To the editor,
Driven mostly by the recent explosion of high-throughput technologies including sequencing, genotyping, and big data analytics, a vast amount of multi-omics and functional annotation data are accumulating for allergic diseases. Yet, these resources are scattered across various databases with different formats, and conducting manual search for genes associated with allergic diseases is a formidable task. As the accumulation of biomedical literature outpaces the ability of most researchers and clinicians to stay abreast of their fields, text mining, which involves automated information extraction by searching documents for text strings and analyzing their frequency and context, is critical. Hence, a comprehensive web-based one-stop bioinformatics tool is needed to efficiently query and retrieve research findings and draw novel hypotheses to understand the unique and shared association (or co-occurrence) of allergic diseases at gene, variant or pathway level. Here we describe AllergyGenDB to systematically generated allergy associated genes (or variants) from over 28 million PubMed biomedical literature citation, and from 93,892 GWAS Catalog and dbGaP SNPs resources,1,2 and distill their functional annotation information from Roadmap Epigenomics, GTEx, and ENCODE data. This accomplishes three things. First, it would serve to validate experiments by demonstrating that known genes occur as predicted. Second, it would rapidly highlight which genes are supported by the literature and which genes are novel in a given context. Third, it would lead novel hypotheses for understanding the unique and shared association (or co-occurrence) of allergic diseases at gene, variant or pathway level.
Disease co-occurrence or overlap at SNPs, genes, pathways and PPI network level and associated functional annotation were determined on-the-fly using Log of the Product of Frequency (LPF) measures.3 The LPF metric measures the co-occurrence of genes or variants and diseases taking into account the number of articles in which the disease and the gene (or variant) are mentioned together. LPF is calculated as follows:
where X is the number of abstracts including both gene (or variant) and disease term, G is the number of abstracts containing the gene (or variant), and T is the number of abstracts containing the disease term. If the LPF measure is zero, the gene or variant is highly associated with the disease and vice-versa. In addition, AllergyGenDB implements the Jaccard index to measure the similarity between diseases based on the number of shared/unique genes/SNPs/pathways.4 Jaccard index (J) is defined as:
where A and B are input gene, SNP or pathway lists; and 0 ≤ J(A, B) ≤ 1. The higher the Jaccard index, the higher similarity between diseases at gene, SNP or pathway level. The co-occurrence of allergic diseases at gene, variant, pathway and network level were determined using Jaccard index following the workflow shown in Fig E1.
AllergyGenDB has both “PubMed” and “Curated Database” portals, as shown in Fig 1. In both scenarios, the AllergyGenDB tool was constructed using disease name(s) followed by genes and variants query. In addition, “PubMed” portal supports custom search based on user-provided sets of literature PMIDs for a specific disorder.
Figure 1.
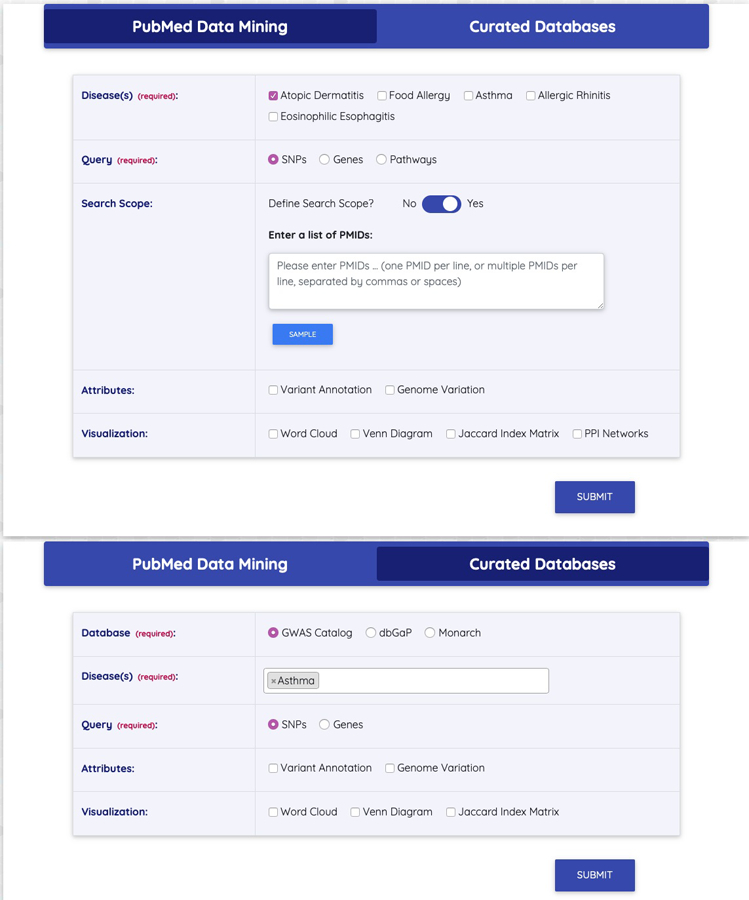
Layout of AllergyGenDB web tool homepage. AllergyGenDB provides two types of gene-disease and disease-disease co-occurrence extraction: PubMed data mining and curated databases (GWAS Catalog, dbGaP and Monarch Initiative).
