Abstract
Aneuploidy occurs within a significant proportion of childhood B-cell acute lymphoblastic leukemia (B-ALL). Some copy number variations (CNV), associated with novel subtypes of childhood B-ALL, have prognostic significance. A total of 233 childhood B-ALL patients were enrolled into this study. Focal copy number alterations of ERG, IKZF1, PAX5, ETV6, RB1, BTG1, EBF1, CDKN2A/2B, and the Xp22.33/Yp11.31 region were assessed by Multiplex Ligation-dependent Probe Amplification (MLPA). The MLPA telomere kit was used to identify aneuploidy through detection of whole chromosome loss or gain. We carried out these procedures alongside measurement of DNA index in order to identify, aneuploidy status in our cohort. MLPA telomere data and DNA index correlated well with aneuploidy status at higher sensitivity than cytogenetic analysis. Three masked hypodiploid patients, undetected by cytogenetics, and their associated copy number neutral loss of heterozygosity (CN-LOH) were identified by STR and SNP arrays. Rearrangements of TCF3, located to 19p, were frequently associated with 19p deletions. Other genetic alterations including iAMP21, IKZF1 deletions, ERG deletions, PAX5AMP, which have clinical significance or are associated with novel subtypes of ALL, were identified. In conclusion, appropriate application of MLPA aids the identifications of CNV and aneuploidy in childhood B-ALL.
Subject terms: Medical research, Molecular medicine
Introduction
As the most common pediatric cancer, acute lymphoblastic leukemia (ALL) accounts for approximately 25% of childhood malignancies. With improved risk-directed treatment and supportive care, the overall 5-year event-free survival rates for this disease now exceed 80% in developed countries1–3. The two major features of risk-directed therapy are based on the genetic alterations of the leukemic cells at diagnosis and the determination of initial treatment response (measured by minimal residual disease, MRD, after induction therapy). The interpretation of MRD levels depends upon the subtype of ALL4,5. Although karyotyping has been the most common approach for detection of numerical chromosomal changes, molecular methods may enhance their detection in childhood B-ALL. Multiplex Ligation-dependent Probe Amplification (MLPA) is a sensitive method based upon the multiplex polymerase chain reaction and capillary electrophoresis that detects multiple copies of around 50 different genomic DNA targets. It has the advantage of lower price and quicker turn-around time than DNA arrays for identification of the important genetic alterations and is now widely used for detection of the important copy number changes in ALL6,7.
Gain or loss of whole chromosomes (aneuploidy) and intrachromosomal amplification of chromosome 21 (iAMP21) accounts for almost 30% of childhood B-ALL identified by traditional methods. High hyperdiploidy with greater than 50 chromosomes comprises up to 30% of childhood B-ALL and most commonly involves gains of chromosomes X, 4, 10, 14, 17 and 218. It is associated with a good outcome, even in patients with induction failure9. Hypodiploidy with less than 44 chromosomes is less common (found in approximately 3% of cases) and is associated with an inferior outcome. Hypodiploid B-ALL can be further divided into three subgroups according to chromosome number. The most common are near-haploidy with 24–31 chromosomes and low-hypodiploidy with 32–39 chromosomes. High-hypodiploidy with 40–43 chromosomes is rare. Low-hypodiploid ALL has a high incidence of TP53 germline mutations10. DNA index (DI) is a well-established method for detection of high hyperdiploidy. The MLPA telomere kit identifies specific gain or loss of individual chromosomes and is suitable for screening for whole chromosome numerical changes11,12. Masked low hypodiploidy, manifesting as doubling of the low hypodiploid chromosome number, can be difficult to diagnose13. Here we show that MLPA and DI are useful in its detection, as confirmed by single-nucleotide polymorphism (SNP) arrays and short tandem repeats (STR). B-ALL patients with iAMP21-ALL were initially shown to have a high relapse risk on standard chemotherapy14,15. It was later demonstrated that treatment on intensive therapy regimens significantly reduced their risk of relapse16–18.
In childhood B-ALL, SNP arrays have successfully identified copy number abnormalities (CNA)19 involving several signaling pathways. For example, deletions of a number of genes within the B-cell differentiation pathway were identified, including PAX5, EFB1 and IKZF119,20. Clinically, IKZF1 alterations have been associated with a poor outcome, particularly in association with Ph-positive (Philadelphia chromosome/BCR-ABL1 positive), and Ph-like ALL (Philadelphia chromosome/BCR-ABL1 negative but the expression profiles were similar to Ph-positive ALL)20–27. Ph-like and iAMP21-ALL have been proposed as novel subtypes of B-ALL in the recent WHO classification of hematologic malignancies, due to their poor prognostic associations28.
In this project, we have used MLPA and DI to study CNA in B-ALL. We show that these approaches are complementary to cytogenetics in improving detection of genetic alterations in childhood B-ALL.
Materials and methods
Patients and protocols
Diagnostic bone marrow (BM) or peripheral blood was obtained from 233 children with B-ALL from January 2002 to July 2018 at the National Taiwan University Hospital. A total of 108 patients were treated on the Taiwan Pediatric Oncology Group TPOG-ALL-2002 protocol, while 125 were treated on TPOG-ALL-2013. Diagnosis of B-ALL was based on BM morphology and the immunophenotype of leukemic cells was determined by flow cytometry. Conventional cytogenetic analysis was carried out as part of the routine work-up29.
The risk-directed TPOG protocols consist of multiple chemotherapeutic agents of different intensities. The treatment protocol was intensified if complete remission was not achieved after initial induction therapy. After 2013, MRD levels were added to risk assignment for therapy. Events were defined as any relapse, death, or secondary malignancy. The Institutional Review Board of National Taiwan University Hospital approved the study and all of the participants or their guardians provided written informed consent in accordance with the Declaration of Helsinki. Details of the protocols and risk group assignment have been published elsewhere27,30,31. We have summarized the risk classification of protocols in the Supplementary information.
