Abstract
Autism spectrum disorder (ASD) is a neurological and developmental disorder characterized by social impairment and restricted interactive and communicative behaviors. It may occur as an isolated disorder or in the context of other neurological, psychiatric, developmental, and genetic disorders. Due to rapid developments in genomics and imaging technologies, imaging genetics studies of ASD have evolved in the last few years. Increased risk for ASD diagnosis is found to be related to many specific single-nucleotide polymorphisms, and the study of genetic mechanisms and noninvasive imaging has opened various approaches that can help diagnose ASD at the nascent level. Identifying risk genes related to structural and functional changes in the brain of ASD patients provide a better understanding of the disease’s neuropsychiatry and can help identify targets for therapeutic intervention that could be useful for the clinical management of ASD patients.
Subject terms: Scientific community, Genomics
Introduction
Autism spectrum disorder (ASD) is a neurological and developmental disorder consisting of a wide range of symptoms and disability that develop in early childhood and persists throughout life. The common symptoms of ASD include limited activities, lower engagement and communication, talking and learning problems, and repetitive behavior. According to the World Health Organization, the global burden of ASD is continuously growing, with a current prevalence rate of 1 in 160 children. Reported prevalence rates vary widely from country to country though. Recent data from the Centers for Disease Control and Prevention showed that about 1 in 68 children in the United States had been identified with some form of ASD, with more than 3 million people affected1. A recent study estimated prevalence of ASD in the United States in 2014–2016 was 2.47% among adolescents and children2. While in the United Kingdom, the annual prevalence rate for children aged 8 years between 2004 and 2010 was 3.8/1000 for boys and 0.8/1000 for girls3. Recent studies have shown that the pooled ASD prevalence estimate in Asia is 0.36%, including data from nine countries (China, Korea, India, Bangladesh, Lebanon, Iran, Israel, Nepal and Sri Lanka)4. The prevalence of ASD in the Middle East region was documented to be 1.4 per 10,000 children in Oman5, 4.3 per 10,000 children in Bahrain6, 1/167 in Saudi Arabia7, and a recent study reported ASD prevalence to be 1.14 % in children aged 6–11 years in Qatar8.
ASD incidence is 4–5 times greater in males than in females9. The exact cause of ASD remains unclear; however, it is thought that both genetic and environmental factors play essential roles. The effect of ASD on society is enormous and multifaceted as it affects not only the child but also the siblings and parents and significantly disturbs the functioning of family routine life. Individuals with ASD are very likely to encounter the criminal justice system, mostly due to a lack of knowledge of their social and communication difficulties. There are also financial pressures associated with the recovery and decreased opportunities for jobs. Various studies have focused on the problems faced by families of autistic children10. It may affect the performance of the child at school, based on the severity of the disorder, and if untreated or undiagnosed, it may continue into adulthood, affecting both personal and professional life. Although there is no standard treatment for ASD, early detection and use of successful therapies can make a difference in the management and improvement of symptoms, and thus the overall quality of life of children with ASD. Before the clinical symptoms occur, therefore, a consensus is reached in treating ASD based on early detection.
Recent cohort studies have reported that hereditary factors can be traced to more than 50% of autism cases11. Historically, the genetic essence of ASD has been identified in family and twin studies12,13. The study of family exome sequencing has found some rare de novo mutations in people with ASD as opposed to healthy controls. It is estimated that ~1000 genes are involved in autism14,15. On the other hand, the main variants associated with autism16 are yet to be identified in genome-wide association studies. By understanding the evidence at gene expression, we will obtain a complete understanding of autism which can aid in the clinical treatment of patients with ASD.
On the other hand, noninvasive imaging provides information on the structural and functional changes of autism in the brain. Studies have shown that connecting genomics to imaging can provide a better understanding of ASD’s pathophysiology17 and can help develop therapeutic approaches to prevent or reverse ASD brain changes. In this review article, as demonstrated in the imaging, we illustrate how genetic variations are correlated with the structural and functional changes in the brain.
Changes in the autistic brain
Pathological findings
Postmortem studies conducted on autistic brains helped to identify the neuroanatomical changes associated with ASD. Smaller cell size and increased density of cells in the hippocampus, limbic system, entorhinal cortex, and amygdala have been observed in all ages of ASD individuals18. Young patients with autism showed abnormally enlarged neurons in the cerebellar nuclei, inferior olive, and vertical limb of the diagonal band of broca18. Another postmortem study found an increased number of neurons in the prefrontal cortex in the brain of ASD children19. Purkinje cell count was found to be decreased in the archicerebellar and neocerebellar cortices of the cerebellar hemisphere in autistic patients20. Another study observed a reduced number of small pale neurons in the brains of adult autistic patients21. A postmortem study of the brain of patients with ASD also reported altered axonal density and the presence of impaired myelin in white matter (WM)22. Overall, the pathological findings suggest a restricted brain development pattern in ASD patients.
Structural brain changes
Brain changes in ASD patients have been primarily studied using magnetic resonance imaging (MRI). MRI is the most widely used noninvasive modality to examine the structural and functional brain changes in different neurological and neurodegenerative disorders. Various MR-based methods are applied to study the morphological, functional, and metabolic changes in the brain of ASD. MRI findings in children with ASD ranging from 2 to 5 years of age revealed that there is an abnormal development of frontal and temporal lobes, lower gray matter (GM), WM, and amygdala volume compared to same-age healthy controls23. A comparison of the brain morphometric features in 3–4 years old ASD children with typically developing (TD) children and developmentally delayed children24 was conducted using high-resolution 3-D MRI. Increased cerebellar volume was found in children with ASD compared to TD, and developmentally delayed children and amygdala enlargement with increased cerebral volume was seen in ASD children24. Another study observed cerebral enlargement in both boys and girls with ASD compared to controls, and boys with regressive autism showed more structural alterations as compared to girls25. Volumetric MR studies demonstrated a 5–10% increase in brain volume of ASD children who were scanned between 18 months and 4 years23,26.
Imaging studies have observed both decreased and increased cortical thickness in patients with ASD. One study, performed on eight autistic subjects, showed decreased cortical thickness in the regions of inferior frontal, occipital, supramarginal, and post- and precentral gyrus, anterior cingulate, prefrontal, parietal, and temporal cortex27. Another study showed increased cortical thickness in the parietal and temporal lobes of autistic children compared to healthy controls28. An MRI study of adults with ASD, who had delayed language development, also called high‐functioning autism (HFA), observed increased cortical thickness in frontal, occipital, temporal, parietal, cingulate, and fusiform gyri while decreased cortical thickness was observed in the post- and paracentral gyrus29. Surface-based morphometry (SBM) analysis is also used to measure cortical thickness in ASD patients. An SBM based study found decreased cortical thickness in the left orbitofrontal and parahippocampal gyrus, left frontal pole, and pars triangularis, while increased cortical thickness was observed in the left precuneus and anterior cingulate cortex30. Another SBM based study reported reduced cortical volume in the left middle temporal gyrus, reduced gyrification index in the left supramarginal gyrus and increased cortical thickness in pars opercularis of inferior frontal gyrus in HFA compared to TD individuals31. The SBM approach was also used to compare the cortical abnormalities in individuals with low‐functioning autism (LFA) and HFA and Asperger’s syndrome with TD controls. In HFA individuals, shape abnormality was observed bilaterally in parietal operculum and ventral postcentral gyrus. LFA individuals showed shape abnormality in the pars opercularis located in the inferior frontal gyrus, and individuals with Asperger’s syndrome showed bilateral abnormalities in the intraparietal sulcus region32. Voxel-based morphometry analysis observed increased GM volume in frontal, parietal, temporal lobes and limbic system and reduced WM volume in limbic system, frontal and temporal lobes of autistic subjects33. Another voxel-based morphometry study showed reduced GM volumes in frontal, temporal, and parietal lobes in HFA individuals compared to healthy controls34.
Diffusion tensor imaging (DTI), a noninvasive MRI technique, exploits the diffusion properties of a water molecule to map the microstructural, architectural, and compositional characteristics of the tissue. Fractional anisotropy (FA) and mean diffusivity are two commonly used DTI metrices in mapping the restricted movement of water molecules in the tissue, where higher FA associates with the greater directionality and tissue integrity35. DTI is widely used to track the changes in the microstructural properties of WM such as axonal density, axonal injury, and myelination in various human brain’ disorders. Provided that ASD is a developmental disorder, DTI can be used to identify variations in WM to better define developmental trajectories in individuals with ASD36. Increased FA was observed in the frontal lobes and corpus callosum regions of children aged 2–3 years with ASD compared to healthy controls37. While, another study showed reduced FA in frontal–posterior tracts in the brain of children with ASD, aged 10–18 years38. DTI studies showed a difference in trajectories of FA in infants aged 6–24 months who were at higher family risk of developing ASD. FA was higher in infants who had ASD diagnosis at the age of 6 months, while FA was found to be lower at the age of 24 months39. One study reported decreased FA in the parietal lobes, temporal lobes, lateral occipital cortex, left anterior cingulate, middle frontal cortex, and in the corticospinal tract of individuals with HFA as compared to the controls34. DTI tractography confirmed abnormality of the WM structures in the regions of the corpus callosum, inferior longitudinal and fronto-occipital fasciculus, and superior longitudinal fasciculus in ASD patients40.
Genes affecting brain morphology
Preclinical studies
While there may be several specific genes affected in autism, there is increasing evidence that these genes converge on a limited number of biological pathways that include changes in cortical development, synapse function, transcription and translation, chromatin modification, and microglial activation41,42 (Table 1).
Table 1.
