Abstract
The interaction of human recombinant DNA topoisomerase 1 (Top1) with linear and circular DNA structures containing a nick or short gap but lacking a specific Top1 recognition site was studied. The effect of key excision repair proteins on formation of the Top1 covalent adduct with the DNA repair intermediates was shown. Partial inhibition of the Top1—DNA-adduct formation upon addition of poly(ADP-ribose) polymerase 1 in the absence of NAD+ was shown, whereas in the presence of NAD+ formation of a high molecular weight product, most likely corresponding to poly(ADP)-ribosylated Top1—DNA adduct, was observed. The data show that the key base excision repair proteins can influence formation of suicide Top1—DNA adducts. Top1 was identified by immunoprecipitation in the bovine testis nuclear extract as the protein forming the main modification product with nick-containing DNA.
Keywords: DNA topoisomerase 1, DNA repair, base excision repair factors
DNA topoisomerases (including topoisomerase 1 (Top1)) are widespread enzymes involved in different cell processes such as DNA replication, transcription, and recombination [1–3]. Topoisomerases are necessary for solution of different topological problems emerging upon replication and transcription of circular closed DNA and influence the rate of these processes. Topoisomerases also provide for maintenance of a certain level of intracellular DNA supercoiling. Topi relaxes supercoiled structures in chromosomal DNA during replication and transcription by creating a nick. Topoisomerases use a tyrosine catalytic residue for nucleophilic attack and disruption of a phosphodiester bond in DNA. Topi establishes a covalent bond with the DNA 3′ end, and in this case hydroxyl is formed at the 5′ end of the break. Such intermediate is usually considered as the Top1—DNA cleavage complex [1–3]. After translocation of cleaved DNA ends relative to each other, Topi restores DNA integrity by catalyzing 5′ hydroxyl ligation in the broken DNA strand. The Top1—DNA cleavage complex creates DNA damage hampering replication and transcription [4–6]. The anticancer drug camptothecin binds covalent DNA—Top1 complex and thus blocks religation of the cleaved DNA strand [7]. Along with camptothecin, many DNA adducts and DNA damages like uracil, 8-oxoguanine, mismatch paired bases, or breaks in DNA located close to the Topi recognition site are able to stabilize the Top1—DNA cleavage complex [8–11]. All these processes can result in incorrect recombination and finally in cell death [12–14].
We showed previously that Topi from the yeast Saccharomyces cerevisiae can interact with DNA structures not containing specific recognition sites for the enzyme [15]. The only necessary condition for formation of Topi covalent adducts with DNA was the presence in one DNA strand of breaks or short gaps. No such products emerged in the case of DNA containing apurinic/apyrimidinic sites (AP sites). However, cleavage of an AP site-containing structures by apurinic/apyrimidinic endonuclease 1 (APE1) resulted in formation of covalent adducts of Top1 with DNA Such products were initially found in S. cerevisiae extracts, and later their existence was confirmed for purified yeast and human Topi [16]. DNA structures containing breaks and gaps in one DNA strand are characteristic intermediates or substrates of DNA repair. This suggests that under certain conditions Top1 can bind such DNA structures by competing with proteins involved in their repair. This might result in inhibition or retardation of DNA repair processes. In this connection, the mechanism of formation of Top1 covalent adducts with DNA structures modeling DNA repair intermediates and the interaction of Top1 with proteins involved in DNA repair is of great interest.
The goal of this work was to investigate the interaction of human Top1 with DNA intermediates of base excision repair, free of Top1-specific sites but having nicks or gaps with different 3′- and 5′-terminal structures, as well as to study the effect of base excision repair proteins on formation of Top1 covalent adduct with DNA.
MATERIALS AND METHODS
The following reagents were used in this work: T4 polynucleotide kinase (5000 U/ml), dNTP (SibEnzyme, Russia), [γ-32P]ATP with specific activity 80 MBq/mol, [α-32P]ATP with specific activity 80 MBq/mmol (Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences), benzonase (Merck, USA), Rainbow molecular mass markers and MicroSpin™ G25 columns (Amersham, USA), dimethyl sulfoxide (DMSO), reagents for electrophoreses and main buffer components (Sigma, USA), polyclonal antibodies to Top1 (Santa Cruz Biotechnology, USA). Other reagents and buffer components from Russian sources were especially pure or pure for analysis.
