but suffers from overfitting (see below), and inclusion of numerous nonapproved biomarkers. The next model included only variables that were entered sequentially into a model based on statistical significance as single markers and resulted in following salivary biomarkers: BNP, CRP, IL-18, sICAM-1, TNF-α, sVCAM-1, E-selectin, Gro-α, IL-6. This “forward selection” model yielded an AUC of 0.91; 95% CI: 0.80–0.97; P = 0.0001.
Fig. 2. ROC plots generated for resulting biomarkers from 4 models, all variables included regardless of statistical or biological significance (“enter,” top left), only significant variables entered sequentially (“forward,” top right), all independent variables first entered and then removed sequentially if not found significant (“backward,” bottom left), and finally, all significant variables entered sequentially with recalculation of the model if a variable is found to become nonsignificant and excluded after inclusion of another independent variable (“stepwise,” bottom right).
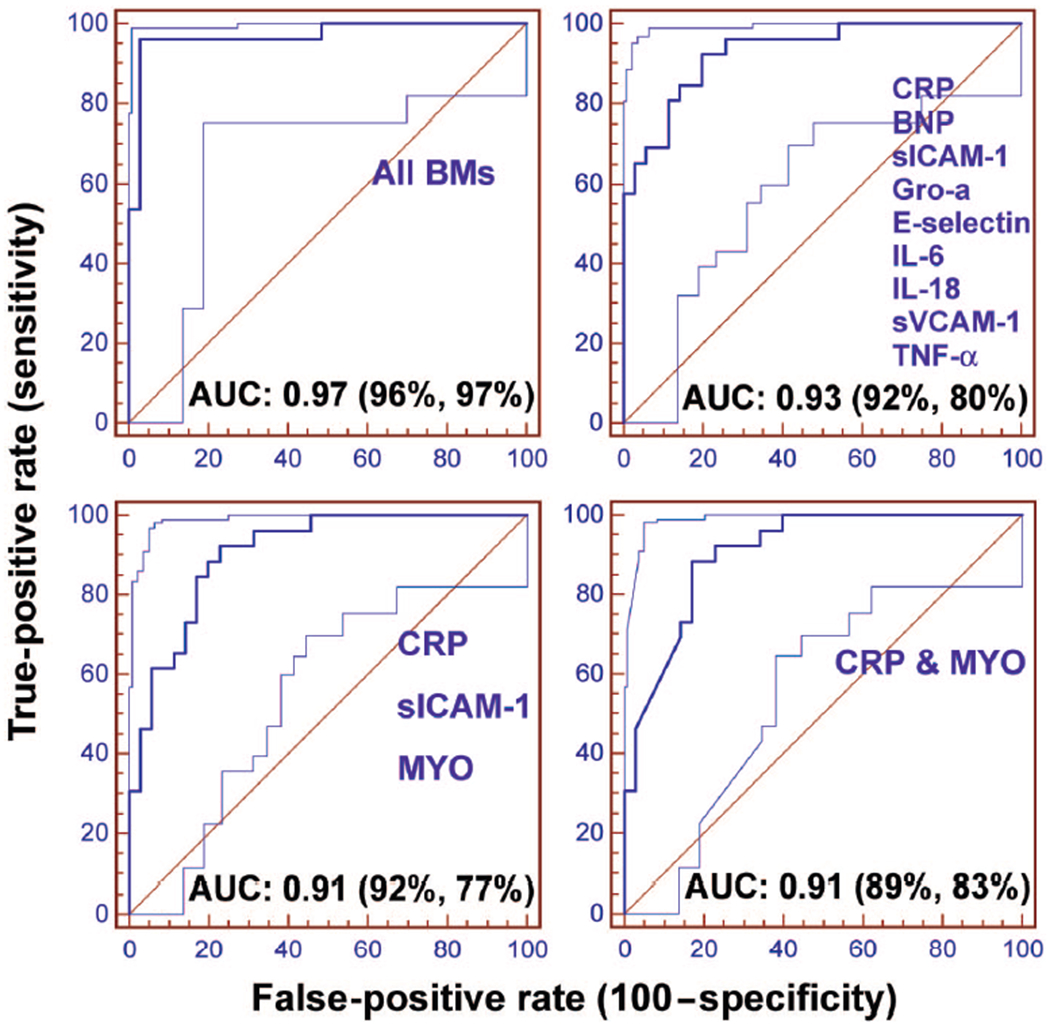
For each ROC plot, the y = x line represents the line of no discrimination. The 95% lower and upper confidence bands are also shown on either side of the ROC curve. The percentages in parentheses list the best-averaged sensitivity and specificity.