ASD-associated genes affecting brain morphology and function.
| Gene (s) | Risk allele/polymorphism | Subjects | Method used | Brain region affected/potentially could be affected | Findings |
|---|---|---|---|---|---|
| ANK2 | ankB mutant mice | DTI | Cortex | Increased interhemispheric asymmetry in the cortex and increased overall connectivity46 | |
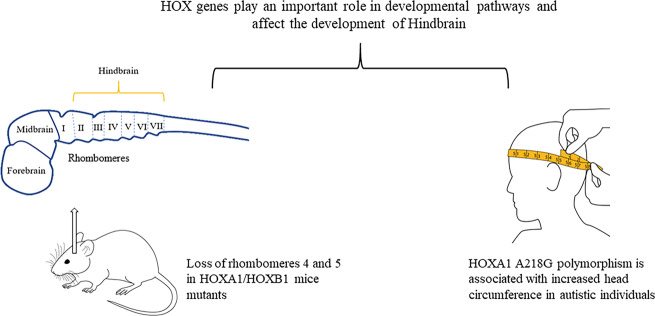
| HOXA1 | HOXA1 A218G polymorphism | Autistic individuals and ethnically matched controls | Family-based association analyses | Could potentially affect hindbrain neural networks that could, in turn, affect brainstem | Enlarged cranial circumference68,143 |
| HOXA1 HOXB1 | Mice mutants of HOXA1/HOXB1 | Immunohistochemistry, RNA in situ hybridization, SEM, cell apoptotic and proliferation assay | Hindbrain | Loss of rhombomere 4 and 5 and loss of second branchial arch in HOXA1/HOXB1 mutant mice53 | |
| PTEN | ASD and macrocephaly | Gene mutation analysis | Cerebral cortex (as one of the subject’s brain MRI showed dilatation of perivascular spaces in the basal ganglia) | Larger head circumference72 | |
| CHD8 | ASD and Developmental delay | Analysis of CHD8 gene expression using Zebrafish model | Forebrain and midbrain | Increased head size55 | |
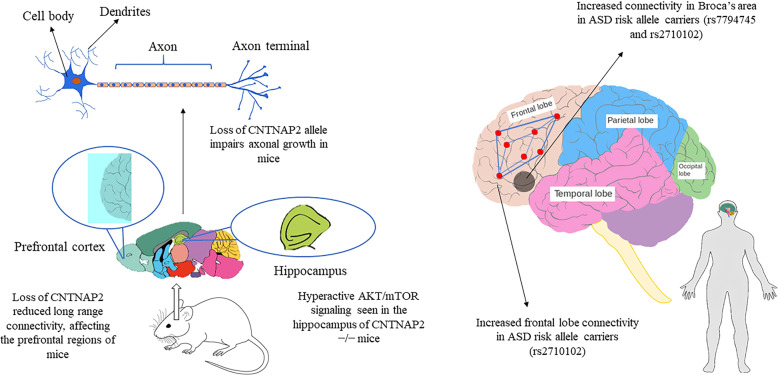
| CNTNAP2 | rs779475 | Healthy controls | DTI, sMRI | Frontal lobe, occipital lobe, cerebellum | Reduction in WM and GM volumes in the cerebellum, frontal and occipital lobes in homozygotes for the risk allele77 |
| rs2710102 | ASD and TD individuals | fMRI (reward-guided implicit learning task) | Frontal cortex | Reduced mPFC activation in nonrisk individuals and increased frontal connectivity in the risk allele carriers117 | |
| rs7794745 T rs2710102 C | Healthy individuals | fMRI (language task) | Prefrontal cortex, temporal cortex | Increased activation of the right inferior frontal gyrus and right lateral temporal cortex in the risk allele carriers118 | |
| c.3709DelG mutation | Individuals with syndromic ASD and healthy controls | MRI | Prefrontal cortex | Increased head circumference and GM volume76 | |
| Forebrain organoid culture | Increased total brain volume in individuals carrying the CNTNAP2 c.3709DelG mutation76 | ||||
| CNTNAP2 knockout mice | Laser-scanning photostimulation, whole-cell recordings, and electron microscopy | Prefrontal cortex | Reduced excitatory and inhibitory synaptic inputs onto L2/3 pyramidal neurons in the mPFC of CNTNAP2 knockout mice with reduced spines and synapses144 | ||
| MET | rs1858830 | Healthy individuals | sMRI | Frontal lobe, temporal lobe, anterior cingulate cortex | Reduction in cortical thickness with increasing C allele dose in temporal gyri, ventral pre- and postcentral gyri, anterior cingulate and in fronto-polar cortex regions78 |
| rs1858830 | ASD and TD individuals | fMRI (observation of emotional faces), resting-state fMRI, DTI | Neocortex | Higher activation of amygdala and striatum and reduced WM integrity and intrinsic connectivity between the posterior cingulate cortex and mPFC regions in MET risk allele carriers79 | |
| SLC6A4 | 5HTTLPR | ASD children | MRI | Cerebrum | 5HTTLPR short allele associated with increasing cerebral GM volumes80 |
| NRXN1 | rs1045881 | ASD and schizophrenia risk allele carriers | sMRI | Frontal lobe | Reduction in frontal WM volumes and altered sensorimotor function115 |
| bi-allelic NRXN1-α del | ASD individual and healthy controls | Single-cell RNA sequencing | Reduced proliferation capability and calcium signaling and high expression of radial glia-like morphology in NRXN1-α del NES cells116 | ||
| OXTR | rs2254298A | Healthy Japanese adults | MRI | Amygdala | Larger bilateral amygdala volume81 |
| rs2254298(G>A) | Healthy females | MRI | Amygdala, anterior cingulate cortex, brainstem | Increased amygdala and GM volume in the brainstem and decreased total GM volume and GM volume in the anterior cingulate cortex in G/A heterozygotes82 | |
| rs53576 | Healthy adults | sMRI and fMRI | Hypothalamus and amygdala | Decrease in hypothalamus GM and amygdala activation and increased functional correlation of hypothalamus and amygdala in minor allele carriers for rs5357683 | |
| rs2254298A | Healthy individuals | MRI | Cerebral cortex | Reduced GM volume in the right insula in males with the risk allele84 | |
| rs2268498 T/C | Healthy individuals | fMRI (fear processing task) | Occipital lobe | T-allele homozygotes showed increased activation of inferior occipital gyrus during recognition of fear expressions86 | |
| rs2268493 | Healthy individuals | fMRI (reward anticipation task) | Nucleus accumbens, amygdala, insula, thalamus, and prefrontal cortical regions | T-allele homozygotes showed reduced activation in the mesolimbic reward circuitry85 | |
| rs401015 | Healthy individuals | fMRI (direct gaze processing task) | Amygdala | Heterozygotes CT variants showed increased amygdala activity87 | |
| rs2254298A | Healthy individuals | VBM, fMRI | Hypothalamus, dorsal anterior cingulate cortex | Decreased hypothalamus GM volume and deactivation of dorsal anterior cingulate gyrus and increased structural coupling of dorsal anterior cingulate gyrus and hypothalamus in A carriers88 | |
| rs53576 | Healthy individuals | VBM, fMRI (mentalizing paradigm) | Amygdala, parietal lobe | Higher brain GM volume in the left amygdala and lower GM volume in superior parietal lobule89 | |
| CDH9 and CDH11 | Mice | In situ hybridization | Cerebellum | High expression of Cdh11 in the central part and Cdh9 in the surrounding areas in lobules VI/VII and Crus I and Crus II regions of the cerebellum145 | |
| RELN | Autistic and healthy individuals (postmortem samples) | SDS-PAGE, Western blotting | Cerebellum | Significant reduction of Reelin protein in the cerebellar region of ASD individuals132 | |
| SLC25A12 | Autistic and healthy individuals (postmortem samples) | qRT-PCR, RNA extraction, in situ hybridization | Prefrontal cortex | Strong expression of SLC25A12 in the BA46 prefrontal cortex of autistic subjects146 | |
| PRKCB1 |
rs3785392 rs3785387 |
Autistic and healthy individuals (postmortem samples) | qRT-PCR, Western blotting, Microarray analysis | Cerebral cortex | Significant reduction of PRKCB1 gene expression in the temporal neocortex of autistic individuals147 |
| TAOK2 | Mice | MRI, DTI | Midbrain, thalamus, hypothalamus and hindbrain regions | Enlarged volumes of the midbrain, hindbrain, hypothalamus, thalamus, cerebellum, and hippocampus and reduced density of fiber tracks in the medial corpus callosum of TAOK2 knockout mice57 | |
| MECP2 | Autistic and healthy individuals (postmortem samples) | Immunofluorescence and laser-scanning cytometry | Frontal cortex, temporal and occipital lobe | Decreased MECP2 expression in the frontal cortex and fusiform gyrus of autistic individuals148 | |
| SHANK3 | SHANK3 transgenic Mice | MRI | Hippocampus, forebrain, and midbrain | Reduction in total brain volume and hippocampal size and increase of basal ganglia in SHANK3 knockout mice as compared to prenatal zinc-deficient mouse model of ASD45 | |
| NL3 | NL3 knockin mice | Ex vivo MRI and DTI | Caudate putamen, substantia nigra, somatosensory cortex, corpus callosum, internal capsule, and cerebral peduncles | Decreased volumes of WM and GM regions in the NL3 knockin mice as compared to wildtype mice43 | |
| AVPR1A | RS1, RS3 | Healthy individuals | fMRI (face-matching task) | Amygdala | RS3 334bp, RS3 longer variants, RS1 312 bp, and RS1 shorter variants risk allele carriers showed stronger activation of left amygdala120 |
|
MECP2 ITGB3 NL3 |
MECP2 308 truncation, ITGB3 knockout, NL3 knockin mouse models |
MRI | Cerebellum | Increased GM and WM volume in crus II lobule and GM volume in paraflocculus in NL3 knockin mice, Expansion of vermis lobules III–X, the anterior lobule, the paraflocculus, and simple, crus I, and crus II hemisphere lobules in the homozygous MECP2 model and reduced cerebellum volume in ITGB3 knockin mice44 | |
| CD38 | rs3796863 | Healthy individuals | fMRI (social, emotional stimuli and gaze processing task) | Temporal cortex | Higher activation of the left fusiform gyrus in homozygous risk allele carriers119 |
| rs3796863 | Healthy individuals | fMRI (social cognition tasks) | Amygdala | Epistasis effect between CD38 and COMT in the amygdala121 | |
| C57BL/6 mice and CD38−/− mice | MRI | Prefrontal cortex | Larger whole brain volume, abnormal cortex development, and impaired synaptic plasticity in the prefrontal cortex of CD38−/− mice108 |
MRI magnetic resonance imaging, DTI diffusion tensor imaging, fMRI functional magnetic resonance imaging, VBM voxel-based morphometry.