Recombinant poly(ADP-ribose) polymerase 1 (PARP1) was the courtesy of M. V. Sukhanova (Institute of Chemical Biology and Fundamental Medicine), DNA of M13mp19 bacteriophage was the courtesy of N. D. Gaiko (Institute of Chemical Biology and Fundamental Medicine), human Topi mutant (Y723F) was the courtesy of M. A. Bjornsti (St. Jude Children’s Research Hospital, Memphis, USA), recombinant APE1, uracilDNAglycosylase, and Stoffel fragment of Taq DNA polymerase were the courtesy of S. N. Khodyreva (Institute of Chemical Biology and Fundamental Medicine), and nuclear extract of bovine testis was the courtesy of R. Prasad (National Institute of Environmental Health Sciences, USA).
Synthetic oligonucleotides used in this work were synthesized by Sigma or in the Institute of Chemical Biology and Fundamental Medicine.
To design DNA duplexes based on phage M13mp19 DNA, the following oligonucleotides were used:
P1, 5′-ATGCAAGCTTGGCGTAATCA-3′, P2, 5′-CCAACGCTCAACAGTAGGGC-3′, P3, 5′-ATTCGGTCGCTGAGGCTTGC-3′.
For design of a single nucleotide gap, the following oligonucleotides were used:
5′-CGGTATCCACCAGGTCTG-3′, 5′-pGACAACGATGAAGCC-3′, 5′-GGCTTCATCGTTGTCGCAGACCTGGTGGAT-ACCG-3′.
A nick was constructed using:
5′-CGGTATCCACCAGGTCTGC-3′, 5′-pGACAACGATGAAGCC-3′, 5′-pFGACAACGATGAAGCC-3′, 5′-OH-GACAACGATGAAGCC-3′, 5′-GGCTTCATCGTTGTCGCAGACCTGGTGGAT- ACCG-3′.
A duplex with U—G in the middle of the strand was constructed using:
5′-GGCTTCATCGTTGTCUCAGACCTGGTGGAT-ACCG-3′, 5′-CGGTATCCACCAGGTCTGGGACAACGAT-GAAGCC-3′.
Preparation of radiolabeled 5′-32P primers
Radiolabel was introduced into the primer 5′-end using T4 polynucleotide kinase as described in [17]. Excess [γ32P]ATP was removed using a MicroSpin™ G25 according to the manufacturer’s protocol. Complementary oligonucleotides were mixed in equimolar amounts and annealed for 3 min at 95°C followed by slow cooling to room temperature.
Modification of Top1 by different DNA structures
Reaction mixtures (10 μl) contained 50 mM Tris-HCl, pH 8.6, 50 mM NaCl, 5 mM MgCl2, 0.2 μM 5′-32P-labeled DNA duplex, and 100 nM hTopl. The reaction mixtures were incubated for 30 min at 37°C. The reaction was stopped by addition of Laemmli buffer and heating for 5 min at 95°C. Reaction products were analyzed as described by Laemmli [18] and visualized by autoradiography using a Molecular Imager FX (BioRad, USA).
DNA was cleaved by Top1
in 20 μl reaction mixture containing 50 mM Tris-HCl, pH 8.6, 50 mM NaCl, 5 mM MgCl2, 0.2 μM 5′-32P-labeled DNA duplex, and 100 nM hTopl. After incubation for 30 min at 37°C, an equal volume of saturated ammonium sulfate solution was added and the final mixture was left overnight at 4°C. The precipitate was collected by centrifugation, washed by (NH4)2SO4 solution (50% saturation), and dissolved in 20 μl buffer (20 mM Tris-HCl, pH 7.6, 5 mM MgCl2). Proteinase K was added to final concentration 250 μg/ml and the resulting mixture was incubated for 20 h at 37°C. DNA fragments were separated by electrophoresis in 20% polyacrylamide gel under denaturing conditions [17]. Aliquots of reaction mixture before and after treatment by proteinase K were analyzed by electrophoresis according to Laemmli [18].