Validation and use-cases:
To demonstrate and validate the usefulness of AllergygenDB, we analyzed the SNPs associated with atopic dermatitis (AD) using PubMed data (Fig E2). Our goal is to determine if the tool can successfully indicate that the top-ranked SNPs (based on PubMed search) are functionality relevant in AD. In this example, we found that the variants in FLG showed the highest co-occurrence with AD. Following this exploration, we showed that these variants have TSS >=0.5 and RegulomeDB <2, indicating their functional relevance in AD. Additional functional studies could investigate the effect of these variants, by mapping to TF, on gene expression. AD-related pathways and their respective enrichment results are shown in Fig E3. Gene word cloud for five major allergic diseases (asthma, atopic dermatitis, food allergy, allergic rhinitis and eosinophilic esophagitis) from literature mining is shown in Fig 2. We assigned gene word cloud weights based on the number of abstract counts. The genes/variants generated using AllergyGenDB were functionally annotated using multiple omics resources (Fig E4). SNP annotations include non-coding variants from RegulomeDB and GWAVA, and regulatory variant from RBP-Var and SNP2TFBS databases and links to VEP and Open Targets Genetics. We annotated genes for eQTL (from GTEx) and gene orthologs in model organisms. Links are provided for multi-omics databases including Ensembl, ENCODE, ClinVar and Roadmap Epigenomics.
Figure 2.

Gene word cloud summary of the five major allergic diseases (asthma, atopic dermatitis, food allergy, allergic rhinitis and eosinophilic esophagitis). Gene word cloud weight was assigned based on the number of abstract counts these genes appeared in literature. Font size is scaled proportional to the weight. Gene word cloud was constructed using D3.js package ‘d3-cloud’.
AllegryGenDB can also be used to explore the shared genes, variants, or pathways among multiple allergic diseases (asthma, atopic dermatitis, food allergy, allergic rhinitis, and eosinophilic esophagitis) in both PubMed and GWAS Catalog. For PubMed mining, there are 3635, 1188, 879, 316, and 76 genes in asthma, atopic dermatitis, allergic rhinitis, food allergy and eosinophilic esophagitis, respectively. The overlapping results (Fig E5A) show that these five diseases have 47 genes in common. Food allergy and eosinophilic esophagitis share 100% genes with asthma, while atopic dermatitis and allergic rhinitis share 88.7% (780/879) and 77.7% (923/1188) of genes with asthma. The Jaccard similarity matrix (Fig E5B) for the five allergic diseases shows that AD and allergic rhinitis genes have the highest Jaccard similarity score of 0.329. The overlap and Jaccard similarity matrix obtained from GWAS Catalog are shown in Fig E5C and Fig E5D, respectively. An interactive visual analysis of the protein-protein interaction networks related to the disease-associated genes is shown in Fig E6. By showing the similarities of genes across different allergic diseases, AllegryGenDB can reveal the shared genetic etiologies. This paves the way to build a combination of shared treatment regimen for allergic diseases based on a set of genetic functions or mutations.
In conclusion, AllergyGenDB is a unique tool to retrieve genes and variants implicated in allergic diseases that would help researchers to validate and support experimental results from literature and develop a new hypothesis. To our knowledge, AllergyGenDB is the first comprehensive omics database tool to identify genes and variants from PubMed or curated database searches that are highly associated with allergic diseases, and rank gene signatures with functional annotations to enable quick and comprehensive visualization. It is developed to provide allergy researchers with an up-to-date online literature mining capability and visualization in a single portal. It is a user-friendly tool with a point-and-click interface and can be accessed freely at https://research.cchmc.org/mershalab/AllergyGenDB/login.php. Hyperlinks are added within the spreadsheet to enable instant review of the genes or PubMed IDs of interest.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grant R01HL132344.
Footnotes
Conflicts of Interest Statement
The authors have no conflicts of interest to declare in relation to this publication.
References
- 1.Chen Y, Zhang X, Zhang GQ, Xu R. Comparative analysis of a novel disease phenotype network based on clinical manifestations. J Biomed Inform. 2015;53:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacArthur J, Bowler E, Cerezo M, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45(D1):D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu YH, Hines LM, Weng HF, et al. Analysis of genomic and proteomic data using advanced literature mining. J Proteome Res. 2003;2(4):405–412. [DOI] [PubMed] [Google Scholar]
- 4.Oerton E, Roberts I, Lewis PSH, Guilliams T, Bender A. Understanding and predicting disease relationships through similarity fusion. Bioinformatics. 2019;35(7):1213–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


