Abstract
Fungi are key players in vital ecosystem services, spanning carbon cycling, decomposition, symbiotic associations with cultivated and wild plants and pathogenicity. The high importance of fungi in ecosystem processes contrasts with the incompleteness of our understanding of the patterns of fungal biogeography and the environmental factors that drive those patterns. To reduce this gap of knowledge, we collected and validated data published on the composition of soil fungal communities in terrestrial environments including soil and plant-associated habitats and made them publicly accessible through a user interface at https://globalfungi.com. The GlobalFungi database contains over 600 million observations of fungal sequences across > 17 000 samples with geographical locations and additional metadata contained in 178 original studies with millions of unique nucleotide sequences (sequence variants) of the fungal internal transcribed spacers (ITS) 1 and 2 representing fungal species and genera. The study represents the most comprehensive atlas of global fungal distribution, and it is framed in such a way that third-party data addition is possible.
Subject terms: Fungal ecology, Biogeography
| Measurement(s) | internal_transcribed_spacer_region |
| Technology Type(s) | digital curation |
| Factor Type(s) | geographic location • sample type • biome • sampling year |
| Sample Characteristic - Organism | Fungi |
| Sample Characteristic - Environment | terrestrial biome |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.12481946
Background & Summary
Fungi play fundamental roles in the ecosystem processes across all terrestrial biomes. As plant symbionts, pathogens or major decomposers of organic matter they substantially influence plant primary production, carbon mineralization and sequestration, and act as crucial regulators of the soil carbon balance1,2. The activities of fungal communities contribute to the production of clean water, food, and air and the suppression of disease-causing soil organisms. Soil fungal biodiversity is thus increasingly recognized to provide services critical to food safety and human health3.
It is of high importance to determine how environmental factors affect the diversity and distribution of fungal communities. So far, only a few studies have focused on fungal distribution and diversity on global scale4–6. Importantly, these single survey studies focused either on a limited number of biomes4,5, fairly narrow groups within the fungal kingdom6, or were restricted only to fungi inhabiting soil. Although individual studies have the advantage of standardized methodology across their whole dataset, their limitation is in the limited sampling efforts in space and time that do not allow general conclusions on distribution of fungal taxa. On the other hand, since the advent of high-throughput-sequencing methods, large amounts of sequencing data on fungi from terrestrial environments accumulated along with metadata across numerous studies and allow interesting analyses when combined7. As an example of this approach, the meta-analysis of 36 papers made it possible to map global diversity of soil fungi collected in >3000 samples and indicated that climate is an important factor for the global distribution of soil fungi8. This approach clearly demonstrated the utility of a meta-study approach to address fungal biogeography, ecology and diversity. In addition, the compilation of these data demonstrated the fact that symbiotic mycorrhizal fungi that aid cultivated and wild plants to access nutrients, are more likely to be affected by rapid changes of climate than other guilds of fungi, including plant pathogens8 and helped to identify which fungi tend to follow alien plants invading new environments9.
Here, we have undertaken a comprehensive collection and validation of data published on the composition of fungal communities in terrestrial environments including soil and plant-associated habitats. This approach enabled us to construct the GlobalFungi database containing, on March 16, 2020, over 110 million unique sequence variants10 (i.e., unique nucleotide sequences) of the fungal nuclear ribosomal internal transcribed spacers (ITS) 1 and 2, covering > 17 000 samples contained in 178 original studies (Fig. 1). The ITS region has been used as molecular marker because it is a universal barcode for fungi11.The dataset of sequence variant frequencies across samples, accompanied by metadata retrieved from published papers and in global climate databases is made publicly available at https://globalfungi.com. To achieve the goal to make published data findable, accessible, interoperable and reusable, the user interface at the above address allows the users to search for individual sequences, fungal species hypotheses12, species or genera, to get a visual representation of their distribution in the environment and to access and download sequence data and metadata. In addition, the user interface also allows authors to submit data from studies not yet covered and in this way to help to build the resource for the community of researchers in systematics, biogeography, and ecology of fungi.
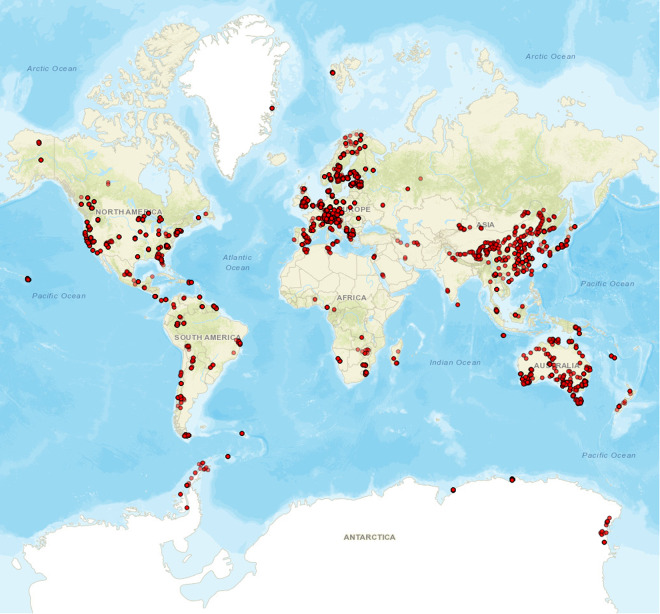
Fig. 1.
Map of locations of samples contained in the GlobalFungi database. Each point represents one or several samples where fungal community composition was reported using high-throughput-sequencing methods targeting the ITS1 or ITS2 marker of fungi. The map was created using the ‘leaflet’ package that uses an open-source JavaScript library for mobile-friendly interactive maps (Leaflet 1.6.0, GNU General Public License).
Methods
Data selection
We explored papers fitting with a main criterion, i.e., high-throughput sequencing for the analysis of fungal communities thanks to the ITS region, and that were published up to the beginning of 2019; in total, we explored 843 papers. The following selection criteria were used for the inclusion of samples (and, consequently, studies) into the dataset: (1) samples came from terrestrial biomes of soil, dead or live plant material (e.g., soil, litter, rhizosphere soil, topsoil, lichen, deadwood, root, and shoot) and were not subject to experimental treatment that artificially modifies the fungal community composition (e.g., temperature or nitrogen increase experiment, greenhouse controlled experiment were excluded); (2) the precise geographic location of each sample was recorded and released using GPS coordinates; (3) the whole fungal community was subject to amplicon sequencing (studies using group-specific primers were excluded); (4) the internal transcribed spacer regions (ITS1, ITS2, or both) were subject to amplification; (5) sequencing data (either in fasta with phred scores reported or fastq format) were publicly available or provided by the authors of the study upon request, and the sequences were unambiguously assigned to samples; (6) the samples could be assigned to biomes according to the Environment Ontology (http://www.ontobee.org/ontology/ENVO)8. In total, 178 publications contained samples that matched our criteria.
Processing of sequencing data
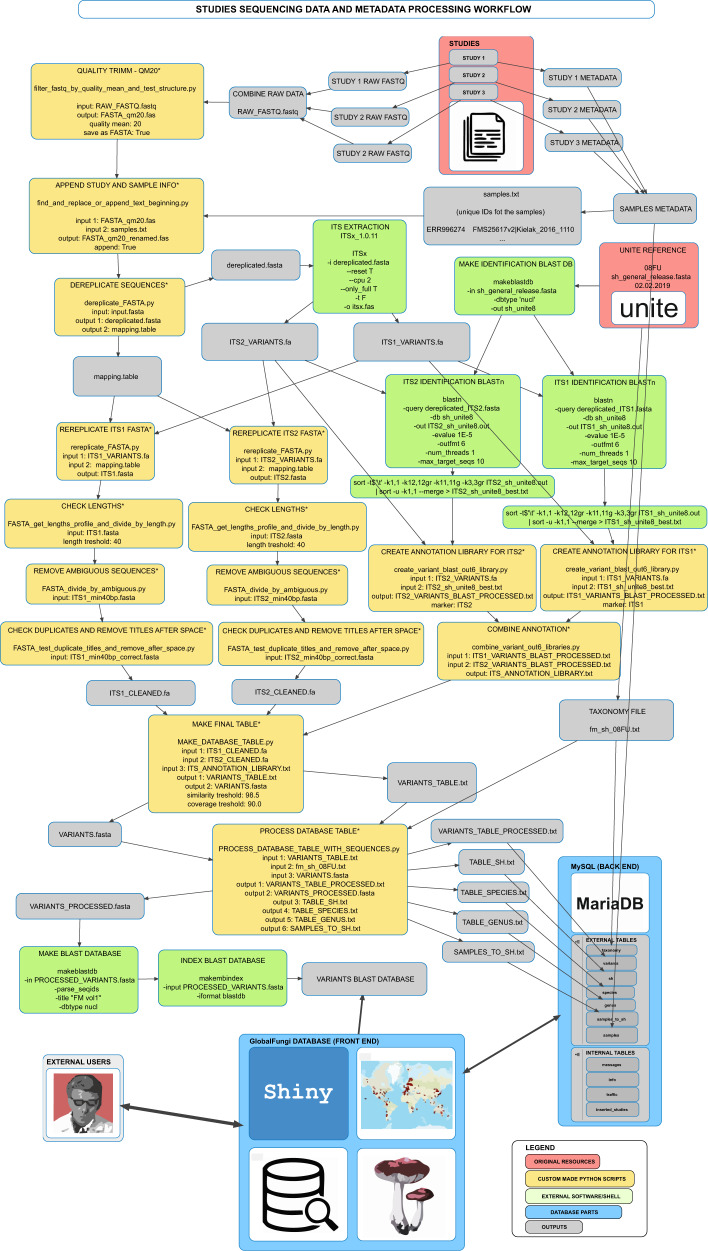
For the processing of data, see Fig. 2 and Code Availability section. Raw datasets from 178 studies, covering 17 242 individual samples were quality filtered by removing all sequences with the mean quality phred scores below 20. Each sequence was labelled using the combination of a sample ID and sequence ID, and the full ITS1 or ITS2 fungal region was extracted using Perl script ITSx v1.0.1113. ITS extraction resulted in a total of 416 291 533 full ITS1 and 231 278 756 full ITS2 sequences. The extracted ITS sequences were classified according to the representative sequence of the closest UNITE species hypothesis (SH) using BLASTn14, using the SH created considering a 98.5% similarity threshold (BLASTDBv5, general release 8.1 from 2.2.201912). A sequence was classified to the best best hit SH only when the following thresholds were met: e-value < 10e−50, sequence similarity > = 98.5%. All representative sequences annotated as nonfungal were discarded. All representative sequences classified to any fungal SH and all unclassified sequences were used to build database library of unique nucleotide sequences (sequence variants). The number of sequence variants accessible through the database is 113 423 871.
Fig. 2.
Processing of raw sequencing data for the GlobalFungi database. Workflow of processing of sequencing data included in the GlobalFungi database.
Sample metadata
Sample metadata were collected from the published papers and/or public repositories where they were submitted by the authors. In some cases, metadata were obtained from the authors of individual studies upon request. The samples were assigned to continents, countries, and specific locations when available, and all sites were categorized into biomes following the classification of Environment Ontology to a maximum achievable depth for each sample. The complete list of metadata included in the database is presented in Table 1.
Table 1.
List of metadata contained in the GlobalFungi database.
| Metadata identifier | Unit | Description of content | Source |
|---|---|---|---|
| Sample ID | unique identifier | generated | |
| Longitude | degrees | Geographical longitude | original paper |
| Latitude | degrees | Geographical latitude | original paper |
| Continent | One of the following: Africa/Antarctica/Asia/Australia/Europe/North America/South America | original paper | |
| Sample type | One of the following: soil/rhizosphere soil/litter/litter + humus/deadwood/lichen/shoot/root | original paper | |
| Biome | One of the following: forest biome/woodland biome/shrubland biome/grassland biome/desert biome/tundra biome/mangrove biome/anthropogenic terrestrial biome/marine biome/freshwater biome/polar desert biome | original paper | |
| Sampling year | Year of sample collection | original paper | |
| Primers | Primers used | original paper | |
| pH | pH | original paper | |
| ITS total | Number of full ITS sequences extracted | generated | |
| MAT (°C) | °C | Mean annual temperature from CHELSA database | CHELSA |
| MAP (mm) | mm | Mean annual precipitation from CHELSA database | CHELSA |
The table lists identifiers, units and sources of metadata contained in the database with the description of their content. The data source “original paper” may also represent additional metadata provided by the authors of the paper.