Genomic DNA extraction
Lymphoblasts were purified from bone marrow or peripheral blood specimens using the Ficoll-Paque centrifugation method, according to the manufacturer’s instructions (GE Healthcare, Piscataway, NJ, USA). Genomic DNA was extracted from leukemic cells using standard phenol/chloroform-based methods. Briefly, 1 million cells were lysed in 10 mM Tris–HCl, 10 mM NaCl, 10 mM EDTA, 20 μg proteinase K, and 0.5% SDS by incubating at 37 °C for 16 h. Total RNA was further removed by adding 500 μg PureLink RNase A (Invitrogen, USA) and incubating for 10 min at 37 °C. An equal volume of phenol–chloroform–isopropanol (25:24:1) was added to lysates and mixed by shaking vigorously, followed by centrifugation at 16,100 × g at 4 °C for 5 min. The upper aqueous phase was transferred to a fresh tube; genomic DNA was then precipitated by adding 2× volume − 80 °C 100% ethanol. The DNA pellet was washed with 75% ethanol and rehydrated with Tris–EDTA buffer. The concentration of DNA was determined using a NanoDrop 1,000 spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA)32.
MLPA analysis
Genomic DNA was analyzed using the SALSA MLPA kit (MRC-Holland, Amsterdam, the Netherlands), according to manufacturer’s instructions. The PCR fragments were separated by capillary electrophoresis on a Life Technologies 3,500 Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA). MLPA data were analyzed using Coffalyser.Net v.140721.1958 (MRC-Holland, Amsterdam, The Netherlands). Probe ratio between 0.75 and 1.3 were considered to be within the normal range. Probe ratio below 0.75 or above 1.3 indicated deletion or gain, respectively. Probe ratio below 0.25 or above 1.8 indicated biallelic deletion or amplification. SALSA MLPA P335 ALL-IKZF1 probemix was used for detection of alterations of EBF1, IKZF1, CDKN2A, CDKN2B, PAX5, ETV6, RB1 and BTG1 genes. SALSA MLPA P327 iAMP21-ERG probemix was used for detecting alterations of ERG gene and iAMP21. SALSA MLPA P329 CRLF2-CSF2RA-IL3RA probemix was used for detecting P2RY8-CRLF2 (PAR1 deletion).
Analysis of ploidy status
Ploidy status was evaluated by SALSA MLPA P036 Subtelomeres Mix 1 probe mix. Whole chromosomal gain or loss was defined when two probes targeting p and q arms of the same chromosome were respectively gained or deleted simultaneously. Chromosome 19p deletions were defined when the probe targeted the p arm of chromosome 19 was deleted while q arm was normal.
DNA index measured by flow cytometry
Freshly prepared or frozen leukemia samples were used for DNA index analysis. Peripheral blood derived from normal healthy individuals was used as controls for diploidy. Mononuclear cells were isolated by Ficoll-Paque (GE Healthcare, Chicago, IL, USA) according to the manufacturer’s instructions. Three cell suspensions were prepared: tube A was a mixture of leukemia cells and normal PBMCs in equal numbers; tubes B and C contained normal PBMCs or leukemia cells alone. Each cell suspension (2 million cells) was fixed with 70% ethanol overnight at – 20 °C. Fixed cells were washed with 1× PBS and then incubated with propidium iodide (50 μg) and RNase (10 μg) for 1 h on ice. Cells were filtered with 100 μm cell strainer and then analyzed by FACSCalibur (BD, Franklin Lakes, NJ, USA). DNA quantity of an individual cell population was determined and DNA index represents the ratio of leukemia sample/normal PBMCs fluorescence calculated from tube A. Tubes B and tube C were used as reference to distinguish the leukemia from PBMC peaks in tube A. Theoretical DNA index (tDI) was calculated using the formula: tDI = chromosome numbers × 0.0202 + 0.067533.
Statistical analysis
Pearson's correlations, the coefficient of determination and p-values were carried out between the results of DI and tDI from MLPA and cytogenetics. Fisher’s exact test was performed to evaluate the enrichment of 19p deletion in TCF3 gene rearranged ALL. The log-rank test compared different survival curves between patients with different major genetic subtypes, patient with or without IKZF1 deletion and patients with or without IKZF1plus. Overall survival (OS) was defined as diagnosis to death. Patients who did not suffer any adverse events within the follow-up period were censored. Event-free survival (EFS) of patients with no response to chemotherapy (refractory), death, and second relapse in induction was set to 0. Univariate and multivariate Cox regression were performed to evaluate hazard ratios (HR) and 95% confidence intervals (CI) of risk factors. All statistical analyses were performed using the Statistical Product and Services Solutions (SPSS) statistical package, v18.0 (IBM, Armonk, NY, USA).
Results
Frequency of copy number abnormalities in children with B-ALL
The demographic, clinical, and laboratory characteristics of 233 children with B-ALL are shown in Table 1. The median age of the cohort was 5.4 years (range < 0.1–17.9 years); 95.3% of the patients were over 1 year of age. The molecular tests performed were those standardized for B-ALL diagnosis including: ETV6-RUNX1, TCF3-PBX1, BCR-ABL1, P2RY8-CRLF2 and KMT2A-AFF1 for 220 samples. Detailed flow diagram of analysis used in this study is demonstrated in Supplementary Fig. S1.
Table 1.
The characteristics of patients in this cohort.
| N | % | |
|---|---|---|
| Sample size | 233 | 100 |
| Gender | ||
| < 1 year | 11 | 4.7 |
| 1–9 years | 160 | 68.7 |
| > 10 years | 62 | 26.6 |
| Sex | ||
| Female | 113 | 48.5 |
| Male | 120 | 51.5 |
| WBC (× 109/l) | ||
| < 50 | 164 | 70.4 |
| 50–100 | 28 | 12.0 |
| > 100 | 38 | 16.3 |
| No data | 3 | 1.3 |
| Protocol | ||
| TPOG 2002 | 108 | 46.4 |
| TPOG 2013 | 125 | 53.6 |
| Early treatment responsea | ||
| MRD positive | 30 | 27.0 |
| MRD negative | 81 | 73.0 |
| Subtype | ||
| Hyperdiploidy | 59 | 25.3 |
| Hypodiploidy | 7 | 3.0 |
| ETV6-RUNX1 | 36 | 15.5 |
| TCF3-PBX1 | 12 | 5.2 |
| Ph+/Ph-like | 16 | 6.9 |
| KMT2A-r | 14 | 6.0 |
| ZNF384/362-r | 9 | 3.9 |
| MEF2D-r | 3 | 1.3 |
| TCF3-HLF | 2 | 0.9 |
| iAMP21 | 4 | 1.7 |
| Other | 71 | 30.5 |
aOnly patients treated with TPOG-2013 were included (n = 111). Fourteen patients were excluded due to missing data; MRD positive: either MRD > 1% at day 15 or > 0.01% at day 35.