Neuroligin-3 (NL3) is a major cell adhesion protein that plays a vital role in the development of the synapse, and it has also been implicated in ASD. An ex vivo MRI study showed a reduction in various WM and GM regions of NL3 knockin mice, with deficits in social and anxiety-related behaviors43. A similar study used MRI to image mouse models with mutations implicated in autism, such as NL3 knockin, MECP2, and integrin β3 (ITGB3) homozygous knockout mouse models to investigate the effects of these mutations on brain morphology44. The study found enlarged WM, and GM volumes of crus II lobule in NL3 mutants increased cerebellar volumes in MECP2 mutants and reduced cerebellar structures in ITGB3 mutants44. A neuroimaging study reported reduced total brain volume and hippocampal size with enlarged basal ganglia structures in SHANK3 knockout mice compared to a prenatal zinc-deficient mouse model of ASD45.
A recent study on mice with ANK2 mutations showed an increased number of excitatory synapses during postnatal development with increased axonal branching, which supports the presence of altered connectivity and penetrant behavioral impairments in mice and humans carrying ASD-related ANK2 mutations46. Mutations in CHD2 have been linked with various neurodevelopmental disorders, including ASD47. CHD2 is shown to play an essential role in brain development as the suppression of CHD2 can inhibit the self-renewing ability of radial glial cells and can increase the generation of intermediate progenitor cells and neurons during the process of neurogenesis that might contribute to abnormal neurodevelopment48. Another study showed that CHD2 knockdown zebrafish exhibited altered locomotor activity, which contributed to epileptic encephalopathy49. These studies show that there is a genetic overlap found between ASD disorder and other neurological disorders such as schizophrenia, epilepsy, and ID50. In addition, knockdown of CHD8 resulted in reduced axon and dendritic growth with delayed neuronal migration in mice51. CHD8 suppression in zebrafish affected the gene expression in neurodevelopmental pathways related to the proliferation of neural progenitor cells52.
One of the studies showed that mice mutants of HOXA1/HOXB1 had several defects, including the loss of rhombomere 4 and 5 and the loss of 2nd branchial arch affecting the development of the hindbrain53. Previous studies have shown the role of 16p11 deletions54 and the CHD8 gene55 in the brain overgrowth in idiopathic ASD. The postnatal development mechanisms such as pruning and dendritic arborization are affected by the overproliferation of cortical progenitor cells. The overproduction of neocortical neurons also contributed to the autistic-like features in mice56. Studies have reported that 16p11 deletions resulted in the alteration of cortical progenitor cell proliferation in mice and increased brain volumes in individuals with syndromic ASD54. One of the genes associated with the 16p11.2 region known as TAOK2 has been found to cause ASD-related cognitive abnormalities57. A neuroimaging study found dosage-dependent abnormalities in TAOK2 heterozygous and knockout mice. MRI of the TAOK2 knockout mice revealed enlarged midbrain, hindbrain, hypothalamus, thalamus, cerebellum, and hippocampus volumes, with a regional delay in the development of the neuronal track in the medial corpus callosum57.
Animal studies have shown that the MET protein regulates early cortical development in the neurobiology of ASD. A study by Qiu et al. showed excitatory hyperconnectivity in neocortical circuits of Met conditional knockout mice58. While another study showed that the reduction of MET and HGF/SF correlated with calbindin+ interneurons in the cortex of urokinase-type plasminogen activator receptor knockout mouse, thereby affecting cortical development59.
Clinical studies
In the clinical setting, several imaging studies were conducted that link specific gene polymorphisms with abnormal brain morphology in ASD patients (Table 1). Gene deletions such as 22q11.2 and 16p11.2 are associated with ASD. These deletions are also found to be linked with other neuropsychiatric disorders, including DiGeorge Syndrome, conotruncal anomaly face syndrome, velocardiofacial syndrome, intellectual disability, and schizophrenia60,61. 22q11.2 deletion in patients having high-risk negative symptoms at risk of developing schizophrenia showed decreased gyrification mainly in the medial occipital and temporal regions, involved in social cognition and early visual processing62. Another study reported increased cortical thickness in the frontal lobe, cerebral cortex, and superior parietal lobes and decreased cortical thickness in the regions of superior temporal gyrus and posterior cingulate cortex in patients with 22q11.2 deletion syndrome63.
Head circumference is abnormally increased in ASD patients64. Many factors can be responsible for the brain overgrowth in ASD, such as altered synapses during the development process, increased neurites or the number of neurons in the brain. HOX genes play an essential role in the development of organs and are responsible for the proper positioning of organism’s segment structures during embryonic development (Fig. 1). Out of all the HOX genes, HOXA1 is a critical gene that is involved in the development of the central nervous system, internal ear, and hyoid bones65,66. HOXA1 polymorphism has been found to induce an increased head growth rate in autistic and TD children67. In addition, autistic individuals with HOXA1 A218G polymorphism displayed an enlarged cranial circumference68, while in autistic patients, one study reported minor contributions of the HOXB1 gene to head circumference69.
Fig. 1. Structural brain changes associated with HOX genes.
HOXA1 A218 gene polymorphism is associated with an increased head circumference in autistic individuals, and HOXA1/HOXB1 mice mutants showed loss of rhombomeres 4 and 5 affecting the hindbrain development53,68.
Mutations in PTEN have also been associated with brain morphological changes in ASD. PTEN mutations have been observed in Cowden syndrome, and macrocephaly is the main feature of patients with Cowden syndrome, and some of the individuals with this syndrome are also found to be autistic70,71. Sequencing of the PTEN gene in ASD patients revealed three heterozygous germline mutations72 and a missense mutation73. An ASD-related syndrome, known as cortical dysplasia-focal epilepsy (CDFE) is a rare neuronal migration disorder, and patients with CDFE exhibit autistic characteristics. A study reported a homozygous mutation in the CNTNAP2 gene in all CDFE patients, which is responsible for an early developmental insult, mainly in the frontal and temporal neocortex74. Another study showed that the impact of CNTNAP2 missense variants on axonal growth as the loss of CNTNAP2 allele contributes to the disruption in axonal growth75. An MRI study investigated the homozygous c.3709DelG mutation in CNTNAP2 in the forebrain organoids generated from human induced pluripotent stem cells that were derived from patients with syndromic ASD. The study found that individuals with c.3709DelG mutation in CNTNAP2 displayed increased head circumference and increased GM volume76. An imaging genetic study found an association of CNTNAP2 rs779475 single-nucleotide polymorphism with WM and GM morphology in healthy controls and observed reduced volume of GM in the regions of frontal and occipital lobes and cerebellum. However, there was a reduction in the WM volume in the regions of right rostral cingulum, right caudal inferior fronto-occipital fasciculus, and the posterior thalamic radiations in homozygotes for the risk allele (rs779475 T/T) as compared to the nonrisk homozygotes and a group of rs779475T heterozygotes77.
Mutations in the chromodomain helicase DNA binding protein 8 (CHD8) gene, which plays a vital role in the chromatin remodeling process, are amongst the most commonly reported mutations in ASD. CHD8 mutations have also been linked with axonal and dendritic growth, and a study showed that CHD8 is highly expressed in a microtubule-associated-protein 2 positive neurons and parvalbumin neurons which plays an essential role in the development of neocortex in the human brain. The findings of the study suggested that CHD8 mutations in humans can result in neuronal deficits that can contribute to ASD pathophysiology51.
Studies have proposed the association of MET proto-oncogene, receptor tyrosine kinase (MET), as a candidate risk gene for ASD. In order to find the association of MET with cortical development, a structural MRI study investigated the relationship between a single-nucleotide polymorphism within the MET promoter (rs1858830, G→C) and the development of cortical thickness in ASD individuals. The study observed cortical thickness reduction in the regions of ventral precentral and postcentral gyri, superior and middle temporal gyri, anterior cingulate cortex, and in the right fronto-polar cortex with increasing C allele dose78. Another study found that MET risk allele carrier (rs1858830) individuals with ASD and TD showed higher activation of amygdala, reduced WM integrity, and reduced connectivity between the posterior cingulate cortex and medial prefrontal cortex (mPFC)79.
A common polymorphism in the serotonin-transporter gene (SLC6A4) involving a serotonin-transporter related polymorphic region (5HTTLPR) results in the generation of short or long alleles. A study by Wassink et al. found that the 5HTTLPR short allele significantly influences the cortical GM volumes in male autistic children80. Several neuroimaging studies have investigated the effects of different oxytocin receptor (OXTR) risk alleles (rs2254298, rs53576, rs226849, rs401015) on brain morphological changes in healthy adults81–89. OXTR risk allele carriers (rs2254298) showed increased amygdala volume81,82,89, decreased GM volume in the hypothalamus83, and right insula84. A study showed that dysregulation in the β-catenin/BRN2 transcriptional cascade was associated with an increased number of neurons and proliferation of neural progenitor cells derived from pluripotent stem cells of ASD individuals and also contributed to increased brain volumes90.
