Abstract
Colon cancer (CC) is considered one of the most common and lethal malignancies occurring both in male and female. Its widespread prevalence demonstrates the need for novel diagnostic and prognostic biomarkers for CC. Emerging evidence has shown that small nucleolar RNAs play critical roles in tumor development. In this study, we investigated the expression profile and functions of SNORD16 in CC. Our data showed that SNORD16, rather than its host gene (RPL4), was upregulated in CC cell lines. Compared to matched adjacent normal tissues, CC tissues showed higher SNORD16 expression levels, and no correlation was found between SNORD16 and RPL4. Patients with high SNORD16 expression levels had a worse prognosis, and multivariate analysis showed the high SNORD16 expression was an independent prognostic factor for CC. In vitro gain- and loss-of-function studies revealed that SNORD16 can promote cell growth, proliferation, migration, and invasion of CC cells by inhibiting apoptosis. These results suggested that SNORD16 has an oncogenic role in CC and might be a novel diagnostic and prognostic biomarker for CC.
Keywords: small nucleolar RNA, SNORD16, colon cancer, oncogene, prognostic biomarker
Introduction
It is estimated that in the United States in 2019, 1 762 450 people will be diagnosed with cancer, 101 420 of which involves colon cancer (CC).1 Combined with rectal cancer, colorectal cancer is considered one of the most common and lethal malignancies in both sexes. Although CC-related mortality has declined over the past few decades, something which has been attributed to improvements in cancer prevention, early diagnosis and treatment, there is still room to improve the overall prognosis of patients with CC.2,3 To do this, there is an urgent need to investigate prognostic biomarkers that can identify high-risk patients and the novel molecular mechanisms of the pathogenesis of CC.
Noncoding RNAs (ncRNAs), especially microRNAs and long noncoding RNAs, have been recognized as important regulatory molecules of biological processes in various diseases, particularly oncogenesis.4-6 Recent studies of small nucleolar RNAs (snoRNAs), another type of ncRNAs, have shown that they can act as novel regulatory RNAs, which has attached attention in field of cancer development.7-9 SnoRNAs are 60 to 300 nucleotides in length, found predominately in the nucleolus, function mainly to guide RNAs for the posttranscriptional modification of ribosomal RNAs (rRNAs), and are categorized into 2 main groups according to the modification type. The box H/ACA snoRNAs (snoRAs) are associated with rRNA pseudouridylation, and the box C/D snoRNAs (snoRDs) are associated with rRNA 2’-O-ribose methylation. Emerging evidence has indicated that snoRNAs also play a critical role in the occurrence and development of various diseases, especially cancer.10-13 SNORA23 was found to be overexpressed in pancreatic ductal adenocarcinoma and correlated with poor patient survival, while SNORA24 was reported to mediate tumor suppression.14,15 Low levels of SNORA24 have been observed in hepatocellular carcinoma (HCC) and are associated with poor patient survival. Zhu et al demonstrated that SNORD89 was highly expressed in ovarian cancer stem cells and could promote the stemness phenotype by regulating the Notch1-c-Myc pathway and could therefore be used as a prognostic biomarker for patients with ovarian cancer.16 Gong et al performed pan-cancer analysis of the expression and clinical relevance of snoRNAs in human cancers through systematic investigation the expression landscape and clinical relevance of snoRNAs in over 10 000 samples across 31 cancer types from The Cancer Genome Atlas (TCGA), and provided a user-friendly data portal.17
In this study, we demonstrated that SNORD16 was overexpressed in CC tissues and inversely correlated with the 5-year overall survival of patients with CC. We also evaluated the relationship between SNORD16 expression and clinicopathological parameters. To further investigate the potential function of SNORD16, a series of knockdown and overexpression studies were performed in vitro. To the best of our knowledge, this is the first study to explore the role of SNORD16 in CC. Our results suggested that SNORD16 might be a novel potential diagnostic and prognostic biomarker for CC.
Materials and Methods
Patients and Samples
In this study, a total of 434 patients with CC from TCGA database were analyzed. When evaluating the relationship between SNORD16 expression and clinicopathological parameters, patients with incomplete clinical data were excluded from the study, leaving a total of 279 patients with a complete data set, including sex, age, TNM stage (American Joint Committee on Cancer), pathological T category, lymph node metastasis, distant metastasis, lymphatic invasion, venous invasion, and history of colon polyps, enrolled in our study. In addition, 20 pairs of matched CC and adjacent normal mucosa tissues were obtained from the department of Colorectal Surgery, Cancer Hospital of China Medical University and used to evaluate SNORD16 expression. Specimen usage was approved by the Ethics Committee, and informed consent was obtained from each patient.
RNA Isolation and Quantitative Real-Time PCR
A total of 1 mL of TRIzol Reagent (Austin, Texas) was added to tissue sample which were then disrupted using the TissueLyser LT (QIAGEN, Hilden, Germany), and total RNA of tissue samples or cultured cells was extracted in accordance with the manufacturer’s instructions. Reverse transcription was performed using the PrimeSript RT reagent kit (Takara, Tokyo, Japan) following the manufacturer’s protocol. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, California) to measure the expression levels of mRNA or snoRNAs. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as internal controls for mRNA and snoRNA, respectively. The 2−ΔΔCt method was used to calculate the relative expression of the target gene. To ensure quantitative accuracy, each sample was run in triplicate. The primer sequences used were as follows: SNORD16 (Forward: 5′-TGCAATGATGTCGTAATTTGCGTC-3′; Reverse: 5′-TTGCTCAGTAAGAATTTTCGTCAA-3′), U6 (Forward: 5′-CTCGCTTCGGCAGCACA-3′; Reverse: 5′-AACGCTTCACGAATTTGCGT-3′), RPL4 (Forward: 5′-CTGCAGGGCCTCTTAACACA-3′; Reverse: 5′-GTGCCACCAAGTGGCTATCT-3′) and GAPDH (Forward: 5′-CATGAGAAGTATGACAACAGCCT-3′; Reverse: 5′-AGTCCTTCCACGATACCAAAGT-3′).
