Abstract
The conserved kinetochore-associated NDC80 complex (composed of Hec1/Ndc80, Nuf2, Spc24, and Spc25) has well-documented roles in mitosis including 1) connecting mitotic chromosomes to spindle microtubules to establish force-transducing kinetochore–microtubule attachments and 2) regulating the binding strength between kinetochores and microtubules such that correct attachments are stabilized and erroneous attachments are released. Although the NDC80 complex plays a central role in forming and regulating attachments to microtubules, additional factors support these processes as well, including the spindle and kinetochore-associated (Ska) complex. Multiple lines of evidence suggest that Ska complexes strengthen attachments by increasing the ability of NDC80 complexes to bind microtubules, especially to depolymerizing microtubule plus ends, but how this is accomplished remains unclear. Using cell-based and in vitro assays, we demonstrate that the Hec1 tail domain is dispensable for Ska complex recruitment to kinetochores and for generation of kinetochore–microtubule attachments in human cells. We further demonstrate that Hec1 tail phosphorylation regulates kinetochore–microtubule attachment stability independently of the Ska complex. Finally, we map the location of the Ska complex in cells to a region near the coiled-coil domain of the NDC80 complex and demonstrate that this region is required for Ska complex recruitment to the NDC80 complex-–microtubule interface.
INTRODUCTION
Successful chromosome segregation during mitosis depends on the formation of stable attachments between chromosomes and spindle microtubules. These attachments are generated at kinetochores, which are macromolecular structures built on centromeric heterochromatin of mitotic chromosomes. Once stable kinetochore–microtubule connections are formed, forces generated by plus-end microtubule dynamics are harnessed for the purpose of congressing chromosomes to the spindle equator and silencing the spindle assembly checkpoint, which prevents anaphase onset until all kinetochores are properly attached to spindle microtubules. The kinetochore-associated NDC80 complex, composed of the proteins Hec1 (also known as Ndc80), Nuf2, Spc24, and Spc25, serves as the core linkage between kinetochores and spindle microtubules (DeLuca and Musacchio, 2012). A direct interaction has been mapped between the “toe” domain of Hec1, which resides in its well-ordered, N-terminal calponin homology (CH) domain, and the microtubule lattice (Ciferri et al., 2008; Wilson-Kubalek et al., 2008; Alushin et al., 2010). This interaction is required for high-affinity NDC80 complex–microtubule interactions in vitro and for kinetochore–microtubule attachment formation in cells from all organisms tested to date (Ciferri et al., 2008; Sundin et al., 2011; Tooley et al., 2011; Lampert et al., 2013; Cheerambathur et al., 2017). The Hec1 protein contains an N-terminal, unstructured “tail” domain that has also been implicated in forming kinetochore–microtubule attachments in cells, although the requirement for the tail domain in this process varies among eukaryotic species (Wimbish and DeLuca, 2020). The Hec1 tail domain in Saccharomyces cerevisiae and Caenorhabditis elegans is dispensable for formation of stable kinetochore–microtubule attachments (Kemmler et al., 2009; Demirel et al., 2012; Cheerambathur et al., 2013; Lampert et al., 2013). In contrast, expression of Hec1 mutants lacking the N-terminal tail domain in mammalian cells has been reported to inhibit the formation of stable attachments (Guimaraes et al., 2008; Miller et al., 2008). The tail domain of Hec1 from all species tested, however, is required for high-affinity binding of NDC80 complexes to microtubules in vitro (Wei et al., 2007; Ciferri et al., 2008; Miller et al., 2008; Alushin et al., 2012; Umbreit et al., 2012; Cheerambathur et al., 2013; Lampert et al., 2013; Zaytsev et al., 2015), suggesting that cellular factors likely compensate for Hec1 tail domain functions to various degrees in different organisms. Why the Hec1 tail domain is required for attachments in mammalian cells, but not in other eukaryotic species, remains an unanswered question.
In addition to generating attachments to spindle microtubules, kinetochores regulate their stability. In early mitosis attachments are labile and undergo rapid turnover, whereas in late mitosis, attachments are stable and long-lived (Zhai et al., 1995; Cimini et al., 2006; DeLuca et al., 2006; Bakhoum et al., 2009). This scheme helps ensure that any erroneous attachments formed in early mitosis are released and corrected and that mature attachments on correctly bi-oriented chromosomes are stabilized. Temporal regulation of attachment strength is primarily achieved through phosphorylation of kinetochore substrates by the Aurora family of kinases (Biggins et al., 1999; Tanaka et al., 2002; Carmena et al., 2012; Krenn and Musacchio, 2015), and the Hec1 N-terminal tail domain is a key component of this phosphoregulatory system (Cheeseman et al., 2006; DeLuca et al., 2006). Nine sites in the Hec1 tail domain have been identified as substrates of Aurora kinases A and B in vitro, and at least five are confirmed to be phosphorylated in cells (Nousiainen et al., 2006; DeLuca et al., 2011, 2018; Kettenbach et al., 2011). In vitro, progressive mutation of these nine target sites to aspartic acid to mimic increasing phosphorylation results in a coordinate decrease in microtubule binding affinity of human NDC80 complexes (Zaytsev et al., 2015). Increasing the number of phosphomimetic substitutions also results in a corresponding decrease in kinetochore–microtubule attachment stability, as evidenced by decreased interkinetochore distances and kinetochore–microtubule bundle densities in mammalian cells (Zaytsev et al., 2014; Etemad et al., 2019; Kuhn and Dumont, 2019). Conversely, expression of Hec1 mutants in which all mapped Aurora kinase target sites are mutated to alanine to prevent phosphorylation results in hyperstabilization of kinetochore–microtubule attachments and defective attachment error correction in mammalian cells (DeLuca et al., 2011; Sundin et al., 2011; Zaytsev et al., 2014; Tauchman et al., 2015; Long et al., 2017; Yoo et al., 2018). A similar phenomenon is observed in embryonic C. elegans cells, where mutation of the four mapped Hec1 tail domain Aurora kinase target sites to alanine results in premature kinetochore–microtubule stabilization (Cheerambathur et al., 2017). One model to explain these results proposes that increased phosphorylation of the Hec1 tail reduces the affinity of the NDC80 complex for microtubules, which in turn decreases kinetochore–microtubule attachment stability.
In addition to the NDC80 complex, the spindle and kinetochore-associated (Ska) complex, a trimer composed of Ska1, Ska2, and Ska3, contributes to the generation and stabilization of kinetochore–microtubule attachments. The Ska complex loads progressively onto kinetochores during mitosis and is required for efficient chromosome congression and for silencing the spindle assembly checkpoint (Hanisch et al., 2006; Daum et al., 2009; Gaitanos et al., 2009; Guimaraes and DeLuca, 2009; Raaijmakers et al., 2009; Theis et al., 2009; Sivakumar et al., 2014, 2016; Auckland et al., 2017). The Ska complex binds both the NDC80 complex and microtubules and stabilizes NDC80 complex–mediated kinetochore–microtubule attachments, likely through its ability to remain bound to depolymerizing microtubule plus ends (Welburn et al., 2009; Jeyaprakash et al., 2012; Schmidt et al., 2013; Abad et al., 2014; Zhang et al., 2017; Helgeson et al., 2018). A major outstanding question is how the Ska complex is recruited to kinetochore-bound NDC80 complexes to promote kinetochore–microtubule attachment stability. Previous studies have suggested that this recruitment is mediated through the Hec1 tail domain (Cheerambathur et al., 2017; Janczyk et al., 2017), the Hec1 loop domain (Zhang et al., 2012, 2017), and the coiled-coil regions of the heterotetrameric complex (Helgeson et al., 2018; Huis in’t Veld et al., 2019); thus the recruitment mechanism remains unresolved.
The Ska complex has also been implicated in regulating kinetochore–microtubule attachment stability. Expression of a nonphosphorylatable Hec1 tail domain mutant in C. elegans embryos resulted in increased kinetochore recruitment of the Ska complex, whereas expression of a phosphomimetic Hec1 tail domain mutant led to the opposite effect (Cheerambathur et al., 2017). Importantly, the increased stability of kinetochore–microtubule attachments observed in cells expressing the nonphosphorylatable mutant version of Hec1 was dependent on the presence of the Ska complex. Thus, in some organisms, rather than directly regulating NDC80 complex–microtubule affinity, phosphorylation of the Hec1 tail likely controls recruitment of Ska complexes, which in turn regulates attachment stability. Whether this mechanism functions in human cells remains to be tested.
Here we investigate how the human Ska complex is recruited to the NDC80 complex in cells and in vitro and how Hec1 tail phosphorylation impacts Ska function. We report that the N-terminal Hec1 tail domain, while required for force generation and attachment regulation, is not explicitly required for either kinetochore–microtubule attachment formation or Ska complex recruitment to kinetochores in human cells. The tail domain is also dispensable for Ska complex–mediated enhancement of NDC80 complex–microtubule binding in vitro. We demonstrate that phosphoregulation of kinetochore–microtubule attachments occurs in the absence of the Ska complex in human cells, providing support for a mechanism whereby Aurora kinase phosphorylation of the Hec1 tail directly modulates kinetochore–microtubule attachment strength. Finally, using two-color fluorescence localization microscopy, we map the location of the Ska complex to a region coincident with the central coiled-coil domain of the NDC80 complex, and consistent with this, we find that this domain of the NDC80 complex is required for the Ska complex to enhance NDC80-microtubule interactions in vitro.