In addition to the metadata provided by the authors of each study, we also extracted bioclimatic variables from the global CHELSA15 and WorldClim 216 databases for each sample based on its GPS location. Since the results based on CHELSA and WorldClim 2 were comparable, we decided to include those from CHELSA, because precipitation patterns are better captured in the CHELSA dataset, in particular for mountain sites15.
For each sequence variant that was classified to SH, fungal species name and genus name was retrieved from the UNITE database12, when available.
Data Records
The raw sequencing reads used to create the database are available at different locations (see Table 2).
Table 2.
List of identifiers and source database of the raw sequencing datasets used.
| Database | Accession Identifiers (in superscripts, respectively: dataset reference, study reference(s)) |
|---|---|
| National Center for Biotechnology Information Sequence Read Archive | SRP00105819,20 SRP00117521,22, SRP00607823,24, SRP01286825,26, SRP01369527,28, SRP01394429,30, SRP01573531,32, SRP01609033,34, SRP02620735,36, SRP02840437,38, SRP03371939,40, SRP03535641,42, SRP04031443,44, SRP04078645,46, SRP04134747,48, SRP04310649,50, SRP0437064,51–53, SRP04398254,55, SRP04466556,57, SRP04516658,59, SRP04558760,61, SRP04574662,63, SRP04593364,65, SRP04604966,67, SRP04803668–70, SRP04885671,72, SRP04954473,74, SRP05103375,76, SRP05222277,78, SRP05271679,80, SRP05595781,82, SRP05743383,84, SRP05754185,86, SRP05850887,88, SRP05855589,90, SRP05885191,92, SRP05928093,94, SRP06083895,96, SRP06117997,98, SRP06130599,100, SRP061904101,102, SRP062647103,104, SRP063711105,106, SRP064158107,108, SRP065817109,110, SRP066030111,112, SRP066284113,114, SRP066331115,116, SRP067301117,118, SRP067367119,120, SRP068514121,122, SRP068608123,124, SRP068620125,126, SRP068654127,128, SRP069065129,130, SRP069742131,132, SRP070568133,134, SRP073070135,136, SRP073265137,138, SRP074055139,140, SRP074496141,142, SRP075989143,144, SRP079403145,146, SRP079521147,148, SRP080210149,150, SRP080428151,152, SRP080680153,154, SRP082472155,156, SRP082976157,158, SRP083394159,160, SRP083434160,161, SRP083901162,163, SRP087715164,165, SRP090261166,167, SRP090335168,169, SRP090490170,171, SRP090651172,173, SRP091741174,175, SRP091855176,177, SRP091867178,179, SRP092609180,181, SRP092777182,183, SRP093592184,185, SRP093928186,187, SRP094708188–190, SRP097883191,192, SRP101553193,194, SRP101605195,196, SRP102378197,198, SRP102417199,200, SRP102775201,202, SRP106137203,204, SRP106774205,206, SRP107174207,208, SRP107743209,210, SRP109164211,212, SRP109773213,214, SRP110522215,216, SRP110810217,218, SRP113348219,220, SRP114697221,222, SRP114821223,224, SRP115350225,226, SRP115464227,228, SRP115599229,230, SRP117302231,232, SRP118875233,234, SRP118960235,236, SRP119174237,238, SRP125864239,240, SRP132277241,242, SRP132591243,244, SRP132598244,245, SRP136886246,247, SRP139483248,249, SRP142723250,251, SRP148813252,253, SRP150527254,255, SRP151262256,257, SRP153934258,259, SRP160913260,261, SRP161632262,263, SRP195764264,265 |
| European Nucleotide Archive Sequence Read Archive | ERP001713266,267, ERP003251268,269, ERP003790270,271, ERP005177272,273, ERP005905274,275, ERP009341276,277, ERP010027278,279, ERP010084280,281, ERP010743282,283, ERP011924284,285, ERP012017286,287, ERP013208288,289, ERP013987290,291, ERP014227292,293, ERP017480294,295, ERP017851296,297, ERP017915298,299, ERP019580300,301, ERP019924302,303, ERP020657304,305, ERP022511306,307, ERP022742308,309, ERP023275310,311, ERP023718312,313, ERP023855314,315, ERP106131316,317, ERP107634318,319, ERP107636319,320, ERP110188321,322, ERP112007323,324 |
| DNA Data Bank of Japan | DRA000926325,326, DRA000937327,328, DRA001737329,330, DRA002424331,332, DRA002469333,334, DRA003024335,336, DRA003730337,338, DRA004913339,340, DRA006519341,342, DRP002783343,344, DRP003138345,346, DRP005365347,348 |
| Dryad Digital Repository | 10.5061/dryad.2fc32349,350, 10.5061/dryad.n82g9351,352, 10.5061/dryad.2343k353,354, 10.5061/dryad.gp302355,356, 10.5061/dryad.cq2rb357,358, 10.5061/dryad.8fn8j359,360, 10.5061/dryad.216tp361,362 |
| GenBank | KAYV00000000.1363,364, KAYU00000000.1364,365, KAYT00000000.1364,366, SAMN02934078367,368, SAMN02934079368,369 |
| Australian Antarctic Data Center database | 10.4225/15/526f42ada05b1370,371 |
| Supplemental Data | Hartmann et al. (2012)Supplementary_Data2,372, Rime et al. (2016)Fungi_SeqsID,373 |
The database contains two data types: sequence variants (individual nucleotide sequences) and samples. For each sequence variant, the following information is stored: sequence variant code, identification of samples where sequence variant occurs and the number of observations, the SH of best hit (when available), fungal species name (when available), fungal genus name (when available). For each sample, metadata information is stored (Table 1). Sequence data and metadata are accessible at Figshare17 (GlobalFungi_ITS_variants.zip, GlobalFungi_metadata.xlsx). All database content is accessible using a public graphical user interface at https://globalfungi.com.
Technical Validation
The technical validation included the screening of the data sources, sequencing data and data reliability. Regarding the data source screening, the data sources (published papers) were screened to fulfil the criteria outlined in the Methods section. The dataset was thoroughly checked for duplicates, and for all records that appeared in multiple publications, only the first original publication of the dataset was considered as a data source. Considering sequence quality, we have only utilized those primer pairs that are generally accepted to target general fungi (see Online-Only Table 1)7,18. Sequences were quality filtered by removing all sequences with the mean quality phred scores below 20 and sequences that did not represent complete ITS1 or ITS2 after extraction or those that were identified as chimeric by the ITS extraction software13 were removed. All representative sequences where the BLASTn search against the UNITE database12 resulted in a nonfungal organism, were discarded.
Online-only Table 1.
List of primer pairs to amplify general fungal communities in the samples included in this Data Descriptor7,17.
| Primer pair | Sequence |
| 1737F / 2043 R | GGAAGTAAAAGTCGTAACAAGG / GCTGCGTTCTTCATCGATGC |
| 18S-F / 5.3S-R | GTAAAAGTCGTAACAAGGTTTC / GTTCAAAGAYTCGATGATTCAC |
| 58A2F / ITS4 | ATCGATGAAGAACGCAG / TCCTCCGCTTATTGATATGC |
| fITS7 / ITS4 | GTGARTCATCGAATCTTTG / TCCTCCGCTTATTGATATGC |
| fITS9 / ITS4 | GAACACAGCGAAATGTGA / TCCTCCGCTTATTGATATGC |
| gITS7 / ITS4 | GTGARTCATCGARTCTTTG / TCCTCCGCTTATTGATATGC |
| gITS7 / ITS4ngs | GTGARTCATCGARTCTTTG / TTCCTSCGCTTATTGATATGC |
| ITS1 / ITS2 | TCCGTAGGTGAACCTGCGG / GCTGCGTTCTTCATCGATGC |
| ITS1 / ITS4 | TCCGTAGGTGAACCTGCGG / TCCTCCGCTTATTGATATGC |
| ITS1F / 58A2R | CTTGGTCATTTAGAGGAAGTAA / CTGCGTTCTTCATCGAT |
| ITS1F / gITS7 | CTTGGTCATTTAGAGGAAGTAA / GTGARTCATCGARTCTTTG |
| ITS1F / ITS2 | CTTGGTCATTTAGAGGAAGTAA / GCTGCGTTCTTCATCGATGC |
| ITS1F / ITS3R | CTTGGTCATTTAGAGGAAGTAA / TCCTCCGCTTATTGATATGC |
| ITS1F / ITS4 | CTTGGTCATTTAGAGGAAGTAA / TCCTCCGCTTATTGATATGC |
| ITS1F_KYO1 / ITS2_KYO1 | CTHGGTCATTTAGAGGAASTAA / CTRYGTTCTTCATCGDT |
| ITS1F_KYO1 / ITS2_KYO2 | CTHGGTCATTTAGAGGAASTAA / TTYRCTRCGTTCTTCATC |
| ITS1F_KYO2 / ITS2_KYO2 | TAGAGGAAGTAAAAGTCGTAA / TTYRCTRCGTTCTTCATC |
| ITS1F_KYO2 / LR3 | TAGAGGAAGTAAAAGTCGTAA / CCGTGTTTCAAGACGGG |
| ITS1FI2 / 5.8 S | GAACCWGCGGARGGATCA / CGCTGCGTTCTTCATCG |
| ITS1FI2 / ITS2 | GAACCWGCGGARGGATCA / GCTGCGTTCTTCATCGATGC |
| ITS1Fngs / ITS2 | GGTCATTTAGAGGAAGTAA / GCTGCGTTCTTCATCGATGC |
| ITS1ngs / ITS2 | TCCGTAGGTGAACCTGC / GCTGCGTTCTTCATCGATGC |
| ITS1-OF / ITS4 | AACT(C)GG(C)CATTTAGAGGAAGT / TCCTCCGCTTATTGATATGC |
| ITS2 / ITS4 | GCTGCGTTCTTCATCGATGC / TCCTCCGCTTATTGATATGC |
| ITS2 / ITS5 | GCTGCGTTCTTCATCGATGC / GGAAGTAAAAGTCGTAACAAGG |
| ITS2F / ITS2R | GCATCGATGAAGAACGC / CCTCCGCTTATTGATATGC |
| ITS3 / ITS4 | GCATCGATGAAGAACGCAGC / TCCTCCGCTTATTGATATGC |
| ITS3_KYO2 / ITS4 | GATGAAGAACGYAGYRAA / TCCTCCGCTTATTGATATGC |
| ITS3_KYO2 / ITS4F | GATGAAGAACGYAGYRAA / CGCTTATTRATATGCTTAAXGXT |
| ITS3_KYO2 / LR_KYO1b | GATGAAGAACGYAGYRAA / MGCWGCATTCCCAAACWA |
| ITS3ngs mix / ITS4ngr mix + ARCH-ITS4-F1 | CA(T)CGATGAAGAACG(C)(A)G / TCCT(S)(C)(G)CTTATTGATATGC |
| ITS3ngs1 to ITS3ngs11 / ITS4ngs | (C)(A)(T)CGATGAAGA(A)CG(C)(A)G / TCCTSCGCTTATTGATATGC |
| ITS3tagmix1 to ITS3tagmix5 / ITS4ngs | CTAGACTCGTCA(T)CGATGAAGAACG(CA)G / TTCCTSCGCTTATTGATATGC |
| ITS4 / fITS9 | TCCTCCGCTTATTGATATGC / GAACACAGCGAAATGTGA |
| ITS4 / ITS3_KYO2 | TCCTCCGCTTATTGATATGC / GATGAAGAACGYAGYRAA |
| ITS4 / ITS5 | TCCTCCGCTTATTGATATGC / GGAAGTAAAAGTCGTAACAAGG |
| ITS4 / ITS9 | TCCTCCGCTTATTGATATGC / GCATTAGAAACTGCTCGTAATG |
| ITS5 / 5.8S_fungi | GGAAGTAAAAGTCGTAACAAGG / CAAGAGATCCGTTGTTGAAAGTT |
| ITS5 / ITS2 | GGAAGTAAAAGTCGTAACAAGG / GCTGCGTTCTTCATCGATGC |
| ITS5 / ITS2_KYO2 | GGAAGTAAAAGTCGTAACAAGG / TTYRCTRCGTTCTTCATC |
| ITS5 / ITS4 | GGAAGTAAAAGTCGTAACAAGG / TCCTCCGCTTATTGATATGC |
| ITS7o / ITS4 | GTGAATCATCRAATYTTTG / TCCTCCGCTTATTGATATGC |
| ITS86F / ITS4 | GTGAATCATCGAATCTTTGAA / TCCTCCGCTTATTGATATGC |
| ITS9 / ITS4 | GCATTAGAAACTGCTCGTAATG / TCCTCCGCTTATTGATATGC |
| ITS9F / ITS4 | CCATCTCATCCCTGCGTGTCTCCGACTCAG / TCCTCCGCTTATTGATATGC |
For data reliability, the geographic location represented by the GPS coordinates was validated first. For each sample set, the geographic location of the sample described in the text of the study was confronted with the location on the map. For those samples where disagreement was recorded (e.g., terrestrial samples positioned in the ocean or located in another region than described in the text), the authors of each study were asked for correction. Studies or samples that could not be reconciled in this way were excluded from the database. The quality of sample metadata was checked and if they were outside the acceptable range (such as content of elements or organic matter > 100%), these invalid metadata were removed.