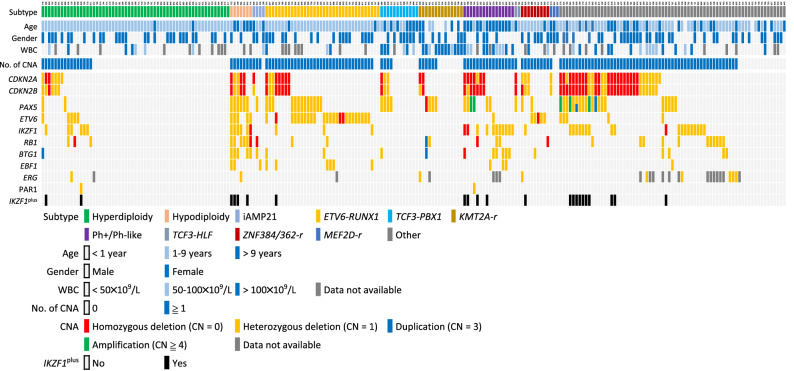
From MLPA testing, overall, 65.7% of the patients (153/233) harbored at least one abnormality (either deletion or amplification) involving the following genes—IKZF1, CDKN2A/2B, PAX5, EBF1, ETV6, BTG1, RB1, ERG or PAR1 region, whereas the remaining 34.3% (80/233) of patients had none of these abnormalities. Simultaneous aberrations in different genes were observed. A heatmap listing these CNA in the entire cohort are given in Fig. 1. Details of the CNA in each major cytogenetic subtype are shown in Supplementary Table S2.
Figure 1.
The heatmap of major subtype of childhood B-cell ALL and the association with CNA detected by MLPA. Ph+ BCR-ABL1 positive, No. number, CN copy number.
DNA index identifies ploidy status in ALL
In 112 samples DNA index analysis was performed; 41 cases showed aneuploidy, of which 35 were high hyperdiploid, 3 were hypodiploid and in 3 cases masked hypodiploidy was indicted, as described below. However, DI cannot identify individual chromosome gain or loss.
MLPA compared to DI and cytogenetics
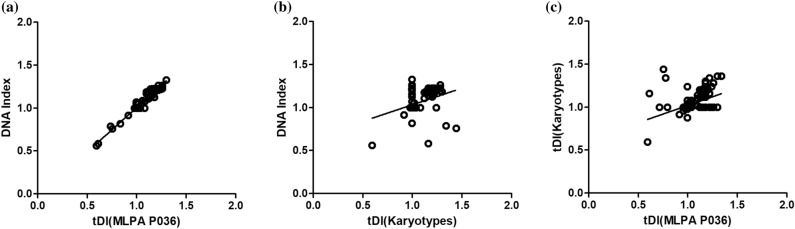
Good quality genomic DNA was available from 204 samples for MLPA analysis using the MLPA P036 kit which identified 57 patients with high hyperdiploidy, 7 with hypodiploidy and 140 with diploidy or near-diploidy. The numerical chromosomal alterations determined by this MLPA P036 kit were compared with the karyotype and DI results. These results showed concordance in number of chromosomes (r = 0.9780, P < 0.0001) for the 111 patients with both MLPA and DI data available (Fig. 2a). There was statistically significant positive correlation between karyotype and DI (r = 0.3308, P = 0.0005) (Fig. 2b), yet lower than MLPA against DI, among 188 patients with karyotype and MLPA data available. The statistically significant positive correlation was also seen between karyotype and MLPA (r = 0.4428, P < 0.0001) (Fig. 2c), but lower than MLPA against DI. We found that 45% (29/64) of patients with high hyperdiploidy or hypodiploidy identified either by DI or MLPA P036 were non-informative. Details of karyotype, DI and MLPA of the cohort are listed in Supplementary Table S3.
Figure 2.
Correlation of the DI and tDI from MLPA P036 and karyotype. (a) DI vs. tDI(MLPA P036) (r = 0.9780, P < 0.0001). (b) DI vs. tDI (Karyotypes) (r = 0.3308, P = 0.0005). (c) tDI (Karyotypes) vs. tDI (MLPA P036) (r = 0.4428, P < 0.0001). tDI theoretical DNA index.
High hyperdiploidy
Among 57 cases with high hyperdiploidy, the majority (94.5%) had gained between 5 and 13 chromosomes (modal chromosome number, MCN, 51–63, inclusive), and the most frequent MCN was 54 chromosomes (Supplementary Fig. S2a). Chromosome gains were non-random and 8 chromosomes accounted for 82% of all gains: 4 (72.7% of cases), 6 (80.7%), 10 (84.2%), 14 (93.0%), 17 (80.7%), 18 (86.0%), 21 (100%), and X (78.9%) (Supplementary Fig. S2b). Gains of chromosomes 5, 8, 9, 11, 12, and 22 represented 15% of the total and were present in between 11 and 35% of cases. Gains of chromosomes 1, 2, 3, 7, 13, 15, 16, 19, and 20 were rare, totaling 3% of chromosomes gained. These patterns of chromosomal gains in these high hyperdiploid cases were similar to previous reports.
The MLPA pattern of iAMP21 and differentiation between iAMP21 and high hyperdiploidy
From their MLPA plots, we identified four patients with iAMP21, as shown in Supplementary Fig. S3. A characteristic chromosome 21 copy number profile has been previously described for cases of iAMP21-ALL from microarray studies and next generation sequencing. It is described as copy number changes from centromere to telomere along chromosome 21, with the highest level of amplification proximal to a telomeric deletion34–36. Tsuchiya et al. reported a case in which RUNX1 was not located within the highest region of amplification of chromosome 2137. In this cohort, RUNX1 was observed within the most highly amplified region of chromosome 21, with the exception of one case (Supplementary Fig. S3). In high hyperdiploid cases, the DI is usually greater than 1.16 and associated with frequent gains of chromosomes 4, 6, 10, 14, 18, 21 and X. We compared the pattern of chromosome 21 gain in high hyperdiploid and iAMP21-ALL in our cohort. SNP arrays analysis was carried out on two iAMP21-ALL samples diagnosed by MLPA (Supplementary Fig. S4). For cases with suspected iAMP21, in the absence of SNP arrays, DI and MLPA P036 and P327 kits can provide the definitive answer.