RESULTS
Phosphorylation of the Hec1 tail affects Ska complex loading to kinetochores
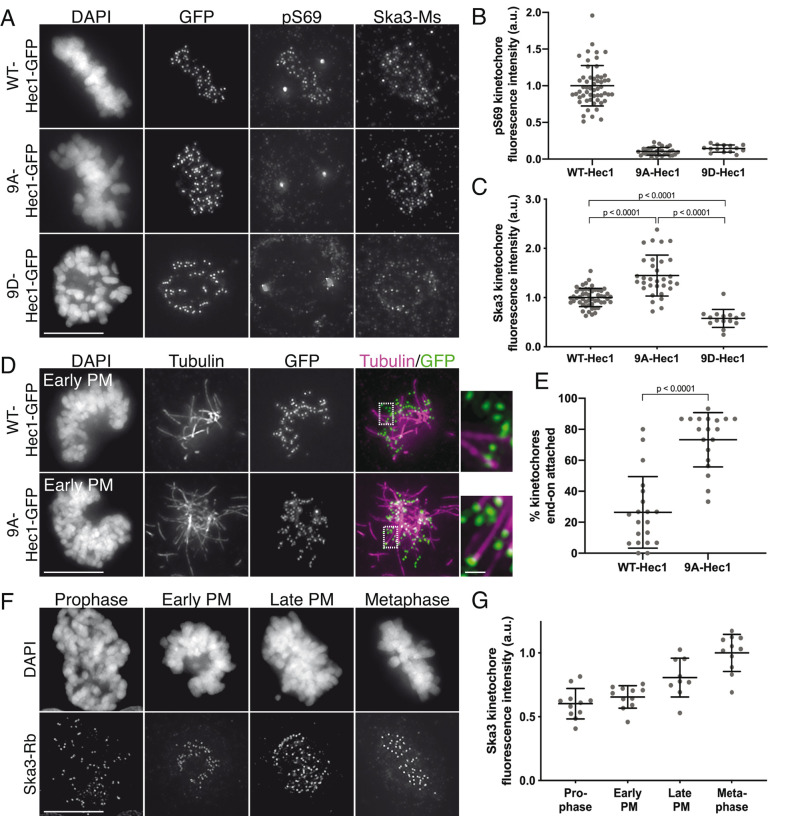
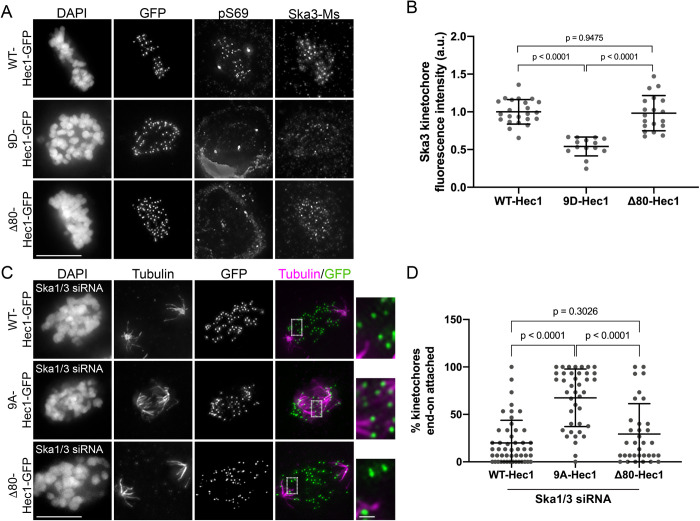
To determine how phosphorylation of the Hec1 tail impacts recruitment of Ska complexes to kinetochores, we expressed mutant versions of green fluorescent protein (GFP)-tagged Hec1 in human cells in which the nine mapped Aurora phosphorylation sites were mutated to either alanine (9A) to prevent phosphorylation or aspartic acid (9D) to mimic constitutive phosphorylation. Western blot analysis revealed that transgene expression levels were variable between Hec1-GFP mutants; thus, we analyzed only cells with similar protein levels based on kinetochore GFP intensity (Supplemental Figure S1, A–C; see also Materials and Methods). We confirmed that expression of the exogenous constructs led to depletion of endogenous Hec1 protein from kinetochores to undetectable levels by staining cells with an antibody to phosphorylated Hec1 Ser-69 (pS69), which does not recognize 9A- or 9D-Hec1 proteins and whose levels do not vary during mitotic progression (DeLuca et al., 2018) (Figure 1, A and B). Similar to the situation described for C. elegans (Cheerambathur et al., 2017), we found that kinetochores in cells expressing 9A-Hec1-GFP were enriched for the Ska complex, while kinetochores in cells expressing 9D-Hec1-GFP exhibited lower levels compared with kinetochores in cells expressing WT-Hec1-GFP (Figure 1, A and C). Similar results were observed in cells expressing Hec1-GFP constructs and depleted of endogenous Hec1 by small interfering RNA (siRNA), confirming that Hec1 transgene expression is sufficient to elicit a dominant phenotype without the additional need for siRNA treatment (Supplemental Figure S1, D–F; Tauchman et al., 2015).
FIGURE 1:
Phosphorylation of the Hec1 tail domain affects kinetochore–microtubule attachment stability and Ska complex loading to kinetochores. (A) Immunofluorescence images of cells expressing WT-, 9A-, and 9D-Hec1-GFP. Cells were fixed and stained using antibodies to Hec1 pS69 and Ska3 (mouse). (B) Quantification of pS69 kinetochore fluorescence intensity from cells expressing WT-, 9A-, and 9D-Hec1-GFP. For each condition, at least 20 kinetochores per cell were measured from at least five cells per experiment from three separate experiments. (C) Quantification of Ska3 kinetochore fluorescence intensity from cells expressing WT-, 9A-, and 9D-Hec1-GFP. For each condition, at least 20 kinetochores per cell were measured from at least five cells per experiment from three separate experiments. Statistical significance was determined by a one-way analysis of variance (ANOVA). (D) Immunofluorescence images of cold-treated cells expressing WT- and 9A-Hec1-GFP. Cells were incubated in ice-cold DMEM for 12 min before fixation, permeabilized, fixed, and stained using antibodies to tubulin. Insets are enlargements of the region indicated by the dashed box. (E) Quantification of end-on attachment in cold-treated cells expressing WT- and 9A-Hec1-GFP. For each condition, at least 15 kinetochores per cell were measured from at least nine cells per experiment from two separate experiments. A Student’s t test was carried out to determine statistical significance. (F) Immunofluorescence images of untreated, control cells in different stages of mitosis fixed and stained with antibodies to Ska3 (rabbit). (G) Quantification of Ska3 kinetochore fluorescence intensity in control cells in progressive stages of mitosis. For each mitotic phase, at least 20 kinetochores were measured from at least four cells per experiment from two separate experiments. On all graphs, each dot represents the average value for all kinetochores from a single cell. Scale bars: 10 and 1 µm for panels and insets, respectively.
Although these results suggest that the phosphorylation state of the tail domain might directly regulate Ska complex recruitment to kinetochores, there is an important caveat to this experiment. Cells expressing 9A-Hec1 mutants generate hyperstable kinetochore–microtubule attachments, in which kinetochore–microtubule bundle densities are increased (Zaytsev et al., 2014), the pulling forces between two sister kinetochores are higher (DeLuca et al., 2011; Yoo et al., 2018), and end-on kinetochore–microtubule attachments are formed earlier than in control cells (Figure 1, D and E). Conversely, cells expressing 9D-Hec1 mutants fail to form stable kinetochore–microtubule attachments during mitosis (DeLuca et al., 2011; Zaytsev et al., 2014). Because the Ska complex loads to kinetochores as microtubule attachments are progressively stabilized (Hanisch et al., 2006; Auckland et al., 2017) (Figure 1, F and G), results from the experiment described above (Figure 1, A–C) do not allow us to differentiate between the two following scenarios: 1) dephosphorylation of the Hec1 tail promotes Ska complex recruitment, and in turn, the Ska complex increases kinetochore–microtubule attachment stability, or 2) dephosphorylation of the Hec1 tail generates stable kinetochore–microtubule attachments, and in turn, stable attachments promote recruitment of the Ska complex to kinetochores.
Phosphorylation of the Hec1 tail does not affect microtubule-independent Ska complex loading to kinetochores
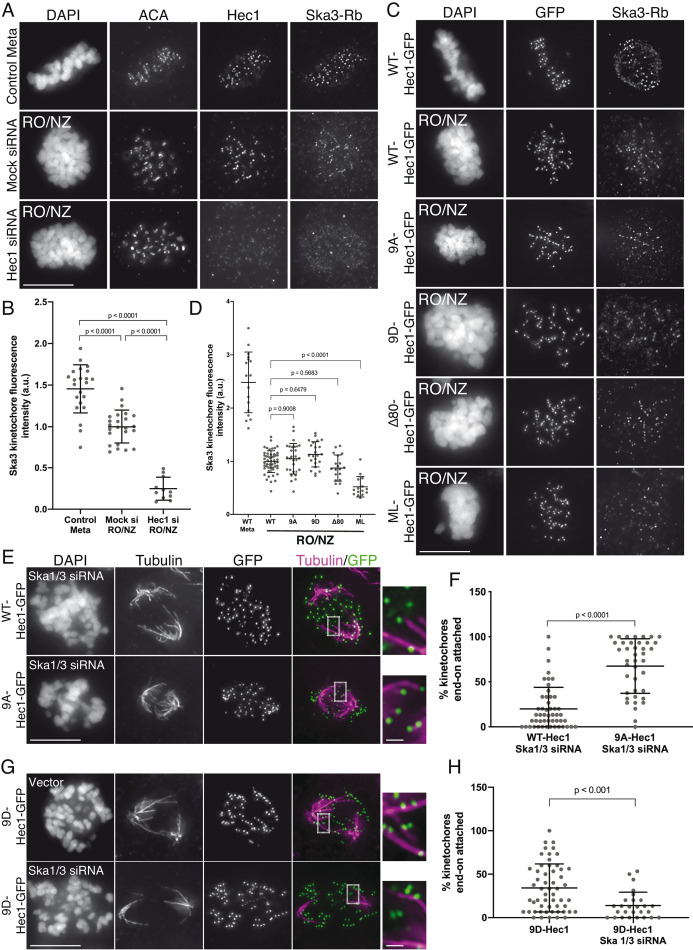
To differentiate between the two possibilities, we measured Ska complex loading to kinetochores in cells expressing WT, 9A-, and 9D-Hec1-GFP in the absence of microtubules. This allowed us to test how mutations in Hec1 affect Ska recruitment without the confounding effects of their impact on kinetochore–microtubule attachment stability. Previous reports have demonstrated that while Ska complexes are maximally loaded onto kinetochores after microtubule attachment, a population of Ska complex localizes to kinetochores in a Hec1-dependent, microtubule-independent manner (Chan et al., 2012; Zhang et al., 2017). Cells transfected with either WT-, 9A-, or 9D-Hec1-GFP were synchronized and arrested in G2 with RO-3306 and then washed out into nocodazole before entry into mitosis. We confirmed that microtubule-independent Ska complex recruitment to kinetochores required the NDC80 complex (Figure 2, A and B) and found that kinetochores in cells expressing WT-, 9D- or 9A-Hec1-GFP all loaded similar levels of the Ska complex (Figure 2, C and D). These results suggest that in the absence of microtubules, the phosphorylation state of the human Hec1 tail domain does not influence Ska complex recruitment to kinetochores. In this experiment, all cells subjected to analysis entered mitosis in the presence of nocodazole, and therefore kinetochores had no contact with microtubules before fixation. Interestingly, when we carried out a similar experiment in an asynchronous population, where nocodazole was added to cells in various stages of mitosis, we found that kinetochores in cells expressing 9A-Hec1-GFP exhibited somewhat higher levels of Ska3 compared with those in cells expressing WT-Hec1-GFP (Supplemental Figure S2, A and B). We speculate that a population of kinetochores in asynchronous cells expressing 9A-Hec1-GFP had previously established kinetochore–microtubule attachments and loaded high levels of the Ska complex to kinetochores before exposure to nocodazole. These results suggest that once Ska complexes are loaded onto kinetochores by microtubule attachment, a subpopulation of the complex remains bound even after microtubule depolymerization.