Identification of bovine testis nuclear extract protein modification products
Reaction mixtures (100 μl) containing 0.2 μM 5′-32P-labeled DNA duplex, nuclear extract proteins (protein concentrations 5–6 mg/ml), and standard buffer components were incubated for 30 min at 37°C. An aliquot of 10 μl was taken, and loading buffer was added. To the remainder 5 μg Top1-specific antibodies or preimmune serum (40 μg/ml) was added. The mixture was incubated for 4 h at 4°C, then 50 μl of Protein G-agarose was added, and the mixture was incubated under mixing for 1 h at 4°C. After centrifugation for 20 min at 14,000 rpm, the supernatant and the agarose-bound protein were removed, and the pellet was washed four times by buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40) with addition of antiprotease cocktail. After the last washing, 50 μl of Laemmli buffer was added to the pellet, and then the mixture was heated for 5 min at 95°C for desorption of protein-bound antibodies. Reaction products were analyzed according to Laemmli [18] and visualized by autoradiography using the Molecular Imager FX.
Preparation of duplexes based on M13mp19 phage DNA
To obtain DNA duplexes, initiating primers (P1. P2, P3) and template (DNA of M13mpl9 phage) were mixed at molar ratio 1:1 with concentrations 0.78 μM in reaction mixture of 65 μl. The sample was placed in a thermostat heated to 95°C and allowed to cool slowly to room temperature. The reaction mixture (100 μl) contained 0.27 μM DNA duplex, 200 units of Stoffel fragment of Taq polymerase, 0.3 mCi [α-32P]ATP, 20 μM dATP, 20 μM dCTP, 20 μM dGTP (total concentration of dTTP and dUTP was 20 μM), and the following standard components: 50 mM Tris-HCl, pH 8.6, 10 mM MgCl2, and 50 mM NaCl. Synthesis was carried out for 60 min at 65 °C and then stopped by addition of EDTAto final concentration 40 mM. The DNA was precipitated by 2.5 volumes of cold 100% ethanol, 0.3 M KOAc, pH 5.5 (kept at −40°C for 40 min). The precipitate was washed with 75% ethanol, air dried at room temperature, and dissolved in 30 μl TE buffer (10 mM Tris-HCl, pH 7.8, 1 mM EDTA). The DNA was separated from low molecular weight components by gel filtration on “spin columns” at 4000 rpm for 2 min in a MiniSpin centrifuge (Eppendorf, Germany).
RESULTS AND DISCUSSION
Modification of human Top1 and its mutant form by different DNA structures
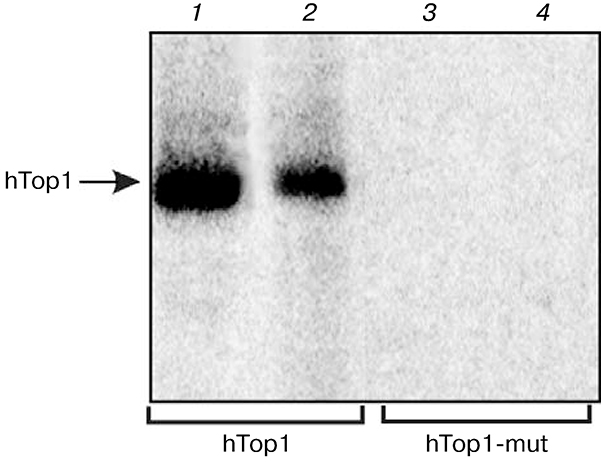
In previous works [15,16] modification of purified human and yeast Top1 by DNA structures containing nicks or short gaps but devoid of specific sequences recognized by Top1 was demonstrated. As is known, Top1 relaxes supercoiled structures in DNA, thus creating a nick. This process involves the catalytic tyrosine residue necessary for nucleophilic attack and disruption of a phosphodiester bond in DNA. To determine whether the tyrosine residue in the Top1 active center takes part in formation of covalent adducts with DNA structures devoid of specific recognition site, we used the enzyme mutant form in which tyrosine in the active center is replaced by phenylalanine (Y723F). Figure 1 shows data on modification of the wild-type hTop1 (1, 2) and its mutant form (hTop1-mut) by DNA structures containing a nick and a single nucleotide gap. As expected, in the absence of tyrosine in the enzyme active center no formation of Top1 covalent adducts with DNA structures was observed (3, 4). Thus, Top1 covalent adducts with DNA containing a single-stranded break or a short gap in the absence of a recognition site are also formed with involvement of the catalytic tyrosine residue in the enzyme active center.
Fig. 1.
Modification of human Topi and its mutant form by different DNA structures. Radiolabeled DNA structures containing a break (1, 3) or a single nucleotide gap (2, 4) were incubated with Top1 (1, 2) or with Topi-mut (3, 4) as described in “Materials and Methods”. Modification products were analyzed in SDS-PAGE.