Cell Culture
HCT116, SW620, and HT29 cell lines were purchased from GeneChem (Shanghai, China) and human normal colon epithelial cells (FHC) cell line was obtained from the American Type Culture Collection (ATCC, Manassas, Virginia). Both HCT116 and SW620 were cultured in RPMI-1640 medium (HyClone; Logan, Utah); HT29 was cultured in Dulbecco’s Modified Eagle Medium (DMEM)/HIGH GLUCOSE (HyClone); FHC was cultured in DMEM: F-12 (ATCC, Manassas). HCT116, SW620, and HT29 culture mediums were supplemented with 10% fetal bovine serum (ExCell Bio; Shanghai, China), and FHC culture medium was supplemented with 10% fetal bovine serum (Gibco, Carlsbad, California). All cells were incubated in a humidified atmosphere containing 5% CO2 at 37°C.
SNORD16 Knockdown and Overexpression
To knockdown SNORD16, 2 specific small interfering RNAs (siRNAs) targeting the SNORD16 RNA sequence were designed and synthesized by Shanghai GenePharma Company (Shanghai, China). Transient transfection of the siRNAs was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, California) following the manufacturer’s instructions. The expression vector GV235 (GeneChem) was used for SNORD16 overexpression and the lentiviral-SNORD16 was also constructed by GeneChem. Transient lentiviral transduction was performed using HiTransG P (GeneChem) according to manufacturer’s guidelines. Quantitative real-time polymerase chain reaction was used to confirm knockdown efficiency and overexpression. The sequence of the SNORD16-siRNAs used were listed as follows: SNORD16-siRNA-1 (Forward: 5′-AGTTGCCTGCTGTCAGTAATT-3′; Reverse: 5′-TTACTGACAGCAGGCAACTTT-3′) and SNORD16-siRNA-2 (Forward: 5′-GCTGGTACAGAAGGTTGACTT-3′; Reverse: 5′-GTCAACCTTCTGTACCAGCTT-3′).
Cell Proliferation Assay and Colony Formation Assay
The Cell Counting Kit-8 (CCK-8) assay (Dojindo, Kumamoto, Japan) and colony formation assay were used to detect the proliferative ability of SNORD16 knocked-down or overexpressed cells. For the CCK-8 assay, 1000 transfected cells were seeded in each well of a 96-well plate and the optical density values were measured at 450 nm every 24 hours using a microplate reader (Sunrise, Tecan, Switzerland). For each group, 5 replicates were performed. For the colony formation assay, 500 transfected cells were plated into wells of a 6-well plate and cultured for 12 days, and medium was changed every 4 days. All cell colonies were fixed with methanol for 1 hour and stained with Giemsa for 3 hours at room temperature. Experiments were performed in triplicate.
Cell Migration and Invasion Assays
For the cell migration assay, transfected/transduced cells (2 × 10 5 cells for siRNA transfection and 1 × 105 for lentiviral transduction) were seeded in the upper chamber of a transwell unit (Costar, Corning, New York) with 100 µL serum-free medium, while the bottom chamber was filled with 600 µL culture medium supplemented with 20% FBS. After 48 hours, cells on the upper surface were washed off, and cells adhering to the lower surface were fixed with 4% paraformaldehyde for 15 minutes, stained with 600 µL 0.1% crystal violet for 2 hours, and imaged using a microscope (Axio Observer A1, Carl Zeiss Shanghai Co, Ltd, Shanghai, China). For the cell invasion assay, same steps were used as for the migration assay, but the transwell units were precoated with 50-µL diluted Matrigel (dilution 1:8; BD Biosciences, San Jose, California) first. Experiments were independently repeated in triplicate.
Cell Apoptosis Assay
Cells were seeded in 24-well plates (5 × 104 cells for siRNA transfection and 4 × 104 cells for lentiviral transduction) and harvested 48 (siRNA) or 72 (lentiviral) hours after transfection/transduction. An Annexin V, FITC Apoptosis Detection Kit (Dojindo, Kumamoto, Japan) was used to detect cell apoptosis, and apoptotic cells were analyzed by flow cytometry (ACEA Bio, San Diego, California).
Western Blot Analysis
Cells were harvested 48 (siRNA) or 72 (lentiviral) hours after transfection/transduction and washed twice with 4°C phosphate-buffered saline. Radioimmunoprecipitation assay buffer (50 mmol Tris-HCl, pH 7.5; 150-mmol NaCl; 0.5% sodium deoxycholate; 1% NP-40) supplemented with phosphatase and protease inhibitors (Roche Applied Science, Indianapolis, Indiana) was used to lyse the cell pellets. Equal amounts of total protein (40 μg) were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto nitrocellulose transfer membranes (Pall Corporation, Pensacola, Florida), then blocked with 5% nonfat milk for 2 hours at room temperature. Then, membranes were incubated with primary antibodies at 4°C for 12 hours. The primary antibodies used included Ribosomal Protein L4 antibody (1:1000, Santa Cruz, California) and GAPDH antibody (1:5000; Proteintech, Wuhan, China). Membranes were then incubated with a corresponding horseradish peroxidase–conjugated secondary antibody for 2 hours at room temperature. The signal intensity of antibody-bound proteins was detected using ImageQuant LAS500 (GE Healthcare, Madison, Wisconsin).