FIGURE 2:
Hec1 tail dephosphorylation does not affect microtubule-independent Ska complex kinetochore loading and stabilizes attachments in the absence of the Ska complex. (A) Immunofluorescence images of untreated cells (top row) or RO3306-synchronized cells released into mitosis in the presence of 10 μM nocodazole (bottom two rows). Cells were stained with ACA (anti-centromere) antibodies and antibodies to Hec1 and Ska3 (rabbit). (B) Quantification of Ska3 kinetochore fluorescence intensity from cells described in panel A. For each condition, at least 20 kinetochores per cell were measured from at least five cells per experiment from two separate experiments. Ska3 intensity was measured only for Hec1 siRNA-treated cells with <20% of endogenous kinetochore-associated Hec1 as determined by staining with an antibody to the CH domain of Hec1 (9G3). A Student’s t test was carried out to determine statistical significance. (C) Immunofluorescence images of cells expressing the indicated Hec1-GFP fusion protein in the absence (top row) or presence of RO3306 synchronization and release into 10 μM nocodazole (remaining rows). Cells were stained with antibodies to Ska3 (rabbit). (D) Quantification of Ska3 kinetochore fluorescence intensity from cold-treated cells described in panel C. For each condition, at least 20 kinetochores per cell were measured from at least five cells per experiment from three separate experiments. Statistical significance was determined by a one-way ANOVA between RO3306-synchronized WT-Hec1-GFP expressing cells and cells expressing the indicated Hec1 fusion proteins. (E) Immunofluorescence images of cold-treated cells expressing WT- and 9A-Hec1-GFP and treated with Ska1 and Ska3 siRNA. Cells were incubated in ice-cold DMEM for 12 min, permeabilized, fixed, and stained using antibodies to tubulin. Insets are enlargements of the regions indicated by the dashed boxes. (F) Quantification of end-on attachment in cells expressing WT- and 9A-Hec1-GFP and treated with Ska1 and Ska3 siRNA. For each condition, at least 15 kinetochores were measured from at least 10 cells from three separate experiments. A Student’s t test was carried out to determine statistical significance. (G) Immunofluorescence images of cells expressing 9D-Hec1-GFP and treated with (bottom panel) or without (top panel) Ska1 and Ska3 siRNA. Cells were incubated in ice-cold DMEM for 12 min, permeabilized, fixed, and stained using antibodies to tubulin. Insets are enlargements of the regions indicated by the dashed boxes. (H) Quantification of end-on attachments in cold-treated cells expressing 9D-Hec1-GFP and treated with or without Ska1 and Ska3 siRNA. For each condition, at least 15 kinetochores were measured per cell from at least nine cells per experiment from at least three separate experiments. A Student’s t test was carried out to determine statistical significance. On all graphs, each dot represents the average value for all kinetochores from a single cell. Scale bars: 10 and 1 µm for panels and insets, respectively.
Hec1 tail phosphorylation contributes to kinetochore–microtubule attachment stability independently of the Ska complex
To investigate the functional dependencies between Hec1 tail dephosphorylation, Ska complex loading, and kinetochore–microtubule attachment stability, we tested whether the stable attachments formed in human cells expressing 9A-Hec1-GFP were dependent on the Ska complex. For these experiments, we depleted the Ska complex from HeLa cells using siRNAs targeting the Ska complex subunits Ska1 and Ska3, which has previously been shown to disrupt kinetochore–microtubule attachments and chromosome alignment (Gaitanos et al., 2009). Immunofluorescence analysis revealed an ∼80% reduction in Ska3 signal at kinetochores of Ska1/Ska3 siRNA-treated cells compared with control cells in metaphase (Supplemental Figure S2, C and D), and most Ska-depleted cells exhibited unaligned chromosomes and defective cold-stable kinetochore–microtubule attachment formation (Supplemental Figure S2, E and F). To determine how the Ska and NDC80 complexes coordinate to form kinetochore–microtubule attachments, we expressed WT- and 9A-Hec1-GFP constructs in Ska-depleted cells, treated the cells with ice-cold media before fixation, stained them with antibodies to tubulin, and scored them for cold-resistant end-on kinetochore–microtubule attachments. Similar to what we observed for Ska-depleted cells alone, cells expressing WT-Hec1-GFP and depleted of Ska1/Ska3 exhibited defects in chromosome alignment and formation of end-on kinetochore–microtubule attachments (Figure 2, E and F). However, in Ska1/Ska3-depleted cells expressing 9A-Hec1-GFP, we observed robust formation of end-on attachments despite cells exhibiting similar defects in chromosome alignment (Figure 2, E and F). These results suggest that Hec1 tail phosphorylation and Ska complex recruitment contribute to regulation of kinetochore–microtubule attachments independently of each other. We reasoned that if this were the case, then the destabilizing effects of expressing a phospho-mimetic Hec1 tail mutant in Ska1/Ska3-depleted cells should be more severe than the effects of expressing a phospho-mimetic Hec1 tail mutant in non–Ska1/Ska3-depleted cells. We found that cells expressing 9D-Hec1-GFP exhibited defects in forming stable, end-on kinetochore–microtubule attachments (average of ∼34% of kinetochores attached per cell), which is consistent with previous studies (Figure 2, G and H; Guimaraes et al., 2008; Zaytsev et al., 2014). In cells depleted of Ska1 and Ska3, expression of 9D-Hec1-GFP indeed resulted in a more penetrant kinetochore–microtubule attachment defect (average of ∼14% of kinetochores attached per cell) (Figure 2, G and H), providing further evidence that in human cells, phosphorylation of the Hec1 tail contributes to kinetochore–microtubule attachment stability independently of the Ska complex. Collectively, these results suggest that the increased loading of Ska complexes to kinetochores in cells expressing 9A-Hec1 is a consequence of increased kinetochore-microtubule attachment stability, rather than a direct effect of blocking Hec1 tail phosphorylation. Similarly, we found that expression of 9A-Hec1-GFP also resulted in higher kinetochore recruitment of Astrin, a subunit of the Astrin-SKAP complex, which has been reported to specifically localize to end-on attached kinetochores (Supplemental Figure S1, G and H; Schmidt et al., 2010; Conti et al., 2019).
Phosphorylation of Hec1 Ser-69 prevents excess Ska loading to kinetochores
In contrast to most Aurora target sites on the Hec1 tail, Ser-69 remains phosphorylated at high levels throughout mitosis, and this modification is important for maintaining proper kinetochore–microtubule attachment dynamics (DeLuca et al., 2018). We next tested whether phosphorylation of this site imparted differences in Ska recruitment compared with a completely dephosphorylated tail. To this end, we expressed 8A-Hec1-GFP in cells, in which eight of the nine mapped Aurora kinase target sites are mutated to alanine and Ser-69 is kept in its wild-type state (DeLuca et al., 2018). In contrast to what we observed for 9A-Hec1, cells expressing 8A-Hec1 recruited normal, wild-type levels of Ska3 in metaphase (Supplemental Figure S3, A and B). In line with the need for high levels of Hec1 tail phosphorylation in early mitosis (Zaytsev et al., 2014), 8A-Hec1 expression resulted in premature stabilization of kinetochore–microtubule attachments (Supplemental Figure S3, C and D). Additionally, and in a manner similar to 9A-Hec1, expression of 8A-Hec1 constructs led to robust kinetochore–microtubule attachment formation in the absence of the Ska complex (Supplemental Figure S3, E and F). These results provide further support for the notion that Hec1 tail dephosphorylation strengthens attachments independently of the Ska complex and suggest that maintenance of wild-type Hec1 tail phosphorylation levels allow normal Ska loading to kinetochores.