DNA containing single-stranded breaks (nicks) and short gaps with different 3′ and 5′ terminus modifications are formed during DNA repair. In particular, such DNA structures are intermediates of base excision repair and also emerge in response to ionizing radiation.
Our study has shown that the intensity of human and yeast Top1 modification was independent of the 3′ and 5′ terminus structure of the break [16]. Since the structure of the break termini including the 3′-terminal phosphate group did not influence the efficiency of formation of the Top1 covalent adduct with DNA, we supposed that a covalent bond is formed at a 3′ terminus of a break formed by the enzyme rather than at the 3′ terminus of the already existing break. It was shown previously that a break or a short gap in certain positions within or close to the recognized sequence can result in inhibition of DNA cleavage by Top1 at the canonical site and generation of additional breaks near this site [9]. It was interesting to determine the character of DNA cleavage in the absence of a recognition sequence. To do this, covalent hTop1 adducts with 5′-32P-labeled primer, the 3′ terminus of which forms a nick, were isolated from the reaction mixture by precipitation with ammonium sulfate. In this case, free DNA remained in solution. The isolated covalent Top1—DNA complex was treated by proteinase K, and DNA products were analyzed by electrophoresis in denaturing polyacrylamide gel.
The results have shown the existence of several DNA products exhibiting lower electrophoretic mobility compared to the initial radiolabeled oligonucleotide (data not shown). Most likely, these products correspond to fragments formed upon DNA cleavage at different positions near the 3′ end of the nick-forming primer. It can also be claimed that the 3′ terminal nucleotide is not involved in formation of the covalent adduct with Top1, because in the case of radiolabel introduction into the 3′ end of the nick no products of covalent DNA attachment to Top1 were observed, similarly to the case using a DNA structure radiolabeled at the 5′ end of the nick. Thus, Top1 cleaves a phosphodiester bond near the 3′ end of the nick and forms a covalent bond with the 3′ end of the newly formed break.
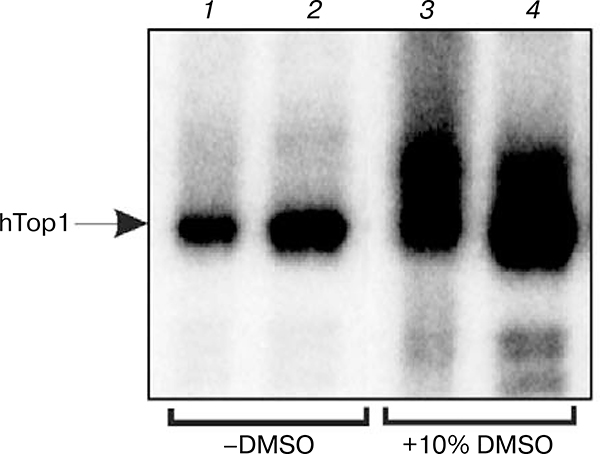
It was shown earlier that the activity of some enzymes increases in the presence of DMSO in low concentrations (5–15%) [19, 20]. To investigate the influence of DMSO on the efficiency of formation of Topi covalent adducts with DNA containing single-stranded breaks and mononucleotide gaps, levels of enzyme modification in the absence and presence of 10% DMSO were compared (Fig. 2). It was shown that in the presence of DMSO the level of Top1 modification increases 2–3-fold (cf. 1, 2 with 3, 4). The effect of DMSO on the enzyme activity can be due to alteration of protein structure in the aqueous-organic medium, which can result in enhancement of its conformational mobility and making reaction easier.
Fig. 2.
Modification of human Top1 in the presence and absence of DMSO. Radiolabeled break-containing DNA structures were incubated with Top1 without DMSO (1, 2) and in the presence of 10% DMSO (3, 4) as described in “Materials and Methods”. Modification products were analyzed by SDS-PAGE.