Hypodiploid cases
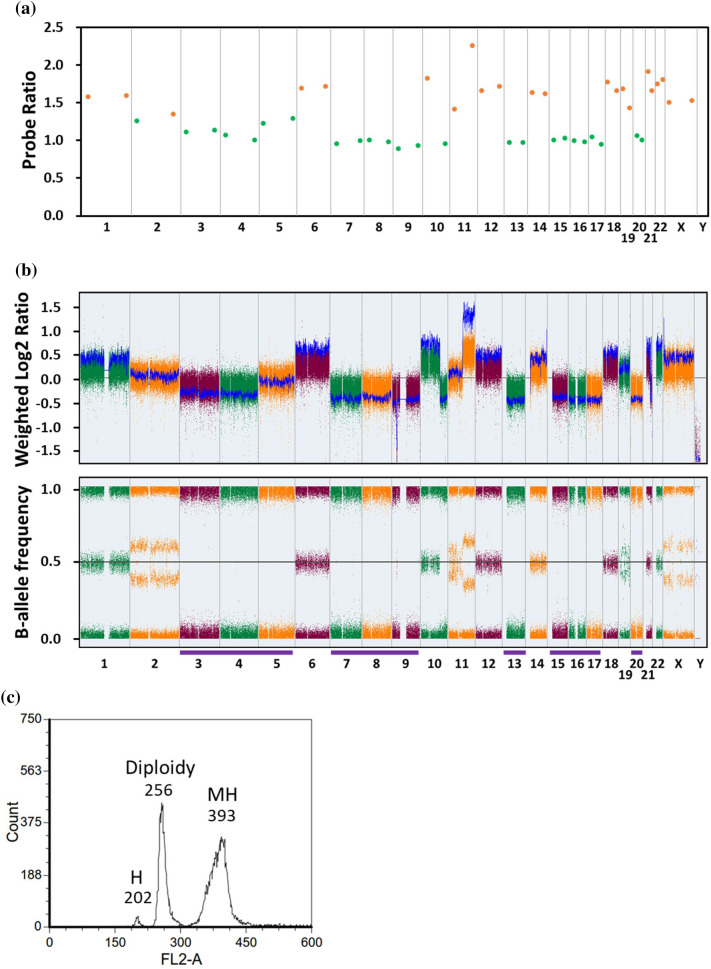
Five patients with low DI were diagnosed with hypodiploidy. Three of them had two peaks in the DI, indicating the presence of hypodiploid clone undergo a doubling of the chromosomes during metaphase. This manifestation is known as masked hypodiploidy. As the diagnosis of masked hypodiploidy requires demonstration of loss of heterozygosity (LOH), these three samples were analyzed by SNP arrays and LOH was seen, as shown in case 984 (Fig. 3). DI showed two peaks: the smaller one (FL2-A value = 202) is the true hypodiploidy and the higher one (FL2-A value = 393) indicates the doubled hypodiploid population. These hypodiploid samples were also tested using MLPA P036 kit. By comparing MLPA with the value of DI, we were able to identify the specific losses and retention of each chromosome number. Thus, we were able to confirm that the masked hypodiploid population originated from doubling of the low hypodiploid one. In Fig. 3, the chromosome gains detected by MLPA P036 corresponded to the retained chromosomes. In contrast, the “normal” chromosomes, for example chromosomes 3, 4, 5, 7, 8, 9, 13, 15, 16, 17 and 20 were shown to be lost. The actual gain or loss of each chromosome cannot be inferred from the DNA index. Using the MLPA P036 kit, we identified another two cases of hypodiploidy (patients 508 and 753) in which LOH was confirmed by STR (see below). Details of these patients are listed in the Table 2.
Figure 3.
Analysis of a case with masked hypodiploidy. (a) The result of MLPA P036. The probe ratio values between 0.75 and 1.3 or greater than 1.3 were indicated with green or orange dots respectively. (b) The result of CytoScan array. Weight log2 ratio and B-allele frequency (BAF) plots are shown. The BAF plot show only two tracts indicate a region with LOH and the whole chromosome LOH is indicated by purple lines. (c) The result of DNA index analysis. H hypodiploidy, MH masked hypodiploidy.
Table 2.
Detailed information of hypodiploidy cases.
| ID | Subtype | Karyotype | MLPA P036 | DNA index | TP53 mutation |
|---|---|---|---|---|---|
| 925 | High-hypodiploidy | 42,X,-4,-9,-13,t(14;17)(q32;p11.2),add(21)(p11.2)[16]/84,idem × 2[3]/46,XY[1] | 42,Y,-4,-9,-13 | 0.91 | R248Q (Unknown) |
| 774 | Low-hypodiploidy | 46,XY[25] | 38,XY,-2,-3,-12,-13,-14,-15,-16,-17 | 0.82 | F341fs (Somatic) |
| 508 | Low-hypodiploidy | 46,XY[20] | 32,XY,-2,-3,-4,-6,-7,-9,-10,-12,-15,-16,-17,-18,-20,-22 | NA | I195F (Germline) |
| 984 | Masked Low-hypodiploidy | 63,XX,-X, + 1,-3,add(3)(q13),del(3)(q?21),-4,-5, + 6,-7,-9,-9,-10, + 12,-13, + 14,-15,-16,-17,-20, + add(21)(q22) × 2, + 22, + 2mar,inc[cp14]/46,XX[6] | 33,XX,-3,-4,-5,-7,-8,-9,-10,-11,-13,-15,-16,-17,-20 | 0.79/1.54 | R273H (Germline) |
| 845 | Masked Low-hypodiploidy | 68,XX,-Y, + 1,-2,-3,-4,del(4)(q21q31),del(5)(q13q33), + 6,-7, + 8, + 9,-10, + 11,-12,-13, + 14,-15,-16,-17,-18, + 19, + 20, + 21, + 22,inc[cp5]/46,XY[20] | 34,Y,-2,-3,-4,-7,-10,-12,-13,-15,-16,-17,-18 | 0.76/1.43 | W53* (Somatic) |
| 952 | Near-haploidy | 26,XY,-1,-2,-3,-4,-5,-6,-7,-8,-9,-11,-12,-13,-14,-15,-16,-17,-18,-19,-20,-22[13] | 26,XY,-1,-2,-3,-4,-5,-6,-7,-8,-9,-11,-12,-13,-14,-15,-16,-17,-18,-19,-20,-22 | 0.56 | Wild-type |
| 753 | Masked Near-haploidy | 52–54,XX, + mar1 ~ mar8[cp4]/46,XX[21] | 27,X,-1,-2,-3,-5,-6,-7,-9,-10,-11,-12,-13,-15,-16,-17,-18,-19,-20,-22 | 1.17 | Wild-type |
A Short Tandem Repeat (STR) is a microsatellite, consisting of a unit of two to thirteen nucleotides repeated hundreds of times on a DNA strand. STR analysis measures the precise number of repeating units. STR is used for confirmation of donor engraftment following stem cell transplantations and this test is available in all medical centers38. Samples of germline (if available) and tumor were sent for STR analysis in order to confirm LOH identified on SNP arrays. We show the interpretation of STR for patient 984 in Supplementary Fig. S5 and all three cases of masked hypodiploidy by STR are shown in Supplementary Table S4. STR provides a simple method to confirm the presence of LOH. Based upon these observations, we have proposed a flowchart for diagnosis of masked hypodiploidy (Supplementary Fig. S6).