The Hec1 tail domain is not required for Ska complex–mediated enhancement of NDC80 complex–microtubule binding
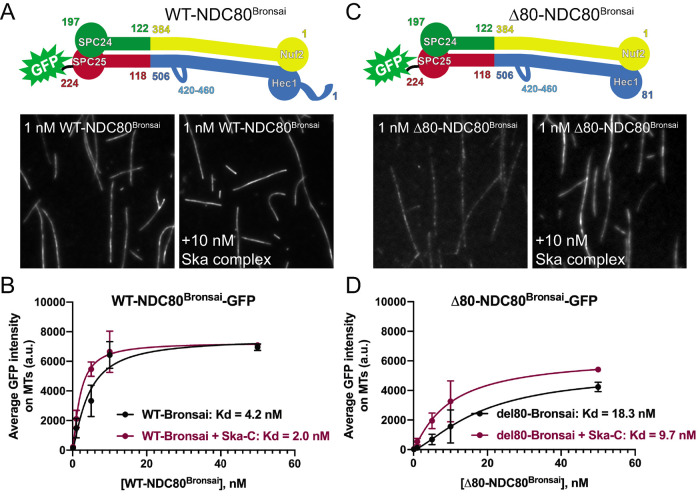
Independent of its phosphorylation state, the tail domain of Hec1 has been implicated in recruiting the Ska complex to the NDC80 complex–microtubule interface in vitro and to kinetochores in human cells (Janczyk et al., 2017). In contrast, other studies have reported that the Hec1 tail is dispensable for the NDC80-Ska complex interaction (Helgeson et al., 2018; Huis in’t Veld et al., 2019). To further investigate these discrepancies, we first asked whether the tail domain is required in vitro for Ska complexes to enhance NDC80 complex–microtubule affinity. Previous studies have shown that purified, recombinant Ska complexes increase the affinity of NDC80 complexes for microtubules in vitro (Schmidt et al., 2012; Helgeson et al., 2018). We therefore measured the microtubule binding affinity of GFP-tagged, recombinantly expressed, purified NDC80 complexes containing WT-Hec1 and Hec1 deleted of its N-terminal 80-amino-acid tail domain (Δ80-Hec1) using a total internal reflection fluorescence (TIRF) microscopy-based assay. For these experiments, we generated NDC80 complexes in which Nuf2 is fused to Spc24 and Hec1 is fused to Spc25-GFP (Figure 3A), termed NDC80Bronsai. These complexes are missing the tetramerization domains from all four subunits but contain the majority of the central coiled-coil region of the complex (Ciferri et al., 2005, 2008), as well as the “loop” domain of Hec1, which is a 40-amino-acid region that briefly disrupts the coiled-coil region (Maiolica et al., 2007). The name represents a hybrid between “NDC80Bonsai,” which is an engineered, truncated version of the NDC80 complex composed of a Nuf2-Spc24 fusion and a Hec1-Spc25-GFP fusion missing the central coiled-coil and tetramerization domains (Ciferri et al., 2008) and “NDC80Broccoli,” which is a dimer of nearly full-length Nuf2 and Hec1 containing the coiled-coil and loop domains (Schmidt et al., 2012). For the binding assays, we incubated increasing concentrations of GFP-labeled NDC80Bronsai complexes with Alexa647-labeled microtubules in the presence or absence of 10 nM recombinantly expressed human Ska complex and measured the average fluorescence intensity along microtubules. WT-NDC80Bronsai complexes robustly bound microtubules, and binding affinity was increased approximately two-fold upon the addition of Ska complexes (Figure 3, A and B). The Δ80-NDC80Bronsai complexes bound to microtubules with significantly lower affinity than the WT complexes (Figure 3, C and D), which is consistent with previously published studies (Ciferri et al., 2008; Miller et al., 2008; Umbreit et al., 2012; Zaytsev et al., 2015). However, addition of purified Ska complex increased the affinity of Δ80-NDC80Bronsai complexes for microtubules by nearly two-fold, similar to the case for WT-NDC80Bronsai (Figure 3D). These results confirm that the Hec1 tail domain is not required for Ska complex–mediated enhancement of microtubule binding by NDC80 complexes in vitro and that Ska complexes are able to compensate for the decreased microtubule binding observed with NDC80 complexes lacking the N-terminal tail domain (Helgeson et al., 2018; Huis in’t Veld et al., 2019). These results also demonstrate that the tetramerization domain of the NDC80 complex is not required for Ska complex binding.
FIGURE 3:
The Hec1 tail domain is not required for Ska complex-mediated enhancement of NDC80 complex–microtubule binding. (A and C) Top: schematics of NDC80Bronsai complexes used in the TIRF-based microtubule binding experiments. Bottom: GFP fluorescence images of NDC80 complexes decorating microtubules in the presence and absence of Ska complex. All images show a single concentration of the NDC80 complex from the experiment (1 nM) with and without added Ska complex (10 nM). (B and D) Binding curves from the microtubule binding assays. Datapoints and curve fits shown in black are from experiments without added Ska complex. Those shown in burgundy are from experiments with added Ska complex. Each point on the curve represents the average fluorescence intensity from three separate experiments. At each concentration, GFP-NDC80 complex fluorescence intensity was measured from at least 40 individual microtubules from at least 10 different TIRF fields per experiment. Scale bars: 10 µm.
We note that one difference between the TIRF-based microtubule binding assays described here and those described in our previous study (Zaytsev et al., 2015) is the choice of assay buffer. When we used standard microtubule binding assay buffers BRB80 (80 mM PIPES, 1 mM MgCl, 1 mM EGTA, pH 6.8) or BRB20 (20 mM PIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.8), purified Ska complexes aggregated in the presence of microtubules (Supplemental Figure S4A). In addition, Ska complexes induced aggregation of NDC80 complexes on microtubules in the presence of BRB80 (Supplemental Figure S4B), which precluded quantitative analysis of fluorescence intensities along microtubules. We therefore developed “SN” buffer (for “Ska-NDC80”) for our assays (20 mM Tris, 50 mM NaCl, pH 7.0), which did not induce aggregation of either Ska or NDC80 complexes (Supplemental Figure S4A).
The Hec1 tail domain is not required for Ska complex recruitment to kinetochores or for kinetochore–MT attachment in human cells
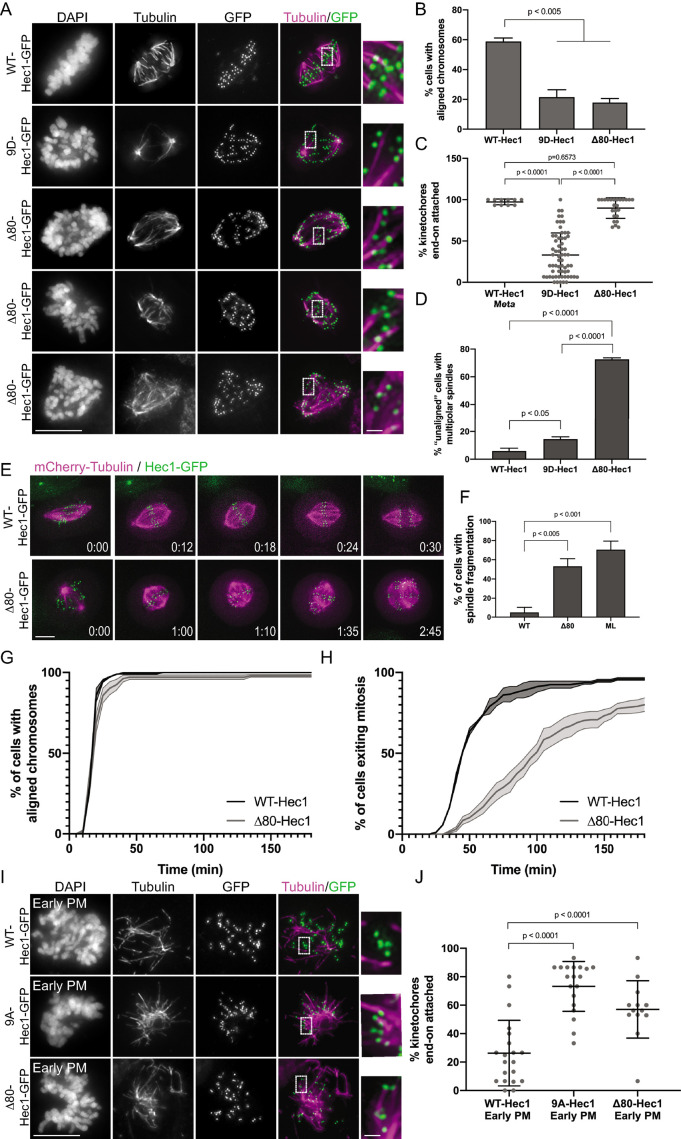
In light of our in vitro results, we next wanted to ask whether the Hec1 tail domain is required for Ska complex recruitment to kinetochores in human cells. For these experiments, we expressed exogenous Δ80-Hec1-GFP in HeLa cells and analyzed only cells with undetectable levels of Hec1-pSer69 at kinetochores. Previous studies in mammalian cells demonstrated that Hec1 tail deletion impacts kinetochore–microtubule attachment stability as evidenced by reductions in interkinetochore distances, decreased cold-resistant microtubule attachments, failure to align chromosomes, and significant mitotic delays (Guimaraes et al., 2008; Miller et al., 2008; Etemad et al., 2015; Janczyk et al., 2017). In line with this, we found that cells expressing Δ80-Hec1-GFP exhibited significant chromosome alignment defects and decreased interkinetochore distances and were unable to silence the spindle assembly checkpoint (Figure 4, A and B, and Supplemental Figure S5, A–C). However, contrary to previous studies, we found that cells expressing Δ80-Hec1-GFP were competent to form cold-resistant kinetochore–microtubule attachments (Figure 4C). This is in contrast to cells expressing 9D-Hec1, which are able neither to properly align chromosomes nor to form stable, cold-resistant kinetochore–microtubule attachments (Figure 4, A–C; DeLuca et al., 2011). Analysis of spindle morphology in Δ80-Hec1 expressing cells revealed that the majority of cells with unaligned chromosomes contained multipolar spindles (Figure 4D). Identical defects in alignment and spindle bipolarity were observed in cells expressing Δ80-Hec1-GFP and depleted of endogenous Hec1 by siRNA (Supplemental Figure S5, D–F). To further investigate this phenotype, we carried out time-lapse imaging of mCherry-tubulin and Hec1-GFP expressing cells. In the case of WT-Hec1-GFP expressing cells, we found that almost all cells formed bipolar spindles and entered anaphase without errors, with only ∼5% of cells undergoing spindle fragmentation before anaphase. Strikingly, while most Δ80-Hec1-GFP expressing cells aligned their chromosomes in a timely manner, cells experienced a metaphase arrest followed by spindle pole fragmentation and subsequent loss of chromosome alignment, resulting in prolonged mitotic delays (Figure 4, E–H). Several non-centrosomal processes, including cohesin fatigue and defective kinetochore force generation, have been suggested to contribute to loss of spindle bipolarity (Daum et al., 2011; Maiato and Logarinho, 2014). Interestingly, live cell imaging revealed that Δ80-Hec1 expressing cells undergo spindle fragmentation with a frequency similar to that of cells expressing a scrambled loop mutant of Hec1 (ML-Hec1; Figure 4F; see also Figure 7 later in this article), which are unable to form stable kinetochore–microtubule attachments and experience dramatic delays in mitotic exit (Varma et al., 2012; Zhang et al., 2012; see Figure 7, H–K, later in this article). Taken with the observation that kinetochores in cells expressing Δ80-Hec1 retain end-on attachments that are under significantly lower tension than kinetochores in cells expressing WT-Hec1 (Figure 4D and Supplemental Figure S5, A and B), these results suggest that the Hec1 tail is dispensable for attachments, but is required for sustaining force at the kinetochore–microtubule interface and timely transit through mitosis.