Effect of PARP1 on Top1 modification efficiency
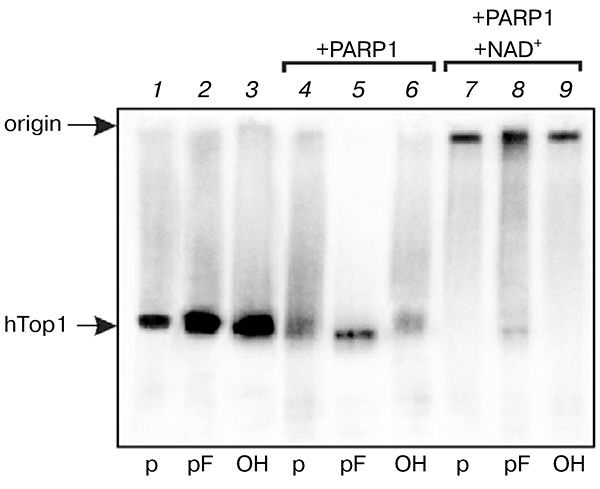
Since single-stranded breaks and gaps emerge during DNA repair processes, especially those associated with base lesions, then Top1 is a potential competitor for proteins carrying out DNA repair, and it should be removed from 3′ end of damaged DNA for following efficient repair, or a complex of repair proteins should prevent the interaction of Top1 with breaks. PARP1 is among the most important proteins that recognize breaks in DNA and coordinate repair processes. Upon interaction with DNA structures containing breaks, PARP1 catalyzes synthesis of poly(ADP)-ribose using NAD+ as a substrate [21, 22]. PARP1 catalyzes both its own poly(ADP)-ribosylation and ribosylation of other proteins involved in interaction with DNA breaks. It was found that PARP1 in the presence of NAD+ removes Top1 from its covalent adducts with DNA formed on DNA containing specific Top1 recognition sites [23]. However, it is not known whether Top1 removal from covalent adducts is due to formation of a negatively charged poly(ADP)-ribose polymer and Top1 ribosylation, or protein—protein interactions of Top1 with PARP1 play a role in this process. It was interesting to elucidate the effect of PARP1 on the stability of Top1—DNA complexes in the presence and absence of NAD+. Purified PARP1 was added to reaction mixtures simultaneously with Top1 or after formation of Top1 covalent complexes with DNA duplexes containing a break with different 5′ ends (phosphate, furan phosphate, or hydroxyl). In all cases partial lowering the level of the Top1—DNA adduct formation in the absence of NAD+ was observed (lanes 4 and 6 in Fig. 3). When PARP1 was added in the presence of NAD+, formation of a high molecular weight product was observed, most likely corresponding to poly(ADP)-ribosylated Top1—DNA adduct (lanes 7–9).
Fig. 3.
Effect of PARP1 on level of human Top1 modification. Radiolabeled DNA structures containing a break with different 5′ ends were incubated with Top1 {1–3). After formation of the Top1—DNA covalent complex, purified PARP1 was added in the absence (4–6) or presence of NAD+ (7–9).
It was shown previously that Top1 covalently joined to DNA can be poly(ADP)-ribosylated by PARP1 [24, 25]. The competition of DNA polymerase β and flapendonuclease 1 with human Top1 for binding to the break-containing DNA structure was also found (data not shown). The results show that key proteins of the base excision repair can influence formation of a suicidal Top1—DNA adduct. This means that efficient functioning of the DNA repair complex hinders formation of Top1 suicidal complexes with DNA breaks, whereas defects in the base repair system can result in emergence of free DNA breaks, potential targets for Top1. Especially severe consequences can be caused by inhibition of the main nick sensor in higher eukaryotic cells — PARP1. Top1—DNA adducts can be formed in cells undergoing intensive influence of damaging agents and cause genetic instability.