19p deletion by MLPA is an indicator of TCF3 translocations in childhood ALL
We identified 7 of 12 cases of TCF3-PBX1 and two cases of TCF3-HLF with 19p loss. This enrichment differs from other subtypes of B-cell ALL (P < 0.0001) (Table 3). TCF3 is an important transcriptional factor with multiple fusion partners in ALL. Samples with 19p deletions without evidence of TCF3-PBX1 or TCF3-HLF fusions may carry TCF3-ZNF384 fusions. TCF3-ZNF384 fusions represent another important subtype of B-cell ALL with a specific immunophenotype showing frequent CD10 loss and CD13 and CD33 expression. From these observations, we observed one sample with 19p loss, loss of CD10, CD13 and CD33 expression in which the TCF3-ZNF384 fusions was identified by RT-PCR (Supplementary Fig. S7). In this series, among a total of 15 samples with 19p loss, 10 of them had TCF3 fusions.
Table 3.
The 19p deletion in TCF3 translocation and non-TCF3 translocation subtypes.
| 19p Normal | 19p Deletion | |
|---|---|---|
| TCF3 translocation | 7 | 10 |
| TCF3-PBX1 | 5 | 7 |
| TCF3-HLF | 0 | 2 |
| TCF3-ZNF384 | 2 | 1 |
| Non-TCF3 translocation | 162 | 5 |
| Hyperdiploidy | 55 | 3 |
| ETV6-RUNX1 | 22 | 1 |
| KMT2A fusions | 11 | 1 |
| Other | 74 | 0 |
Fisher’s exact test of comparing TCF3 translocation with non-TCF3 translocation subtypes, P < 0.0001.
Novel subtypes of ALL, intragenic amplifications of PAX5 (PAX5AMP), IKZF1-plus and ERG deletions
Recently two papers have reported two novel high-risk subtypes of childhood ALL, PAX5AMP and IKZF1plus7,39. There were 23 IKZF1plus patients and 5 patients with PAX5AMP in this cohort. Nine patients (9/233 = 3.9%) were identified with ERG deletions. These ERG deletions were associated with different subtypes of ALL (Fig. 1).
Survival analysis
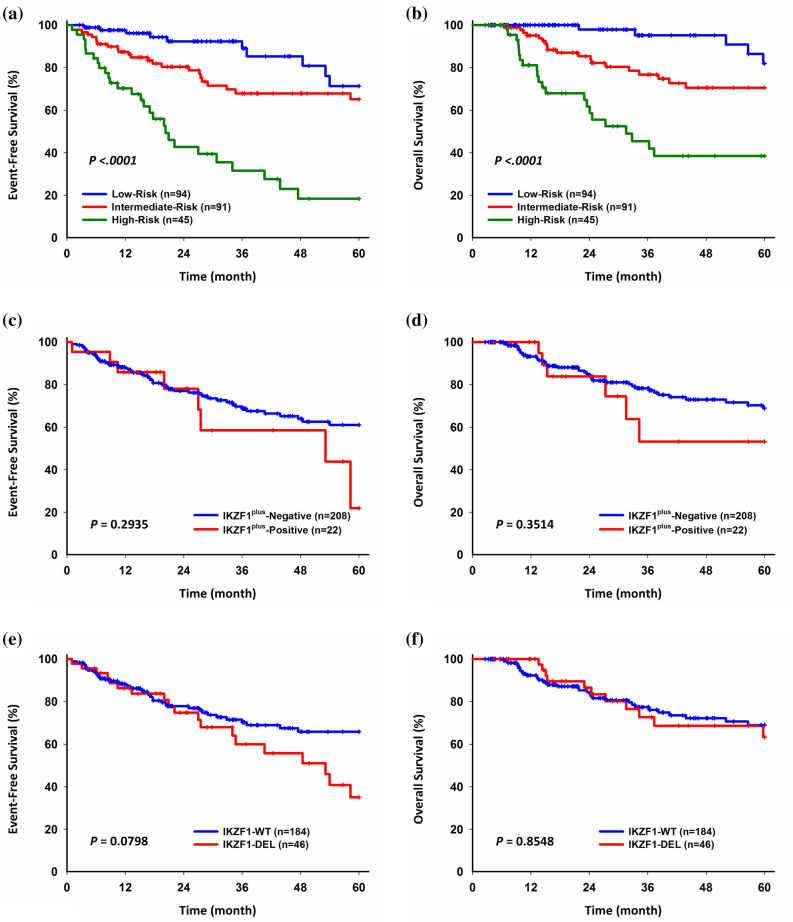
Among patients with the major cytogenetic alterations, two with TCF3-HLF relapsed and died within 5 years from diagnosis. Patients with high-risk subtypes (Ph-positive/-like, hypodiploidy, MEF2D-r, KMT2A-r, TCF3-HLF, iAMP21) had inferior 5-year EFS (P < 0.0001) and OS (P < 0.0001) (Fig. 4a, b). The overall outcome was slightly inferior compared to previous TPOG reports, likely due to many of them being referred from other hospitals after relapse30. All patients with iAMP21 were not detected at diagnosis. There is a trend that patients with IKZF1plus had inferior 5 year-EFS and OS than patients without IKZF1plus, but it did not reach statistical significance (Fig. 4c, d). Patients with IKZF1 deletions had inferior 5-year EFS and 5-year OS, but it also did not reach statistical significance (Fig. 4e, f). In the Cox multivariate regression model, IKZF1 deletions were not a strong predictor of poor outcome (Supplementary Table S5).