Usage Notes
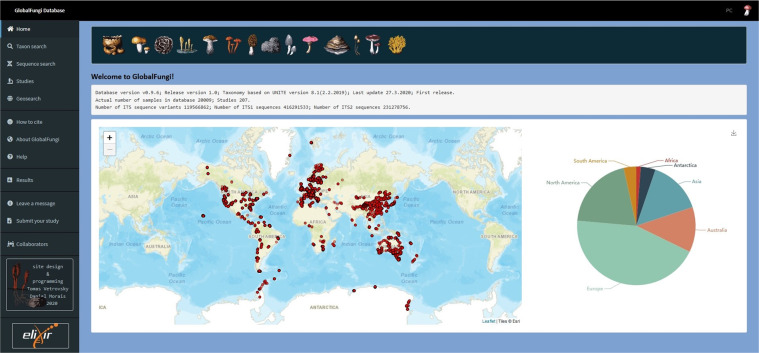
The user interface at https://globalfungi.com enables the users to access the database in several ways (Fig. 3). In the taxon search, it is possible to search for genera or species of fungi or for the 98.5% SH species hypotheses of UNITE, contained in the general release 8.1 from 2.2.2019. The search results open the options to download the corresponding SH or the corresponding sequence variants. It is also possible to view a breakdown of samples by type, biome, mean annual temperature, mean annual precipitation, pH, and continents. The results also contain an interactive map of the taxon distribution with relative abundances of sequences of the taxon across samples and a list of samples with metadata. Several modes of filtering of results are available as well.
Fig. 3.
User interface to access the GlobalFungi database.
In the sequence search, it is possible to search for multiple nucleotide sequences by choosing if the result will be the exact match or a BLAST result. The BLAST option gives the possibility to retrieve the sequence variant best hit in the database, or, when only one sequence is submitted, it is possible to display multiple ranked high score hits among the sequence variants.
It is also possible to open individual studies and access their content. Finally, in the Geosearch, users can select a group of samples on the map, with a range of tools, and retrieve data for these samples (such as the FASTA file with all occurring sequence variants).
Importantly, the database is intended to grow, both by the continuing activity of the authors and by using the help of the scientific community. For that, the “Submit your study” section of the web interface enabling the submission of studies not yet represented is available to users. The submission tool guides the submitting person through the steps where details about the publication, samples, sample metadata and sequences are sequentially submitted. The submitted data will be used to update the database twice a year after processing and validation by the authors. Thus, users submitting their data, besides a precious contribution to mycological progress, will benefit from making their data accessible to the international scientific community in an easily accessible form and increasing the visibility of their results. Users can also maximize their visibility by approving to add their name and affiliation to the online list of collaborators and/or to the GlobalFungi Group Author’ list that will be mentioned in future publications describing the database content, its development, or metastudies using the whole database.
Among the possible uses of the GlobalFungi Database, fungal ecologists will be able to link fungal diversity data with the panel of collected metadata, which should allow them to determine the environmental factors driving the fungal diversity. This kind of study can be done at different geographic levels, from country scale up to the entire world, and for all the fungal communities or by focusing on some ecosystem compartments. This should lead to a better understanding of the biogeography of the fungal diversity. Větrovský et al.8 brought interesting findings by doing this for soil fungal communities at the scale of the globe. The evolutionary biologists could study, for example, the effect of global change on the fungal diversity by comparing the natural versus anthropogenic biomes. In addition to focus on the fungal diversity, some studies could trigger specific fungi. Thus, mycologists could determine the biogeography of one specific fungal species. They could also determine the composition of the fungal communities associated with the focused species and detect some potential recurrent fungal associations. The GlobalFungi Database could also speed up the progress in fungal taxonomy by highlighting the existence of a high number of fungal sequences not currently assigned to species along with environmental metadata promoting thus the interest in their description.
Acknowledgements
We acknowledge funding from the Czech Science Foundation Grant No 18–26191S. ELIXIR CZ research infrastructure project LM2015047 by the Ministry of Education, Youth and Sports of the Czech Republic is acknowledged for hosting the database. All corresponding authors of published studies that provided additional information on the samples included in the database are gratefully acknowledged.
Online-only Table
Author contributions
T.V., P.K. and P.B. jointly conceived the study. C.L. and coordinated data acquisition, T.V., D.M., M.K. and P.B. designed the database, T.V. and D.M. developed the database and created the user interface. C.A.G., S.A.H., B.D.B., K.B., V.B., F.D., R.Z.H., M.J., J.K., C.L., S.L., R.L.M., T. Mar., T. Maš., L. Me., L. Mi., T. Mi., S.M., D.N., I.O., S.P.C., M.Š., K.Š., V.T., M.U., L.V., J.V. and L.Ž. identified data sources, processed and analysed sequencing information, collated and analysed metadata. P.B. drafted the manuscript along with T.V., C.L. and P.K. All authors wrote and reviewed the manuscript.
Code availability
The workflow included several custom made python scripts (labelled by star in the Fig. 2) which are accessible here: https://github.com/VetrovskyTomas/GlobalFungi.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tomáš Větrovský, Daniel Morais, Petr Kohout, Clémentine Lepinay.
References
- 1.Crowther TW, et al. Quantifying global soil carbon losses in response to warming. Nature. 2016;540:104–108. doi: 10.1038/nature20150. [DOI] [PubMed] [Google Scholar]
- 2.Peay KG, Kennedy PG, Talbot JM. Dimensions of biodiversity in the Earth mycobiome. Nature Rev. Microbiol. 2016;14:434–447. doi: 10.1038/nrmicro.2016.59. [DOI] [PubMed] [Google Scholar]
- 3.Wall DH, Nielsen UN, Six J. Soil biodiversity and human health. Nature. 2015;528:69–76. doi: 10.1038/nature15744. [DOI] [PubMed] [Google Scholar]
- 4.Tedersoo L, et al. Global diversity and geography of soil fungi. Science. 2014;346:1256688. doi: 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- 5.Bahram M, et al. Structure and function of the global topsoil microbiome. Nature. 2018;560:233–237. doi: 10.1038/s41586-018-0386-6. [DOI] [PubMed] [Google Scholar]
- 6.Egidi E, et al. A few Ascomycota taxa dominate soil fungal communities worldwide. Nature Commun. 2019;10:2369. doi: 10.1038/s41467-019-10373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nilsson RH, et al. Mycobiome diversity: high-throughput sequencing and identification of fungi. Nature Rev. Microbiol. 2019;17:95–109. doi: 10.1038/s41579-018-0116-y. [DOI] [PubMed] [Google Scholar]
- 8.Větrovský T, et al. A meta-analysis of global fungal distribution reveals climate-driven patterns. Nature Commun. 2019;10:5142. doi: 10.1038/s41467-019-13164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlk, L. et al. Early successional ectomycorrhizal fungi are more likely to naturalize outside their native range than other ectomycorrhizal fungi. New Phytol., 10.1111/nph.16557 (2020). [DOI] [PubMed]
- 10.Thompson LR, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoch CL, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nilsson RH, et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019;47:D259–D264. doi: 10.1093/nar/gky1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bengtsson-Palme J, et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Meth. Ecol. Evol. 2013;4:914–919. [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Karger DN, et al. Data Descriptor: Climatologies at high resolution for the earth’s land surface areas. Scientific Data. 2017;4:170122. doi: 10.1038/sdata.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fick SE, Hijmans RJ. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017;37:4302–4315. [Google Scholar]
- 17.Baldrian P, 2020. GlobalFungi: Global database of fungal records from high-throughput-sequencing metabarcoding studies. figshare. [DOI] [PMC free article] [PubMed]
- 18.Anslan S, et al. Great differences in performance and outcome of high-throughput sequencing data analysis platforms for fungal metabarcoding. Mycokeys. 2018;39:29–40. doi: 10.3897/mycokeys.39.28109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.2010. NCBI Sequence Read Archive. SRP001058
- 20.Jumpponen A, Jones KL. Seasonally dynamic fungal communities in the Quercus macrocarpa phyllosphere differ between urban and nonurban environments. New Phytol. 2010;186:496–513. doi: 10.1111/j.1469-8137.2010.03197.x. [DOI] [PubMed] [Google Scholar]
- 21.2010. NCBI Sequence Read Archive. SRP001175
- 22.Jumpponen A, Jones KL, Mattox JD, Yaege C. Massively parallel 454-sequencing of fungal communities in Quercus spp ectomycorrhizas indicates seasonal dynamics in urban and rural sites. Mol. Ecol. 2010;19:41–53. doi: 10.1111/j.1365-294X.2009.04483.x. [DOI] [PubMed] [Google Scholar]
- 23.2011. NCBI Sequence Read Archive. SRP006078
- 24.Mello A, et al. ITS-1 versus ITS-2 pyrosequencing: a comparison of fungal populations in truffle grounds. Mycologia. 2011;103:1184–1193. doi: 10.3852/11-027. [DOI] [PubMed] [Google Scholar]
- 25.2012. NCBI Sequence Read Archive. SRP012868
- 26.Ihrmark K, et al. New primers to amplify the fungal ITS2 region - evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012;82:666–677. doi: 10.1111/j.1574-6941.2012.01437.x. [DOI] [PubMed] [Google Scholar]
- 27.2012. NCBI Sequence Read Archive. SRP013695
- 28.Zimmerman NB, Vitousek PM. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc. Natl. Acad. Sci. USA. 2012;109:13022–13027. doi: 10.1073/pnas.1209872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.2016. NCBI Sequence Read Archive. SRP013944
- 30.Uroz S, et al. Specific impacts of beech and Norway spruce on the structure and diversity of the rhizosphere and soil microbial communities. Sci. Rep. 2016;6:27756. doi: 10.1038/srep27756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.2015. NCBI Sequence Read Archive. SRP015735
- 32.Gao C, et al. Community assembly of ectomycorrhizal fungi along a subtropical secondary forest succession. New Phytol. 2015;205:771–785. doi: 10.1111/nph.13068. [DOI] [PubMed] [Google Scholar]
- 33.2015. NCBI Sequence Read Archive. SRP016090
- 34.Clemmensen KE, et al. Carbon sequestration is related to mycorrhizal fungal community shifts during long-term succession in boreal forests. New Phytol. 2015;205:1525–1536. doi: 10.1111/nph.13208. [DOI] [PubMed] [Google Scholar]
- 35.2014. NCBI Sequence Read Archive. SRP026207
- 36.De Beeck MO, et al. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One. 2014;9:e97629. doi: 10.1371/journal.pone.0097629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.2015. NCBI Sequence Read Archive. SRP028404
- 38.De Beeck MO, et al. Impact of metal pollution on fungal diversity and community structures. Environ. Microbiol. 2015;17:2035–2047. doi: 10.1111/1462-2920.12547. [DOI] [PubMed] [Google Scholar]
- 39.2015. NCBI Sequence Read Archive. SRP033719
- 40.Chaput DL, Hansel CM, Burgos WD, Santelli CM. Profiling microbial communities in manganese remediation systems treating coal mine drainage. Appl. Environ. Microbiol. 2015;81:2189–2198. doi: 10.1128/AEM.03643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.2015. NCBI Sequence Read Archive. SRP035356
- 42.Sterkenburg E, Bahr A, Brandström Durling M, Clemmensen KE, Lindahl BD. Changes in fungal communities along a boreal forest soil fertility gradient. New Phytol. 2015;207:1145–1158. doi: 10.1111/nph.13426. [DOI] [PubMed] [Google Scholar]