FIGURE 4:
The Hec1 tail domain is not required for the formation of stable end-on kinetochore–microtubule attachments in cells. (A) Immunofluorescence images of cold-treated cells expressing WT-, 9D-, and Δ80-Hec1-GFP. Cells were incubated in ice-cold DMEM for 12 min, permeabilized, fixed, and stained using antibodies to tubulin. Insets are enlargements of the regions indicated by the dashed boxes. Three examples of cells expressing Δ80-Hec1-GFP are shown. (B) Quantification of chromosome alignment in cells expressing WT-, 9D-, and Δ80-Hec1-GFP. For each condition, chromosome alignment was assessed in at least 100 cells per experiment from two separate experiments. Cells were scored as “aligned” if they had a metaphase plate with <5 chromosomes off the plate. Statistical significance was determined by a one-way ANOVA. (C) Quantification of end-on attachment in cells expressing WT-, 9D-, and Δ80-Hec1-GFP and cold-treated prior to fixation. For each condition, at least 15 kinetochores per cell were measured from at least 10 cells per experiment from two separate experiments. Statistical significance was determined by a one-way ANOVA. (D) Quantification of multipolarity observed in cells expressing WT-, 9D- and Δ80-Hec1-GFP. Cells with unaligned chromosomes were scored for containing bi- vs multipolar spindles, and the percent of cells with multipolar spindles is shown. For each condition, at least 100 cells per experiment were analyzed from two separate experiments. Statistical significance was determined by a one-way ANOVA. (E) Still images from time-lapse experiments of cells expressing Hec1-GFP and mCherry-tubulin. Time from nuclear envelope breakdown (NEBD) is denoted on bottom right corner of each image (hours:minutes). (F) Quantification of spindle pole fragmentation frequency quantified from time-lapse imaging experiments. Cells were scored as undergoing fragmentation events if loss of spindle bipolarity was observed during time-lapse imaging as determined from the mCherry-tubulin signal. Quantifications shown are averages from two (WT-, ML-) or four (Δ80-Hec1) independent experiments. Statistical significance was determined by a one-way ANOVA. (G) Quantification of chromosome alignment efficiency in cells from the experiment shown in panel E. Cell fate was tracked after mitotic entry (as determined by NEBD) for 3 h, and cells were scored as “aligned” upon metaphase plate formation (as determined by Hec1-GFP fluorescence). Data for WT- and Δ80-Hec1 are from 175 cells from two independent experiments and 165 cells from four independent experiments, respectively. (H) Quantification of mitotic exit timing in cells from the experiment shown in panel E. Cell fate was tracked after mitotic entry (as determined by NEBD) for 3 h, and cells were scored as “exiting mitosis” upon anaphase entry. Data for WT- and Δ80-Hec1 are from 175 cells from two independent experiments and 165 cells from four independent experiments, respectively. (I) Immunofluorescence images of cold-treated, early prometaphase cells expressing WT, 9A-, and Δ80-Hec1-GFP. Cells were incubated in ice-cold DMEM for 12 min, permeabilized, fixed, and stained with antibodies to tubulin. Insets are enlargements of the regions indicated by the dashed boxes. (J) Quantification of end-on attachment in early prometaphase cells expressing WT-, 9A-, and Δ80- Hec1-GFP. The WT- and 9A-Hec1 data shown are from the experiment presented in Figure 2. For each condition, at least 15 kinetochores per cell were measured from at least six cells per experiment from at least two separate experiments. Statistical significance was determined by a one-way ANOVA. On all dot plots, each dot represents the average value for all kinetochores from a single cell. Scale bars: 10 and 1 µm for panels and insets, respectively.
FIGURE 7:
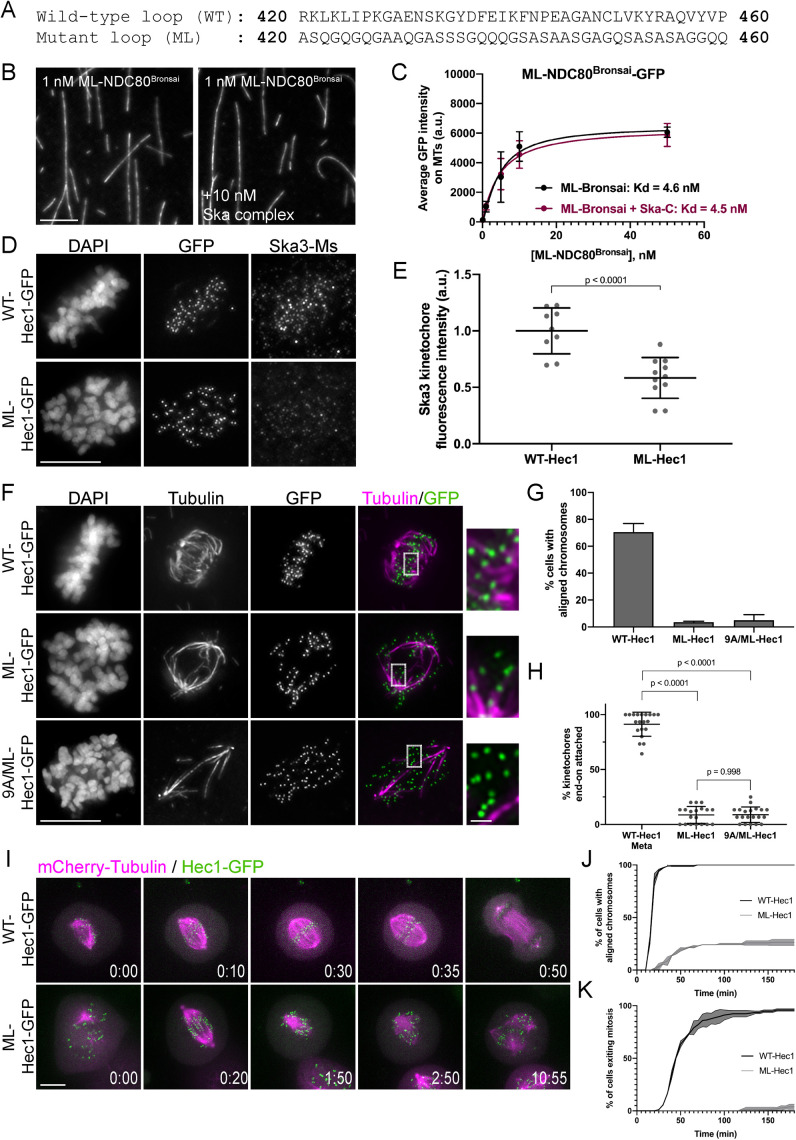
The Hec1 loop domain contributes to Ska complex recruitment to kinetochores and generation of kinetochore–microtubule attachments. (A) Sequence of the wild-type (WT) and mutated (ML) loop region in Hec1. (B) GFP fluorescence images of the ML-NDC80Bronsai complex decorating microtubules in the presence and absence of the Ska complex. Images show a single concentration of the ML-NDC80Bronsai complex from the experiment (1 nM) with and without added Ska complex (10 nM). (C) Binding curves from the microtubule binding assays. Datapoints and curve fits shown in black are from experiments without added Ska complex. Those shown in burgundy are from experiments with added Ska complex. Each point on the curve represents the average fluorescence intensity at that concentration from three separate experiments. For each concentration, fluorescence intensities of GFP-NDC80 complexes were measured on at least 40 individual microtubules from at least 10 different TIRF fields per experiment. (D) Immunofluorescence images of cells expressing WT- and ML-Hec1-GFP. Cells were fixed and stained using antibodies to Ska3 (mouse). (E) Quantification of Ska3 kinetochore fluorescence intensity from cells expressing WT- and ML-Hec1-GFP. For each condition, at least 20 kinetochores per cell were measured from at least four cells per experiment from two separate experiments. (F) Immunofluorescence images of cold-treated cells expressing WT-, ML-, and 9A/ML-Hec1-GFP. Cells were incubated in ice-cold DMEM for 12 min, permeabilized, fixed, and stained with antibodies to tubulin. Insets are enlargements of the regions indicated by the dashed boxes. (G) Quantification of chromosome alignment in cells expressing WT-, ML-, and 9A/ML-Hec1-GFP. For each condition, chromosome alignment was assessed in at least 100 cells per experiments from two separate experiments. Cells were scored as “aligned” if they contained a metaphase plate with <5 chromosomes off the plate. (H) Quantification of end-on attachment in cells expressing WT-, ML-, and 9A/ML-Hec1-GFP. Statistical significance was determined using a one-way ANOVA. For each condition, at least 15 kinetochores per cell were measured from at least nine cells per experiment from two separate experiments. (I) Still images from time-lapse experiments of cells expressing Hec1-GFP and mCherry-tubulin. Time from nuclear envelope breakdown (NEBD) denoted at bottom right corner of each image (hours:minutes). (J) Quantification of chromosome alignment efficiency in cells from the experiment shown in Figure I. Cell fate was tracked after mitotic entry (as determined by NEBD) for 3 h, and cells were scored as “aligned” upon metaphase plate formation (as determined by Hec1-GFP fluorescence). Data for WT- and ML-Hec1 are from 175 cells from two independent experiments and 165 cells from two independent experiments, respectively. (K) Quantification of mitotic exit timing in cells from the experiment shown in Figure I. Cell fate was tracked after mitotic entry (as determined by NEBD) for 3 h, and cells were scored as “exiting mitosis” upon anaphase entry. Data for WT- and ML-Hec1 are from 175 cells from two independent experiments and 165 cells from two independent experiments, respectively. Scale bars: 10 and 1 µm for panels and insets, respectively.
Given that cells expressing Δ80-Hec1 form cold-stable attachments, we hypothesized that these attachments are mediated through the Hec1 CH domain and have lost their ability to be negatively regulated through Aurora kinase phosphorylation. To test this hypothesis, we analyzed cold-resistant end-on attachments in early prometaphase cells shortly after nuclear envelope breakdown. Similar to 9A-Hec1-GFP expressing cells, and in contrast to WT-Hec1-GFP expressing cells, early prometaphase cells expressing Δ80-Hec1-GFP formed end-on kinetochore–microtubule attachments that resisted cold depolymerization (Figure 4, I and J), suggesting that the Hec1 tail domain is required for temporal regulation of attachments. Collectively, these data support a role for the Hec1 tail in force generation and attachment regulation at the kinetochore, but suggest that it is not required for kinetochore–microtubule attachment formation.