Figure 4.
Five-year EFS and OS analysis. (a, b) Major genetic subtypes. (c, d) IKZF1plus-Positive vs. IKZF1plus-Negative. (e, f) IKZF1-DEL vs. IKZF1-WT. EFS event-free survival, OS overall survival, DEL deletion, WT wild type.
Discussion
In this retrospective study, the MLPA P036 subtelomeres probemix kit provided accurate detection of aneuploidy in childhood B-cell ALL and good correlation with the results from DI. MLPA and DI are superior to traditional cytogenetics, due to the shorter turn-around time, irrespective of mitotic index and improved sensitivity. Detections of specific gains or losses of each chromosome assist the differential diagnosis of hyperdiploidy from iAMP21. In addition, DI is helpful for diagnosis of masked hypodiploidy and LOH should be confirmed by SNP arrays. STR provides a simple method, available in most medical centers in Taiwan, to document LOH in these masked hypodiploid cases. Around 1.7% (4/233) of B-ALL patients had iAMP21. We also identified some of the novel ALL subtypes, including PAX5AMP, and IKZF1plus7,39. TCF3 rearrangements were frequently associated with 19p deletions.
High hyperdiploidy accounts for around 20 ~ 25 percentage of childhood B-cell ALL40. In this cohort, the most frequent modal chromosome number was 54 followed by 55. The most frequent gains included chromosomes 4, 6, 10, 18, 16, 17, 18, 21 and X, in agreement with previous reports8,40,41. This incidence of high hyperdiploidy was lower in Taiwan than Caucasian populations30,42,43. Using DI and the MLPA P036 kit, the incidence was around 27% in this cohort. In this study, 45% of high hyperdiploid patients were not detected by cytogenetics, manifesting as normal karyotype. In previous TPOG ALL 2002 report, hyperdiploidy accounted for 13.6% in B-ALL (n = 1,209). The incidence was much lower than that of this report. The reason for this discrepancy might be the relative smaller case numbers in this study. For cases without metaphases or normal karyotype, DI and MLPA can be successfully used for diagnosis of high hyperdiploidy11.
iAMP21-ALL is a novel subtype of B-ALL proposed by WHO14,15,28,33. The initial gold standard for diagnosis was FISH using probes directed to the RUNX1 gene, but array-CGH or SNP arrays are now the main method for diagnosis33. One MLPA kit can successfully identify iAMP21 due to the density of probes along the long arm of chromosome 21. We identified 4 cases with iAMP21 by MLPA. In these cases, the level of gain was variable along the length of chromosome 21 with the ratio being more than 3.0, higher than in cases where chromosome 21 is gained as part of a high hyperdiploidy karyotype in which the probe ratio for every probe in the kit being ~ 1.5–2.0. These data correlated with other gains, especially of chromosomes 4, 6, 10, 18, 16, 17, 18 and X. If gains of chromosomes X, 4, 6, 10, 14, 17 and 18 are detected at the same time as gains of 21, it is most likely that the patient has high hyperdiploidy rather than iAMP21-ALL.
Masked hypodiploidy can be difficult to diagnose. Another study used a similar MLPA approach to identify the aneuploidy status of relapsed B-cell ALL12. Three patients with high hyperdiploidy had the highest number of chromosomal gains (median 11). Gains of the classical high hyperdiploidy pattern were less frequent, but gains of non-classical chromosomes, especially 1, 5, 11, 19 and 22, accounted for 49% of all gains in these patients. All three patient relapse samples carried TP53 mutations, two of which were present in the germline. In all three cases, no underlying hypodiploid clone was detected by DI or cytogenetic analyses, making diagnosis difficult. A recent report by Carroll et al. demonstrated that a considerable proportion (25% or higher) of hypodiploidy in children with B-ALL may have been overlooked in previous studies due to the presence of only a doubled hypodiploid population, mistakenly interpreted as typical high hyperdiploidy associated with a favorable risk44. In this cohort, the chromosome number in high hyperdiploidy was mostly in the range of 52 ~ 59, which could overlap with masked hypodiploidy. For masked hypodiploid cases, the MLPA P036 kit results, alongside DNA index, can detect the specific gain or loss of each chromosome. LOH can also be confirmed by STR.
TCF3, located to 19p, is rearranged with several genes in childhood ALL. The most frequent is TCF3-PBX1 and rarely the poor risk TCF3-HLF2. We observed 19p loss in all TCF3-PBX1 and TCF3-HLF cases. TCF3 has also been identified to be rearranged with ZNF384, a novel fusion recently identified45–48. In cases with 19p deletions without TCF3-PBX1 or TCF3-HLF detected by RT-PCR or cytogenetics, 19p deletions may point to other TCF3 fusions. TCF3-ZNF384 fusions are also frequently associated with CD10 loss, with the presence of CD13 and CD3345,48,49. These two characteristics are useful for its identification by RT-PCR.
In our cohort, patients with iAMP21 and KMT2A fusions had an inferior 5-year EFS and OS in comparison to patients with ETV6-RUNX1 or high hyperdiploidy. Patients with hypodiploidy also had an inferior 5-year EFS and OS, although most of them were not identified at the time of diagnosis. The outcome for patients with iAMP21-ALL may be improved if detected at diagnosis, so that they may be treated with more intensive chemotherapy. No events were seen in patients with PAX5AMP, while patents with IKZF1plus showed a trend towards inferior EFS and OS, although the P-value was not significant. IKZF1 deletions showed a trend towards poorer clinical outcomes, as observed in a number of other studies22,27,50. Due to the relative small case numbers in this study, larger studies are indicated in Taiwan in order to evaluate the clinical impact of these genetic alterations in Taiwan.
In conclusion, MLPA and DNA index together can rapidly provide reliable information for identification of aneuploidy of childhood B-ALL. Using these methods, diagnosis of aneuploidy in Taiwan might be improved particularly among those cases currently classified within unknown subtype of B-cell ALL, and especially those without metaphases or normal karyotype. STR provides a simple method to demonstrate LOH if masked hypodiploidy is suspected. Other important abnormalities such as IKZF1 deletions, IKZF1plus and ERG deletions can also be identified by MLPA. These tools are helpful for the diagnosis of some important subtype of ALL.