RNA Sequencing (RNA-Seq) and Bioinformatics Analysis
RNA from SNORD16 overexpressed group and control group were utilized for RNA-seq (each group containing 3 replicate). RNA-Seq library construction and next-generation sequencing were performed by LC Biotech (Hangzhou, China), and Paired-end sequencing was performed with an Illumina X10 sequencer. The differentially expressed genes were selected with a false discovery rate of <0.05 and log2 (fold change) >1 or log2 (fold change) < −1. Other bioinformatics analysis, such as Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment were also performed by LC Biotech (Hangzhou, China).
Statistical Analysis
Data are presented as the mean ± standard deviation, and all experiments were performed at least in triplicate. The Wilcoxon rank correlation test and Mann-Whitney U tests were employed to analyze the difference in SNORD16 expression between CC and adjacent normal mucosa samples. Spearman correlation analysis was used to evaluate the correlation between the expression of SNORD16 and RPL4. The association between SNORD16 expression and clinicopathological parameters was analyzed using a χ2 test. Survival curves were plotted using the Kaplan-Meier method and the log-rank test was applied to analyze the difference survival groups using the SNORD16 expression median value as the cutoff. Uni- and multivariate Cox proportional hazard models were used to evaluate the relationship between SNORD16 expression and survival. A Student t test was used to analyze the differences between cell lines, the expression changes after transfection/transduction, and cell function experiments. Statistical analyses were performed using SPSS software version 24.0 (SPSS, Chicago, Illinois) or GraphPad Prism version 7.0 (GraphPad Software, La Jolla, California), and P < .05 was considered statistically significant.
Results
SNORD16 Is Overexpressed in CC
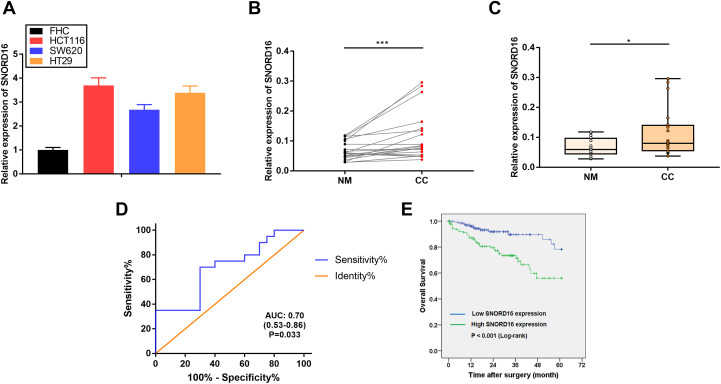
We first investigated the differences in SNORD16 expression levels between FHC and colorectal cancer cells (HCT116, SW620, and HT29) by qPT-PCR. When normalized to FHC, SNORD16 expression was increased by more than 2-fold in all 3 colorectal cancer cell lines (Figure 1A). To further investigate the expression profile of SNORD16, 20 pairs of matched CC and adjacent normal mucosa tissues were used. As shown in Figure 1B and C, at the transcript level, SNORD16 was significantly overexpressed in tumor tissues when compared with to normal mucosa tissues, which was consistent with the results obtained from cell lines. Furthermore, the receiver operating characteristic curve analysis indicated that SNORD16 expression could discriminate between CC and normal mucosa tissues (area under the curve: 0.70, 95% confidence interval: 0.53-0.86, P = .033), suggesting that SNORD16 could be used in CC diagnosis (Figure 1D). Taken together, these results show that SNORD16 was overexpressed in CC and has a potential to be a diagnostic biomarker.
Figure 1.
SNORD16 expression in colon cancer cell lines, cancer tissues, and its clinical significance. A, SNORD16 expression levels in colon cancer (CC) cell lines (HCT116, SW620, and HT29) compared to normal colon epithelial cell line (FHC) via quantitative real-time polymerase chain reaction (qRT-PCR). B and C, SNORD16 is overexpressed in CC compared to matched adjacent normal mucosa tissues, expression normalized to U6 (P < .001 and <.05, Wilcoxon rank correlation test and Mann-Whitney U tests, respectively). D, Receiver operating characteristic (ROC) curve analysis indicated that SNORD16 expression levels distinguished CC tissues from matched adjacent normal mucosa tissues (P = .033, areas under the curve (AUC): 0.70). E, Kaplan-Meier analysis of overall survival. Patients with high SNORD16 expression exhibited worse survival (P < .001, log-rank test). The median expression level of SNORD16 was used as the cutoff.
The Association Between SNORD16, Clinicopathological Parameters, and Prognosis
We stratified patients with CC into 2 groups according to SNORD16 expression levels (median value) and evaluated the relationship between SNORD16 expression and clinicopathological features (sex, age, TNM stage, pathological T category, lymph node metastasis, distant metastasis, lymphatic invasion, venous invasion, history of colon polyps, and survival time). As shown in Table 1, SNORD16 expression significantly associated with age (P = .023), lymphatic invasion (P < .001), and a patient history of colon polyps (P < .001). Patients who were older, positive for both lymphatic invasion and colon polyps are more likely to have higher levels of SNORD16 expression.
Table 1.