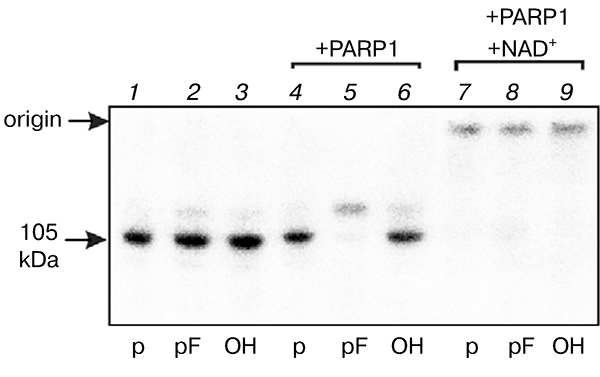
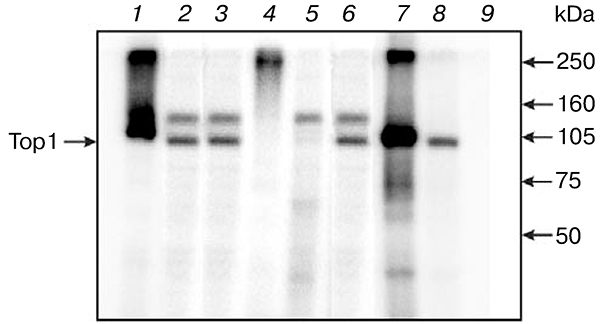
As noted earlier, initially products of Top1 modification by DNA structures having no specific sites for recognition by this enzyme but containing a nick or a short gap were detected in extracts of yeasts known to have no analogs of nick-sensor proteins such as PARP1 in higher eukaryotes. To reveal the possibility of Top1 modification in the higher eukaryote cell extracts, we carried out modification of proteins of bovine testis nuclear extract by different DNA structures. Like in the case of S. cerevisiae extract, the use of nick-containing DNA revealed a modification product of ~ 105 kDa corresponding by its electrophoretic mobility to Topi (Fig. 4). Addition of purified PARP1 to the extract resulted in decreased level of Top1 covalent adduct with DNA, especially pronounced in the case of DNA duplex containing a tetrahydrofuran phosphate residue at the 5′ end of single-stranded break. In this case PARP1 modification was also observed (lane 5 in Fig. 4). Addition of PARP1 to nuclear extract in the presence of NAD+ results in appearance of modification products with low electrophoretic mobility due to (ADP)-ribosylation of the protein.
Fig. 4.
Effect of PARP1 on level of modification of bovine testis nuclear extract proteins. Radiolabeled DNA structures containing a break with different 5′ ends were incubated with the nuclear extract proteins (1–3). After formation of covalent Top1—DNA complex, purified PARP1 was added in the absence (4–6) or presence of NAD+ (7–9).
Identification of Top1 in nuclear extracts
To identify the protein forming the main modification product in the bovine testis nuclear extract with nick-containing DNA, we used an immunoprecipitation technique that allows target protein adsorption on Protein G-agarose in the form of a complex with specific antibodies (Fig. 5). No binding to the resin of labeled nuclear extract products was observed when preimmune serum was used (9), whereas in the case of specific antibodies modification products were bound to the resin (7, 8). The data indicate that the nuclear extract protein of ~ 105 kDa corresponds to the Top1 modification product.
Fig. 5.
Identification of products of modification of bovine testis nuclear extract proteins by break-containing DNA structure. After incubation of extract proteins with radiolabeled DNA structure, the reaction mixture was divided into two parts. One was incubated in the presence of antibodies to Top1, while the other part was incubated with preimmune serum. Then Protein G-agarose was added. The resin-bound protein was eluted with hot loading buffer containing 2% SDS. For control, similar experiments were carried out with purified human Top1 (1, 4, 7). Lanes: 1–3) products of reaction mixture separation; 7–9) products obtained by separation of reaction mixture after elution of preimmune serum containing samples (9) and antibodies to Top1 (7, 8); 4–6) products of separation of proteins not bound to the resin.
To increase selectivity of Top1 modification in cell and nuclear extracts by the break- and gap-containing DNA duplexes, circular DNA containing AP sites statistically distributed in one strand were constructed and used. Extended DNA mimicking intermediates of base excision repair, like those containing statistically distributed AP sites or breaks in the sugar-phosphate backbone, seem a convenient tool for solution of a number of prob lems. Long DNAs imitate natural structures to a higher extent compared to short DNA duplexes. Another advantage of long DNA over short DNA duplexes is the decreased concentration of DNA termini in the reaction mixture. This is important in investigation in cell extracts, because the latter contain a great number of proteins exhibiting high affinity to terminal DNA regions (such as PARP1, Ku antigen) [26–28]. Their efficient binding can prevent interaction with DNA of other proteins of the extract, in particular of Top1, due to competition, and thus to lower yields of covalent adducts with target proteins. Thus, it was shown that Ku-antigen binding in HeLa cell extract with DNA substrates of base excision repair inhibits this reaction pathway [29].