Supplementary information
Acknowledgements
This work was supported by grants from the Ministry of Science and Technology, Taiwan (MOST-103-2314-B-002-199 and MOST-107-2314-B-002-173-MY2 to YLY). The authors are grateful for the PI of TPOG ALL protocol, Dr. HC Liu for his valuable feedback. The authors express their gratitude to all of the patients who participated in this study, and their parents. The authors also acknowledge the efforts of the TPOG and the Childhood Cancer Foundation in Taiwan.
Author contributions
C.-H.Y., T.-K.L., C.-C.H., S.-W.L. and Y.-L.Y. designed the study, analyzed the data and wrote the manuscript. C.-H.Y., T.-K.L. and Y.-N.S. acquired, processed patient specimens and performed experiments. C.-Y.L. and H.-Y.C. performed the analysis. K.-H.L., S.-T.J., M.-Y.L., S.-H.C., K.-H.W., S.-C.W., H.-H.C., M,-J.L. and D.-T.L. provided clinical samples and data. CJ.H. provided the techniques. The manuscript was written by C.-H.Y., T.-K. L., C.-C.H., S.-W.L. and Y.-L.Y. and was reviewed and edited by all authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chih-Hsiang Yu and Tze-Kang Lin.
Contributor Information
Chia-Cheng Hung, Email: double@sofivagenomics.com.tw.
Shu-Wha Lin, Email: mtshuwha@ntu.edu.tw.
Yung-Li Yang, Email: yangyl92@ntu.edu.tw.
Supplementary information
is available for this paper at 10.1038/s41598-020-68311-9.
References
- 1.Pui CH, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N. Engl. J. Med. 2009;360:2730–2741. doi: 10.1056/NEJMoa0900386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pui CH, Nichols KE, Yang JJ. Somatic and germline genomics in paediatric acute lymphoblastic leukaemia. Nat. Rev. Clin. Oncol. 2019;16:227–240. doi: 10.1038/s41571-018-0136-6. [DOI] [PubMed] [Google Scholar]
- 3.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N. Engl. J. Med. 2015;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 4.Pui CH, et al. Clinical impact of minimal residual disease in children with different subtypes of acute lymphoblastic leukemia treated with Response-Adapted therapy. Leukemia. 2016 doi: 10.1038/leu.2016.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Connor D, et al. Genotype-specific minimal residual disease interpretation improves stratification in pediatric acute lymphoblastic leukemia. J. Clin. Oncol. 2018;36:34–43. doi: 10.1200/JCO.2017.74.0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwab CJ, et al. Evaluation of multiplex ligation-dependent probe amplification as a method for the detection of copy number abnormalities in B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2010;49:1104–1113. doi: 10.1002/gcc.20818. [DOI] [PubMed] [Google Scholar]
- 7.Stanulla M, et al. IKZF1plus defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric B-cell precursor acute lymphoblastic leukemia. J. Clin. Oncol. 2018;36:1240–1249. doi: 10.1200/jco.2017.74.3617. [DOI] [PubMed] [Google Scholar]
- 8.Moorman AV, et al. Outcome heterogeneity in childhood high-hyperdiploid acute lymphoblastic leukemia. Blood. 2003;102:2756–2762. doi: 10.1182/blood-2003-04-1128. [DOI] [PubMed] [Google Scholar]
- 9.Oudot C, et al. Prognostic factors for leukemic induction failure in children with acute lymphoblastic leukemia and outcome after salvage therapy: The FRALLE 93 study. J. Clin. Oncol. 2008;26:1496–1503. doi: 10.1200/jco.2007.12.2820. [DOI] [PubMed] [Google Scholar]
- 10.Holmfeldt L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vázquez-Reyes A, et al. Aneuploidy identification in pre-B acute lymphoblastic leukemia patients at diagnosis by Multiplex Ligation-dependent Probe Amplification (MLPA) Leuk. Res. 2017;59:117–123. doi: 10.1016/j.leukres.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Groeneveld-Krentz S, et al. Aneuploidy in children with relapsed B-cell precursor acute lymphoblastic leukaemia: Clinical importance of detecting a hypodiploid origin of relapse. Br. J. Haematol. 2019;185:266–283. doi: 10.1111/bjh.15770. [DOI] [PubMed] [Google Scholar]
- 13.Safavi S, Paulsson K. Near-haploid and low-hypodiploid acute lymphoblastic leukemia: Two distinct subtypes with consistently poor prognosis. Blood. 2017;129:420–423. doi: 10.1182/blood-2016-10-743765. [DOI] [PubMed] [Google Scholar]
- 14.Harewood L, et al. Amplification of AML1 on a duplicated chromosome 21 in acute lymphoblastic leukemia: A study of 20 cases. Leukemia. 2003;17:547–553. doi: 10.1038/sj.leu.2402849. [DOI] [PubMed] [Google Scholar]
- 15.Robinson HM, Harrison CJ, Moorman AV, Chudoba I, Strefford JC. Intrachromosomal amplification of chromosome 21 (iAMP21) may arise from a breakage-fusion-bridge cycle. Genes Chromosomes Cancer. 2007;46:318–326. doi: 10.1002/gcc.20412. [DOI] [PubMed] [Google Scholar]
- 16.Harrison CJ, et al. An international study of intrachromosomal amplification of chromosome 21 (iAMP21): Cytogenetic characterization and outcome. Leukemia. 2014;28:1015–1021. doi: 10.1038/leu.2013.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heerema NA, et al. Intrachromosomal amplification of chromosome 21 is associated with inferior outcomes in children with acute lymphoblastic leukemia treated in contemporary standard-risk children's oncology group studies: A report from the children's oncology group. J. Clin. Oncol. 2013;31:3397–3402. doi: 10.1200/jco.2013.49.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moorman AV, et al. Risk-directed treatment intensification significantly reduces the risk of relapse among children and adolescents with acute lymphoblastic leukemia and intrachromosomal amplification of chromosome 21: A comparison of the MRC ALL97/99 and UKALL2003 trials. J. Clin. Oncol. 2013;31:3389–3396. doi: 10.1200/jco.2013.48.9377. [DOI] [PubMed] [Google Scholar]