We were somewhat surprised at the ability of cells expressing Δ80-Hec1-GFP to retain end-on attachments after cold treatment, since it was previously observed that the tail domain contributes to the formation and/or maintenance of kinetochore–microtubule attachments in both human and marsupial cells (Guimaraes et al., 2008; Miller et al., 2008; Etemad et al., 2015; Janczyk et al., 2017). To confirm that this was not a cell type–specific phenomenon, we expressed WT- and Δ80-Hec1-GFP constructs in human RPE1 cells and found that, similar to what was observed in HeLa cells, RPE1 cells expressing Δ80-Hec1-GFP were competent to form cold-resistant end-on kinetochore–microtubule attachments (Supplemental Figure S6A). Interestingly, we found that location of the GFP had a major impact on the ability of Δ80-Hec1 expressing cells to form kinetochore–microtubule attachments. HeLa cells expressing either C- or N-terminally GFP-tagged WT-Hec1 constructs formed stable, end-on attachments, as previously reported (Guimaraes et al., 2008; Miller et al., 2008; DeLuca et al., 2011; Mattiuzzo et al., 2011; Etemad et al., 2015; Janczyk et al., 2017). In contrast, HeLa cells expressing C-terminally GFP-tagged Δ80-Hec1 formed end-on kinetochore–microtubule attachments, while those expressing N-terminally GFP-tagged Δ80-Hec1 did not (Supplemental Figure S6B). Similar results were found in RPE1 cells (Supplemental Figure S6D). We quantified cold-resistant attachment stability in HeLa cells expressing either C- or N- terminally GFP-tagged Δ80-Hec1 and confirmed that while cells expressing C-terminally tagged Δ80-Hec1 were able to form cold-stable, end-on attachments, cells expressing N-terminally tagged Δ80-Hec1 were not (Supplemental Figure S6, C and D).
After characterizing the phenotype of cells expressing Δ80-Hec1-GFP, we returned to our original question of whether the Hec1 tail is required for Ska complex recruitment to kinetochores. Immunofluorescence analysis revealed no significant difference in Ska3 levels at kinetochores in metaphase cells expressing WT- versus Δ80-Hec1-GFP (Figure 5, A and B). Similarly, microtubule-independent Ska3 recruitment to kinetochores also remained high in cells expressing Δ80-Hec1-GFP (Figure 2, C and D), suggesting that the tail domain is dispensable for both microtubule-dependent and -independent Ska complex recruitment. Because purified Ska complexes compensated for the weak binding affinity of Δ80-NDC80 complexes in vitro (Figure 3), we asked whether formation of kinetochore–microtubule attachments in cells expressing Δ80-Hec1-GFP required the presence of an intact Ska complex. We depleted Ska1 and Ska3 from HeLa cells, expressed either WT- or Δ80-Hec1-GFP, incubated the cells in cold media, and measured the abundance of end-on kinetochore–microtubule attachments. Kinetochore–microtubule attachments failed to form in Ska1/Ska3-depleted cells expressing either WT- or Δ80-Hec1-GFP (Figure 5, C and D), which is in contrast to cells expressing 8A- and 9A-Hec1-GFP (Figure 2, E and F; Supplemental Figure S3, E and F). Thus, tail-less NDC80 complexes, similar to WT complexes, require the Ska complex to form attachments to microtubules. Collectively, our results suggest that the tail domain of Hec1 is not explicitly required for either Ska complex recruitment to kinetochores or formation of stable kinetochore–microtubule attachments, but it likely plays a role in force generation at the attachment interface in human cells.
FIGURE 5:
The Hec1 tail domain is dispensable for Ska complex recruitment to kinetochores and is required for kinetochore–microtubule attachments in the absence of the Ska complex. (A) Immunofluorescence images of cells expressing WT, 9D-, and Δ80-Hec1-GFP. Cells were fixed and stained using antibodies to Hec1 pS69 and Ska3 (mouse). (B) Quantification of Ska3 kinetochore fluorescence intensity from cells expressing WT, 9D-, and Δ80-Hec1-GFP. For each condition, at least 20 kinetochores per cell were measured from at least five cells per experiment from three separate experiments. Statistical significance was determined by a one-way ANOVA. (C) Immunofluorescence images of cold-treated cells expressing WT-, 9A-, and Δ80-Hec1-GFP and treated with Ska1 and Ska3 siRNA. Cells were incubated in ice-cold DMEM for 12 min, permeabilized, fixed, and stained with antibodies to tubulin. Insets are enlargements of the regions indicated by the dashed boxes. (D) Quantification of end-on attachment in cells expressing WT-, 9A-, and Δ80-Hec1-GFP and treated with Ska1 and Ska3 siRNA. The WT- and 9A-Hec1 data shown are from the experiment presented in Figure 2. For each condition, at least 15 kinetochores per cell were measured from at least 10 cells per experiment from three separate experiments. Statistical significance was determined by a one-way ANOVA. On all dot plots, each dot represents the average value for all kinetochores from a single cell. Scale bars: 10 and 1 µm for panels and insets, respectively.
The Ska complex is recruited to the internal coiled-coil domain region of the NDC80 complex to enhance NDC80-MT binding
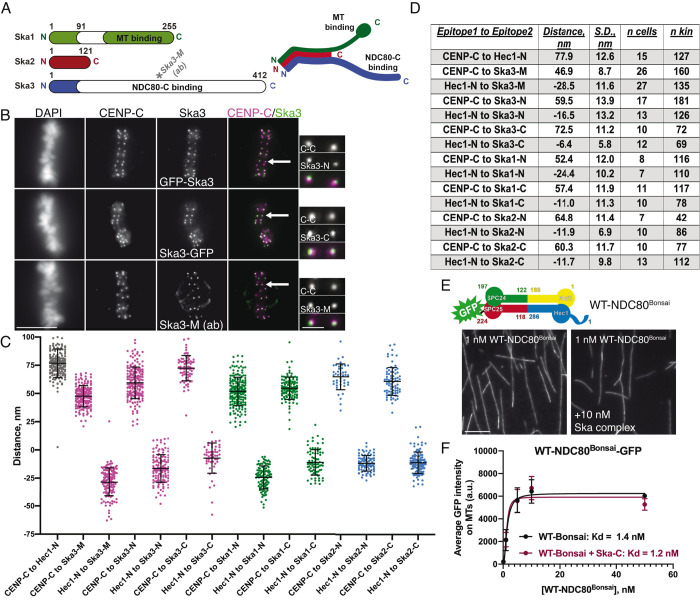
To home in on the Ska complex recruitment domain within the NDC80 complex, we carried out two-color fluorescence localization mapping of Ska complex components at metaphase kinetochores (Wan et al., 2009). Since the C-terminal half of Ska3 contains the putative NDC80 binding site, we first mapped the distance between a Ska3 antibody that recognizes amino acids 226–253 (Figure 6A; “Ska3-M,” for “middle”) and both CENP-C (inner kinetochore) and the N-terminus of Hec1 (outer kinetochore). These measurements revealed that amino acids 226–253 of Ska3 reside ∼47 nm outside of CENP-C and ∼29 nm inside of the N-terminus of Hec1 (Figure 6, A–D), suggesting that a region encompassed by the NDC80 complex–binding domain is localized near the internal, coiled-coil region of the NDC80 complex. Reconstituted, purified human Ska complexes have been shown to exist as either monomers or dimers of the Ska1, Ska2, and Ska3 trimer, which are formed through oligomerization of the N-termini of each protein to form a three-helix bundle (Jeyaprakash et al., 2012; Helgeson et al., 2018). Ska1’s C-terminus contains a winged-helix domain that has microtubule binding activity, and Ska3 contains a predominantly unstructured C-terminal region that is responsible for interaction with the NDC80 complex (Jeyaprakash et al., 2012; Abad et al., 2014, 2016). To better understand how the Ska complex components are organized at the kinetochore–microtubule interface, we carried out further paired fluorescence localization mapping using N- and C-terminal GFP tags on the Ska complex components. The N-terminal GFP tags on Ska1, Ska2, and Ska3 all mapped to a similar domain within the kinetochore, which was 52–65 nm outside of CENP-C and 12–24 nm inside the CH domain of Hec1 (Figure 6, C and D). This is not surprising, since the N-termini of Ska1, 2, and 3 form a well-folded, relatively compact oligomerization domain (Jeyaprakash et al., 2012). Furthermore, we found that all C-terminal domains of Ska1, Ska2, and Ska3 also mapped to a region inside the Hec1 CH domain. However, we note that the C-terminal GFP tag on Ska3 was localized very close to this region, with a mapped distance of ∼73 nm outside of CENP-C and ∼6 nm inside the Hec1 CH domain (Figure 6, B–D). This suggests that the unstructured domain of Ska3 may extend substantially along the length of the coiled-coil domain of the NDC80 complex. These experiments were carried out using a 2D analysis of kinetochore domain localization (Wan et al., 2009), and we note that our reported average distance between CENP-C and the CH domain of Hec1 is consistent with previously reported 2D and 3D measurements (Figure 6, C and D; Wan et al., 2009; Suzuki et al., 2018; Roscioli et al., 2019).
FIGURE 6:
The Ska complex localizes to kinetochores at the central coiled-coil domain of the NDC80 complex. (A) Left: schematic showing the domain architecture of the Ska complex components. White regions indicate predicted disordered domains (Jeyaprakash et al., 2012). The Ska3 antibody directed to amino acids 226–253 is indicated on the schematic and represents the “Ska3-Middle Domain” (“Ska3-M”). Right: schematic of a single Ska complex (one copy of each subunit), showing the trimerization domains located in the N-termini of Ska1, Ska2, and Ska3, the microtubule binding domain of Ska1 (green), and the proposed NDC80 complex–binding region in Ska3 (blue). (B) Immunofluorescence images of metaphase cells expressing N- and C-terminally GFP-tagged Ska3 and stained with antibodies to inner kinetochore protein CENP-C (top two rows) and immunofluorescence images of a metaphase cell stained with antibodies to Ska3-M (rabbit) and CENP-C (bottom row). Arrows point to the kinetochore pairs shown in the insets. (C) Plots of the mean distance between the indicated kinetochore proteins/protein domains. Measurements with “Hec1-N” were carried out with an antibody to the CH domain in the N-terminus of Hec1 (9G3). “N” and “C” epitopes for each of the Ska complex components are N- and C-terminal GFP moieties, respectively. Each point on the graph represents a distance measurement for a pair of sister kinetochores. (D) Summary of data presented in panel C. Positive values indicate that epitope 1 was mapped inside epitope 2. Negative values indicate that epitope 1 was mapped outside of epitope 2. The numbers of cells (n cells) and kinetochore pairs (n kin) are indicated. (E) Top: schematic of the NDC80Bonsai complex. Bottom: GFP fluorescence images of the NDC80Bonsai complex decorating microtubules in the presence and absence of Ska complex. Images show a single concentration of the NDC80Bonsai complex from the experiment (1 nM) with and without added Ska complex (10 nM). (F) Binding curves from the microtubule binding assays. Datapoints and curve fits shown in black are from experiments without added Ska complex. Those shown in burgundy are from experiments with added Ska complex. Each point on the curve represents the average fluorescence intensity at that concentration from three separate experiments. For each concentration, fluorescence intensities of GFP-NDC80 complexes were measured on at least 40 individual microtubules from at least 10 different TIRF fields per experiment. Scale bars: 10 and 1 µm for panels and insets, respectively.