Functional brain changes
Resting-state fMRI (rsfMRI) is a powerful tool that is being used for mapping the neuronal activity of various neurological disorders. rsfMRI studies have revealed abnormalities in the neuronal network in ASD patients that help in understanding the underlying neurobiology of ASD. An rsfMRI study observed altered functional connectivity between the cerebral cortex and dentate nucleus in individuals with ASD, which is suggestive of impaired interaction between the cerebellum and social cortical regions of the brain91. Another study observed reduced integration of the default mode network with the mPFC and angular gyrus regions in the ASD group92. The mPFC region is mainly involved in decision making, and one of its primary function is encoding task-related information in the working memory93, while angular gyrus is involved in semantic processing, default mode network, social cognition, and memory retrieval and attention94. Another rsfMRI study showed that the underconnectivity in the temporal sulcus, which is a human voice processing region, in ASD children in addition to a decreased connectivity in reward-related regions in the brain of ASD children. While task-based fMRI showed altered network connectivity in multiple brain regions that control social and emotional processing in ASD95. A sentence comprehension task-based study showed decreased connectivity in cortical areas of ASD individuals, which signifies the low congruence of brain regions that are essential for integrating information96. Other task-based studies have also reported decreased connectivity in the fronto-parietal regions that are involved in working memory (n-back working memory task)97, facial processing (emotional facial processing task)98, response inhibition (simple inhibition/1-back inhibition task)99 and visuomotor coordination (measuring BOLD signal)100 in ASD individuals. Task-based study based on social processing found hypoactivation in social brain nodes in the regions of the mPFC, frontal gyrus, insula, temporal sulcus, amygdala, and the fusiform gyrus101.
Task-based fMRI studies focused on cognitive control reported fronto-striatal dysfunction in ASD, specifically in frontal gyrus, basal ganglia, and anterior cingulate cortex regions. Task-based studies that involved examining brain responses associated with figures of speech like puns102 and examining the neural activity during the detection of senseless sentences103 have also shown communication impairments such as decreased brain synchrony in language processing regions in ASD102,103. Task-based studies focusing on reward processing showed impairments in mesolimbic and meso cortical brain regions in ASD. Both over connectivity and underconnectivity between frontal and posterior regions was observed in task-based functional connectivity study104. An fMRI study revealed that ASD individuals had reduced activation of higher-order social (fusiform, cingulate, amygdala, gyrus, insula) and executive processing (prefrontal cortex) of the brain and higher activation of lower-order structures that are involved in the mediation of primary motor and sensory processing during emotional task performance105. While some task-based studies such as source recognition task106 and emotional face processing task107 have reported increased connectivity between the brain regions of ASD individuals106,107.
Limitations of functional imaging in ASD
Neuroimaging can help bridge the gap between genes, environment, and different ASD behavioral phenotypes. Although functional imaging techniques such as fMRI have revolutionized the understanding of various mental disorders, the fMRI technique also contains several limitations. The big challenge in current fMRI-based methods is the impact of age/gender and the development of scanning techniques for the diagnosis of ASD across heterogeneous populations. One of the limitations of fMRI is that it has no role in measuring the direct neuronal activity instead it provides changes in the signal based on the blood oxygen level, which may lead to the false-positive or -negative signal specially in patients with abrupt behavioral changes such as ASD. Secondly, an individual’s performance on a specific task must be considered in a task-based fMRI and must be well-thought-out when analyzing the fMRI data. The fMRI data are also analyzed as group averages; therefore, it is not straightforward to diagnose individual autistic patients. This could be par by introducing an individualized diagnostic model based on artificial intelligence.
Furthermore, the data obtained from fMRI may be compromised to some extent due to head motion artifacts that alter voxel and stable state magnetization leading to distortion of anatomical locations and may show false regions associated with brain activity. In addition, fMRI data can only demonstrate that a particular region of the brain is involved in a specific cognitive function but cannot determine whether it is a cause or a consequence of that specific cognitive function. The lack of longitudinal data also makes it challenging to identify the abnormal developmental course in ASD during infancy. Nevertheless, the integration of multiple sites and data sources and the increase in the number of subjects and intra-variability among subjects can improve the feasibility of the fMRI technique in ASD.
Genes affecting brain functions
Preclinical studies
Reduced levels of oxytocin have been found in ASD patients, and CD38 is a vital enzyme that controls oxytocin production. A CD38−/− mice study showed abnormal development of the cortex and impaired synaptic plasticity in the prefrontal cortex region with impaired social and emotional responses108 (Table 1). Genetic variations in the 1A gene of the arginine vasopressin receptor (AVPR1A) were linked to autism. Impaired social recognition and reduced anxiety behaviors were observed in knockout mice of AVPR1A, while overexpression of AVPR1A in mice led to increased social memory109,110. There is also an inversion of cortical layering due to neuronal migration defects in the Reelin (RELN) knockout mice111, resulting in various behavioral abnormalities, such as aggressive behavior and memory and learning impairments112. CNVs of the cytoplasmic FMR1-interacting protein 1 (CYFIP1) was found to be associated with ASD and other neuropsychiatric disorders such as schizophrenia. CYFIP1 plays an essential part in regional brain connectivity and corpus callosum function. A study found that the CYFIP1-heterozygous mice displayed reduced bilateral functional connectivity across the entire brain and defects in the WM architecture. In these mice, reduced myelination is also observed in the callosal axons, an altered presynaptic feature that contributes to the motor and sensorimotor abnormalities113.
Another rsfMRI study showed decreased local and long-range functional connectivity in prefrontal and midbrain of mice with the homozygous loss of CNTNAP2 and may contribute to the development of autistic-like features and other neurodevelopment disorders114.
Clinical studies
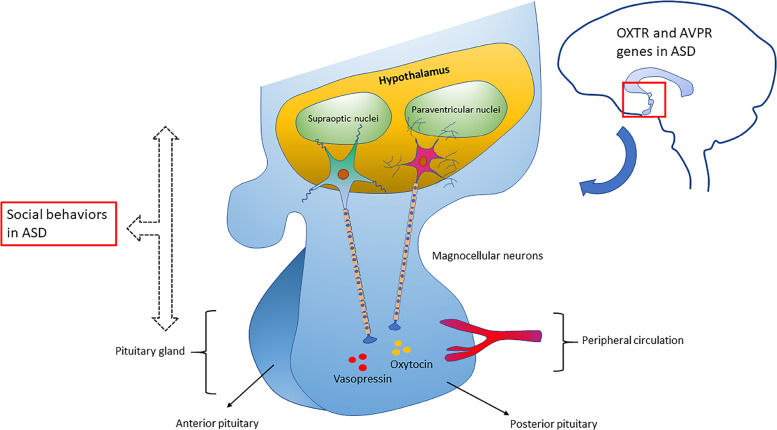
Imaging genetics studies have shown the involvement of ASD risk genes in altering the brain circuits that control reward and language processing, and social behavior (Table 1). Neurexin-1 (NRXN1) is one of the ASD risk genes that are found to influence the brain structure and functions, and its polymorphisms are found to be associated with structural alterations in the prefrontal–thalamic circuitry in healthy individuals, conferring a potential risk of developing ASD or schizophrenia115. Single-cell analysis of neural stem cells from an autistic patient with bi-allelic NRXN1-α deletion observed a phenotypic shift toward radial glia-like cell with impaired maturation action potential and reduced calcium signaling116. On the other hand, CNTNAP2 polymorphisms are found to be associated with altered brain connectivity in regions that are involved in reward and language development117 (Fig. 2). Healthy individuals carrying the risk alleles for ASD and language impairment (rs7794745 T, rs2710102 C) showed increased activation in the right inferior frontal gyrus and right lateral temporal cortex118. Mutations in the genes involved in the oxytocin/vasopressin system may also contribute to ASD risk as they influence the function of brain regions such as the amygdala and hypothalamus that are associated with emotional and social processing119,120 (Fig. 3). Studies have found an association between a common genetic variant in CD38 (rs3796863), autism, and the impact of oxytocin levels on brain response to social stimuli119,121. An fMRI study observed increased activation of the left fusiform gyrus in homozygotes for CD38 genetic variant rs3796863119. The MET gene variants are also found to be involved in altering the connectivity and integrity of WM in the temporo-parieto-occipital regions that are involved in high neurological functions such as working memory, language processing, and face and object recognition, all that may predispose an individual to ASD79. Another study reported that the loss of CNTNAP2 contributed to the reduction in long-range connectivity in the brain regions of mice, mainly affecting the prefrontal regions that act as connectivity hubs in the brain122 and this hypoconnectivity in the fronto-posterior region lead to impaired social behaviors, predisposing to autism risk114. The OXTR polymorphism rs1042778 is found to be associated with two common clinical phenotypes of ASD, such as aggression and panic in male ASD patients. As compared to GG homozygotes, the T-allele carriers were at higher risk of possessing aggressive and panic behaviors123. Some of the task-based fMRI studies in healthy individuals carrying the OXTR risk alleles showed increased activation of inferior occipital gyrus during a fear processing task86, decreased activation in the mesolimbic reward circuitry in a reward anticipation task85 and increased amygdala activity during a direct gaze processing task87. One study found elevated levels of AVP with reduced levels of apelin in ASD patients that point to the vasopressinergic dysfunction in autism124. Another study proposed that the shorter alleles of RS1 polymorphism lead to reduced transcription of AVPR1A that may increase the susceptibility to ASD125. An fMRI study found an association between ASD and AVPR1A genetic variants and reported differential activation of amygdala during a face-matching task in RS1 and RS3 risk allele carriers120.