- 19.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 20.Mullighan CG, et al. BCR–ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 21.Alexander TB, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature. 2018;562:373–379. doi: 10.1038/s41586-018-0436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullighan CG, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Engl. J. Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat. Genet. 2016;48:1481–1489. doi: 10.1038/ng.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen IM, et al. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: A Children's Oncology Group study. Blood. 2012;119:3512–3522. doi: 10.1182/blood-2011-11-394221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts KG, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waanders E, et al. Integrated use of minimal residual disease classification and IKZF1 alteration status accurately predicts 79% of relapses in pediatric acute lymphoblastic leukemia. Leukemia. 2010 doi: 10.1038/leu.2010.275. [DOI] [PubMed] [Google Scholar]
- 27.Yang YL, et al. IKZF1 deletions predict a poor prognosis in children with B-cell progenitor acute lymphoblastic leukemia: A multicenter analysis in Taiwan. Cancer Sci. 2011;102:1874–1881. doi: 10.1111/j.1349-7006.2011.02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arber DA, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 29.Tien HF, et al. Correlation of cytogenetic results with immunophenotype, genotype, clinical features, and ras mutation in acute myeloid leukemia. A study of 235 Chinese patients in Taiwan. Cancer Genet. Cytogenet. 1995;84:60–68. doi: 10.1016/0165-4608(95)00084-4. [DOI] [PubMed] [Google Scholar]
- 30.Li M-J, et al. Treatment for childhood acute lymphoblastic leukemia in Taiwan: Taiwan Pediatric Oncology Group ALL-2002 study emphasizing optimal reinduction therapy and central nervous system preventive therapy without cranial radiation. Pediatr. Blood Cancer. 2017;64:234–241. doi: 10.1002/pbc.26142. [DOI] [PubMed] [Google Scholar]
- 31.Liang DC, et al. Long-term results of Taiwan Pediatric Oncology Group studies 1997 and 2002 for childhood acute lymphoblastic leukemia. Leukemia. 2010;24:397–405. doi: 10.1038/leu.2009.248. [DOI] [PubMed] [Google Scholar]
- 32.Yu C-H, et al. TP53 alterations in relapsed childhood acute lymphoblastic leukemia. Cancer Sci. 2020;111:229–238. doi: 10.1111/cas.14238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rachieru-Sourisseau P, et al. DNA Index in childhood acute lymphoblastic leukaemia: A karyotypic method to validate the flow cytometric measurement. Int. J. Lab. Hematol. 2010;32:288–298. doi: 10.1111/j.1751-553X.2009.01189.x. [DOI] [PubMed] [Google Scholar]
- 34.Harrison CJ. Blood Spotlight on iAMP21 acute lymphoblastic leukemia (ALL), a high-risk pediatric disease. Blood. 2015;125:1383–1386. doi: 10.1182/blood-2014-08-569228. [DOI] [PubMed] [Google Scholar]
- 35.Strefford JC, et al. Complex genomic alterations and gene expression in acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8167–8172. doi: 10.1073/pnas.0602360103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuchinskaya E, et al. Tiling-resolution array-CGH reveals the pattern of DNA copy number alterations in acute lymphoblastic leukemia with 21q amplification: The result of telomere dysfunction and breakage/fusion/breakage cycles? Leukemia. 2007;21:1327. doi: 10.1038/sj.leu.2404628. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchiya KD, Davis B, Gardner RA. Is intrachromosomal amplification of chromosome 21 (iAMP21) always intrachromosomal? Cancer Genet. 2017;218:10–14. doi: 10.1016/j.cancergen.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Butler JM. Genetics and genomics of core short tandem repeat loci used in human identity testing. J. Forensic Sci. 2006;51:253–265. doi: 10.1111/j.1556-4029.2006.00046.x. [DOI] [PubMed] [Google Scholar]
- 39.Schwab C, et al. Intragenic amplification of PAX5: A novel subgroup in B-cell precursor acute lymphoblastic leukemia? Blood Adv. 2017;1:1473–1477. doi: 10.1182/bloodadvances.2017006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison CJ, Foroni L. Cytogenetics and molecular genetics of acute lymphoblastic leukemia. Rev. Clin. Exp. Hematol. 2002;6:91–113. doi: 10.1046/j.1468-0734.2002.00069.x. [DOI] [PubMed] [Google Scholar]
- 41.Paulsson K, Johansson B. High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2009;48:637–660. doi: 10.1002/gcc.20671. [DOI] [PubMed] [Google Scholar]
- 42.Liang D-C, et al. Frequencies of ETV6–RUNX1 fusion and hyperdiploidy in pediatric acute lymphoblastic leukemia are lower in far east than west. Pediatr. Blood Cancer. 2010;55:430–433. doi: 10.1002/pbc.22628. [DOI] [PubMed] [Google Scholar]
- 43.Yang YL, et al. Multiplex reverse transcription-polymerase chain reaction as diagnostic molecular screening of 4 common fusion chimeric genes in Taiwanese children with acute lymphoblastic leukemia. J. Pediatr. Hematol. Oncol. 2010;32:e323–330. doi: 10.1097/MPH.0b013e3181ed1655. [DOI] [PubMed] [Google Scholar]
- 44.Carroll AJ, et al. Masked hypodiploidy: Hypodiploid acute lymphoblastic leukemia (ALL) mimicking hyperdiploid ALL in children: A report from the Children's Oncology Group. Cancer Genet. 2019;238:62–68. doi: 10.1016/j.cancergen.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirabayashi S, et al. ZNF384-related fusion genes consist of a subgroup with a characteristic immunophenotype in childhood B-cell precursor acute lymphoblastic leukemia. Haematologica. 2016 doi: 10.3324/haematol.2016.151035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J-F, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc. Natl. Acad. Sci. 2018;115:E11711–E11720. doi: 10.1073/pnas.1814397115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu YF, et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine. 2016;8:173–183. doi: 10.1016/j.ebiom.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian M, et al. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res. 2017;27:185–195. doi: 10.1101/gr.209163.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shago M, Abla O, Hitzler J, Weitzman S, Abdelhaleem M. Frequency and outcome of pediatric acute lymphoblastic leukemia with ZNF384 gene rearrangements including a novel translocation resulting in an ARID1B/ZNF384 gene fusion. Pediatr. Blood Cancer. 2016;63:1915–1921. doi: 10.1002/pbc.26116. [DOI] [PubMed] [Google Scholar]
- 50.Kuiper RP, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24:1258–1264. doi: 10.1038/leu.2010.87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.