- 43.2014. NCBI Sequence Read Archive. SRP040314
- 44.Talbot JM, et al. Endemism and functional convergence across the North American soil mycobiome. Proc. Natl. Acad. Sci. USA. 2014;111:6341–6346. doi: 10.1073/pnas.1402584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.2015. NCBI Sequence Read Archive. SRP040786
- 46.Saravesi K, et al. Moth outbreaks alter root-associated fungal communities in subarctic mountain birch forests. Microb. Ecol. 2015;69:788–797. doi: 10.1007/s00248-015-0577-8. [DOI] [PubMed] [Google Scholar]
- 47.2015. NCBI Sequence Read Archive. SRP041347
- 48.Liu J, et al. Soil carbon content drives the biogeographical distribution of fungal communities in the black soil zone of northeast China. Soil Biol. Biochem. 2015;83:29–39. [Google Scholar]
- 49.2015. NCBI Sequence Read Archive. SRP043106
- 50.Hoppe B, et al. Linking molecular deadwood-inhabiting fungal diversity and community dynamics to ecosystem functions and processes in Central European forests. Fungal Divers. 2015;77:367–379. [Google Scholar]
- 51.2017. NCBI Sequence Read Archive. SRP043706
- 52.Hiiesalu I, Bahram M, Tedersoo L. Plant species richness and productivity determine the diversity of soil fungal guilds in temperate coniferous forest and bog habitats. Mol. Ecol. 2017;26:4846–4858. doi: 10.1111/mec.14246. [DOI] [PubMed] [Google Scholar]
- 53.Tedersoo L, et al. Tree diversity and species identity effects on soil fungi, protists and animals are context dependent. ISME J. 2016;10:346–362. doi: 10.1038/ismej.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.2015. NCBI Sequence Read Archive. SRP043982
- 55.Jarvis SG, Woodward S, Taylor AF. Strong altitudinal partitioning in the distributions of ectomycorrhizal fungi along a short (300 m) elevation gradient. New Phytol. 2015;206:1145–1155. doi: 10.1111/nph.13315. [DOI] [PubMed] [Google Scholar]
- 56.2016. NCBI Sequence Read Archive. SRP044665
- 57.Nacke H, et al. Fine spatial scale variation of soil microbial communities under European Beech and Norway Spruce. Front. Microbiol. 2016;7:2067. doi: 10.3389/fmicb.2016.02067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.2015. NCBI Sequence Read Archive. SRP045166
- 59.Rincón A, et al. Compartmentalized and contrasted response of ectomycorrhizal and soil fungal communities of Scots pine forests along elevation gradients in France and Spain. Environ. Microbiol. 2015;17:3009–3024. doi: 10.1111/1462-2920.12894. [DOI] [PubMed] [Google Scholar]
- 60.2016. NCBI Sequence Read Archive. SRP045587
- 61.Bahram M, et al. Stochastic distribution of small soil eukaryotes resulting from high dispersal and drift in a local environment. ISME J. 2016;10:885–896. doi: 10.1038/ismej.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.2014. NCBI Sequence Read Archive. SRP045746
- 63.Walker DM, et al. A metagenomics-based approach to the top-down effect on the detritivore food web: a salamanders influence on fungal communities within a deciduous forest. Ecol. Evol. 2014;4:4106–4116. doi: 10.1002/ece3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.2015. NCBI Sequence Read Archive. SRP045933
- 65.Zhang T, Wei XL, Zhang YQ, Liu HY, Yu LY. Diversity and distribution of lichen-associated fungi in the Ny-Alesund Region (Svalbard, High Arctic) as revealed by 454 pyrosequencing. Sci. Rep. 2015;5:14850. doi: 10.1038/srep14850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.2016. NCBI Sequence Read Archive. SRP046049
- 67.Oh SY, Fong JJ, Park MS, Lim YW. Distinctive feature of microbial communities and bacterial functional profiles in Tricholoma matsutake dominant soil. PLoS One. 2016;11:e0168573. doi: 10.1371/journal.pone.0168573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.2016. NCBI Sequence Read Archive. SRP048036
- 69.Yang T, et al. Carbon constrains fungal endophyte assemblages along the timberline. Environ. Microbiol. 2016;18:2455–2469. doi: 10.1111/1462-2920.13153. [DOI] [PubMed] [Google Scholar]
- 70.Yang T, Sun H, Shen C, Chu H. Fungal assemblages in different habitats in an Erman’s Birch forest. Front. Microbiol. 2016;7:1368. doi: 10.3389/fmicb.2016.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.2015. NCBI Sequence Read Archive. SRP048856
- 72.Elliott DR, Caporn SJ, Nwaishi F, Nilsson RH, Sen R. Bacterial and fungal communities in a degraded ombrotrophic peatland undergoing natural and managed re-vegetation. PLoS One. 2015;10:e0124726. doi: 10.1371/journal.pone.0124726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.2015. NCBI Sequence Read Archive. SRP049544
- 74.Goldmann K, Schöning I, Buscot F, Wubet T. Forest management type influences diversity and community composition of soil fungi across temperate forest ecosystems. Front. Microbiol. 2015;6:1300. doi: 10.3389/fmicb.2015.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.2016. NCBI Sequence Read Archive. SRP051033
- 76.Roy-Bolduc A, Laliberté E, Boudreau S, Hijri M. Strong linkage between plant and soil fungal communities along a successional coastal dune system. FEMS Microbiol. Ecol. 2016;92:fiw156. doi: 10.1093/femsec/fiw156. [DOI] [PubMed] [Google Scholar]
- 77.2017. NCBI Sequence Read Archive. SRP052222
- 78.Fernández-Martínez MA, et al. Microbial succession dynamics along glacier forefield chronosequences in Tierra del Fuego (Chile) Polar Biol. 2017;40:1939–1957. [Google Scholar]
- 79.2015. NCBI Sequence Read Archive. SRP052716
- 80.Leff JW, et al. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. USA. 2015;112:10967–10972. doi: 10.1073/pnas.1508382112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.2015. NCBI Sequence Read Archive. SRP055957
- 82.Tedersoo L, et al. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. Mycokeys. 2015;10:1–43. [Google Scholar]
- 83.2016. NCBI Sequence Read Archive. SRP057433
- 84.Wang W, Zhai Y, Cao L, Tan H, Zhang R. Endophytic bacterial and fungal microbiota in sprouts, roots and stems of rice (Oryza sativa L.) Microbiol. Res. 2016;188:1–8. doi: 10.1016/j.micres.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 85.2016. NCBI Sequence Read Archive. SRP057541
- 86.Waring BG, Adams R, Branco S, Powers JS. Scale-dependent variation in nitrogen cycling and soil fungal communities along gradients of forest composition and age in regenerating tropical dry forests. New Phytol. 2016;209:845–854. doi: 10.1111/nph.13654. [DOI] [PubMed] [Google Scholar]
- 87.2016. NCBI Sequence Read Archive. SRP058508
- 88.Glassman SI, Levine CR, DiRocco AM, Battles JJ, Bruns TD. Ectomycorrhizal fungal spore bank recovery after a severe forest fire: some like it hot. ISME J. 2016;10:1228–1239. doi: 10.1038/ismej.2015.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.2016. NCBI Sequence Read Archive. SRP058555
- 90.De Gannes V, et al. Microbial community structure and function of soil following ecosystem conversion from native forests to Teak plantation forests. Front. Microbiol. 2016;7:1976. doi: 10.3389/fmicb.2016.01976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.2018. NCBI Sequence Read Archive. SRP058851
- 92.Bach EM, Williams RJ, Hargreaves SK, Yang F, Hofmockel KS. Greatest soil microbial diversity found in micro-habitats. Soil Biol. Biochem. 2018;118:217–226. [Google Scholar]
- 93.2016. NCBI Sequence Read Archive. SRP059280
- 94.Roy‐Bolduc A, Laliberté E, Hijri M. High richness of ectomycorrhizal fungi and low host specificity in a coastal sand dune ecosystem revealed by network analysis. Ecol. Evol. 2016;6:349–362. doi: 10.1002/ece3.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.2016. NCBI Sequence Read Archive. SRP060838
- 96.He F, et al. Changes in composition and diversity of fungal communities along Quercus mongolica forests developments in Northeast China. Appl. Soil Ecol. 2016;100:162–171. [Google Scholar]
- 97.2016. NCBI Sequence Read Archive. SRP061179
- 98.Valverde A, et al. Specific microbial communities associate with the rhizosphere of Welwitschia mirabilis, a living fossil. PLoS One. 2016;11:e0153353. doi: 10.1371/journal.pone.0153353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.2017. NCBI Sequence Read Archive. SRP061305
- 100.Yao F, et al. Microbial taxa distribution is associated with ecological trophic cascades along an elevation gradient. Front. Microbiol. 2017;8:2071. doi: 10.3389/fmicb.2017.02071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.2015. NCBI Sequence Read Archive. SRP061904
- 102.Veach AM, Dodds WK, Jumpponen A. Woody plant encroachment, and its removal, impact bacterial and fungal communities across stream and terrestrial habitats in a tallgrass prairie ecosystem. FEMS Microbiol. Ecol. 2015;91:fiv109. doi: 10.1093/femsec/fiv109. [DOI] [PubMed] [Google Scholar]
- 103.2016. NCBI Sequence Read Archive. SRP062647
- 104.Newsham KK, et al. Relationship between soil fungal diversity and temperature in the maritime Antarctic. Nat. Clim. Change. 2016;6:182. [Google Scholar]
- 105.2017. NCBI Sequence Read Archive. SRP063711
- 106.Poosakkannu A, Nissinen R, Männistö M, Kytöviita MM. Microbial community composition but not diversity changes along succession in arctic sand dunes. Environ. Microbiol. 2017;19:698–709. doi: 10.1111/1462-2920.13599. [DOI] [PubMed] [Google Scholar]
- 107.2017. NCBI Sequence Read Archive. SRP064158
- 108.Tian J, et al. Patterns and drivers of fungal diversity along an altitudinal gradient on Mount Gongga, China. J. Soil. Sediment. 2017;17:2856–2865. [Google Scholar]
- 109.2017. NCBI Sequence Read Archive. SRP065817
- 110.Zhang W, Lu Z, Yang K, Zhu J. Impacts of conversion from secondary forests to larch plantations on the structure and function of microbial communities. Appl. Soil Ecol. 2017;111:73–83. [Google Scholar]
- 111.2016. NCBI Sequence Read Archive. SRP066030
- 112.Porter TM, Shokralla S, Baird D, Golding GB, Hajibabaei M. Ribosomal DNA and plastid markers used to sample fungal and plant communities from wetland soils reveals complementary biotas. PLoS One. 2016;11:e0142759. doi: 10.1371/journal.pone.0142759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.2017. NCBI Sequence Read Archive. SRP066284
- 114.Wang M, et al. Influence of Peanut cultivars and environmental conditions on the diversity and community composition of Pod Rot soil fungi in China. Mycobiology. 2017;45:392–400. doi: 10.5941/MYCO.2017.45.4.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.2017. NCBI Sequence Read Archive. SRP066331
- 116.Delgado‐Baquerizo M, et al. Soil microbial communities drive the resistance of ecosystem multifunctionality to global change in drylands across the globe. Ecol. Lett. 2017;20:1295–1305. doi: 10.1111/ele.12826. [DOI] [PubMed] [Google Scholar]
- 117.2017. NCBI Sequence Read Archive. SRP067301
- 118.Cross H, et al. Fungal diversity and seasonal succession in ash leaves infected by the invasive ascomycete Hymenoscyphus fraxineus. New Phytol. 2017;213:1405–1417. doi: 10.1111/nph.14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.2016. NCBI Sequence Read Archive. SRP067367
- 120.Zhang T, Wang NF, Liu HY, Zhang YQ, Yu LY. Soil pH is a key determinant of soil fungal community composition in the Ny-Alesund region, Svalbard (High Arctic) Front. Microbiol. 2016;7:227. doi: 10.3389/fmicb.2016.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.2016. NCBI Sequence Read Archive. SRP068514
- 122.Gehring CA, et al. Cheatgrass invasion alters the abundance and composition of dark septate fungal communities in sagebrush steppe. Botany. 2016;94:481–491. [Google Scholar]