Next, all independent variables were first entered into model and then removed sequentially if not found significant. This “backward selection” model lead to inclusion of following salivary biomarkers: CRP, sICAM-1, and MYO, yielding an AUC of 0.93; 95% CI: 0.84–0.98; P = 0.0001. Finally, all significant biomarkers were entered sequentially and model was recalculated after exclusion of any variable found nonsignificant upon inclusion of another independent variable. This “stepwise” method indicated that salivary CRP and MYO had large effects and yielded an AUC of 0.91; 95% CI: 0.80–0.97; P = 0.0001.
Even with acquisition of robust protein measurements across 84 patients in this case-control study, other factors including outlier samples, sample stability, and measurement inaccuracies potentially can influence main conclusions related to utility of various salivary biomarkers. To acquire a better understanding of the potential diagnostic capabilities of these salivary biomarkers, composite data were examined from a number of perspectives. Thus, an aggregate ranking system, as summarized in the right panel of Fig. 1, was created for each salivary biomarker by using the following 3 factors: (a) AUC obtained for single biomarkers (AUC in Fig. 1), leading to ranking r1, (b) the value of the ratio of median diseased over median non-AMI controls (r2 in Fig. 1), (c) the P value assessing the statistical significance of the difference between the medians of diseased and control populations (r3 in Fig. 1), and (d) aggregate rankings averaging prior 3 factors (R in Fig. 1).
The aggregate score projects the following list of top 10 biomarkers and is considered to yield most valuable information for diagnosis of AMI from a single salivary biomarker perspective: CRP (#1), sICAM-1 (#2), sCD40L (#3), MPO (#4), MMP-9 (#5), TNF-α (#6), MYO (#7), IL-1β (#8), adiponectin (#9), and RANTES (#10).
The results of ROC analysis of combination panels involving combinations of these 10 biomarkers are listed in Table 1. Salivary CRP-MPO and CRP-MYO, as well as trio panel involving CRP-MPO-MYO yielded similar AUCs of 0.82, 0.85, and 0.85, respectively. Next, we explored utility of saliva assessment as a companion test to ECG to capture those NSTEMI patients that are not diagnosed in initial ECG screen (i.e., 39% of AMI patients in this case-control study). A new set of ROC curves were built, based on use of the same panels as previously described, except that salivary biochemistry data is combined with ECG screening information (i.e., a value of 1 is input for STEMI patients, and 0 is used for NSTEMI patients), which is an independent variable.
Table 1.
AUC, SE, P, AUC 95% CI, and best averaged sensitivity and specificity for various salivary biomarkers and biomarkers combinations.