Fig. 2. Specific studies in humans and mice with ASD showing functional brain changes correlated with CNTNAP2 gene.
The CNTNAP2 gene is associated with increased frontal lobe connectivity117 and increased connectivity in ASD risk allele carriers within Broca’s area118. Studies on mice show that the loss of CNTNAP2 gene impairs axonal growth75, reduces functional connectivity in the prefrontal cortex114, and hyperactive AKT/mTOR pathway149.
Fig. 3. Oxytocin and vasopressin (OT-AVP) pathway associated with social behavior in ASD.
In the hypothalamus, magnocellular neurons originating from paraventricular and supraoptic nuclei release oxytocin and vasopressin in the posterior pituitary which then goes into the peripheral circulation. Any deficiencies in oxytocin or vasopressin levels contribute to ASD’s social behavioral impairments.
Reduced expression of three genes namely metaxin 2 (MTX2), neurofilament light polypeptide (NEFL), and solute carrier family 25, member 27 (SLC25A27) was found in the anterior cingulate gyrus, the motor cortex, and thalamus regions of autistic patients. NEFL plays a vital role in the maintenance and assembly of the axonal cytoskeleton126, while SLC25A27 is expressed in the central nervous system and has been found to play an essential role in neuronal cell differentiation127, apoptosis inhibition128, and reduction of reactive oxygen species129.
One of the critical regulators of neuronal migration is RELN glycoprotein, shown to play a vital role in the neuron layering. Aberrant reelin signaling has been reported in many studies in individuals with ASD130–132. RELN mutations disrupt neuronal migration and connectivity and cerebellar hypoplasia133. A significant reduction in the RELN protein has also been observed in the cerebellum of autistic individuals132. Another gene T-Box Brain Protein 1 that encodes a brain-specific T-box transcription factor plays an essential role in the neuronal development, axonal migration, and the development of the cerebral cortex and amygdala134. This gene has been identified as an ASD causative gene135 and is involved in the differentiation of neurons during the early development of neocortex136.
Diagnostic models of ASD
Diagnostic models based on MR-derived features
MRI-based diagnostic models are used for the behavioral assessment of autistic patients. These diagnostic model studies involve three steps, including extraction of properties from MR images, construction of diagnostic model using statistical models followed by evaluation and validation by researchers. Several studies have focussed on MRI-based diagnostic models for the detection and classification of ASD30,137. Diagnostic model performance is strongly influenced by types of entities selected as components of the model. For example, a study showed the comparison of diagnostic models based on regional thickness derived from surface morphometry (SBM) with diagnostic models based on volumetric morphometry involving four different classification methods and classification based on thickness was found to be more efficient and predictive of ASD compared to classification based on volume30. rsfMRI pipelines have been used to extract predictive biomarkers in autism by constructing participant-specific connectomes and then comparing these connectomes across participants to learn connectivity patterns that may identify ASD individuals138. The results suggested that rsfMRI data collected from different sites could reveal robust functional connectivity biomarkers of ASD.
Diagnostic models based on imaging genetics
Imaging genetics in ASD has proven useful, and pathways that include common genetic variation in TD individuals at risk of developing ASD have been characterized. Prenatal transcription regulation and synapse formation in the developing brain is impacted by the genes associated with ASD (Table 1). Alteration in frontal WM connectivity and structure and disturbance in the frontal, temporal, and occipital circuits involved in visual and language processing was found to be associated with NRXN superfamily genes115. Neuropeptide signaling and emotional functioning was found to be influenced by the oxytocin and arginine vasopressin receptor genes via structural and functional modification in the amygdala–hypothalamus circuitry77. One study showed a relationship between frontal lobe connectivity and common genetic variants in CNTNAP2 using a functional neuroimaging study117 and the study found that ASD and TD individuals who were nonrisk allele carriers showed more reduction in the activation of mPFC during an fMRI task as compared to risk allele carriers117. Another study showed decreased functional connectivity in the prefrontal cortex, cortical spinal tract, corpus callosum, and decreased integrity of WM in children and adolescents carrying MET rs1858830, C risk allele. Such studies suggest that the genes affect the brain regions that are involved in social and emotional processing139.
More detailed brain analysis combined with visualization, will promote the elucidation of the relationship between genes and altered neuronal circuits. Also, researching the time-dependent impacts of ASD-involved genetic variants that are the cause of neurodevelopmental modification can aid in the progress of ASD imaging genetics. In addition, the plasticity that is altered in different neuropsychiatric disorders such as ASD can be measured with high resolution by combining electroencephalography or fMRI with transcranial magnetic stimulation. Advances in noninvasive brain imaging techniques and scans during rest or task may help in infant neuroimaging that are at a higher risk to develop ASD. With studies investigating genetic effects on brain development using high-resolution imaging, studying brain-behavior connections and the impact of new therapies on brain development, further translational progress will happen in the future. By neuroimaging endophenotypes, a greater understanding of the impact of ASD risk genes on the brain circuitry139 can be achieved. Ultimately, imaging genetics in ASD might be promising for the clinical management of ASD-affected patients.
Challenges and future perspectives
The field of imaging genetics has exponentially grown in recent decades from its candidate gene studies to large-scale longitudinal studies, cross-modal investigations, and translational animal models of various psychiatric disorders. In addition, imaging genetics has begun integrating transcriptomic data and analytical methods for assessing pathway enrichment, such as the score system for pathway regulation. Of addition to translational animal research and pharmacological intervention in vitro and in vivo, imaging genetics can also provide an insight into various behavioral and genetic factors that contribute to the risk of ASDs.
One of the challenges facing imaging genetics is the conceptualization of endophenotypes, which states that endophenotypes are heritable and associated with psychiatric disorders and may impede research on brain-based associations by limiting imaging genetic research to genes previously associated with a psychiatric disorder140. It is important to properly replicate the studies, particularly those with false-positive results, to address the impact of a genetic variation in a disease, and this problem can be solved by correcting genome-wide associations with large sample size imaging phenotypes.
Another challenge in ASD neuroimaging research is the discrepancy in the brain connectivity findings due to motion artifacts. The motion artifacts in the rsfMRI data can misrepresent group differences in connectivity metrics which can lead to incorrect interpretation of the altered brain connectivity data in ASD. Other methodological variables such as pipeline type, field of view, and dataset can significantly impact (in terms of over-or underconnectivity) the functional connectivity MRI studies in ASD141. Also, the differences in age and severity of the ASD cohort may lead to disparity in findings, and so large-scale studies are required that concentrate on exploring the developmental trajectories that are crucial to understanding the neurobiology underlying ASD. Such large-scale study databases can be obtained from various consortia, such as autism brain imaging data exchange and National Autism Research Database (NDAR). The heterogeneity of the ASD phenotypes, therefore, leads to the difficulties faced in ASD neuroimaging genetics as there is a high degree of variability in frequency, the number of behavioral symptoms reported, and the associated characteristics in ASD. In addition, the brain responses produced as a result of a particular task stimulus are inadequate to determine if these responses result from a neural processing disorder or are due to the introduction of an extrinsic factor.
In addition, more development in data analytical methods such as machine learning algorithms or software can allow data from multiple modalities to be incorporated into a heterogeneous population, and can also recognize and detect digital biomarkers142 that can be used for the clinical management of ASD patients.
Conclusion
To explain the origin of ASD’s underlying pathophysiology, the study of genetic alterations and the relationship between genes and environmental factors is crucial. Imaging genetics studies have revealed important information about the pathophysiology of ASD. One of the key aims of imaging genetics studies in ASD is to elucidate neural pathways that give rise to phenotypical heterogeneity. Enhanced risk of ASD is shown to be related to several different single-nucleotide polymorphisms, and the stratification of neuroimaging data by specific genetic risk factors occurring in ASD may help to understand the neurobiology underlying this disorder. Such studies will help to elucidate ASD from various perspectives and subsequently will pave the way for personalized treatment for patients with ASD.
Acknowledgements
This study was supported by a PI grant from Sidra Medicine (5071012001) to M.H. A.A.B. is supported by Sidra Medicine internal grant (5011041002).
Author contributions
S.H., S.N, A.A.B.: prepared the scientific material, wrote the paper, generated tables and figures. S.Y., M.W., P.B., K.F., R.R., M.P.F.: critical revision and editing of the scientific contents. M.H. conceived of and designed the review contents, and contributed to paper writing and editing. All authors read and approved the final paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sheema Hashem, Sabah Nisar
References
- 1.Baio J, et al. Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2014. MMWR Surveill. Summ. 2018;67:1–23. doi: 10.15585/mmwr.ss6706a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu G, Strathearn L, Liu B, Bao W. Prevalence of autism spectrum disorder among US children and adolescents, 2014–2016. JAMA. 2018;319:81–82. doi: 10.1001/jama.2017.17812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor B, Jick H, MacLaughlin D. Prevalence and incidence rates of autism in the UK: time trend from 2004–2010 in children aged 8 years. BMJ Open. 2013;3:e003219. doi: 10.1136/bmjopen-2013-003219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu S, et al. Prevalence of autism spectrum disorder in Asia: a systematic review and meta-analysis. Psychiatry Res. 2020;284:112679. doi: 10.1016/j.psychres.2019.112679. [DOI] [PubMed] [Google Scholar]
- 5.Al-Farsi YM, et al. Brief report: prevalence of autistic spectrum disorders in the Sultanate of Oman. J. Autism Dev. Disord. 2011;41:821–825. doi: 10.1007/s10803-010-1094-8. [DOI] [PubMed] [Google Scholar]
- 6.Al-Ansari A, Mahmood M. Epidemiology of autistic disorder in Bahrain: prevalence and obstetric and familial characteristics. East. Mediterr. Health J. 2013;19:769–774. [PubMed] [Google Scholar]
- 7.Aljarallah, A., Alwaznah, T., Alansari, S. & Alhazmi, M. A. Study of Autism and Developmental Disorders in Saudi Children (King Abdulaziz City for Science and Technology, Riyadh, Saudi Arabia, 2007).