The mapping experiments suggested that the Ska complex is recruited to the central coiled-coil region of the NDC80 complex. To further investigate a role for this region in Ska complex binding, we carried out microtubule binding experiments in the presence and absence of purified Ska complexes using NDC80Bonsai, a truncated NDC80 complex missing most of the central coiled-coil region and the loop domain (Ciferri et al., 2008). Indeed, we found that while the NDC80Bonsai complexes bound robustly to microtubules, the affinity of NDC80Bonsai complexes for microtubules was not increased with the addition of Ska complexes (Figure 6, E and F). These findings are consistent with recent results from the Liu and Musacchio labs, which demonstrate that NDC80Bonsai complexes are unable to bind to purified Ska complexes (Zhang et al., 2017; Huis in’t Veld et al., 2019). Additionally, we found that in BRB80 buffer, addition of the Ska complex did not induce clustering of NDC80Bonsai complexes on microtubules, in contrast to NDC80Bronsai complexes (Supplemental Figure S3B), further supporting the idea that the central coiled-coil region of the NDC80 complex is required for Ska complex association.
We next generated a version of NDC80Bronsai in which the amino acids that make up the “loop” region of Hec1 (amino acids 420–460; Maiolica et al., 2007) were substituted with alternative amino acids predicted to form a flexible motif (Varma et al., 2012) (ML-NDC80Bronsai, Figure 7A). We then tested whether the microtubule binding affinity of this mutant version of the complex was increased by addition of purified Ska complexes. We found that while ML-NDC80Bronsai bound to microtubules with an affinity similar to that of WT-NDC80Bronsai, the addition of purified Ska complex had no significant effect on its microtubule binding affinity (Figure 7, B and C). These results suggest that mutation of the loop domain either prevents the Ska complex from directly interacting with the NDC80 complex or precludes a conformation that promotes formation of a NDC80/Ska/microtubule complex.
We then asked whether the loop domain was required for Ska complex recruitment to kinetochores in cells. For this purpose, we expressed the Hec1 loop mutant (ML-Hec1-GFP) in HeLa cells and found that Ska3 levels were significantly reduced at kinetochores compared with kinetochores from cells expressing WT-Hec1-GFP (Figure 7, D and E). We also found, consistent with previously published results, that end-on kinetochore–microtubule attachments failed to form and chromosome alignment was abolished in cells expressing ML-Hec1-GFP (Figure 7, F–H) (Varma et al., 2012; Zhang et al., 2012). Live cell imaging revealed that cells expressing ML-Hec1 failed to align chromosomes and exit mitosis and that these cells exhibited high levels of spindle fragmentation (Figures 7, I–K, and 4F). Given that Ska complexes maximally load to kinetochores with end-on attachments, again we could not distinguish between two possibilities: 1) the Hec1 loop domain promotes Ska complex recruitment, and in turn, the Ska complex is required for end-on attachment formation; or 2) the Hec1 loop domain is required for generation of stable kinetochore–microtubule attachments, and in turn, stable attachments promote Ska complex loading. We therefore measured Ska complex loading to kinetochores in the absence of microtubules and found that cells expressing ML-Hec1-GFP exhibited reduced levels of Ska3 at kinetochores (Figure 2, C and D), suggesting that an intact loop domain is required for efficient Ska complex recruitment to kinetochores.
Both chromosome alignment and kinetochore–microtubule attachment formation were severely impaired in cells expressing ML-Hec1-GFP. We therefore tested whether these defects were exclusively due to loss of Ska complex recruitment to kinetochores. Experiments in Figure 2 demonstrated that cells expressing 9A-Hec1-GFP formed hyperstable kinetochore–microtubule attachments, and this phenotype was independent of the Ska complex (Figure 2, E and F). These results indicate that attachment defects arising from Ska1/Ska3 depletion can be compensated for by the strong attachments generated in cells expressing 9A-Hec1-GFP. We therefore reasoned that if a mutated loop domain results in attachment defects solely due to loss of Ska complex recruitment, then preventing phosphorylation of the tail domain should rescue this defect. Thus, we generated a hybrid mutant containing a 9A tail domain and the mutant loop sequence (9A/ML-Hec1-GFP). We found that in cells expressing 9A/ML-Hec1-GFP, stable kinetochore–microtubule attachments failed to form and chromosome alignment was severely defective, similar to what we observed in cells expressing ML-Hec1-GFP (Figure 7, F–H). These results suggest that although the loop domain may participate in recruiting the Ska complex to kinetochores, it likely plays an additional, non-Ska complex–dependent role in generating kinetochore–microtubule attachments, perhaps through recruitment of other kinetochore proteins. It is also possible that mutation of the loop domain results in changes in NDC80 complex architecture at kinetochores that preclude formation of end-on, stable kinetochore–microtubule attachments.
A variety of loop mutations (including mutants with the loop sequence scrambled, reversed, or deleted altogether) have been reported to result in severe defects in chromosome alignment and end-on attachment formation when expressed in human cells (Varma et al., 2012; Zhang et al., 2012; Figure 7). To narrow down the region of the loop required for wild-type function, we generated systematic alanine substitutions of short stretches of 5–6 amino acids within the Hec1 loop (Supplemental Figure S7A) and expressed these mutants in cells. Expression of several, but not all, of these mutants mimicked the phenotype observed in cells expressing the ML-Hec1-GFP mutant and led to severe chromosome alignment defects (Supplemental Figure S7, A and B). We noted that mutating regions within the loop that resulted in substantial changes in local net charge produced the strongest chromosome misalignment phenotypes, while mutating regions with low net charge density resulted in no observable defects (Supplemental Figure S7, A–C). This suggests that the distributed charge of the loop region is likely critical for formation of kinetochore–microtubule attachments, potentially through forming interactions with the Ska complex and/or other kinetochore-associated proteins such as Cdt1 (Varma et al., 2012).
DISCUSSION
Hec1 tail phosphorylation affects kinetochore–microtubule attachments independently of the Ska complex
The positively charged, N-terminal tail domain of Ndc80/Hec1 is a target of Aurora kinases, and it has been suggested that phosphorylation of the tail directly reduces the affinity of NDC80 complexes for the negatively charged microtubule lattice, which in turn reduces kinetochore–microtubule attachment strength. It is also possible that phosphorylation of the Hec1 tail domain indirectly affects kinetochore–microtubule attachment strength by regulating the recruitment of additional kinetochore-associated microtubule-binding proteins. One possible candidate for imparting this regulation is the Ska complex, which loads to kinetochores progressively during mitosis and contributes to the stabilization of kinetochore–microtubule attachments. We found here that cells expressing a nonphosphorylatable Hec1 mutant (9A-Hec1) recruited increased levels of Ska complex components to kinetochores in human cells, similar to what has been reported in C. elegans (Cheerambathur et al., 2017). However, we found that stable kinetochore-–microtubule attachments were not dependent on the Ska complex in cells expressing 9A-Hec1, in which tail phosphorylation is completely blocked, or in cells expressing 8A-Hec1, where Ser-69 is left intact and remains phosphorylated throughout mitosis (DeLuca et al., 2018). Furthermore, we demonstrated that the phosphorylation state of the tail domain did not affect the levels of Ska complexes recruited to kinetochores in the absence of microtubules. Finally, we showed that kinetochore–microtubule attachments were additively destabilized in cells expressing a mutant version of Hec1 containing a phospho-mimetic tail (9D-Hec1) and depleted of the Ska complex. Collectively, these results suggest that, despite coincidental timing, Hec1 tail dephosphorylation and Ska complex recruitment to kinetochores likely contribute to kinetochore-microtubule attachment stabilization independently.
It is noteworthy to mention that the metaphase recruitment of Ska3 to kinetochores differed between 9A-Hec1 and 8A-Hec1 expressing cells. Previous studies have demonstrated that complete dephosphorylation of the Hec1 tail results in hyperstable kinetochore–microtubule attachment formation as evidenced by increases in interkinetochore distance and kinetochore-fiber intensity, lagging chromosomes in anaphase, and dampened metaphase chromosome oscillations (Guimaraes et al., 2008; DeLuca et al., 2011, 2018; Zaytsev et al., 2014; Long et al., 2017). Strikingly, leaving Ser-69 unperturbed while blocking phosphorylation at all other Aurora sites (8A-Hec1) was sufficient to restore chromosome oscillation kinetics to wild-type levels (DeLuca et al., 2018). Similarly, we showed here that cells expressing 8A-Hec1 recruited wild-type levels of Ska3 to kinetochores, while cells expressing 9A-Hec1 recruited significantly higher levels. Despite this, and similar to the case for 9A-Hec1, cells expressing 8A-Hec1 formed cold-stable attachments prematurely and retained robust attachments in the absence of the Ska complex, supporting the model that Hec1 tail dephosphorylation modulates kinetochore–microtubule attachment strength in a Ska complex–independent manner.
Also of note are the somewhat divergent mechanisms for Ska complex recruitment and NDC80 complex–mediated kinetochore–microtubule attachment regulation described in this study compared with those described in C. elegans. In C. elegans, but not in human cells, depletion of the Ska complex rescued hyperstable kinetochore–microtubule attachment formation caused by expression of a nonphosphorylatable Hec1 tail domain mutant (Cheerambathur et al., 2017). Furthermore, while a pool of attachment-independent Ska complex localizes to kinetochores in human cells, C. elegans Ska complex localizes to kinetochores at detectable levels only upon chromosome alignment. These results suggest that in C. elegans, dephosphorylation of the Hec1 tail domain promotes Ska complex association to kinetochores, which in turn promotes stabilization of kinetochore–microtubule attachments. As such, the mechanism for Ska complex recruitment to kinetochores is likely not completely conserved from C. elegans to humans.