| Strategy | Biomarker (BM) panela | AUC | SE | 95% CI | P | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|
| Aggregate based on R | BM 1 | 0.78 | 0.051 | 0.679–0.865 | 0.0001 | 68.3 | 73.7 |
| Aggregate based on R | BMs 1–2 | 0.77 | 0.052 | 0.668–0.856 | 0.0001 | 68.3 | 76.7 |
| Aggregate based on R | BMs 1–3 | 0.78 | 0.051 | 0.681–0.866 | 0.0001 | 65.1 | 79.1 |
| Aggregate based on R | BMs 1–4 | 0.81 | 0.048 | 0.709–0.887 | 0.0001 | 73.2 | 79.1 |
| Aggregate based on R | BMs 1–5 | 0.81 | 0.048 | 0.705–0.884 | 0.0001 | 73.2 | 79.1 |
| Aggregate based on R | BMs 1–6 | 0.82 | 0.047 | 0.718–0.893 | 0.0001 | 85.4 | 65.1 |
| Aggregate based on R | BMs 1–7 | 0.85 | 0.045 | 0.752–0.920 | 0.0001 | 89.5 | 68.3 |
| Aggregate based on R | BMs 1–8 | 0.85 | 0.044 | 0.752–0.921 | 0.0001 | 89.5 | 70.7 |
| Aggregate based on R | BMs 1–9 | 0.85 | 0.044 | 0.752–0.921 | 0.0001 | 89.5 | 70.7 |
| Aggregate based on R | BMs 1–10 | 0.87 | 0.042 | 0.775–0.935 | 0.0001 | 89.5 | 70.7 |
| All biomarkers | All 21 biomarkers | 0.97 | 0.025 | 0.889–0.996 | 0.0001 | 96.2 | 97.1 |
| BPSFAb | CRP MPO | 0.82 | 0.047 | 0.720–0.895 | 0.0001 | 90.2 | 62.8 |
| BPSFA | CRP MYO | 0.85 | 0.044 | 0.756–0.923 | 0.0001 | 92.1 | 73.2 |
| BPSFA | CRP MPO MYO | 0.85 | 0.045 | 0.746–0.916 | 0.0001 | 92.1 | 68.3 |
| All biomarkers + ECG | All 21 biomarkers + ECG | 1.00 | 0.000 | 0.941–1.000 | 0.0001 | 100.0 | 100.0 |
| BPSFA + ECG | CRP MPO & ECG | 0.95 | 0.026 | 0.872–0.983 | 0.0001 | 90.2 | 90.7 |
| BPSFA + ECG | CRP MYO & ECG | 0.94 | 0.028 | 0.866–0.982 | 0.0001 | 100.0 | 73.2 |
| BPSFA + ECG | CRP MPO MYO & ECG | 0.94 | 0.028 | 0.866–0.982 | 0.0001 | 81.6 | 92.7 |
| Reduced training set | CRP MYO | 0.88 | 0.048 | 0.766–0.953 | 0.0001 | 96.3 | 71.4 |
| Reduced training set | CRP MYO & ECG | 0.96 | 0.028 | 0.869–0.993 | 0.0001 | 92.6 | 85.7 |
| Testing set | CRP MYO | 0.85 | 0.084 | 0.641–0.958 | 0.0001 | 90.2 | 69.2 |
| Testing set | CRP MYO & ECG | 0.89 | 0.073 | 0.693–0.977 | 0.0001 | 81.8 | 92.3 |
BMs are ranked and combinations assembled according to aggregate score (R) listed in Figure 1: CRP (1), sICAM-1 (2), sCD40L (3), MPO (4), MMP-9 (5), TNF-α (6), MYO (7), IL-1β (8), adiponectin (9), and RANTES (10).
BPSFA, Biomarkers with precedent in serum that are FDA approved.
Inclusion of information from the ECG in the test panels increased AUC in most combinations of biomarkers. A focus on 2 panels with minimal numbers of biomarkers was chosen to demonstrate the utility of the combination ECG-salivary test. Logistic regression and ROC analysis of the established panel of CRP-MPO-MYO in conjunction with ECG yielded an AUC of 0.94 (0.87–0.98 95% CI, P = 0.0001) (Table 1). This same panel demonstrated 82% sensitivity and 90% specificity in differentiating cardiac disease patients from controls. CRP-MYO provided a similar AUC and displayed 100% sensitivity and 73% specificity (Table 1). These values exceed those of ECG by itself, which gave a sensitivity of only 61% in this study. This limited dataset was split into training and test sets to explore in a preliminary manner the ruggedness of the procedure for selecting classification models. Logistic regression coefficients were recalculated and models were established from the data obtained from 55 patients (approximately two-thirds of the total sample population) used in the training set. The CRP and MYO model was challenged with the remaining one-third of the samples as external data, alone and as complemented by ECG.