- 8.Alshaban F, et al. Prevalence and correlates of autism spectrum disorder in Qatar: a national study. J. Child Psychol. Psychiatry. 2019;60:1254–1268. doi: 10.1111/jcpp.13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werling DM, Geschwind DH. Sex differences in autism spectrum disorders. Curr. Opin. Neurol. 2013;26:146–153. doi: 10.1097/WCO.0b013e32835ee548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pisula E, Porębowicz-Dörsmann A. Family functioning, parenting stress and quality of life in mothers and fathers of Polish children with high functioning autism or Asperger syndrome. PLoS One. 2017;12:e0186536. doi: 10.1371/journal.pone.0186536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaugler T, et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallmayer J, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandin S, et al. The familial risk of autism. JAMA. 2014;311:1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss LA, et al. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fakhoury M. Imaging genetics in autism spectrum disorders: linking genetics and brain imaging in the pursuit of the underlying neurobiological mechanisms. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2018;80:101–114. doi: 10.1016/j.pnpbp.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 18.Kemper TL, Bauman M. Neuropathology of infantile autism. J. Neuropathol. Exp. Neurol. 1998;57:645–652. doi: 10.1097/00005072-199807000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Courchesne E, et al. Neuron number and size in prefrontal cortex of children with autism. JAMA. 2011;306:2001–2010. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- 20.Arin DM, Bauman ML, Kemper TL. The distribution of Purkinje cell loss in the cerebellum in autism. Neurology. 1991;41:307. [Google Scholar]
- 21.Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. Int. J. Dev. Neurosci. 2005;23:183–187. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Zikopoulos B, Barbas H. Changes in prefrontal axons may disrupt the network in autism. J. Neurosci. 2010;30:14595–14609. doi: 10.1523/JNEUROSCI.2257-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Sparks BF, et al. Brain structural abnormalities in young children with autism spectrum disorder. Neurology. 2002;59:184. doi: 10.1212/wnl.59.2.184. [DOI] [PubMed] [Google Scholar]
- 25.Nordahl CW, et al. Brain enlargement is associated with regression in preschool-age boys with autism spectrum disorders. Proc. Natl. Acad. Sci. USA. 2011;108:20195–20200. doi: 10.1073/pnas.1107560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Courchesne E, et al. Unusual brain growth patterns in early life in patients with autistic disorder. Neurology. 2001;57:245. doi: 10.1212/wnl.57.2.245. [DOI] [PubMed] [Google Scholar]
- 27.Piven J, An MRI. study of the corpus callosum in autism. Am. J. Psychiatry. 1997;154:1051–1056. doi: 10.1176/ajp.154.8.1051. [DOI] [PubMed] [Google Scholar]
- 28.Hardan AY, et al. study of increased cortical thickness in autism. Am. J. Psychiatry. 2006;163:1290–1292. doi: 10.1176/appi.ajp.163.7.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyde KL, Samson F, Evans AC, Mottron L. Neuroanatomical differences in brain areas implicated in perceptual and other core features of autism revealed by cortical thickness analysis and voxel-based morphometry. Hum. Brain Mapp. 2010;31:556–566. doi: 10.1002/hbm.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiao Y, et al. Predictive models of autism spectrum disorder based on brain regional cortical thickness. NeuroImage. 2010;50:589–599. doi: 10.1016/j.neuroimage.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Libero LE, DeRamus TP, Deshpande HD, Kana RK. Surface-based morphometry of the cortical architecture of autism spectrum disorders: volume, thickness, area, and gyrification. Neuropsychologia. 2014;62:1–10. doi: 10.1016/j.neuropsychologia.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Nordahl CW, et al. Cortical folding abnormalities in autism revealed by surface-based morphometry. J. Neurosci. 2007;27:11725–11735. doi: 10.1523/JNEUROSCI.0777-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen R, Jiao Y, Herskovits EH. Structural MRI in autism spectrum disorder. Pediatr. Res. 2011;69:63R–68R. doi: 10.1203/PDR.0b013e318212c2b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller S, et al. Convergent findings of altered functional and structural brain connectivity in individuals with high functioning autism: a multimodal MRI study. PLoS One. 2013;8:e67329. doi: 10.1371/journal.pone.0067329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Bihan D. Looking into the functional architecture of the brain with diffusion MRI. Nat. Rev. Neurosci. 2003;4:469–480. doi: 10.1038/nrn1119. [DOI] [PubMed] [Google Scholar]
- 36.Travers BG, et al. Diffusion tensor imaging in autism spectrum disorder: a review. Autism Res. 2012;5:289–313. doi: 10.1002/aur.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben Bashat D, et al. Accelerated maturation of white matter in young children with autism: a high b value DWI study. NeuroImage. 2007;37:40–47. doi: 10.1016/j.neuroimage.2007.04.060. [DOI] [PubMed] [Google Scholar]
- 38.Sahyoun CP, Belliveau JW, Mody M. White matter integrity and pictorial reasoning in high-functioning children with autism. Brain Cognit. 2010;73:180–188. doi: 10.1016/j.bandc.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolff JJ, et al. Differences in white matter fiber tract development present from 6 to 24 months in infants with autism. Am. J. Psychiatry. 2012;169:589–600. doi: 10.1176/appi.ajp.2011.11091447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jou RJ, et al. Diffusion tensor imaging in autism spectrum disorders: preliminary evidence of abnormal neural connectivity. Aust. N. Z. J. Psychiatry. 2011;45:153–162. doi: 10.3109/00048674.2010.534069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geschwind DH, State MW. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 2015;14:1109–1120. doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar M, et al. High resolution magnetic resonance imaging for characterization of the neuroligin-3 knock-in mouse model associated with autism spectrum disorder. PLoS One. 2014;9:e109872. doi: 10.1371/journal.pone.0109872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steadman PE, et al. Genetic effects on cerebellar structure across mouse models of autism using a magnetic resonance imaging atlas. Autism Res. 2014;7:124–137. doi: 10.1002/aur.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoen, M. et al. Shank3 transgenic and prenatal zinc-deficient autism mouse models show convergent and individual alterations of brain structures in MRI. Front. Neural Circuits13, 10.3389/fncir.2019.00006 (2019). [DOI] [PMC free article] [PubMed]
- 46.Yang R, et al. ANK2 autism mutation targeting giant ankyrin-B promotes axon branching and ectopic connectivity. Proc. Natl. Acad. Sci. USA. 2019;116:15262–15271. doi: 10.1073/pnas.1904348116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamar K-MJ, Carvill GL. Chromatin remodeling proteins in epilepsy: lessons from CHD2-associated epilepsy. Front. Mol. Neurosci. 2018;11:208–208. doi: 10.3389/fnmol.2018.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen T, Ji F, Yuan Z, Jiao J. CHD2 is required for embryonic neurogenesis in the developing cerebral cortex. Stem Cells. 2015;33:1794–1806. doi: 10.1002/stem.2001. [DOI] [PubMed] [Google Scholar]
- 49.Suls A, et al. De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am. J. Hum. Genet. 2013;93:967–975. doi: 10.1016/j.ajhg.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, et al. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol. Psychiatry. 2016;21:290–297. doi: 10.1038/mp.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Q, et al. Autism-associated CHD8 deficiency impairs axon development and migration of cortical neurons. Mol. Autism. 2018;9:65. doi: 10.1186/s13229-018-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugathan A, et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. USA. 2014;111:E4468–E4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rossel M, Capecchi MR. Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development. 1999;126:5027. doi: 10.1242/dev.126.22.5027. [DOI] [PubMed] [Google Scholar]
- 54.Qureshi AY, et al. Opposing brain differences in 16p11.2 deletion and duplication carriers. J. Neurosci. 2014;34:11199–11211. doi: 10.1523/JNEUROSCI.1366-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernier R, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang W-Q, et al. Overproduction of upper-layer neurons in the neocortex leads to autism-like features in mice. Cell Rep. 2014;9:1635–1643. doi: 10.1016/j.celrep.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 57.Richter M, et al. Altered TAOK2 activity causes autism-related neurodevelopmental and cognitive abnormalities through RhoA signaling. Mol. Psychiatry. 2019;24:1329–1350. doi: 10.1038/s41380-018-0025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiu S, Anderson CT, Levitt P, Shepherd GMG. Circuit-specific intracortical hyperconnectivity in mice with deletion of the autism-associated Met receptor tyrosine kinase. J. Neurosci. 2011;31:5855–5864. doi: 10.1523/JNEUROSCI.6569-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Powell EM, Mars WM, Levitt P. Hepatocyte growth factor/scatter factor is a motogen for interneurons migrating from the ventral to dorsal telencephalon. Neuron. 2001;30:79–89. doi: 10.1016/s0896-6273(01)00264-1. [DOI] [PubMed] [Google Scholar]
- 60.Bassett AS, et al. Practical guidelines for managing patients with 22q11.2 deletion syndrome. J. Pediatr. 2011;159:332–339. doi: 10.1016/j.jpeds.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moreno-De-Luca A, et al. The role of parental cognitive, behavioral, and motor profiles in clinical variability in individuals with chromosome 16p11.2 deletions. JAMA Psychiatry. 2015;72:119–126. doi: 10.1001/jamapsychiatry.2014.2147. [DOI] [PubMed] [Google Scholar]
- 62.Mihailov A, et al. Morphological brain changes associated with negative symptoms in patients with 22q11.2 deletion syndrome. Schizophr. Res. 2017;188:52–58. doi: 10.1016/j.schres.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 63.Schmitt JE, et al. Aberrant cortical morphometry in the 22q11.2 deletion syndrome. Biol. Psychiatry. 2015;78:135–143. doi: 10.1016/j.biopsych.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fombonne E, Rogé B, Claverie J, Courty S, Frémolle J. Microcephaly and macrocephaly in autism. J. Autism Dev. Disord. 1999;29:113–119. doi: 10.1023/a:1023036509476. [DOI] [PubMed] [Google Scholar]
- 65.Torres M, Giráldez F. The development of the vertebrate inner ear. Mech. Dev. 1998;71:5–21. doi: 10.1016/s0925-4773(97)00155-x. [DOI] [PubMed] [Google Scholar]
- 66.Persico, A. M. in Neural Circuit Development and Function in the Brain (eds Rubenstein, John L. R. & Rakic, Pasko) 651–694 (Academic Press, Cambridge, 2013).