Extended DNA mimicking intermediates of base excision repair were obtained on the basis of circular ssDNA of M13mpl9 phage. Preliminary analysis showed the absence of specific Top1 recognition sites from the M13mpl9 phage DNA sequences. Synthesis was carried out using three primers in order to increase the completeness of single-stranded DNA transformation to double-stranded and thus to exclude the probability of emergence of single-stranded regions. The M13mpl9 DNA regions complementary to P1, P2, and P3 regions were at a distance of 2345–2470 bases from each other. DNA was synthesized using the Stoffel fragment of Taq DNA polymerase. Besides natural deoxynucleoside 5′-triphos-phates, dUTP and [α-32P]dATP were introduced into the system, which made it possible to obtain DNA with statistically distributed dUMP and [α-32P]dAMP residues in the same strand.
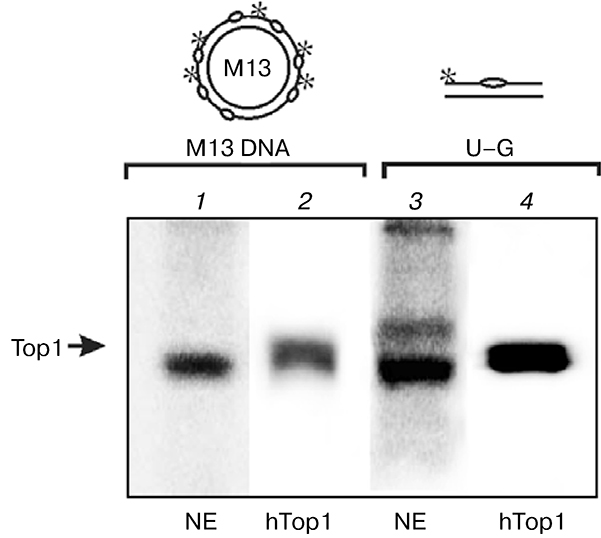
DNA structures with breaks were obtained on the basis of synthesized double-stranded circular DNA using purified enzymes or endogenous enzymes of the bovine testis nuclear extract. Statistically distributed uracil residues were eliminated using uracil-DNA glycosylase, which resulted in formation of AP sites in positions corresponding to uracils. Single-stranded breaks were formed in response to purified APE1 or APE of cell extracts. So the obtained double-stranded circular DNAs with single-stranded breaks were used as probes for covalent bridging of the bovine testis nuclear extract proteins or recombinant human Top1 as control. Besides circular M13mpl9 phage DNA, the 34-mer DNA duplex containing uracil in position 16 of one of the duplex strands (U—G) was used for comparison as damaged DNA structure. Covalent joining of a sufficiently large circular DNA sharply decreases electrophoretic mobility of DNA—protein adducts; therefore, samples containing phage DNA were treated with benzonase before application on the gel. As seen in Fig. 6, in the case of nuclear extract protein modification using M13mpl9 DNA is more selective than that by short oligonucleotide (cf. 1 and 3). Most likely, PARP1 is also modified when the 34-mer DNA duplex is used.
Fig. 6.
Increase in selectivity of Topi modification in nuclear extract using M13mpl9 phage DNA. Radiolabeled M13mpl9 phage DNA (1, 2) or 34-mer DNA duplex (3, 4) were incubated with nuclear extract (NE) proteins (1, 3) or with Top1 (2, 4). Modification products were analyzed by SDS-PAGE.
Thus, the substrate-mimicking DNA or base excision repair intermediates can be used for Top1 modification in eukaryotic cell extracts. Moreover, estimation of the level of this enzyme modification in different cell extracts using break-containing DNA can be useful for prediction of the conditions of repair systems in these cells.
Acknowledgments
This work was supported by the Russian Foundation for Basic Research (grants 07-04-00389 and 09-0491320), HRJRG-102, the Russian Academy of Sciences program “Molecular and Cell Biology”, program “Leading Research Schools” (652.2008), FTsPK (State contract 2009-1.1-201-018-002), and CRDF-BRHE (RNP.2.2.2.3.10031).
Abbreviations
- AP site
apurinic/apyrimidinic site
- APE1
apurinic/apyrimidinic endonuclease 1
- DMSO
dimethyl sulfoxide
- PARP1
poly(ADP-ribose) polymerase 1
- Top1
DNA topoisomerase 1
Footnotes
The authors are grateful to Prof. S. Boiteux for useful advice and discussion of results.