- 123.2016. NCBI Sequence Read Archive. SRP068608
- 124.Li Y, et al. Changes of soil microbial community under different degraded gradients of alpine meadow. Agric. Ecosyst. Environ. 2016;222:213–222. [Google Scholar]
- 125.2016. NCBI Sequence Read Archive. SRP068620
- 126.Zhou J, et al. Temperature mediates continental-scale diversity of microbes in forest soils. Nat. Commun. 2016;7:12083. doi: 10.1038/ncomms12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.2016. NCBI Sequence Read Archive. SRP068654
- 128.Cox F, Newsham KK, Bol R, Dungait JA, Robinson CH. Not poles apart: Antarctic soil fungal communities show similarities to those of the distant Arctic. Ecol. Lett. 2016;19:528–536. doi: 10.1111/ele.12587. [DOI] [PubMed] [Google Scholar]
- 129.2017. NCBI Sequence Read Archive. SRP069065
- 130.Bergottini VM, et al. Exploring the diversity of the root-associated microbiome of Ilex paraguariensis St. Hil. (Yerba Mate) Appl. Soil Ecol. 2017;109:23–31. [Google Scholar]
- 131.2017. NCBI Sequence Read Archive. SRP069742
- 132.Moussa TA, Al-Zahrani HS, Almaghrabi OA, Abdelmoneim TS, Fuller MP. Comparative metagenomics approaches to characterize the soil fungal communities of western coastal region, Saudi Arabia. PLoS One. 2017;12:e0185096. doi: 10.1371/journal.pone.0185096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.2016. NCBI Sequence Read Archive. SRP070568
- 134.Goldmann K, et al. Divergent habitat filtering of root and soil fungal communities in temperate beech forests. Sci. Rep. 2016;11:31439. doi: 10.1038/srep31439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.2016. NCBI Sequence Read Archive. SRP073070
- 136.Liu C, et al. The influence of soil properties on the size and structure of bacterial and fungal communities along a paddy soil chronosequence. Eur. J. Soil Biol. 2016;76:9–18. [Google Scholar]
- 137.2017. NCBI Sequence Read Archive. SRP073265
- 138.Smith ME, et al. Investigating niche partitioning of ectomycorrhizal fungi in specialized rooting zones of the monodominant leguminous tree Dicymbe corymbosa. New Phytol. 2017;215:443–453. doi: 10.1111/nph.14570. [DOI] [PubMed] [Google Scholar]
- 139.2016. NCBI Sequence Read Archive. SRP074055
- 140.Bissett A, et al. Introducing BASE: the Biomes of Australian Soil Environments soil microbial diversity database. GigaScience. 2016;5:21. doi: 10.1186/s13742-016-0126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.2016. NCBI Sequence Read Archive. SRP074496
- 142.Vannette RL, Leopold DR, Fukami T. Forest area and connectivity influence root-associated fungal communities in a fragmented landscape. Ecology. 2016;97:2374–2383. doi: 10.1002/ecy.1472. [DOI] [PubMed] [Google Scholar]
- 143.2017. NCBI Sequence Read Archive. SRP075989
- 144.Zhou X, et al. Rhizospheric fungi and their link with the nitrogen-fixing Frankia harbored in host plant Hippophae rhamnoides L. J. Basic Microbiol. 2017;57:1055–1064. doi: 10.1002/jobm.201700312. [DOI] [PubMed] [Google Scholar]
- 145.2017. NCBI Sequence Read Archive. SRP079403
- 146.Glassman SI, Wang IJ, Bruns TD. Environmental filtering by pH and soil nutrients drives community assembly in fungi at fine spatial scales. Mol. Ecol. 2017;26:6960–6973. doi: 10.1111/mec.14414. [DOI] [PubMed] [Google Scholar]
- 147.2018. NCBI Sequence Read Archive. SRP079521
- 148.Cline LC, Schilling JS, Menke J, Groenhof E, Kennedy PG. Ecological and functional effects of fungal endophytes on wood decomposition. Funct. Ecol. 2018;32:181–191. [Google Scholar]
- 149.2016. NCBI Sequence Read Archive. SRP080210
- 150.Johansen RB, et al. A native and an invasive dune grass share similar, patchily distributed, root-associated fungal communities. Fungal Ecol. 2016;23:141–155. [Google Scholar]
- 151.2017. NCBI Sequence Read Archive. SRP080428
- 152.Zhang S, Chen X, Zhong Q, Huang Z, Bai Z. Relations among epiphytic microbial communities from soil, leaves and grapes of the grapevine. Front. Life Sci. 2017;10:73–83. [Google Scholar]
- 153.2017. NCBI Sequence Read Archive. SRP080680
- 154.Fernandez CW, et al. Ectomycorrhizal fungal response to warming is linked to poor host performance at the boreal-temperate ecotone. Glob. Change Biol. 2017;23:1598–1609. doi: 10.1111/gcb.13510. [DOI] [PubMed] [Google Scholar]
- 155.2017. NCBI Sequence Read Archive. SRP082472
- 156.Zhang Z, et al. Fungal communities in ancient peatlands developed from different periods in the Sanjiang Plain, China. PLoS One. 2017;12:e0187575. doi: 10.1371/journal.pone.0187575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.2017. NCBI Sequence Read Archive. SRP082976
- 158.Gomes SI, Merckx VS, Saavedra S. Fungal-host diversity among mycoheterotrophic plants increases proportionally to their fungal-host overlap. Ecol. Evol. 2017;7:3623–3630. doi: 10.1002/ece3.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.2017. NCBI Sequence Read Archive. SRP083394
- 160.Zhou X, et al. Conversion from long-term cultivated wheat field to Jerusalem artichoke plantation changed soil fungal communities. Sci. Rep. 2017;7:41502. doi: 10.1038/srep41502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.2017. NCBI Sequence Read Archive. SRP083434
- 162.2017. NCBI Sequence Read Archive. SRP083901
- 163.Gomes SI, Aguirre‐Gutiérrez J, Bidartondo MI, Merckx VS. Arbuscular mycorrhizal interactions of mycoheterotrophic Thismia are more specialized than in autotrophic plants. New Phytol. 2017;213:1418–1427. doi: 10.1111/nph.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.2017. NCBI Sequence Read Archive. SRP087715
- 165.Tian H, et al. Changes in soil microbial communities after 10 years of winter wheat cultivation versus fallow in an organic-poor soil in the Loess Plateau of China. PLoS One. 2017;12:e0184223. doi: 10.1371/journal.pone.0184223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.2016. NCBI Sequence Read Archive. SRP090261
- 167.Gourmelon V, et al. Environmental and geographical factors structure soil microbial diversity in New Caledonian ultramafic substrates: a metagenomic approach. PLoS One. 2016;11:e0167405. doi: 10.1371/journal.pone.0167405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.2017. NCBI Sequence Read Archive. SRP090335
- 169.Younginger BS, Ballhorn DJ. Fungal endophyte communities in the temperate fern Polystichum munitum show early colonization and extensive temporal turnover. Am. J. Bot. 2017;104:1188–1194. doi: 10.3732/ajb.1700149. [DOI] [PubMed] [Google Scholar]
- 170.2017. NCBI Sequence Read Archive. SRP090490
- 171.Kamutando CN, et al. Soil nutritional status and biogeography influence rhizosphere microbial communities associated with the invasive tree Acacia dealbata. Sci. Rep. 2017;7:6472. doi: 10.1038/s41598-017-07018-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.2017. NCBI Sequence Read Archive. SRP090651
- 173.Anthony MA, Frey SD, Stinson KA. Fungal community homogenization, shift in dominant trophic guild, and appearance of novel taxa with biotic invasion. Ecosphere. 2017;8:e01951. [Google Scholar]
- 174.2017. NCBI Sequence Read Archive. SRP091741
- 175.Ge ZW, Brenneman T, Bonito G, Smith ME. Soil pH and mineral nutrients strongly influence truffles and other ectomycorrhizal fungi associated with commercial pecans (Carya illinoinensis) Plant Soil. 2017;418:493–505. [Google Scholar]
- 176.2018. NCBI Sequence Read Archive. SRP091855
- 177.Mirmajlessi SM, et al. Survey of soil fungal communities in Strawberry fields by Illumina amplicon sequencing. Eurasian Soil Sci. 2018;51:682–691. [Google Scholar]
- 178.2016. NCBI Sequence Read Archive. SRP091867
- 179.Harrison JG, Forister ML, Parchman TL, Koch GW. Vertical stratification of the foliar fungal community in the world’s tallest trees. Am. J. Bot. 2016;103:2087–2095. doi: 10.3732/ajb.1600277. [DOI] [PubMed] [Google Scholar]
- 180.2019. NCBI Sequence Read Archive. SRP092609
- 181.Semenova‐Nelsen TA, Platt WJ, Patterson TR, Huffman J, Sikes BA. Frequent fire reorganizes fungal communities and slows decomposition across a heterogeneous pine savanna landscape. New Phytol. 2019;224:916–927. doi: 10.1111/nph.16096. [DOI] [PubMed] [Google Scholar]
- 182.2017. NCBI Sequence Read Archive. SRP092777
- 183.Dean SL, et al. A study of Glycine max (soybean) fungal communities under different agricultural practices. Plant Gene. 2017;11:8–16. [Google Scholar]
- 184.2017. NCBI Sequence Read Archive. SRP093592
- 185.Kyaschenko J, Clemmensen KE, Hagenbo A, Karltun E, Lindahl BD. Shift in fungal communities and associated enzyme activities along an age gradient of managed Pinus sylvestris stands. ISME J. 2017;11:863–874. doi: 10.1038/ismej.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.2017. NCBI Sequence Read Archive. SRP093928
- 187.Tian J, et al. Ecological succession pattern of fungal community in soil along a retreating glacier. Front. Microbiol. 2017;8:1028. doi: 10.3389/fmicb.2017.01028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.2017. NCBI Sequence Read Archive. SRP094708
- 189.Oono R, Rasmussen A, Lefèvre E. Distance decay relationships in foliar fungal endophytes are driven by rare taxa. Environ. Microbiol. 2017;19:2794–2805. doi: 10.1111/1462-2920.13799. [DOI] [PubMed] [Google Scholar]
- 190.Oono R. A confidence interval analysis of sampling effort, sequencing depth, and taxonomic resolution of fungal community ecology in the era of high-throughput sequencing. PLoS One. 2017;12:e0189796. doi: 10.1371/journal.pone.0189796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.2017. NCBI Sequence Read Archive. SRP097883
- 192.Marín C, et al. Functional land-use change effects on soil fungal communities in Chilean temperate rainforests. J. Soil Sci. Plant Nut. 2017;17:985–1002. [Google Scholar]
- 193.2017. NCBI Sequence Read Archive. SRP101553
- 194.Siles JA, Margesin R. Seasonal soil microbial responses are limited to changes in functionality at two Alpine forest sites differing in altitude and vegetation. Sci. Rep. 2017;7:2204. doi: 10.1038/s41598-017-02363-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.2018. NCBI Sequence Read Archive. SRP101605
- 196.Kazartsev I, Shorohova E, Kapitsa E, Kushnevskaya H. Decaying Picea abies log bark hosts diverse fungal communities. Fungal Ecol. 2018;33:1–12. [Google Scholar]
- 197.2017. NCBI Sequence Read Archive. SRP102378
- 198.Peay KG, et al. Convergence and contrast in the community structure of bacteria, fungi and archaea along a tropical elevation-climate gradient. FEMS Microbiol. Ecol. 2017;93:fix045. doi: 10.1093/femsec/fix045. [DOI] [PubMed] [Google Scholar]
- 199.2018. NCBI Sequence Read Archive. SRP102417
- 200.Coleine C, et al. Antarctic cryptoendolithic fungal communities are highly adapted and dominated by Lecanoromycetes and Dothideomycetes. Front. Microbiol. 2018;9:1392. doi: 10.3389/fmicb.2018.01392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.2018. NCBI Sequence Read Archive. SRP102775
- 202.Park MS, et al. Diversity of fungi associated with roots of Calanthe orchid species in Korea. J. Microbiol. 2018;56:49–55. doi: 10.1007/s12275-018-7319-9. [DOI] [PubMed] [Google Scholar]
- 203.2018. NCBI Sequence Read Archive. SRP106137
- 204.Glynou K, Nam B, Thines M, Maciá‐Vicente JG. Facultative root-colonizing fungi dominate endophytic assemblages in roots of nonmycorrhizal Microthlaspi species. New Phytol. 2018;217:1190–1202. doi: 10.1111/nph.14873. [DOI] [PubMed] [Google Scholar]