- 67.Muscarella LA, et al. HOXA1 gene variants influence head growth rates in humans. Am. J. Med. Genet. Part B: Neuropsychiatr. Genet. 2007;144B:388–390. doi: 10.1002/ajmg.b.30469. [DOI] [PubMed] [Google Scholar]
- 68.Conciatori M, et al. Association between the HOXA1 A218G polymorphism and increased head circumference in patients with autism. Biol. Psychiatry. 2004;55:413–419. doi: 10.1016/j.biopsych.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 69.Muscarella LA, et al. Candidate gene study of HOXB1 in autism spectrum disorder. Mol. Autism. 2010;1:9. doi: 10.1186/2040-2392-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pilarski R, Eng C. Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J. Med. Genet. 2004;41:323–326. doi: 10.1136/jmg.2004.018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goffin A, Hoefsloot LH, Bosgoed E, Swillen A, Fryns J-P. PTEN mutation in a family with Cowden syndrome and autism. Am. J. Med. Genet. 2001;105:521–524. doi: 10.1002/ajmg.1477. [DOI] [PubMed] [Google Scholar]
- 72.Butler MG, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buxbaum JD, et al. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144B:484–491. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strauss K, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N. Engl. J. Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 75.Canali G, et al. Genetic variants in autism-related CNTNAP2 impair axonal growth of cortical neurons. Hum. Mol. Genet. 2018;27:1941–1954. doi: 10.1093/hmg/ddy102. [DOI] [PubMed] [Google Scholar]
- 76.de Jong, J. et al. Cortical overgrowth in a preclinical forebrain organoid model of CNTNAP2-associated autism spectrum disorder. (2019). [DOI] [PMC free article] [PubMed]
- 77.Tan GCY, Doke TF, Ashburner J, Wood NW, Frackowiak RSJ. Normal variation in fronto-occipital circuitry and cerebellar structure with an autism-associated polymorphism of CNTNAP2. NeuroImage. 2010;53:1030–1042. doi: 10.1016/j.neuroimage.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hedrick A, et al. Autism risk gene MET variation and cortical thickness in typically developing children and adolescents. Autism Res. 2012;5:434–439. doi: 10.1002/aur.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rudie JD, et al. Autism-associated promoter variant in MET impacts functional and structural brain networks. Neuron. 2012;75:904–915. doi: 10.1016/j.neuron.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wassink TH, et al. Cerebral cortical gray matter overgrowth and functional variation of the serotonin transporter gene in autism. Arch. Gen. Psychiatry. 2007;64:709–717. doi: 10.1001/archpsyc.64.6.709. [DOI] [PubMed] [Google Scholar]
- 81.Inoue H, et al. Association between the oxytocin receptor gene and amygdalar volume in healthy adults. Biol. Psychiatry. 2010;68:1066–1072. doi: 10.1016/j.biopsych.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 82.Furman DJ, Chen MC, Gotlib IH. Variant in oxytocin receptor gene is associated with amygdala volume. Psychoneuroendocrinology. 2011;36:891–897. doi: 10.1016/j.psyneuen.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tost H, et al. A common allele in the oxytocin receptor gene (OXTR) impacts prosocial temperament and human hypothalamic-limbic structure and function. Proc. Natl. Acad. Sci. USA. 2010;107:13936–13941. doi: 10.1073/pnas.1003296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saito Y, et al. Neural correlate of autistic-like traits and a common allele in the oxytocin receptor gene. Soc. Cognit. Affect. Neurosci. 2014;9:1443–1450. doi: 10.1093/scan/nst136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Damiano CR, et al. Association between the oxytocin receptor (OXTR) gene and mesolimbic responses to rewards. ecular. Mol. Autism. 2014;5:7. doi: 10.1186/2040-2392-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O’Connell G, et al. Association of genetic variation in the promoter region of OXTR with differences in social affective neural processing. J. Behav. Brain Sci. 2012;2:60. [Google Scholar]
- 87.Montag C, Sauer C, Reuter M, Kirsch P. An interaction between oxytocin and a genetic variation of the oxytocin receptor modulates amygdala activity toward direct gaze: evidence from a pharmacological imaging genetics study. Eur. Arch. Psychiatry Clin. Neurosci. 2013;263:169–175. doi: 10.1007/s00406-013-0452-x. [DOI] [PubMed] [Google Scholar]
- 88.Tost H, et al. Neurogenetic effects of OXTR rs2254298 in the extended limbic system of healthy Caucasian adults. Biol. Psychiatry. 2011;70:e37–e39. doi: 10.1016/j.biopsych.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 89.Schneider-Hassloff H, et al. Oxytocin receptor polymorphism and childhood social experiences shape adult personality, brain structure and neural correlates of mentalizing. NeuroImage. 2016;134:671–684. doi: 10.1016/j.neuroimage.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 90.Marchetto MC, et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol. Psychiatry. 2017;22:820–835. doi: 10.1038/mp.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Olivito G, et al. Resting-state functional connectivity changes between dentate nucleus and cortical social brain regions in autism spectrum disorders. Cerebellum. 2017;16:283–292. doi: 10.1007/s12311-016-0795-8. [DOI] [PubMed] [Google Scholar]
- 92.Joshi G, et al. Integration and segregation of default mode network resting-state functional connectivity in transition-age males with high-functioning autism spectrum disorder: a proof-of-concept study. Brain Connect. 2017;7:558–573. doi: 10.1089/brain.2016.0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baddeley A. Working memory: looking back and looking forward. Nat. Rev. Neurosci. 2003;4:829–839. doi: 10.1038/nrn1201. [DOI] [PubMed] [Google Scholar]
- 94.Seghier ML. The angular gyrus: multiple functions and multiple subdivisions. Neuroscientist. 2013;19:43–61. doi: 10.1177/1073858412440596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abrams DA, et al. Underconnectivity between voice-selective cortex and reward circuitry in children with autism. Proc. Natl. Acad. Sci. USA. 2013;110:12060–12065. doi: 10.1073/pnas.1302982110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Just MA, Cherkassky VL, Keller TA, Minshew NJ. Cortical activation and synchronization during sentence comprehension in high-functioning autism: evidence of underconnectivity. Brain. 2004;127:1811–1821. doi: 10.1093/brain/awh199. [DOI] [PubMed] [Google Scholar]
- 97.Koshino H, et al. Functional connectivity in an fMRI working memory task in high-functioning autism. NeuroImage. 2005;24:810–821. doi: 10.1016/j.neuroimage.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 98.Kleinhans NM, et al. Abnormal functional connectivity in autism spectrum disorders during face processing. Brain. 2008;131:1000–1012. doi: 10.1093/brain/awm334. [DOI] [PubMed] [Google Scholar]
- 99.Kana RK, Keller TA, Minshew NJ, Just MA. Inhibitory control in high-functioning autism: decreased activation and underconnectivity in inhibition networks. Biol. Psychiatry. 2007;62:198–206. doi: 10.1016/j.biopsych.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Villalobos ME, Mizuno A, Dahl BC, Kemmotsu N, Müller R-A. Reduced functional connectivity between V1 and inferior frontal cortex associated with visuomotor performance in autism. NeuroImage. 2005;25:916–925. doi: 10.1016/j.neuroimage.2004.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blakemore S-J. The social brain in adolescence. Nat. Rev. Neurosci. 2008;9:267–277. doi: 10.1038/nrn2353. [DOI] [PubMed] [Google Scholar]
- 102.Kana RK, Wadsworth HM. “The archeologist’s career ended in ruins”: hemispheric differences in pun comprehension in autism. NeuroImage. 2012;62:77–86. doi: 10.1016/j.neuroimage.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 103.Catarino A, et al. An fMRI investigation of detection of semantic incongruities in autistic spectrum conditions. Eur. J. Neurosci. 2011;33:558–567. doi: 10.1111/j.1460-9568.2010.07503.x. [DOI] [PubMed] [Google Scholar]
- 104.Dichter GS. Functional magnetic resonance imaging of autism spectrum disorders. Dialog. Clin. Neurosci. 2012;14:319–351. doi: 10.31887/DCNS.2012.14.3/gdichter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ameis SH, Szatmari P. Imaging-genetics in autism spectrum disorder: advances, translational impact, and future directions. Front. Psychiatry. 2012;3:46. doi: 10.3389/fpsyt.2012.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Noonan SK, Haist F, Müller R-A. Aberrant functional connectivity in autism: evidence from low-frequency BOLD signal fluctuations. Brain Res. 2009;1262:48–63. doi: 10.1016/j.brainres.2008.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Safar K, Wong SM, Leung RC, Dunkley BT, Taylor MJ. Increased functional connectivity during emotional face processing in children with autism spectrum disorder. Front. Hum. Neurosci. 2018;12:408. doi: 10.3389/fnhum.2018.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martucci LL, et al. A multiscale analysis in CD38(-/-) mice unveils major prefrontal cortex dysfunctions. FASEB J. 2019;33:5823–5835. doi: 10.1096/fj.201800489R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Egashira N, et al. Impaired social interaction and reduced anxiety-related behavior in vasopressin V1a receptor knockout mice. Behav. Brain Res. 2007;178:123–127. doi: 10.1016/j.bbr.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 110.Bielsky IF, Hu S-B, Szegda KL, Westphal H, Young LJ. Profound impairment in social recognition and reduction in anxiety-like behavior in vasopressin V1a receptor knockout mice. Neuropsychopharmacology. 2004;29:483–493. doi: 10.1038/sj.npp.1300360. [DOI] [PubMed] [Google Scholar]