Correlations Between SNORD16 Expression and Clinicopathological Parameters.
| Characteristics | Case | SNORD16 Expressiona | χ2 | P | |
|---|---|---|---|---|---|
| Low | High | ||||
| Sex | 0.766 | .811 | |||
| Female | 143 | 70 | 73 | ||
| Male | 136 | 69 | 67 | ||
| Age (year) | 5.483 | .023 b | |||
| ≤ 70 | 147 | 83 | 64 | ||
| > 70 | 132 | 56 | 76 | ||
| TNM stage (AJCC) | 1.629 | .228 | |||
| Stage I/II | 158 | 84 | 74 | ||
| Stage III/IV | 121 | 55 | 66 | ||
| Pathological T category | 0.001 | 1.000 | |||
| T1/T2 | 52 | 26 | 26 | ||
| T3/T4 | 227 | 113 | 114 | ||
| Lymph node metastasis | 1.077 | .333 | |||
| Negative | 160 | 84 | 76 | ||
| Positive | 119 | 55 | 64 | ||
| Distant metastasis | 1.239 | .329 | |||
| Negative | 234 | 120 | 114 | ||
| Positive | 45 | 19 | 26 | ||
| Lymphatic invasion | 14.321 | <.001b | |||
| Negative | 174 | 102 | 72 | ||
| Positive | 105 | 37 | 68 | ||
| Venous invasion | 2.688 | .125 | |||
| Negative | 211 | 111 | 100 | ||
| Positive | 68 | 28 | 40 | ||
| History of colon polyps | 14.117 | <.001b | |||
| Negative | 185 | 107 | 78 | ||
| Positive | 94 | 32 | 62 | ||
Abbreviations: AJCC, American Joint Committee on cancer.
a Cutoff threshold of SNORD16 expression is median value in all patients in this cohort.
b P < .05.
The Kaplan-Meier method and log-rank tests were applied to analyze the difference between the high and low SNORD16 expression groups. Results revealed that patients with high SNORD16 expression had a worse overall survival (Figure 1E; P < .001), and the 5-year survival rate of high and low SNORD16 expression groups was 56.0% and 78.4%, respectively. To further investigate the prognostic value of SNORD16, uni- and multivariate Cox proportional hazards model were applied to analyze the prognostic factors that affect the overall survival of patients with CC (Table 2). As expected, age, TNM stage, pathological T category, lymph node metastasis, distant metastasis, lymphatic invasion, venous invasion, and SNORD16 expression were significantly associated with overall survival in univariate analysis. However, multivariate analysis indicated that only age, TNM stage, and SNORD16 expression were independent prognostic factors for overall CC survival. We also evaluated the prognostic value of SNORD16 in other cancer types using the user-friendly data portal, snoRNA in cancer (http://bioinfo.life.hust.edu.cn/SNORic).17 High SNORD16 expression associated with poor prognosis in adrenocortical carcinoma (ACC), esophageal carcinoma (ESCA), kidney renal clear cell carcinoma (KIRC), pancreatic adenocarcinoma (PAAD), and thyroid carcinoma (THCA; Figure S1). SNORD16 expression was not associated with overall survival in rectum adenocarcinoma (READ). Taken together, these results indicated that in patients with CC, SNORD16 upregulation contributed to poor prognosis and that SNORD16 might be used as a potential prognostic biomarker for CC.
Table 2.
Cox Regression for Overall Survival Analysis.
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age (≤ 70 vs > 70) | 1.810 | 1.015-3.229 | .045a | 2.052 | 1.121-3.756 | .020a |
| TNM Stage (stage I/II vs stage III/IV) | 4.475 | 2.327-8.606 | <.001a | 12.443 | 2.213-69.960 | .004a |
| Pathological T category (T1/T2 vs T3/T4) | 10.021 | 1.379-72.845 | .023a | 5.297 | 0.701-40.034 | .106 |
| Lymph node metastasis (negative vs positive) | 3.677 | 1.972-6.855 | <.001a | 0.252 | 0.054-1.174 | .079 |
| Distant metastasis (negative vs positive) | 3.544 | 1.960-6.407 | <.001a | 1.476 | 0.740-2.945 | .269 |
| Lymphatic invasion (negative vs positive) | 2.302 | 1.300-4.075 | .004a | 1.111 | 0.520-2.375 | .786 |
| Venous invasion (negative vs positive) | 1.947 | 1.077-3.522 | .028a | 1.384 | 0.657-2.916 | .393 |
| History of colon polyps (negative vs positive) | 0.853 | 0.448-1.626 | .630 | |||
| SNORD16 expression (low vs high)b | 3.012 | 1.588-5.713 | .001a | 2.697 | 1.381-5.266 | .004 a |
Abbreviation: CI, confidence interval.
a P < .05.
b Cutoff threshold of SNORD16 expression is median value in all patients in this cohort.
SNORD16, Rather Than Its Host Gene RPL4, Is Overexpressed in CC
We postulated that SNORD16 might have a functional relationship with RPL4, because it is located at the intron of the RPL4 gene. In this study, RPL4 mRNA expression was detected in cell lines and clinical specimens. Results (Figure 2A-C) showed that RPL4 expression was not upregulated in either cancer cells or CC tissues. RPL4 expression cannot be used to discriminate between CC and normal mucosa tissues and was not correlated with SNORD16 expression in 20 pairs of clinical specimens (Figure S2). We further verified these results using the TCGA database and found that there was no correlation between SNORD16 and RPL4 expression (Figure 2D). Furthermore, RPL4 mRNA and protein expression levels were also not affected when SNORD16 expression was either up- or downregulated (Figure 2E-G). Together, these observations suggested that SNORD16, rather than its host gene, was overexpressed in CC and did not regulate expression of its host gene.
Figure 2.