- 205.2018. NCBI Sequence Read Archive. SRP106774
- 206.Saitta A, Anslan S, Bahram M, Brocca L, Tedersoo L. Tree species identity and diversity drive fungal richness and community composition along an elevational gradient in a Mediterranean ecosystem. Mycorrhiza. 2018;28:39–47. doi: 10.1007/s00572-017-0806-8. [DOI] [PubMed] [Google Scholar]
- 207.2017. NCBI Sequence Read Archive. SRP107174
- 208.Almario J, et al. Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proc. Natl. Acad. Sci. USA. 2017;114:E9403–E9412. doi: 10.1073/pnas.1710455114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.2017. NCBI Sequence Read Archive. SRP107743
- 210.Fernández‐Mendoza F, Fleischhacker A, Kopun T, Grube M, Muggia L. ITS1 metabarcoding highlights low specificity of lichen mycobiomes at a local scale. Mol. Ecol. 2017;26:4811–4830. doi: 10.1111/mec.14244. [DOI] [PubMed] [Google Scholar]
- 211.2017. NCBI Sequence Read Archive. SRP109164
- 212.Varenius K, Lindahl BD, Dahlberg A. Retention of seed trees fails to lifeboat ectomycorrhizal fungal diversity in harvested Scots pine forests. FEMS Microbiol. Ecol. 2017;93:fix105. doi: 10.1093/femsec/fix105. [DOI] [PubMed] [Google Scholar]
- 213.2017. NCBI Sequence Read Archive. SRP109773
- 214.He D, et al. Diversity and co-occurrence network of soil fungi are more responsive than those of bacteria to shifts in precipitation seasonality in a subtropical forest. Soil Biol. Biochem. 2017;115:499–510. [Google Scholar]
- 215.2017. NCBI Sequence Read Archive. SRP110522
- 216.Mendoza JR, Kok CR, Stratton J, Bianchini A, Hallen-Adams HE. Understanding the mycobiota of maize from the highlands of Guatemala, and implications for maize quality and safety. Crop Prot. 2017;101:5–11. [Google Scholar]
- 217.2017. NCBI Sequence Read Archive. SRP110810
- 218.Miura T, Sánchez R, Castañeda LE, Godoy K, Barbosa O. Is microbial terroir related to geographic distance between vineyards? Environ. Microbiol. Rep. 2017;9:742–749. doi: 10.1111/1758-2229.12589. [DOI] [PubMed] [Google Scholar]
- 219.2018. NCBI Sequence Read Archive. SRP113348
- 220.Zhang J, et al. Distinct large-scale biogeographic patterns of fungal communities in bulk soil and soybean rhizosphere in China. Sci. Total Environ. 2018;644:791–800. doi: 10.1016/j.scitotenv.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 221.2017. NCBI Sequence Read Archive. SRP114697
- 222.Sharma-Poudyal D, Schlatter D, Yin C, Hulbert S, Paulitz T. Long-term no-till: A major driver of fungal communities in dryland wheat cropping systems. PLoS One. 2017;12:e0184611. doi: 10.1371/journal.pone.0184611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.2018. NCBI Sequence Read Archive. SRP114821
- 224.Ren C, et al. Differential responses of soil microbial biomass, diversity, and compositions to altitudinal gradients depend on plant and soil characteristics. Sci. Total Environ. 2018;610:750–758. doi: 10.1016/j.scitotenv.2017.08.110. [DOI] [PubMed] [Google Scholar]
- 225.2018. NCBI Sequence Read Archive. SRP115350
- 226.Schneider-Maunoury L, et al. Is Tuber melanosporum colonizing the roots of herbaceous, non-ectomycorrhizal plants? Fungal Ecol. 2018;31:59–68. [Google Scholar]
- 227.2018. NCBI Sequence Read Archive. SRP115464
- 228.Sapkota R, Nicolaisen M. Cropping history shapes fungal, oomycete and nematode communities in arable soils and affects cavity spot in carrot. Agric. Ecosyst. Environ. 2018;257:120–131. [Google Scholar]
- 229.2018. NCBI Sequence Read Archive. SRP115599
- 230.Schroeder JW, et al. Community composition and diversity of Neotropical root‐associated fungi in common and rare trees. Biotropica. 2018;50:694–703. [Google Scholar]
- 231.2018. NCBI Sequence Read Archive. SRP117302
- 232.Fan K, Weisenhorn P, Gilbert JA, Chu H. Wheat rhizosphere harbors a less complex and more stable microbial co-occurrence pattern than bulk soil. Soil Biol. Biochem. 2018;125:251–260. [Google Scholar]
- 233.2018. NCBI Sequence Read Archive. SRP118875
- 234.Montagna M, et al. Differential biodiversity responses between kingdoms (plants, fungi, bacteria and metazoa) along an Alpine succession gradient. Mol. Ecol. 2018;27:3671–3685. doi: 10.1111/mec.14817. [DOI] [PubMed] [Google Scholar]
- 235.2018. NCBI Sequence Read Archive. SRP118960
- 236.Schön ME, Nieselt K, Garnica S. Belowground fungal community diversity and composition associated with Norway spruce along an altitudinal gradient. PLoS One. 2018;13:e0208493. doi: 10.1371/journal.pone.0208493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.2017. NCBI Sequence Read Archive. SRP119174
- 238.Thiem D, Piernik A, Hrynkiewicz K. Ectomycorrhizal and endophytic fungi associated with Alnus glutinosa growing in a saline area of central Poland. Symbiosis. 2017;75:17–28. doi: 10.1007/s13199-017-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.2016. NCBI Sequence Read Archive. SRP125864
- 240.Barnes CJ, Maldonado C, Frøslev TG, Antonelli A, Rønsted N. Unexpectedly high beta-diversity of root-associated fungal communities in the Bolivian Andes. Front. Microbiol. 2016;7:1377. doi: 10.3389/fmicb.2016.01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.2018. NCBI Sequence Read Archive. SRP132277
- 242.Schlatter DC, Kahl K, Carlson B, Huggins DR, Paulitz T. Fungal community composition and diversity vary with soil depth and landscape position in a no-till wheat-based cropping system. FEMS Microbiol. Ecol. 2018;94:fiy098. doi: 10.1093/femsec/fiy098. [DOI] [PubMed] [Google Scholar]
- 243.2018. NCBI Sequence Read Archive. SRP132591
- 244.Rasmussen PU, et al. Multiscale patterns and drivers of arbuscular mycorrhizal fungal communities in the roots and root-associated soil of a wild perennial herb. New Phytol. 2018;220:1248–1261. doi: 10.1111/nph.15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.2018. NCBI Sequence Read Archive. SRP132598
- 246.2012. NCBI Sequence Read Archive. SRP136886
- 247.Guo J, et al. Soil fungal assemblage complexity is dependent on soil fertility and dominated by deterministic processes. New Phytol. 2019;226:232–243. doi: 10.1111/nph.16345. [DOI] [PubMed] [Google Scholar]
- 248.2019. NCBI Sequence Read Archive. SRP139483
- 249.Song H, et al. Tropical forest conversion to rubber plantation in southwest China results in lower fungal beta diversity and reduced network complexity. FEMS Microbiol. Ecol. 2019;95:fiz092. doi: 10.1093/femsec/fiz092. [DOI] [PubMed] [Google Scholar]
- 250.2018. NCBI Sequence Read Archive. SRP142723
- 251.Rogers TJ, et al. Exploring variation in phyllosphere microbial communities across four hemlock species. Ecosphere. 2018;9:e02524. [Google Scholar]
- 252.2018. NCBI Sequence Read Archive. SRP148813
- 253.Schlegel M, Queloz V, Sieber TN. The endophytic mycobiome of European Ash and Sycamore Maple leaves – geographic patterns, host specificity and influence of Ash Dieback. Front. Microbiol. 2018;9:2345. doi: 10.3389/fmicb.2018.02345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.2019. NCBI Sequence Read Archive. SRP150527
- 255.Truong C, et al. Ectomycorrhizal fungi and soil enzymes exhibit contrasting patterns along elevation gradients in southern Patagonia. New Phytol. 2019;222:1936–1950. doi: 10.1111/nph.15714. [DOI] [PubMed] [Google Scholar]
- 256.2018. NCBI Sequence Read Archive. SRP151262
- 257.Jiao S, et al. Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome. 2018;6:146. doi: 10.1186/s40168-018-0526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.2018. NCBI Sequence Read Archive. SRP153934
- 259.Marasco R, et al. Rhizosheath microbial community assembly of sympatric desert speargrasses is independent of the plant host. Microbiome. 2018;6:215. doi: 10.1186/s40168-018-0597-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.2018. NCBI Sequence Read Archive. SRP160913
- 261.Bickford WA, Goldberg DE, Kowalski KP, Zak DR. Root endophytes and invasiveness: no difference between native and non‐native Phragmites in the Great Lakes region. Ecosphere. 2018;9:e02526. [Google Scholar]
- 262.2018. NCBI Sequence Read Archive. SRP161632
- 263.Si P, et al. Rhizosphere microenvironments of eight common deciduous fruit trees were shaped by microbes in Northern China. Front. Microbiol. 2018;9:3147. doi: 10.3389/fmicb.2018.03147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.2019. NCBI Sequence Read Archive. SRP195764
- 265.Purahong W, Wu YT, Chen CT, Mapook A. Characterization of the Castanopsis carlesii deadwood mycobiome by Pacbio sequencing of the full-length fungal nuclear ribosomal internal transcribed spacer (ITS) Front. Microbiol. 2019;10:983. doi: 10.3389/fmicb.2019.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.2014. European Nucleotide Archive. ERP001713
- 267.Geml J, et al. The contribution of DNA metabarcoding to fungal conservation: diversity assessment, habitat partitioning and mapping red-listed fungi in protected coastal Salix repens communities in the Netherlands. PLoS One. 2014;9:e99852. doi: 10.1371/journal.pone.0099852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.2013. European Nucleotide Archive. ERP003251
- 269.Schmidt PA, et al. Illumina metabarcoding of a soil fungal community. Soil Biol. Biochem. 2013;65:128–132. [Google Scholar]
- 270.2015. European Nucleotide Archive. ERP003790
- 271.van der Wal A, Ottosson E, De Boer W. Neglected role of fungal community composition in explaining variation in wood decay rates. Ecology. 2015;96:124–133. doi: 10.1890/14-0242.1. [DOI] [PubMed] [Google Scholar]
- 272.2015. European Nucleotide Archive. ERP005177
- 273.Muller LA, Hilger HH. Insights into the effects of serpentine soil conditions on the community composition of fungal symbionts in the roots of Onosma echioides. Soil Biol. Biochem. 2015;81:1–8. [Google Scholar]
- 274.2015. European Nucleotide Archive. ERP005905
- 275.Sun H, et al. Fungal community shifts in structure and function across a boreal forest fire chronosequence. Appl. Environ. Microbiol. 2015;81:7869–7880. doi: 10.1128/AEM.02063-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.2015. European Nucleotide Archive. ERP009341
- 277.Rajala T, Tuomivirta T, Pennanen T, Mäkipää R. Habitat models of wood-inhabiting fungi along a decay gradient of Norway spruce logs. Fungal Ecol. 2015;18:48–55. [Google Scholar]
- 278.2017. European Nucleotide Archive. ERP010027
- 279.Purahong W, et al. Characterization of unexplored deadwood mycobiome in highly diverse subtropical forests using culture-independent molecular technique. Front. Microbiol. 2017;8:574. doi: 10.3389/fmicb.2017.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.2016. European Nucleotide Archive. ERP010084
- 281.van der Wal A, Gunnewiek PJK, Cornelissen JHC, Crowther TW, de Boer W. Patterns of natural fungal community assembly during initial decay of coniferous and broadleaf tree logs. Ecosphere. 2016;7:e01393. [Google Scholar]
- 282.2016. European Nucleotide Archive. ERP010743
- 283.Reese AT, et al. Urban stress is associated with variation in microbial species composition-but not richness-in Manhattan. ISME J. 2016;10:751–760. doi: 10.1038/ismej.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.2016. European Nucleotide Archive. ERP011924
- 285.Kielak AM, Scheublin TR, Mendes LW, Van Veen JA, Kuramae EE. Bacterial community succession in Pine-wood decomposition. Front. Microbiol. 2016;7:231. doi: 10.3389/fmicb.2016.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.2016. European Nucleotide Archive. ERP012017