The CRP-MYO biomarker panel yielded similar AUC for the full training set (0.85) and the reduced training set (0.88), with similar sensitivity and specificity values (see Table 1). In addition, consistent increases in AUC were observed for this panel when it was used in conjunction with ECG (AUC = 0.96, 93% sensitivity, 86% specificity). When evaluated in the testing set, AUC was 0.89. The logit equation obtained from the logistic regression analysis was:
In a final step, we translated a saliva multiplexed test relevant for AMI screening to an LOC system that could be used for POC testing (Fig. 3) and enabled detection of CRP, MYO, IL-1β, and MPO in fluorescent multiplex assays performed on both AMI and control patients. Note that high fidelity signals for these salivary biomarkers are extracted with the nano-biochip approach whereby the high surface area of the beads, high local concentration of capture antibody, and pressure drive transport of samples and reagents afford an integrated approach that allows for these salivary biomarker measurements to be completed in POC settings.
Fig. 3. Multiplex lab-on-a-chip (LOC) for AMI screening.
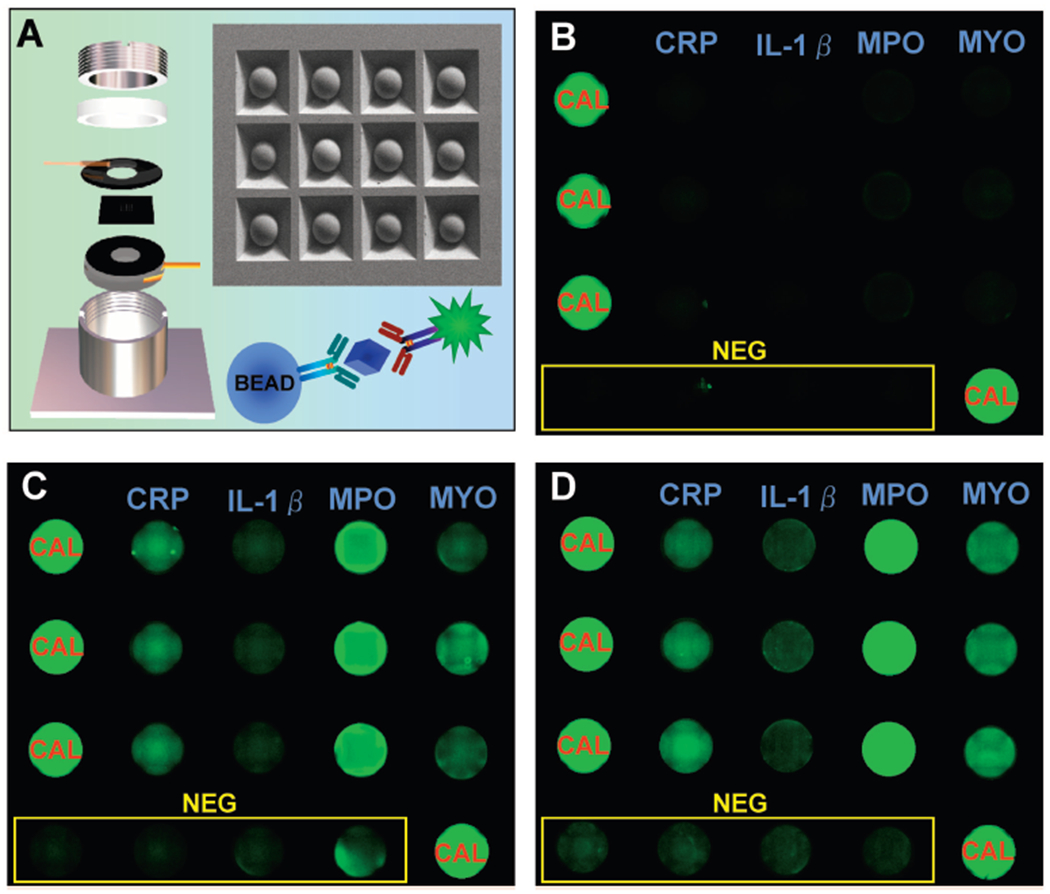
(A), A scanning electron micrograph of the silicon microchip is shown with the LOC fluidic compartment on the left. An immuno-schematic depicts the sandwich type immunoassay detection modality and the analyte of interest (CRP, IL-1β, MYO, or MPO antigens are represented in blue). Examples of fluorescence micrographs of a LOC multiplex assay for CRP, IL-1β, MYO, and MPO are shown for non-AMI control (B), NSTEMI (C), and STEMI (D) patients. NEG, negative; CAL, calibrator.