SNORD16, rather than RPL4 (its host gene), is overexpressed in colon cancer. A, RPL4 expression levels in colon cancer (CC) cell lines (HCT116, SW620 and HT29) compared with normal colon epithelial cells (FHC) via quantitative real-time polymerase chain reaction (qRT-PCR). B and C, No significantly difference in RPL4 expression was found between CC and matched adjacent normal mucosa tissues (P > .05, Wilcoxon rank correlation test and Mann-Whitney U tests, respectively). RPL4 expression was normalized to GAPDH. D, Spearman correlation analysis indicating no correlation between SNORD16 and RPL4 expression (P = .86). (E-G) RPL4 expression is unaffected by SNORD16 up- or downregulation at either the transcriptional or posttranscriptional level.
SNORD16 Promotes Proliferation and Colony Formation
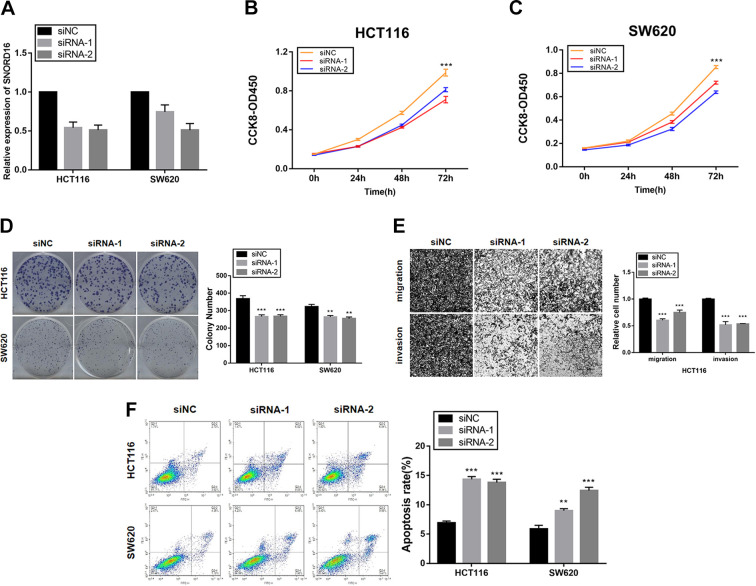
To investigate the function of SNORD16 in CC, HCT116 and SW620 cells were transduced with lentiviral-SNORD16 and lentiviral-control, respectively. After 72 hours post-transduction, cells were harvested to detect SNORD16 expression using qRT-PCR. Results revealed that SNORD16 was successfully upregulated in both HCT116 and SW620 cells (Figure 3A). Cell Counting Kit-8 and colony formation experiments (Figure 3B-D) showed that cells overexpressing SNORD16 exhibited increased cell proliferation and colony formation ability. To knockdown SNORD16 expression in HCT116 and SW620 cells, 2 independent SNORD16-targeting siRNAs were used to perform loss-of-function analysis. After 24 hours post-transfection, cells were harvested to detect SNORD16 expression using qRT-PCR. Results showed that SNORD16 was successfully downregulated in both HCT116 and SW620 cells (Figure 4A). Cell Counting Kit-8 and colony formation assays showed that knocking down SNORD16 expression significantly suppressed cell growth and clonogenicity (Figure 4B-D). These results suggested that SNORD16 promoted proliferation and colony formation in HCT116 and SW620 cells.
Figure 3.
Effect of SNORD16 overexpression on colon cancer in vitro. A, SNORD16 expression levels in HCT116 and SW620 cells after transduction with LV-NC and LV-SNORD16. B and C, Cell Counting Kit-8 (CCK-8) assays indicate that SNORD16 overexpression promotes cell proliferation in both HCT116 and SW620 cells. D, SNORD16 overexpression in HCT116 and SW620 increases colony formation. E, SNORD16 overexpression enhances the migration and invasion capacity of HCT116 cells. F, SNORD16 overexpression significantly reduces apoptosis in both HCT116 and SW620 cells treated with H2O2 (400 μM for 5 hours). *P < .05; **P < .01; ***P < .001.
Figure 4.
Effect of SNORD16 knockdown on colon cancer in vitro. A, The knockdown efficiency of SNORD16 in HCT116 and SW620 cells after transfection with siNC, siRNA-1, and siRNA-2. B and C, Cell Counting Kit-8 (CCK-8) assays indicate that SNORD16 knockdown inhibits the cell proliferation in both HCT116 and SW620 cells. D, SNORD16 knockdown decreases colony formation in HCT116 and SW620. E, SNORD16 knockdown reduces the migration and invasion capacity of HCT116 cells. F, SNORD16 knockdown induces apoptosis in both HCT116 and SW620 cells. *P < .05; **P < .01; ***P < .001.
SNORD16 Promotes Migration and Invasion
The positive correlation between SNORD16 expression and lymphatic (P < .001) or venous invasion (P = .125) prompted us to investigate whether it influences cell migration and invasion ability. In vitro transwell migration and invasion assays showed that HCT116 cells overexpressing SNORD16 exhibited increased migration and invasion compared to the control group (Figure 3E). Conversely, knocking down SNORD16 in HCT116 cells significantly suppressed migration and invasion (Figure 4E). Taken together, these results showed that SNORD16 not only promoted cell proliferation and colony formation but also significantly promoted CC cell migration and invasion, observations that were consistent with our clinical data.
SNORD16 Knockdown Induces Cell Apoptosis
To investigate whether SNORD16’s cell proliferation and metastatic function correlated with apoptosis, an Annexin V, FITC Apoptosis Detection Kit was used to detect apoptotic cells. As shown in Figure 4F, SNORD16 inhibition in both HCT116 and SW620 cells resulted in a marked increase in the percentage of apoptotic cells. In CC cells, SNORD16 exhibited an antiapoptotic effect after H2O2 treatment (Figure 3F). These results indicated that SNORD16 promoted the proliferation and metastatic ability by inhibiting apoptosis.