- 287.Santalahti, M., Sun, H., Jumpponen, A., Pennanen, T. & Heinonsalo, J. Vertical and seasonal dynamics of fungal communities in boreal Scots pine forest soil. FEMS Microbiol. Ecol. 92, fiw170 (2016). [DOI] [PubMed]
- 288.2016. European Nucleotide Archive. ERP013208
- 289.Frey B, et al. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016;92:fiw018. doi: 10.1093/femsec/fiw018. [DOI] [PubMed] [Google Scholar]
- 290.2017. European Nucleotide Archive. ERP013987
- 291.Wilhelm RC, et al. A metagenomic survey of forest soil microbial communities more than a decade after timber harvesting. Sci. Data. 2017;4:170092. doi: 10.1038/sdata.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.2016. European Nucleotide Archive. ERP014227
- 293.Lanzén A, et al. Multi-targeted metagenetic analysis of the influence of climate and environmental parameters on soil microbial communities along an elevational gradient. Sci. Rep. 2016;6:28257. doi: 10.1038/srep28257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.2018. European Nucleotide Archive. ERP017480
- 295.Purahong W, et al. Increasing N deposition impacts neither diversity nor functions of deadwood‐inhabiting fungal communities, but adaptation and functional redundancy ensure ecosystem function. Environ. Microbiol. 2018;20:1693–1710. doi: 10.1111/1462-2920.14081. [DOI] [PubMed] [Google Scholar]
- 296.2017. European Nucleotide Archive. ERP017851
- 297.Yang T, et al. Soil fungal diversity in natural grasslands of the Tibetan Plateau: associations with plant diversity and productivity. New Phytol. 2017;215:756–765. doi: 10.1111/nph.14606. [DOI] [PubMed] [Google Scholar]
- 298.2017. European Nucleotide Archive. ERP017915
- 299.Nguyen D, et al. Foliar fungi of Betula pendula: impact of tree species mixtures and assessment methods. Sci. Rep. 2017;7:41801. doi: 10.1038/srep41801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.2017. European Nucleotide Archive. ERP019580
- 301.Tu B, et al. Microbial diversity in chinese temperate steppe: unveiling the most influential environmental drivers. FEMS Microbiol. Ecol. 2017;93:fix031. doi: 10.1093/femsec/fix031. [DOI] [PubMed] [Google Scholar]
- 302.2017. European Nucleotide Archive. ERP019924
- 303.Yang T, et al. Fungal community assemblages in a high elevation desert environment: absence of dispersal limitation and edaphic effects in surface soil. Soil Biol. Biochem. 2017;115:393–402. [Google Scholar]
- 304.2017. European Nucleotide Archive. ERP020657
- 305.van der Wal A, Gunnewiek PK, de Hollander M, de Boer W. Fungal diversity and potential tree pathogens in decaying logs and stumps. Forest Ecol. Manag. 2017;406:266–273. [Google Scholar]
- 306.2019. European Nucleotide Archive. ERP022511
- 307.Alzarhani AK, et al. Are drivers of root-associated fungal community structure context specific? ISME J. 2019;13:1330–1344. doi: 10.1038/s41396-019-0350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.2017. European Nucleotide Archive. ERP022742
- 309.van der Wal A, Gunnewiek PK, de Boer W. Soil-wood interactions: Influence of decaying coniferous and broadleaf logs on composition of soil fungal communities. Fungal Ecol. 2017;30:132–134. [Google Scholar]
- 310.2018. European Nucleotide Archive. ERP023275
- 311.Purahong W, et al. Determinants of deadwood-inhabiting fungal communities in temperate forests: molecular evidence from a large scale deadwood decomposition experiment. Front. Microbiol. 2018;9:2120. doi: 10.3389/fmicb.2018.02120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.2018. European Nucleotide Archive. ERP023718
- 313.Sun R, et al. Tillage changes vertical distribution of soil bacterial and fungal communities. Front. Microbiol. 2018;9:699. doi: 10.3389/fmicb.2018.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.2018. European Nucleotide Archive. ERP023855
- 315.Santalahti M, et al. Reindeer grazing alter soil fungal community structure and litter decomposition related enzyme activities in boreal coniferous forests in finnish lapland. Appl. Soil Ecol. 2018;132:74–82. [Google Scholar]
- 316.2018. European Nucleotide Archive. ERP106131
- 317.Gałązka A, Grządziel J. Fungal genetics and functional diversity of microbial communities in the soil under long-term monoculture of Maize using different cultivation techniques. Front. Microbiol. 2018;9:76. doi: 10.3389/fmicb.2018.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.2019. European Nucleotide Archive. ERP107634
- 319.Ramirez KS, et al. Range-expansion effects on the belowground plant microbiome. Nat. Ecol. Evol. 2019;3:604. doi: 10.1038/s41559-019-0828-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.2019. European Nucleotide Archive. ERP107636
- 321.2019. European Nucleotide Archive. ERP110188
- 322.George PB, et al. Divergent national-scale trends of microbial and animal biodiversity revealed across diverse temperate soil ecosystems. Nat. Commun. 2019;10:1107. doi: 10.1038/s41467-019-09031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.2019. European Nucleotide Archive. ERP112007
- 324.Álvarez-Garrido L, Viñegla B, Hortal S, Powell JR, Carreira JA. Distributional shifts in ectomycorrizhal fungal communities lag behind climate-driven tree upward migration in a conifer forest-high elevation shrubland ecotone. Soil Biol. Biochem. 2019;137:107545. [Google Scholar]
- 325.2014. DNA Data Bank of Japan. DRA000926
- 326.Yamamoto S, et al. Spatial segregation and aggregation of ectomycorrhizal and root-endophytic fungi in the seedlings of two Quercus species. PLoS One. 2014;9:e96363. doi: 10.1371/journal.pone.0096363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.2014. DNA Data Bank of Japan. DRA000937
- 328.Kadowaki K, et al. Detection of the horizontal spatial structure of soil fungal communities in a natural forest. Popul. Ecol. 2014;56:301–310. [Google Scholar]
- 329.2016. DNA Data Bank of Japan. DRA001737
- 330.Izuno A, et al. Structure of phyllosphere fungal communities in a tropical dipterocarp plantation: A massively parallel next-generation sequencing analysis. Mycoscience. 2016;57:171–180. [Google Scholar]
- 331.2016. DNA Data Bank of Japan. DRA002424
- 332.Matsuoka S, Kawaguchi E, Osono T. Temporal distance decay of similarity of ectomycorrhizal fungal community composition in a subtropical evergreen forest in Japan. FEMS Microbiol. Ecol. 2016;92:fiw061. doi: 10.1093/femsec/fiw061. [DOI] [PubMed] [Google Scholar]
- 333.2016. DNA Data Bank of Japan. DRA002469
- 334.Izuno A, Kanzaki M, Artchawakom T, Wachrinrat C, Isagi Y. Vertical structure of phyllosphere fungal communities in a tropical forest in Thailand uncovered by high-throughput sequencing. PLoS One. 2016;11:e0166669. doi: 10.1371/journal.pone.0166669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.2016. DNA Data Bank of Japan. DRA003024
- 336.Matsuoka S, Mori AS, Kawaguchi E, Hobara S, Osono T. Disentangling the relative importance of host tree community, abiotic environment and spatial factors on ectomycorrhizal fungal assemblages along an elevation gradient. FEMS Microbiol. Ecol. 2016;92:fiw044. doi: 10.1093/femsec/fiw044. [DOI] [PubMed] [Google Scholar]
- 337.2016. DNA Data Bank of Japan. DRA003730
- 338.Toju H, Yamamoto S, Tanabe AS, Hayakawa T, Ishii HS. Network modules and hubs in plant-root fungal biomes. J. R. Soc. Interface. 2016;13:20151097. doi: 10.1098/rsif.2015.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.2017. DNA Data Bank of Japan. DRA004913
- 340.Shen Z, et al. Banana Fusarium Wilt disease incidence is influenced by shifts of soil microbial communities under different monoculture spans. Microb. Ecol. 2017;75:739–750. doi: 10.1007/s00248-017-1052-5. [DOI] [PubMed] [Google Scholar]
- 341.2018. DNA Data Bank of Japan. DRA006519
- 342.Matsuoka S, Ogisu Y, Sakoh S, Hobara S, Osono T. Taxonomic, functional, and phylogenetic diversity of fungi along primary successional and elevational gradients near Mount Robson, British Columbia. Polar Sci. 2018;21:165–171. [Google Scholar]
- 343.2015. DNA Data Bank of Japan. DRP002783
- 344.Fukasawa Y, Matsuoka S. Communities of wood-inhabiting fungi in dead pine logs along a geographical gradient in Japan. Fungal Ecol. 2015;18:75–82. [Google Scholar]
- 345.2016. DNA Data Bank of Japan. DRP003138
- 346.Toju H, Tanabe AS, Ishii HS. Ericaceous plant-fungus network in a harsh alpine-subalpine environment. Mol. Ecol. 2016;25:3242–3257. doi: 10.1111/mec.13680. [DOI] [PubMed] [Google Scholar]
- 347.2019. DNA Data Bank of Japan. DRP005365
- 348.Shigyo N, Umeki K, Hirao T. Seasonal dynamics of soil fungal and bacterial communities in cool-temperate montane forests. Front. Microbiol. 2019;10:1944. doi: 10.3389/fmicb.2019.01944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Semenova TA, 2014. Data from: Long-term experimental warming alters community composition of ascomycetes in Alaskan moist and dry arctic tundra. Dryad. [DOI] [PubMed]
- 350.Geml J, et al. Long-term warming alters richness and composition of taxonomic and functional groups of arctic fungi. FEMS Microbiol. Ecol. 2015;91:fiv095. doi: 10.1093/femsec/fiv095. [DOI] [PubMed] [Google Scholar]
- 351.Oriol G, 2017. Data from: Abrupt changes in the composition and function of fungal communities along an environmental gradient in the High. Arctic. Dryad. [DOI] [PubMed]
- 352.Grau O, et al. Abrupt changes in the composition and function of fungal communities along an environmental gradient in the high Arctic. Mol. Ecol. 2017;26:4798–4810. doi: 10.1111/mec.14227. [DOI] [PubMed] [Google Scholar]
- 353.Mundra S, 2015. Data from: Arctic fungal communities associated with roots of Bistorta vivipara do not respond to the same fine-scale edaphic gradients as the above-ground vegetation. Dryad. [DOI] [PubMed]
- 354.Mundra S, et al. Arctic fungal communities associated with roots of Bistorta vivipara do not respond to the same fine-scale edaphic gradients as the aboveground vegetation. New Phytol. 2015;205:1587–1597. doi: 10.1111/nph.13216. [DOI] [PubMed] [Google Scholar]
- 355.Rime T, 2014. Data from: Vertical distribution of the soil microbiota along a successional gradient in a glacier forefield. Dryad. [DOI] [PubMed]
- 356.Rime T, et al. Vertical distribution of the soil microbiota along a successional gradient in a glacier forefield. Mol. Ecol. 2015;24:1091–1108. doi: 10.1111/mec.13051. [DOI] [PubMed] [Google Scholar]
- 357.Semenova TA, 2017. Data from: Compositional and functional shifts in arctic fungal communities in response to experimentally increased snow depth. Dryad. [DOI]
- 358.Semenova TA, et al. Compositional and functional shifts in arctic fungal communities in response to experimentally increased snow depth. Soil Biol. Biochem. 2016;100:201–209. [Google Scholar]
- 359.Geml J, 2014. Data from: Large-scale fungal diversity assessment in the Andean Yungas forests reveals strong community turnover among forest types along an altitudinal gradient. Dryad. [DOI] [PubMed]
- 360.Wicaksono CY, et al. Contracting montane cloud forests: a case study of the Andean alder (Alnusacuminata) and associated fungi in the Yungas. Biotropica. 2017;49:141–152. [Google Scholar]
- 361.Yao F, 2013. Data from: Substantial compositional turnover of fungal communities in an alpine ridge-to-snowbed gradient. Dryad. [DOI] [PubMed]
- 362.Yao F, et al. Substantial compositional turnover of fungal communities in an alpine ridge-to-snowbed gradient. Mol. Ecol. 2013;22:5040–5052. doi: 10.1111/mec.12437. [DOI] [PubMed] [Google Scholar]