Discussion
To our knowledge, this study is the first to investigate the use of oral fluid of AMI patients to evaluate biomarkers involved in the cardiovascular disease cascade. These salivary biomarkers were combined into panels that were tested for their capacity to distinguish AMI patients from non-AMI controls. Interestingly, evaluation of serum markers of cTnI, CK-MB, BNP, and MYO for expression levels in UWS yielded only modest ratios and discriminating ability for distinguishing AMI patients from controls (see Table 1 and online Supplemental Table 1). Careful analysis of these saliva samples revealed that although these biomarkers are expressed at measurable levels in extreme phenotypes, in typical samples for diseased patients the concentrations of these biomarkers can fall below the limit of detection for the Beckman Access instrument when measured at the initial time interval available for this study (i.e., within 48 h). Because troponins are the dominant serum biomarkers for classification of AMI patients, in future studies we will investigate salivary troponins more carefully, measuring them at earlier time points using next-generation and more sensitive salivary nano-biochip sensor systems.
From an initial 21 biomarkers, we found that measurement of salivary CRP and MYO serves as a minimal reliable panel. Both of these biomarkers have been cleared by the FDA for clinical use in serum, an important consideration for intended application, although for other indications than saliva AMI screens. Several combinations (Table 1), including up- and downregulated biomarkers (data not shown) can be used to differentiate between the AMI and control groups with high statistical significance, but panels with only 2 or 3 select biomarkers are often found to perform as well or better than more inclusive panels.
Panels involving CRP, MYO, and MPO, when used as companion tests for ECG, were found to yield excellent diagnostic accuracies, as measured by ROC analyses in which they yielded an AUC of 0.96 that is comparable to the best serum multimarker panels reported previously (0.98 for BNP, MYO, cTnI, and CK-MB) (8). With sensitivity values in the range of 90%–100% for MYO-CRP and MYO-CRP-MPO panels, these new rapid salivary biochemistry tests would seem to be excellent supplements to standard ECGs. There is ample reported evidence indicating that chest pain evaluation might incorporate a multimarker approach that includes some combination of myoglobin, CK-MB, troponin, and inflammatory markers in the diagnostic strategy (8,35–38). However, in this proof-of-feasibility study, we collected samples 12–48 h (median 30) after the onset of AMI-related chest pain.The relatively late time of sample acquisition in the clinical course of AMI is a limitation of this initial study, as is the use of nonchest pain patients for the control samples. Also, caution must be used when interpreting the results of logistic regression for larger panels (i.e., >5 biomarkers) because resulting improvements in AUC become increasingly associated with overfitting of the logistic regression model owing to imbalance between the number of patients in each studied group and the number of biomarkers. In contrast, smaller panels are robust and appear to hold up to challenges of external data. To address these issues, further studies involving more patients are now in progress. In these studies we aim to delineate the kinetic profile of these biomarkers in saliva collected from chest pain patients at earlier time points. Furthermore, the development of ultrasensitive saliva LOC assays for troponin, currently the gold standard for AMI diagnostic evaluation in serum, will allow us to reevaluate the contributions and combinations from these biomarkers in a similar manner to that used in the present study. Just as in the initial biomarker validation phases, our LOC studies also document measurable signal differences in protein fingerprint patterns of these 2 patient groups. Bead-based immunoassay systems display strong analytical performance characteristics (typical intraassay variance of 4%–8% and interassay variance of 6%–10%) (26, 27, 31). Correlation studies completed with FDA-approved instruments for serum CRP yield R2 values of 0.98.
Ongoing studies aim at refining this test for the detection of AMI in chest pain patients at the point of need. With the integrated LOC system, sample handling and analyte separation, detection, and analysis can be completed on disposable nano-biochips intended for use in conjunction with a portable analyzer, as previously reported (29, 39), which could help reduce the time to diagnosis and therapies.