REFERENCES
- 1.Chen AY, and Liu L E (1994) Annu. Rev. Pharmacol. Toxicol, 34, 191–218. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC (1996) Annu. Rev. Biochem, 65, 635–692. [DOI] [PubMed] [Google Scholar]
- 3.Wang JC (2002) Nat. Rev. Mol. Cell. Biol, 3, 430–440. [DOI] [PubMed] [Google Scholar]
- 4.Holm C, Covey JM, Kerrigan D, and Pommier Y (1989) Cancer Res, 49, 6365–6368. [PubMed] [Google Scholar]
- 5.Hsiang YH, Lihou MG, and Liu LF (1989) Cancer Res, 49, 5077–5082. [PubMed] [Google Scholar]
- 6.Wu J, and Liu LF (1997) Nucleic Acids Res, 25, 4181–4186.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svejstrup JQ, Christiansen K, Gromova II, Andersen AH, and Westergaard O (1991) J. Mol. Biol, 222, 669–678. [DOI] [PubMed] [Google Scholar]
- 8.Pourquier P, Ueng L-M, Kohlhagen G, Mazumder A, Gupta M, Kohn KW, and Pommier Y (1997) J. Biol. Chem, 272, 7792–7796. [DOI] [PubMed] [Google Scholar]
- 9.Pourquier R, Pilon AA, Kohlhagen G, Mazumder A, Sharma A, and Pommier Y (1997) J. Biol. Chem, 272, 26441–26447. [DOI] [PubMed] [Google Scholar]
- 10.Pourquier P, Ueng L-M, Fertala J, Wang D, Park H, Essigman JM, Bjornsti MA, and Pommier Y (1999) J. Biol. Chem, 274, 8516–8523. [DOI] [PubMed] [Google Scholar]
- 11.Cline SD, and Hanawalt PC (2003) Nat. Rev. Mol. Cell. Biol, 4, 361–372. [DOI] [PubMed] [Google Scholar]
- 12.Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, Zhang H, and Kohn KW (2003) Mutat. Res, 532, 173–203. [DOI] [PubMed] [Google Scholar]
- 13.Pommier Y (2006) Nat. Rev. Cancer, 6, 789–802. [DOI] [PubMed] [Google Scholar]
- 14.Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, Agama K, Nitiss JL, and Redon C (2006) Prog. Nucleic Acid Res. Mol. Biol, 81, 179–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebedeva N, Auffret van der Kemp P, Bjornsti MA, Lavrik O, and Boiteux S (2006) DNA Repair, 5, 799–809. [DOI] [PubMed] [Google Scholar]
- 16.Lebedeva N, Rechkunova N,. Boiteux S, and Lavrik O (2008) IUBMB Life, 60, 130–134. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch EF, and Maniatis T (1989) Molecular Cloning, Cold Spring Harbor Laboratory Press, Cold Spring Flarbor, N. Y. [Google Scholar]
- 18.Laemmli UK (1970) Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 19.Kujo C, and Ohshima T (1998)Appl. Environ. Microbiol, 64,2152–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lonhienne T, Gerday C, and Feller G (2000) Biochim. Biophys. Acta, 1543, 1–10. [DOI] [PubMed] [Google Scholar]
- 21.Shall S, and de Murcia G (2000) Mutat. Res, 460, 1–15. [DOI] [PubMed] [Google Scholar]
- 22.Lindahl T, Satoh MS, Poirier GG, and Klungland A (1995) Trends Biochem. Sci, 20, 405–411. [DOI] [PubMed] [Google Scholar]
- 23.Malanga M, and Althaus FR (2004) J. Biol. Chem, 279, 5244–5248. [DOI] [PubMed] [Google Scholar]
- 24.Ferro AM, Higgins NP, and Olivera BM (1983) J. Biol. Chem, 258, 6000–6003. [PubMed] [Google Scholar]
- 25.Yung TM, Sato S, and Satoh MS (2004) J. Biol. Chem, 279, 39686–39696. [DOI] [PubMed] [Google Scholar]
- 26.Ame JC, Spenlehauer C, and de Murcia G (2004) Bioessays, 26, 882–893. [DOI] [PubMed] [Google Scholar]
- 27.Doherty AJ, and Jackson SP (2001) Curr. Biol, 11, R920–R924. [DOI] [PubMed] [Google Scholar]
- 28.Ilina ES, Lavrik OI, and Khodyreva SN (2008) Biochim. Biophys. Acta, 1784, 1777–1785. [DOI] [PubMed] [Google Scholar]
- 29.Frit R, Calsou P, Chen DJ, and Salles B (1998) Mol. Biol, 4, 963–973. [DOI] [PubMed] [Google Scholar]