- 363.Schappe T, 2017. Uncultured fungus internal transcribed spacer 1, targeted locus study. GenBank. KAYV00000000.1
- 364.Schappe T, et al. The role of soil chemistry and plant neighbourhoods in structuring fungal communities in three Panamanian rainforests. J. Ecol. 2017;105:569–579. [Google Scholar]
- 365.Schappe T, 2017. Uncultured fungus internal transcribed spacer 1, targeted locus study. GenBank. KAYU00000000.1
- 366.Schappe T, 2017. Uncultured fungus internal transcribed spacer 1, targeted locus study. GenBank. KAYT00000000.1
- 367.Vaz AB, 2017. MIMS Environmental/Metagenome sample from biofilm metagenome. BioSample. SAMN02934078
- 368.Vaz AB, et al. Using Next-Generation Sequencing (NGS) to uncover diversity of wood-decaying fungi in neotropical atlantic forests. Phytotaxa. 2017;295:1–21. [Google Scholar]
- 369.Vaz AB, 2017. MIMS Environmental/Metagenome sample from biofilm metagenome. BioSample. SAMN02934079
- 370.Siciliano S, 2014. Polar soil bacterial and fungal biodiversity survey. Australian Antarctic Data Centre. [DOI]
- 371.Ji M, et al. Microbial diversity at Mitchell Peninsula, Eastern Antarctica: a potential biodiversity “hotspot”. Polar Biol. 2016;39:237–249. [Google Scholar]
- 372.Hartmann M, et al. Significant and persistent impact of timber harvesting on soil microbial communities in Northern coniferous forests. ISME J. 2012;6:2199–2218. doi: 10.1038/ismej.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Rime T, Hartmann M, Frey B. Potential sources of microbial colonizers in an initial soil ecosystem after retreat of an alpine glacier. ISME J. 2016;10:1625–1641. doi: 10.1038/ismej.2015.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Baldrian P, 2020. GlobalFungi: Global database of fungal records from high-throughput-sequencing metabarcoding studies. figshare. [DOI] [PMC free article] [PubMed]
- 2010. NCBI Sequence Read Archive. SRP001058
- 2010. NCBI Sequence Read Archive. SRP001175
- 2011. NCBI Sequence Read Archive. SRP006078
- 2012. NCBI Sequence Read Archive. SRP012868
- 2012. NCBI Sequence Read Archive. SRP013695
- 2016. NCBI Sequence Read Archive. SRP013944
- 2015. NCBI Sequence Read Archive. SRP015735
- 2015. NCBI Sequence Read Archive. SRP016090
- 2014. NCBI Sequence Read Archive. SRP026207
- 2015. NCBI Sequence Read Archive. SRP028404
- 2015. NCBI Sequence Read Archive. SRP033719
- 2015. NCBI Sequence Read Archive. SRP035356
- 2014. NCBI Sequence Read Archive. SRP040314
- 2015. NCBI Sequence Read Archive. SRP040786
- 2015. NCBI Sequence Read Archive. SRP041347
- 2015. NCBI Sequence Read Archive. SRP043106
- 2017. NCBI Sequence Read Archive. SRP043706
- 2015. NCBI Sequence Read Archive. SRP043982
- 2016. NCBI Sequence Read Archive. SRP044665
- 2015. NCBI Sequence Read Archive. SRP045166
- 2016. NCBI Sequence Read Archive. SRP045587
- 2014. NCBI Sequence Read Archive. SRP045746
- 2015. NCBI Sequence Read Archive. SRP045933
- 2016. NCBI Sequence Read Archive. SRP046049
- 2016. NCBI Sequence Read Archive. SRP048036
- 2015. NCBI Sequence Read Archive. SRP048856
- 2015. NCBI Sequence Read Archive. SRP049544
- 2016. NCBI Sequence Read Archive. SRP051033
- 2017. NCBI Sequence Read Archive. SRP052222
- 2015. NCBI Sequence Read Archive. SRP052716
- 2015. NCBI Sequence Read Archive. SRP055957
- 2016. NCBI Sequence Read Archive. SRP057433
- 2016. NCBI Sequence Read Archive. SRP057541
- 2016. NCBI Sequence Read Archive. SRP058508
- 2016. NCBI Sequence Read Archive. SRP058555
- 2018. NCBI Sequence Read Archive. SRP058851
- 2016. NCBI Sequence Read Archive. SRP059280
- 2016. NCBI Sequence Read Archive. SRP060838
- 2016. NCBI Sequence Read Archive. SRP061179
- 2017. NCBI Sequence Read Archive. SRP061305
- 2015. NCBI Sequence Read Archive. SRP061904
- 2016. NCBI Sequence Read Archive. SRP062647
- 2017. NCBI Sequence Read Archive. SRP063711
- 2017. NCBI Sequence Read Archive. SRP064158
- 2017. NCBI Sequence Read Archive. SRP065817
- 2016. NCBI Sequence Read Archive. SRP066030
- 2017. NCBI Sequence Read Archive. SRP066284
- 2017. NCBI Sequence Read Archive. SRP066331
- 2017. NCBI Sequence Read Archive. SRP067301
- 2016. NCBI Sequence Read Archive. SRP067367
- 2016. NCBI Sequence Read Archive. SRP068514
- 2016. NCBI Sequence Read Archive. SRP068608
- 2016. NCBI Sequence Read Archive. SRP068620
- 2016. NCBI Sequence Read Archive. SRP068654
- 2017. NCBI Sequence Read Archive. SRP069065
- 2017. NCBI Sequence Read Archive. SRP069742
- 2016. NCBI Sequence Read Archive. SRP070568
- 2016. NCBI Sequence Read Archive. SRP073070
- 2017. NCBI Sequence Read Archive. SRP073265
- 2016. NCBI Sequence Read Archive. SRP074055
- 2016. NCBI Sequence Read Archive. SRP074496
- 2017. NCBI Sequence Read Archive. SRP075989
- 2017. NCBI Sequence Read Archive. SRP079403
- 2018. NCBI Sequence Read Archive. SRP079521
- 2016. NCBI Sequence Read Archive. SRP080210
- 2017. NCBI Sequence Read Archive. SRP080428
- 2017. NCBI Sequence Read Archive. SRP080680
- 2017. NCBI Sequence Read Archive. SRP082472
- 2017. NCBI Sequence Read Archive. SRP082976
- 2017. NCBI Sequence Read Archive. SRP083394
- 2017. NCBI Sequence Read Archive. SRP083434
- 2017. NCBI Sequence Read Archive. SRP083901
- 2017. NCBI Sequence Read Archive. SRP087715
- 2016. NCBI Sequence Read Archive. SRP090261
- 2017. NCBI Sequence Read Archive. SRP090335
- 2017. NCBI Sequence Read Archive. SRP090490
- 2017. NCBI Sequence Read Archive. SRP090651
- 2017. NCBI Sequence Read Archive. SRP091741
- 2018. NCBI Sequence Read Archive. SRP091855
- 2016. NCBI Sequence Read Archive. SRP091867
- 2019. NCBI Sequence Read Archive. SRP092609
- 2017. NCBI Sequence Read Archive. SRP092777
- 2017. NCBI Sequence Read Archive. SRP093592
- 2017. NCBI Sequence Read Archive. SRP093928
- 2017. NCBI Sequence Read Archive. SRP094708
- 2017. NCBI Sequence Read Archive. SRP097883
- 2017. NCBI Sequence Read Archive. SRP101553
- 2018. NCBI Sequence Read Archive. SRP101605
- 2017. NCBI Sequence Read Archive. SRP102378
- 2018. NCBI Sequence Read Archive. SRP102417
- 2018. NCBI Sequence Read Archive. SRP102775
- 2018. NCBI Sequence Read Archive. SRP106137
- 2018. NCBI Sequence Read Archive. SRP106774
- 2017. NCBI Sequence Read Archive. SRP107174
- 2017. NCBI Sequence Read Archive. SRP107743
- 2017. NCBI Sequence Read Archive. SRP109164
- 2017. NCBI Sequence Read Archive. SRP109773
- 2017. NCBI Sequence Read Archive. SRP110522
- 2017. NCBI Sequence Read Archive. SRP110810
- 2018. NCBI Sequence Read Archive. SRP113348
- 2017. NCBI Sequence Read Archive. SRP114697
- 2018. NCBI Sequence Read Archive. SRP114821
- 2018. NCBI Sequence Read Archive. SRP115350
- 2018. NCBI Sequence Read Archive. SRP115464
- 2018. NCBI Sequence Read Archive. SRP115599
- 2018. NCBI Sequence Read Archive. SRP117302
- 2018. NCBI Sequence Read Archive. SRP118875
- 2018. NCBI Sequence Read Archive. SRP118960
- 2017. NCBI Sequence Read Archive. SRP119174
- 2016. NCBI Sequence Read Archive. SRP125864
- 2018. NCBI Sequence Read Archive. SRP132277
- 2018. NCBI Sequence Read Archive. SRP132591
- 2018. NCBI Sequence Read Archive. SRP132598
- 2012. NCBI Sequence Read Archive. SRP136886
- 2019. NCBI Sequence Read Archive. SRP139483
- 2018. NCBI Sequence Read Archive. SRP142723
- 2018. NCBI Sequence Read Archive. SRP148813
- 2019. NCBI Sequence Read Archive. SRP150527
- 2018. NCBI Sequence Read Archive. SRP151262
- 2018. NCBI Sequence Read Archive. SRP153934
- 2018. NCBI Sequence Read Archive. SRP160913
- 2018. NCBI Sequence Read Archive. SRP161632
- 2019. NCBI Sequence Read Archive. SRP195764
- 2014. European Nucleotide Archive. ERP001713
- 2013. European Nucleotide Archive. ERP003251
- 2015. European Nucleotide Archive. ERP003790
- 2015. European Nucleotide Archive. ERP005177
- 2015. European Nucleotide Archive. ERP005905
- 2015. European Nucleotide Archive. ERP009341
- 2017. European Nucleotide Archive. ERP010027
- 2016. European Nucleotide Archive. ERP010084
- 2016. European Nucleotide Archive. ERP010743
- 2016. European Nucleotide Archive. ERP011924
- 2016. European Nucleotide Archive. ERP012017
- 2016. European Nucleotide Archive. ERP013208
- 2017. European Nucleotide Archive. ERP013987
- 2016. European Nucleotide Archive. ERP014227
- 2018. European Nucleotide Archive. ERP017480
- 2017. European Nucleotide Archive. ERP017851
- 2017. European Nucleotide Archive. ERP017915
- 2017. European Nucleotide Archive. ERP019580
- 2017. European Nucleotide Archive. ERP019924
- 2017. European Nucleotide Archive. ERP020657
- 2019. European Nucleotide Archive. ERP022511
- 2017. European Nucleotide Archive. ERP022742
- 2018. European Nucleotide Archive. ERP023275
- 2018. European Nucleotide Archive. ERP023718
- 2018. European Nucleotide Archive. ERP023855
- 2018. European Nucleotide Archive. ERP106131
- 2019. European Nucleotide Archive. ERP107634
- 2019. European Nucleotide Archive. ERP107636
- 2019. European Nucleotide Archive. ERP110188
- 2019. European Nucleotide Archive. ERP112007
- 2014. DNA Data Bank of Japan. DRA000926
- 2014. DNA Data Bank of Japan. DRA000937
- 2016. DNA Data Bank of Japan. DRA001737
- 2016. DNA Data Bank of Japan. DRA002424
- 2016. DNA Data Bank of Japan. DRA002469
- 2016. DNA Data Bank of Japan. DRA003024
- 2016. DNA Data Bank of Japan. DRA003730
- 2017. DNA Data Bank of Japan. DRA004913
- 2018. DNA Data Bank of Japan. DRA006519
- 2015. DNA Data Bank of Japan. DRP002783
- 2016. DNA Data Bank of Japan. DRP003138
- 2019. DNA Data Bank of Japan. DRP005365
- Semenova TA, 2014. Data from: Long-term experimental warming alters community composition of ascomycetes in Alaskan moist and dry arctic tundra. Dryad. [DOI] [PubMed]
- Oriol G, 2017. Data from: Abrupt changes in the composition and function of fungal communities along an environmental gradient in the High. Arctic. Dryad. [DOI] [PubMed]
- Mundra S, 2015. Data from: Arctic fungal communities associated with roots of Bistorta vivipara do not respond to the same fine-scale edaphic gradients as the above-ground vegetation. Dryad. [DOI] [PubMed]
- Rime T, 2014. Data from: Vertical distribution of the soil microbiota along a successional gradient in a glacier forefield. Dryad. [DOI] [PubMed]
- Semenova TA, 2017. Data from: Compositional and functional shifts in arctic fungal communities in response to experimentally increased snow depth. Dryad. [DOI]
- Geml J, 2014. Data from: Large-scale fungal diversity assessment in the Andean Yungas forests reveals strong community turnover among forest types along an altitudinal gradient. Dryad. [DOI] [PubMed]
- Yao F, 2013. Data from: Substantial compositional turnover of fungal communities in an alpine ridge-to-snowbed gradient. Dryad. [DOI] [PubMed]
- Schappe T, 2017. Uncultured fungus internal transcribed spacer 1, targeted locus study. GenBank. KAYV00000000.1
- Schappe T, 2017. Uncultured fungus internal transcribed spacer 1, targeted locus study. GenBank. KAYU00000000.1
- Schappe T, 2017. Uncultured fungus internal transcribed spacer 1, targeted locus study. GenBank. KAYT00000000.1
- Vaz AB, 2017. MIMS Environmental/Metagenome sample from biofilm metagenome. BioSample. SAMN02934078
- Vaz AB, 2017. MIMS Environmental/Metagenome sample from biofilm metagenome. BioSample. SAMN02934079
- Siciliano S, 2014. Polar soil bacterial and fungal biodiversity survey. Australian Antarctic Data Centre. [DOI]
Data Availability Statement
The workflow included several custom made python scripts (labelled by star in the Fig. 2) which are accessible here: https://github.com/VetrovskyTomas/GlobalFungi.