Potential Mechanism of SNORD16’s Biological Functions
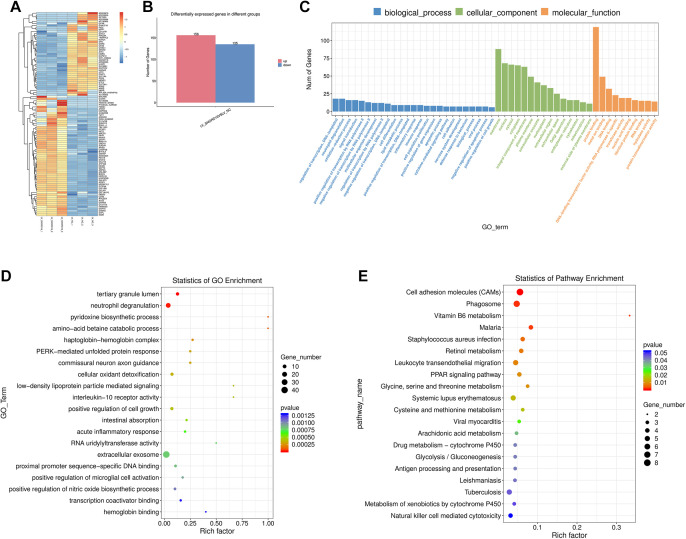
To investigate potential downstream targets of SNORD16 in CC cells, RNA-seq analysis was performed on HCT116 cells treated with LV-NC versus LV-SNORD16. A total of 156 genes upregulated genes and 135 downregulated genes were observed when overexpressing SNORD16 (Figure 5B). Based on the GO analysis, SNORD16 is involved in biological processes, including oxidation-reduction, cell differentiation, cell proliferation, cell adhesion, and apoptosis (Figure 5C and D). In addition, KEGG pathway analysis identified several signaling pathways influenced by SNORD16 overexpression (Figure 5E). Intriguingly, these biological processes and signaling pathways matched the above results. For example, cell adhesion molecule pathways are thought to be involved in regulating the cell adhesion, differentiation, and invasion of cancer cells, and our data showed that SNORD16 expression significantly associated with lymphatic invasion. Data obtained by our in vitro gain- and loss-of-function studies suggested that SNORD16 promoted cell growth, proliferation, migration, and invasion of CC cells by inhibiting apoptosis, which was in accordance with the GO analysis. Therefore, SNORD16 could promote CC cell proliferation and the metastatic ability by inhibiting apoptosis through multiple cancer-related pathways.
Figure 5.
RNA sequencing and bioinformatics analysis. A, Heat map of the SNORD16-regulated genes in HCT116 cells. (B) Barplot of differentially expressed genes. (C-D) Gene ontology (GO) enrichment analysis of SNORD16-regulated genes. E, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of SNORD16-regulated genes.
Discussion
Colon cancer, combined with rectum cancer, is considered one of the most common and lethal malignancies occurring in both sexes.1 The understanding of the pathogenesis of colorectal cancer has improved with advances made in the discovery of new oncogenes and anti-oncogenes.18-20 Despite these advances, the underlying mechanism of these diseases remains largely unknown. Recent evidence has shown that snoRNAs play an important regulatory role in the occurrence and development of various diseases, especially cancer.21-23 Considering the potential value of snoRNAs as diagnostic and/or prognostic biomarkers and therapeutic targets, we investigated the potential role of SNORD16 in CC.
In this study, we demonstrated that SNORD16 was overexpressed in CC tissues when compared to adjacent normal mucosa, which was consistent with the results obtained from cell lines. Receiver operating characteristic analyses indicated that SNORD16 expression discriminated CC tissues from adjacent normal mucosa, thereby suggesting that SNORD16 can be utilized for CC diagnosis. Furthermore, SNORD16 expression was significantly associated with patient age, lymphatic invasion, and history of colon polyps. Patients with CC with high SNORD16 expression had a worse prognosis, and SNORD16 expression was confirmed to be an independent prognostic factor for CC. In addition, the prognostic value of SNORD16 in other cancer types was investigated. High SNORD16 expression associated with poor prognosis in ACC, ESCA, KIRC, PAAD, and THCA but did not associate with overall survival in READ. These results indicated that SNORD16 might act as a CC oncogene.
In several studies, it was indicated that both snoRNAs and its host gene could play a critical role in regulating the malignant phenotype of cancer. For example, Askarian-Amiri et al found that Zfas1 is a snoRNA host gene that was downregulated in breast tumors when compared to normal tissue and that suppressing the expression of Zfas1 increased cellular proliferation and differentiation without altering the levels of host snoRNAs.24 Both SNORD126 and its host gene CCNB1IP1 were overexpressed in HCC samples; however, no significant correlation was observed. Neither up- nor downregulation of SNORD126 changed the expression of CCNB1IP1, thereby indicating that the host gene of SNORD126 may not participate in SNORD126-mediated cellular malignancy.23 Because SNORD16 is located at the intron of RPL4, we proposed that it might have a relationship with RPL4. The expression profile of RPL4 was determined, and no correlation was found between the expression of SNORD16 and its host gene. Furthermore, RPL4 mRNA and protein expression levels were not influenced by SNORD16 being up- or downregulated. These observations suggest that the function of SNORD16 might be independent of its host gene. The in vitro of SNORD16 in CC cell lines was investigated using a series of gain- and loss-of-function analysis. As shown in previous studies on SNORA21 and SNORA42 in CRC, our results showed that decreasing SNORD16 expression inhibited cell growth, proliferation, migration, and invasion capacities of CC cells by inducing apoptosis.25,26 Conversely, upregulated SNORD16 expression promoted CC cell growth, proliferation, migration, and invasion capacities. To further clarify the biological function and to investigate the potential downstream targets of SNORD16, RNA-seq analysis was conducted on HCT116 cells that were treated with LV-NC versus LV-SNORD16. Intriguingly, we found that RNA-seq data were in accordance with our clinical observations and function experiments. SNORD16 is involved in biological processes, including oxidation-reduction, cell differentiation, cell proliferation, cell adhesion, and apoptosis. Kyoto Encyclopedia of Genes and Genomes pathway analysis suggested that SNORD16 is involved in several signaling pathways, including cell adhesion molecule pathways, which were thought to be involved in regulating the cell adhesion, differentiation, and invasion of cancer cells and matched our in vitro function data. Based on the results mentioned above, SNORD16 could promote CC cell proliferation, and the metastatic ability by inhibiting apoptosis through multiple cancer-related pathways.