- 111.Trommsdorff M, et al. Reeler/disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 112.Salinger W, Ladrow P, Wheeler C. Behavioral phenotype of the reeler mutant mouse: effects of reln gene dosage and social isolation. Behav. Neurosci. 2004;117:1257. doi: 10.1037/0735-7044.117.6.1257. [DOI] [PubMed] [Google Scholar]
- 113.Domínguez-Iturza N, et al. The autism- and schizophrenia-associated protein CYFIP1 regulates bilateral brain connectivity and behaviour. Nat. Commun. 2019;10:3454. doi: 10.1038/s41467-019-11203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liska, A. et al. Homozygous loss of autism-risk gene CNTNAP2 results in reduced local and long-range prefrontal functional connectivity. Cold Spring Harbor Labs J. 060335. 10.1101/060335. (2016). [DOI] [PubMed]
- 115.Voineskos AN, et al. Neurexin-1 and frontal lobe white matter: an overlapping intermediate phenotype for schizophrenia and autism spectrum disorders. PLoS One. 2011;6:e20982. doi: 10.1371/journal.pone.0020982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lam M, et al. Single cell analysis of autism patient with bi-allelic NRXN1-alpha deletion reveals skewed fate choice in neural progenitors and impaired neuronal functionality. Exp. Cell Res. 2019;383:111469. doi: 10.1016/j.yexcr.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 117.Scott-Van Zeeland AA, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci. Transl. Med. 2010;2:56ra80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Whalley HC, et al. Genetic variation in CNTNAP2 alters brain function during linguistic processing in healthy individuals. Am. J. Med. Genet. Part B: Neuropsychiatr. Genet. 2011;156:941–948. doi: 10.1002/ajmg.b.31241. [DOI] [PubMed] [Google Scholar]
- 119.Sauer C, Montag C, Wörner C, Kirsch P, Reuter M. Effects of a common variant in the CD38 gene on social processing in an oxytocin challenge study: possible links to autism. Neuropsychopharmacology. 2012;37:1474–1482. doi: 10.1038/npp.2011.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meyer-Lindenberg A, et al. Genetic variants in AVPR1A linked to autism predict amygdala activation and personality traits in healthy humans. Mol. Psychiatry. 2009;14:968–975. doi: 10.1038/mp.2008.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sauer C, Montag C, Reuter M, Kirsch P. Imaging oxytocin × dopamine interactions: an epistasis effect of CD38 and COMT gene variants influences the impact of oxytocin on amygdala activation to social stimuli. Front. Neurosci. 2013;7:45. doi: 10.3389/fnins.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liska A, Galbusera A, Schwarz AJ, Gozzi A. Functional connectivity hubs of the mouse brain. NeuroImage. 2015;115:281–291. doi: 10.1016/j.neuroimage.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 123.de Oliveira Pereira Ribeiro L, et al. Evidence for association between OXTR gene and ASD clinical phenotypes. J. Mol. Neurosci. 2018;65:213–221. doi: 10.1007/s12031-018-1088-0. [DOI] [PubMed] [Google Scholar]
- 124.Boso M, et al. Reduced plasma apelin levels in patients with autistic spectrum disorder. Arch. Med. Res. 2007;38:70–74. doi: 10.1016/j.arcmed.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 125.Tansey KE, et al. Functionality of promoter microsatellites of arginine vasopressin receptor 1A (AVPR1A): implications for autism. Mol. Autism. 2011;2:3. doi: 10.1186/2040-2392-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Muma NA, Hoffman PN. Neurofilaments are intrinsic determinants of axonal caliber. Micron. 1993;24:677–683. [Google Scholar]
- 127.Smorodchenko A, et al. Comparative analysis of uncoupling protein 4 distribution in various tissues under physiological conditions and during development. Biochim. Biophys. Acta—Biomembr. 2009;1788:2309–2319. doi: 10.1016/j.bbamem.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 128.Zhang M, et al. Overexpression of uncoupling protein 4 promotes proliferation and inhibits apoptosis and differentiation of preadipocytes. Life Sci. 2006;79:1428–1435. doi: 10.1016/j.lfs.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 129.Liu D, et al. Mitochondrial UCP4 mediates an adaptive shift in energy metabolism and increases the resistance of neurons to metabolic and oxidative stress. NeuroMol. Med. 2006;8:389–413. doi: 10.1385/NMM:8:3:389. [DOI] [PubMed] [Google Scholar]
- 130.Bonora E, et al. Analysis of reelin as a candidate gene for autism. Mol. Psychiatry. 2003;8:885–892. doi: 10.1038/sj.mp.4001310. [DOI] [PubMed] [Google Scholar]
- 131.Persico AM, et al. Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol. Psychiatry. 2001;6:150–159. doi: 10.1038/sj.mp.4000850. [DOI] [PubMed] [Google Scholar]
- 132.Fatemi SH, et al. Reelin signaling is impaired in autism. Biol. Psychiatry. 2005;57:777–787. doi: 10.1016/j.biopsych.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 133.Hong SE, et al. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat. Genet. 2000;26:93–96. doi: 10.1038/79246. [DOI] [PubMed] [Google Scholar]
- 134.Huang T-N, Hsueh Y-P. Brain-specific transcriptional regulator T-brain-1 controls brain wiring and neuronal activity in autism spectrum disorders. Front. Neurosci. 2015;9:406. doi: 10.3389/fnins.2015.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang T-N, et al. Haploinsufficiency of autism causative gene Tbr1 impairs olfactory discrimination and neuronal activation of the olfactory system in mice. Mol. Autism. 2019;10:5. doi: 10.1186/s13229-019-0257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ecker C, et al. Describing the brain in autism in five dimensions-magnetic resonance imaging-assisted diagnosis of autism spectrum disorder using a multiparameter classification approach. J. Neurosci. 2010;30:10612–10623. doi: 10.1523/JNEUROSCI.5413-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Abraham A, et al. Deriving reproducible biomarkers from multi-site resting-state data: an Autism-based example. NeuroImage. 2017;147:736–745. doi: 10.1016/j.neuroimage.2016.10.045. [DOI] [PubMed] [Google Scholar]
- 139.Hernandez LM, Rudie JD, Green SA, Bookheimer S, Dapretto M. Neural signatures of autism spectrum disorders: insights into brain network dynamics. Neuropsychopharmacology. 2015;40:171–189. doi: 10.1038/npp.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bogdan R, et al. Imaging genetics and genomics in psychiatry: a critical review of progress and potential. Biol. Psychiatry. 2017;82:165–175. doi: 10.1016/j.biopsych.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nair, A. et al. Impact of methodological variables on functional connectivity findings in autism spectrum disorders. Hum. Brain Mapp. 35. 10.1002/hbm.22456 (2014). [DOI] [PMC free article] [PubMed]
- 142.Coravos A, Khozin S, Mandl KD. Developing and adopting safe and effective digital biomarkers to improve patient outcomes. NPJ Digit. Med. 2019;2:14. doi: 10.1038/s41746-019-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Domínguez-del-Toro E, et al. Generation of a novel functional circuit in Hoxa1 mutant mice. J. Neurosci. 2001;21:5637–5642. doi: 10.1523/JNEUROSCI.21-15-05637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lazaro MT, et al. Reduced prefrontal synaptic connectivity and disturbed oscillatory population dynamics in the CNTNAP2 model of autism. Cell Rep. 2019;27:2567–2578. doi: 10.1016/j.celrep.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang C, Pan Y-H, Wang Y, Blatt G, Yuan X-B. Segregated expressions of autism risk genes Cdh11 and Cdh9 in autism-relevant regions of developing cerebellum. Mol. Brain. 2019;12:40. doi: 10.1186/s13041-019-0461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lepagnol-Bestel AM, et al. SLC25A12 expression is associated with neurite outgrowth and is upregulated in the prefrontal cortex of autistic subjects. Mol. Psychiatry. 2008;13:385–397. doi: 10.1038/sj.mp.4002120. [DOI] [PubMed] [Google Scholar]
- 147.Lintas C, et al. Involvement of the PRKCB1 gene in autistic disorder: significant genetic association and reduced neocortical gene expression. Mol. Psychiatry. 2009;14:705–718. doi: 10.1038/mp.2008.21. [DOI] [PubMed] [Google Scholar]
- 148.Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1:e1–e11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xing X, et al. Hyperactive Akt-mTOR pathway as a therapeutic target for pain hypersensitivity in Cntnap2-deficient mice. Neuropharmacology. 2020;165:107816. doi: 10.1016/j.neuropharm.2019.107816. [DOI] [PubMed] [Google Scholar]