Supplementary Material
Acknowledgments:
Funding provided by NIH through the National Institute of Dental and Craniofacial Research (UO1 DE15017 and U01 DE017793), General Clinical Research Center at University of Kentucky (M01-RR02602), and National Center for Research Resources (P20 RR020145).
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Authors’ Disclosures of Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest:
Employment or Leadership: J. Spertus, CV Outcomes; S.R. Steinhubl, the Medicines Company.
Consultant or Advisory Role: J. Spertus, St. Jude Medical, Otsuka, and United Healthcare; J.T. McDevitt, LabNow.
Stock Ownership: S.R. Steinhubl, the Medicines Company; J.T. McDevitt, LabNow.
Honoraria: None declared.
Research Funding: C. Miller, NIH; J. Spertus, Amgen, Roche Diagnostics, Atherotech, BMS/Sanofi Aventis, Lilly, Johnson & Johnson, and Medtronic; J.T. McDevitt, NIH–National Institute of Dental and Craniofacial Research, and LabNow.
Expert Testimony: None declared.
Publisher's Disclaimer: Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent or reflect views of the NIH or the Federal Government.
Nonstandard abbreviations: AMI, acute myocardial infarction; ECG, electrocardiogram; STEMI, ST elevation myocardial infarction; NSTEMI, non-STEMI; MYO, myoglobin; CK-MB, creatine kinase-MB; cTnT, cardiac troponin T; cTnI, cardiac troponin I; POC, point of care; LOC, lab-on-a-chip; CRP, C-reactive protein; UWS, unstimulated whole saliva; BNP, brain natriuretic peptide; IL, interleukin; MCP-1, monocyte chemoattractant protein-1; MPO, myeloperoxidase; sCD40L, soluble cluster of differentiation ligand; TNF-α, tumor necrosis factor-α; RANTES, regulated on activation, normal T expressed and secreted; sVCAM-1, soluble vascularization cellular adhesion molecule-1; ENA-78, epithelial cell–derived neutrophil-activating peptide 78; Gro-α, growth related protein-α; sICAM-1, soluble intercellular adhesion molecule-1; MMP-9, matrix metalloprotease-9; AUC, area under the curve.
References
- 1.Rosamond W, Flegal K, Friday G, Furie K, Go A, Greenlund K, et al. Heart disease and stroke statistics, 2007 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 2007; 115:E69–E171. [DOI] [PubMed] [Google Scholar]
- 2.Ting HH, Krumholz HM, Bradley EH, Cone DC, Curtis JP, Drew BJ, et al. Implementation and integration of prehospital ECGs into systems of care for acute coronary syndrome: a scientific statement from the American Heart Association Interdisciplinary Council on Quality of Care and Outcomes Research, Emergency Cardiovascular Care Committee, Council on Cardiovascular Nursing, and Council on Clinical Cardiology. Circulation 2008;118:1066–79. [DOI] [PubMed] [Google Scholar]
- 3.Christenson RH, ed. The National Academy of Clinical Biochemistry presents laboratory medicine practice guidelines: biomarkers of acute coronary syndromes and heart failure. TheAmerican Association for Clinical Chemistry; 2007. http://www.aacc.org/SiteCollectionDocuments/NACB/LMPG/ACS_PDF_online.pdf (Accessed July 2009). [Google Scholar]
- 4.NACB Writing Group M, Morrow DA, Cannon CP, Jesse RL, Newby LK, Ravkilde J, et al. National Academy of Clinical Biochemistry laboratory medicine practice guidelines: clinical characteristics and utilization of biochemical markers in acute coronary syndromes. Clin Chem 2007;53:552–74. [DOI] [PubMed] [Google Scholar]
- 5.Thygesen K, Alpert JS, White HD; Joint ESC/ACCF/ AHA/WHF Task Force for the Redefinition of Myocardial Infarction, Jaffe AS, Apple FS, et al. Universal definition of myocardial infarction. Circulation 2007;116:2634–53. [DOI] [PubMed] [Google Scholar]