Ionizing radiation reacts both directly and indirectly (through a variety of free radicals generated by the radiolysis of water) with several types of biological macromolecules in tissues, including DNA, proteins, and lipids to cause complex biological effects or lethal cellular injury.27 Hydrogen peroxide is one of the reactive oxygen species (ROS) that plays an important role in ionizing radiation killing cells (indirect effects). Our results have demonstrated that CC cells exhibited significantly resistance to hydrogen peroxide–induced apoptosis after overexpressing SNORD16, which indicated that SNORD16-overexprssion might have a potential radiation resistant role through inhibiting the indirect effects of ionizing radiation. RNA-seq analysis showed that SNORD16 is involved in biological processes, including oxidation-reduction and apoptotic. In addition, we found that CHAC1, which induced cell apoptosis via ROS generation,28 was significantly downregulated 7.1-fold after overexpressing SNORD16 in HCT116 cells (Table S1). Therefore, we speculated that the overexpression of SNORD16 in CC cells resulted in CHAC1 suppression, reduction in ROS generation, and resistance to hydrogen peroxide-induced apoptosis. Thus, SNORD16 might have a potential role in predicting the radiosensitivity of CC.
Our study has some limitations. Firstly, only 20 pairs of clinical samples were collected and used in this study and a larger sample size is essential to confirm our results. Secondly, in vivo experiments need to be performed to confirm our in vitro results. Lastly, the specific oncogenic mechanism of SNORD16 in CC also needs further verification using corresponding experiments. Despite these limitations, our study was the first to identify the oncogenic role of SNORD16 in CC and we proposed that it might be a novel diagnostic and prognostic biomarker for CC, as well as a novel therapeutic target.
Conclusions
In summary, our results showed that SNORD16 is overexpressed in CC tissues and that patients with high SNORD16 expression had a worse prognosis. SNORD16 can promote cell growth, proliferation, migration, and invasion of CC cells by inhibiting apoptosis. Our study was the first to identify the oncogenic role of SNORD16 in CC, and our results suggested that SNORD16 might be a novel potential diagnostic and prognostic biomarker for CC.
Supplemental Material
Supplemental Material, Figure_S1 for Small Nucleolar RNA, C/D Box 16 (SNORD16) Acts as a Potential Prognostic Biomarker in Colon Cancer by Jun-yan He, Xin Liu, Zhen-hua Qi, Qi Wang, Wen-qing Lu, Qing-tong Zhang, Shu-ya He and Zhi-dong Wang in Dose-Response
Supplemental Material, Figure_S2 for Small Nucleolar RNA, C/D Box 16 (SNORD16) Acts as a Potential Prognostic Biomarker in Colon Cancer by Jun-yan He, Xin Liu, Zhen-hua Qi, Qi Wang, Wen-qing Lu, Qing-tong Zhang, Shu-ya He and Zhi-dong Wang in Dose-Response
Supplemental Material, Table_S1(RNA-seq_results) for Small Nucleolar RNA, C/D Box 16 (SNORD16) Acts as a Potential Prognostic Biomarker in Colon Cancer by Jun-yan He, Xin Liu, Zhen-hua Qi, Qi Wang, Wen-qing Lu, Qing-tong Zhang, Shu-ya He and Zhi-dong Wang in Dose-Response
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China [31770913 to Z.W.].