- 6.Apple FS, Ler R, Chung AY, Berger MJ, Murakami MM. Point-of-care i-STAT cardiac troponin I for assessment of patients with symptoms suggestive of acute coronary syndrome. Clin Chem 2006;52: 322–5. [DOI] [PubMed] [Google Scholar]
- 7.Wilper AP, Woolhandler S, Lasser KE, McCormick D, Cutrona SL, Bor DH, Himmelstein DU. Waits to see an emergency department physician: U.S. trends and predictors, 1997–2004. Health Aff (Millwood) 2008;27(2):w84–95. [DOI] [PubMed] [Google Scholar]
- 8.Hollander JE, Peacock WF, Shofer FS, Green GB, Lowenstein CJ, Blankenberg S, et al. A cardiac multimarker index performs better than standard markers to diagnose acute myocardial infarction. Acad Emerg Med 2005;12(5 Suppl 1):33a.15635135 [Google Scholar]
- 9.Sluss PM. Cardiac markers: current technologies for their measurement at points of care. Point of Care 2006;5:38–46. [Google Scholar]
- 10.Wu AHB. Point-of-care testing for conventional cardiac markers. Point of Care 2006;5:20–4. [Google Scholar]
- 11.Di Serio F, Lovero R, Leone M, De Sario R, Ruggieri V, Varraso L, Pansini N. Integration between the tele-cardiology unit and the central laboratory: methodological and clinical evaluation of point-of- care testing cardiac marker in the ambulance. Clin Chem Lab Med 2006;44:768–73. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgibbon F, Meenan BJ, Brown A, Dixon D. User perspectives of cardiac marker point-of-care testing for hospital-based chest pain diagnosis. Point of Care 2008;7:47–53. [Google Scholar]
- 13.Apple FS, Smith SW, Pearce LA, Murakami MM. Assessment of the multiple-biomarker approach for diagnosis of myocardial infarction in patients presenting with symptoms suggestive of acute coronary syndrome. Clin Chem 2009;55:93–100. [DOI] [PubMed] [Google Scholar]
- 14.Denny P, Hagen FK, Hardt M, Liao LJ, Yan WH, Arellanno M, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res 2008;7:1994–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siqueira WL, Salih E, Wan DL, Helmerhorst EJ, Oppenheim FG. Proteome of human minor salivary gland secretion. J Dent Res 2008;87:445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Streckfus CF, Bigler L, Dellinger T, Kuhn M, Chouinard N, Dai XL. The expression of the c-erbB-2 receptor protein in glandular salivary secretions. J Oral Pathol Med 2004;33:595–600. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res 2004;10: 8442–50. [DOI] [PubMed] [Google Scholar]
- 18.Herr AE, Hatch AV, Throckmorton DJ, Tran HM, Brennan JS, Giannobile WV, Singh AK. Microfluidic immunoassays as rapid saliva-based clinical diagnostics. Proc Nat Acad Sci 2007;104:5268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufman E, Lamster IB. The diagnostic application of saliva: a review. Crit Rev Oral Biol Med 2002;13:197–212. [DOI] [PubMed] [Google Scholar]
- 20.Mandel ID. Salivary diagnosis: more than a lick and a promise. J Am Dent Assoc 1993;124:85–7. [DOI] [PubMed] [Google Scholar]
- 21.Malamud D Saliva as a diagnostic fluid. Br Med J 1992;305:207–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavigne JJ, Savoy S, Clevenger MB, Ritchie JE, McDoniel B, Yoo SJ, et al. Solution-based analysis of multiple analytes by a sensor array: toward the development of an “electronic tongue.” J Am Chem Soc 1998;120:6429–30. [Google Scholar]
- 23.Goodey A, Lavigne JJ, Savoy SM, Rodriguez MD, Curey T, Tsao A, et al. Development of multianalyte sensor arrays composed of chemically derivatized polymeric microspheres localized in micromachined cavities. J Am Chem Soc 2001;123: 2559–70. [DOI] [PubMed] [Google Scholar]
- 24.Christodoulides N, Tran M, Floriano PN, Rodriguez M, Goodey A, Ali M, et al. A microchip-based multianalyte assay system for the assessment of cardiac risk. Anal Chem 2002;74:3030–6. [DOI] [PubMed] [Google Scholar]
- 25.Ali MF, Kirby R, Goodey AP, Rodriguez MD, Ellington AD, Neikirk DP, McDevitt JT. DNA hybridization and discrimination of single-nucleotide mismatches using chip-based microbead arrays. Anal Chem 2003;75:4732–9. [DOI] [PubMed] [Google Scholar]
- 26.Christodoulides N, Floriano PN, Acosta SA, Michael-Ballard K, Weigum SE, Dharshan P, et al. Towards the development of a lab-on-a-chip dual function white blood cell and C-reactive protein analysis method for the assessment of inflammation and cardiac risk. Clin Chem 2005;51:2391–5. [DOI] [PubMed] [Google Scholar]
- 27.Christodoulides N, Mohanty S, Miller CS, Langub MC, Floriano PN, Dharshan P, et al. Application of microchip assay system for the measurement of C-reactive protein in human saliva. Lab Chip 2005;5:261–9. [DOI] [PubMed] [Google Scholar]
- 28.Floriano PN, Christodoulides N, Romanovicz DK, Bernard B, Simmons GW, Cavell M, McDevitt JT. Membrane-based on-line optical analysis system for rapid detection of bacteria and spores. Biosens Bioelectron 2005;20:2079–88. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez WR, Christodoulides N, Floriano PN, Graham S, Mohanty S, Dixon M, et al. A microchip CD4 counting method for HIV monitoring in resource-poor settings. Plos Medicine 2005;2: 663–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christodoulides N, Dharshan P, Wong J, Floriano PN, Neikirk DP, McDevitt JT. A microchip-based assay for interleukin-6 In: Floriano PN, ed. Microchip-based assay systems, methods and applications. Vol. 385 Totowa (NJ): Humana Press; 2007. p 131–44. [DOI] [PubMed] [Google Scholar]
- 31.Christodoulides N, Floriano PN, Miller CS, Ebersole JL, Mohanty S, Dharshan P, et al. Lab-on-a-chip methods for point-of-care measurements of salivary biomarkers of periodontitis. Oral-Based Diagn 2007;1098:411–28. [DOI] [PubMed] [Google Scholar]
- 32.Weigum SE, Floriano PN, Christodoulides N, McDevitt JT. Cell-based sensor for analysis of EGFR biomarker expression in oral cancer. Lab Chip 2007;7:995. [DOI] [PubMed] [Google Scholar]
- 33.Apple FS, Wu AHB, Mair J, Ravkilde J, Panteghini M, Tate J, et al. Future biomarkers for detection of ischemia and risk stratification in acute coronary syndrome. Clin Chem 2005;51:810–24. [DOI] [PubMed] [Google Scholar]
- 34.Carreiro-Lewandowski E Update on cardiac biomarkers. Lab Med 2006;37:597–605. [Google Scholar]
- 35.Apple FS, Murakami MM, Pearce LA, Herzog CA. Multi-biomarker risk stratification of N-terminal pro-B-type natriuretic peptide, high-sensitivity C-reactive protein, and cardiac troponin T and I in end-stage renal disease for all-cause death. Clin Chem 2004;50:2279–85. [DOI] [PubMed] [Google Scholar]
- 36.Baldus S, Heeschen C, Meinertz T, Zeiher AM, Eiserich JP, Munzel T, et al. Myeloperoxidase serum levels predict risk in patients with acute coronary syndromes. Circulation 2003;108: 1440–5. [DOI] [PubMed] [Google Scholar]
- 37.Blomkalns AL, Gibler WB. Chest pain unit concept: rationale and diagnostic strategies. Cardiol Clin 2005;23:411–21. [DOI] [PubMed] [Google Scholar]
- 38.Sabatine MS, Morrow DA, de Lemos JA, Gibson CM, Murphy SA, Rifai N, et al. Multimarker approach to risk stratification in non-ST elevation acute coronary syndromes: simultaneous assessment of troponin I, C-reactive protein, and B-type natriuretic peptide. Circulation 2002;105:1760–3. [DOI] [PubMed] [Google Scholar]
- 39.Cohen J Monitoring treatment: at what cost? Science (Wash DC) 2004;304:1936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