ORCID iD: Zhi-dong Wang  https://orcid.org/0000-0003-0594-1638
https://orcid.org/0000-0003-0594-1638
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Siegel RL, Miller KD. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 2. Taieb J, Andre T, Auclin E. Refining adjuvant therapy for non-metastatic colon cancer, new standards and perspectives. Cancer Treat Rev. 2019;75:1–11. [DOI] [PubMed] [Google Scholar]
- 3. Van Cutsem E, Cervantes A, Adam R, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27(8):1386–1422. [DOI] [PubMed] [Google Scholar]
- 4. Harrandah AM, Mora RA, Chan EKL. Emerging microRNAs in cancer diagnosis, progression, and immune surveillance. Cancer Lett. 2018;438:126–132. [DOI] [PubMed] [Google Scholar]
- 5. Lian H, Xie P, Yin N, et al. Linc00460 promotes osteosarcoma progression via miR-1224-5p/FADS1 axis. Life Sci. 2019;233:116757. [DOI] [PubMed] [Google Scholar]
- 6. Bach DH, Lee SK. Long noncoding RNAs in cancer cells. Cancer Lett. 2018;419:152–166. [DOI] [PubMed] [Google Scholar]
- 7. Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38(5):485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat rev Cancer. 2012;12(2):84–88. [DOI] [PubMed] [Google Scholar]
- 9. Siprashvili Z, Webster DE, Johnston D, et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet. 2016;48(1):53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bortolin-Cavaille ML, Cavaille J. The SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) gene clusters at the imprinted Prader-Willi locus generate canonical box C/D snoRNAs. Sci Rep. 2012;40(14):6800–6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cavaille J. Box C/D small nucleolar RNA genes and the Prader-Willi syndrome: a complex interplay. Wiley Interdiscip Rev: RNA. 2017;8:e1417. [DOI] [PubMed] [Google Scholar]
- 12. Cao P, Yang A, Wang R, et al. Germline duplication of SNORA18L5 increases risk for HBV-related hepatocellular carcinoma by altering localization of ribosomal proteins and decreasing levels of p53. Gastroenterology. 2018;155(2):542–556. [DOI] [PubMed] [Google Scholar]
- 13. Xu G, Yang F, Ding CL, et al. Small nucleolar RNA 113-1 suppresses tumorigenesis in hepatocellular carcinoma. Molecular cancer. 2014;13:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cui L, Nakano K, Obchoei S, et al. Small nucleolar noncoding RNA snora23, up-regulated in human pancreatic ductal adenocarcinoma, regulates expression of spectrin repeat-containing nuclear envelope 2 to promote growth and metastasis of xenograft tumors in mice. Gastroenterology. 2017;153(1):292–306.e2. [DOI] [PubMed] [Google Scholar]
- 15. McMahon M, Contreras A, Holm M, et al. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. eLife. 2019;8:e48847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu W, Niu J, He M, et al. SNORD89 promotes stemness phenotype of ovarian cancer cells by regulating Notch1-c-Myc pathway. J Transl Med. 2019;17(1):259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gong J, Li Y, Liu CJ, et al. A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep. 2017;21(7):1968–1981. [DOI] [PubMed] [Google Scholar]
- 18. Wang L, Li B, Zhang L, et al. miR-664a-3p functions as an oncogene by targeting Hippo pathway in the development of gastric cancer. Cell Proliferat. 2019;52(3):e12567. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19. Wang H, Chen W, Jin M, et al. CircSLC3A2 functions as an oncogenic factor in hepatocellular carcinoma by sponging miR-490-3p and regulating PPM1F expression. Mol Cancer. 2018;17(1):165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu B, Pan S, Xiao Y, Liu Q, Xu J, Jia L. LINC01296/miR-26a/GALNT3 axis contributes to colorectal cancer progression by regulating O-glycosylated MUC1 via PI3K/AKT pathway. J Exp & Clin Cancer Res. 2018;37(1):316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang H, Ma P, Liu P, Chen B, Liu Z. Small nucleolar RNA U2_19 promotes hepatocellular carcinoma progression by regulating Wnt/beta-catenin signaling. Biochem Bioph Res Commun. 2018;500(2):351–356. [DOI] [PubMed] [Google Scholar]
- 22. Wu L, Chang L, Wang H, Ma W, Peng Q, Yuan Y. Clinical significance of C/D box small nucleolar RNA U76 as an oncogene and a prognostic biomarker in hepatocellular carcinoma. Clin Res Hepatol Gas. 2018;42(1):82–91. [DOI] [PubMed] [Google Scholar]
- 23. Fang X, Yang D, Luo H, et al. SNORD126 promotes HCC and CRC cell growth by activating the PI3K-AKT pathway through FGFR2. J Mol Cell Biol. 2017;9(3):243–255. [DOI] [PubMed] [Google Scholar]
- 24. Askarian-Amiri ME, Crawford J, French JD, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17(5):878–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoshida K, Toden S, Weng W, et al. SNORA21—an oncogenic small nucleolar RNA, with a prognostic biomarker potential in human colorectal cancer. EBioMedicine. 2017;22:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okugawa Y, Toiyama Y, Toden S, et al. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. 2017;66(1):107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kalman NS, Zhao SS, Anscher MS, et al. Current status of targeted radioprotection and radiation injury mitigation and treatment agents: a critical review of the literature. Int J Radiat Oncol, Biol, Phys. 2017;98(3):662–682. [DOI] [PubMed] [Google Scholar]
- 28. Sun Y, Atmadibrata B, Yu D, et al. Upregulation of LYAR induces neuroblastoma cell proliferation and survival. Cell Death Differ. 2017;24(9):1645–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Figure_S1 for Small Nucleolar RNA, C/D Box 16 (SNORD16) Acts as a Potential Prognostic Biomarker in Colon Cancer by Jun-yan He, Xin Liu, Zhen-hua Qi, Qi Wang, Wen-qing Lu, Qing-tong Zhang, Shu-ya He and Zhi-dong Wang in Dose-Response
Supplemental Material, Figure_S2 for Small Nucleolar RNA, C/D Box 16 (SNORD16) Acts as a Potential Prognostic Biomarker in Colon Cancer by Jun-yan He, Xin Liu, Zhen-hua Qi, Qi Wang, Wen-qing Lu, Qing-tong Zhang, Shu-ya He and Zhi-dong Wang in Dose-Response
Supplemental Material, Table_S1(RNA-seq_results) for Small Nucleolar RNA, C/D Box 16 (SNORD16) Acts as a Potential Prognostic Biomarker in Colon Cancer by Jun-yan He, Xin Liu, Zhen-hua Qi, Qi Wang, Wen-qing Lu, Qing-tong Zhang, Shu-ya He and Zhi-dong Wang in Dose-Response