The Hec1 tail is dispensable for kinetochore–microtubule attachments in cells
Consistent with our findings here, chromosome alignment errors and decreased interkinetochore distances have been previously observed in mammalian cells expressing tail-less, Δ80-Hec1 mutants (Guimaraes et al., 2008; Miller et al., 2008; Etemad et al., 2015; Janczyk et al., 2017). These phenotypes have been widely attributed to loss of stable kinetochore–microtubule attachments (Guimaraes et al., 2008; Miller et al., 2008). Contrary to this, we found that Δ80-Hec1 expression did not prevent formation of cold-resistant, stable kinetochore–microtubule attachments in either HeLa or RPE1 cells. Instead, we report that cells lacking the tail domain formed kinetochore–microtubule attachments prematurely, presumably due to a lack of Aurora kinase–mediated regulation. In such a scenario, cells expressing tail-less Hec1 are unable to negatively regulate the initial formation of end-on kinetochore–microtubule attachments, and the NDC80 complex is able to bind spindle microtubule plus ends through strong Hec1 CH domain–mediated interactions, which would otherwise be kept labile by a highly phosphorylated Hec1 tail domain. Despite this early accumulation of attachments, however, we found that cells expressing Δ80-Hec1-GFP exhibited decreased interkinetochore distances, suggesting that attachments are unable to produce sufficient forces to generate wild-type tension across sister kinetochore pairs.
Maintenance of a bipolar mitotic spindle requires a balance of forces within the spindle, some of which are derived from chromosomes and their attachments to spindle microtubules (Manning and Compton, 2007; Maiato and Logarinho, 2014). Fragmentation of spindles leading to multipolarity can be caused by alterations in forces generated at the kinetochore–microtubule interface and by loss of sister chromatid cohesion (Daum et al., 2011; Stevens et al., 2011; Maiato and Logarinho, 2014). In the case of cells expressing tail-less Hec1, we observed a high incidence of multipolarity, similar to what we observed in cells expressing the Hec1 loop mutation, in which kinetochore-microtubule attachments failed to form altogether. Cells expressing Δ80-Hec1 were able to form cold-stable end-on attachments, but these attachments did not generate wild-type force and were not sufficient to allow cells to exit mitosis with normal timing. Thus, defective force generation and/or cohesin fatigue may contribute to the high incidence of multipolarity observed in Δ80-Hec1 expressing cells. Alternatively, the Hec1 tail may have an uncharacterized role at the centrosome, perhaps involving the Hec1-Hice1 interaction, which is required for maintenance of spindle bipolarity (Wu et al., 2009). Future studies will be required to fully characterize the causes of multipolarity in Δ80-Hec1 expressing cells.
It is noteworthy to mention that a recent in vitro study has also implicated the Hec1 tail domain in force generation at the kinetochore–microtubule interface: Huis in’t Veld et al. (2019) artificially trimerized NDC80 complexes on the surface of beads and measured the ability of NDC80 complex trimers to resist force from an optical trap. NDC80 complex trimers lacking the Hec1 tail—despite binding to microtubules with high affinity—detached from depolymerizing microtubules in both the presence and absence of applied force, whereas wild-type NDC80 trimers remained bound under these conditions (Huis in’t Veld et al., 2019). These results led the authors to conclude that the Hec1 tail is critical for force coupling at the kinetochore–microtubule interface.
An important distinction between our study and several previously published studies is the requirement for the Hec1 tail in formation of kinetochore–microtubule attachments. Notable experimental differences may explain the conflicting results. For example, in one previous study, N-terminally tagged Hec1 constructs were used (Etemad et al., 2015). In our current study, when we tagged WT-Hec1 with GFP on either the C- or N-terminus, we found that both constructs were competent to support formation of kinetochore–microtubule attachments. However, while cells expressing C-terminally tagged ∆80-Hec1-GFP were able to form attachments, those expressing N-terminally tagged GFP-∆80-Hec1 were not (Supplemental Figure S6, B-–D). Thus, replacing the N-terminal tail of Hec1 with a GFP moiety results in a failure to form kinetochore–microtubule attachments. In other published studies, researchers used ∆80-Hec1 mutants lacking a GFP tag to rescue depletion of endogenous Hec1 and reported failure to form kinetochore–microtubule attachments (Miller et al., 2008; Janczyk et al., 2017). In our study, we found that transfection of equal amounts of Hec1 transgenic DNA led to significant differences in protein expression, with ∆80-Hec1 being expressed at ∼50% lower levels than WT-Hec1 (Supplemental Figure S1, A and B). In the case of siRNA silence-rescue experiments, this difference in expression would likely result in an incomplete rescue and defects in kinetochore–microtubule attachments. In most of our experiments, we did not treat cells with Hec1 siRNA, but rather measured endogenous Hec1 depletion from kinetochores using an antibody to the constitutively phosphorylated residue Ser-69 on the Hec1 tail. In this experimental scheme, we analyzed only cells with similar kinetochore GFP levels (Supplemental Figure S1C; see also Materials and Methods), thus avoiding any discrepancies associated with incomplete rescue. Finally, a previous study from our lab expressed C-terminally tagged ∆80-Hec1 mutants in PtK1 cells and reported defective kinetochore–microtubule attachment (Guimaraes et al., 2008). This study was done using PtK1 cells stably expressing photo-activatable (PA)-GFP-tubulin, and it is possible that the expression levels of this tubulin construct specifically compromised the kinetochore–microtubule interface in a manner that sensitized cells to Hec1 tail loss. Alternatively, it is possible that PtK1 cells have a requirement for this domain of Hec1 different from that of human cells. Expressing ∆80-Hec1 in otherwise unperturbed PtK1 cells will be important to address this question.
Our results describing the formation of end-on attachments in human cells expressing tail-less Hec1 are consistent with results from both budding yeast and C. elegans, where the Ndc80 tail domain is not strictly required for kinetochore–microtubule attachment (Kemmler et al., 2009; Demirel et al., 2012; Cheerambathur et al., 2013; Lampert et al., 2013). However, similar to the scenario in human cells, the tail domain does play some role at the kinetochore–microtubule interface in these organisms. For example, in budding yeast the tail domain becomes required for cell survival upon perturbation of the Dam1 complex, a kinetochore-associated complex found in yeasts but not higher eukaryotes, which contributes to generating stable kinetochore–microtubule attachments (Demirel et al., 2012; Lampert et al., 2013). In addition, a study using a tension sensor inserted near the N-terminus of budding yeast Hec1 demonstrated that while the tail domain is not required for kinetochore–microtubule attachment formation per se in cells, its deletion results in reduced tension at the NDC80 complex–microtubule interface (Suzuki et al., 2016). Taken with the result that the Hec1 tail is required for load-bearing attachments of NDC80 complexes to microtubules (Huis in’t Veld et al., 2019), the available data suggest that this function of the Hec1 tail domain is generally conserved from yeast to humans.
The Ska complex compensates for Hec1 tail domain function
Studies using NDC80 complexes purified from various organisms have demonstrated that the Hec1 tail domain is required for high-affinity NDC80 complex–microtubule binding in vitro (Cheeseman et al., 2006; Wei et al., 2007; Alushin et al., 2012; Umbreit et al., 2012; Cheerambathur et al., 2013; Lampert et al., 2013; Zaytsev et al., 2015). In the case of human NDC80 complexes, addition of the Ska complex compensates for deletion of the Hec1 tail in a number of in vitro NDC80 complex–microtubule interaction assays. Helgeson et al. (2018) carried out optical trapping experiments using NDC80 complex–coated beads to demonstrate that while NDC80 complexes lacking the Hec1 tail generated weak attachments to microtubules that could be disrupted under low rupture forces, addition of soluble Ska complexes significantly strengthened these attachments. Consistent with these findings, Huis in’t Veld et al. (2019) found that addition of the Ska complex to trimerized NDC80 complexes lacking the Hec1 tail enabled these complexes to track depolymerizing microtubules, a property not observed in the absence of the Ska complex. The notion that the Ska complex can functionally compensate for Hec1 tail deletion is reminiscent of studies carried out in budding yeast with the Dam1 complex, which has been suggested to be a functional orthologue of the Ska complex (Welburn et al., 2009). Analogous to the experiments described above for human Ska and NDC80 complexes, Dam1 is able to enhance the affinity of tail-less budding yeast NDC80 complexes for microtubules in vitro (Lampert et al., 2010; Tien et al., 2010; Lampert et al., 2013). Consistently, deletion of the Ndc80/Hec1 tail is not lethal in budding yeast (Kemmler et al., 2009; Demirel et al., 2012; Lampert et al., 2013), but deletion or mutation of Dam1 sensitizes cells to loss of the Ndc80/Hec1 tail, resulting in cell death due to cell division defects (Demirel et al., 2012; Lampert et al., 2013; Suzuki et al., 2016). In line with these results, we found that cells expressing Δ80-Hec1-GFP were able to form cold-stable kinetochore–microtubule attachments, but this required the presence of the Ska complex. Collectively, our in vitro and cell-based results suggest that the Ska complex can compensate for the Hec1 tail’s role in forming stable kinetochore–microtubule attachments in human cells.
The Ska complex is recruited to the internal coiled-coil domain of the NDC80 complex rather than the Hec1 tail domain
Ska complex loading to kinetochores requires the NDC80 complex (Gaitanos et al., 2009; Welburn et al., 2009; Chan et al., 2012; Zhang et al., 2012), and the two complexes directly interact (Zhang et al., 2017; Helgeson et al., 2018; Huis in’t Veld et al., 2019). Although Ska3 is known to mediate the interaction, its binding site on the NDC80 complex remains unresolved (Zhang et al., 2018). Our results demonstrate that the Hec1 tail is not required for Ska complex–mediated enhancement of NDC80-microtubule interactions or for Ska complex localization to kinetochores in cells. We note that these results are inconsistent with a previous report from Janczyk et al. (2017), where it was shown that mutations in the Hec1 tail abolish Ska recruitment to the NDC80 complex–microtubule interface in vitro and to kinetochores in cells. It is unclear why our results differ from theirs, although a potential explanation is that the tail mutations made in the Janczyk et al. study impacted overall kinetochore architecture in a manner that precluded Ska recruitment independently of the Hec1 tail. To map the Ska complex kinetochore recruitment domain, we used two-color colocalization imaging and found that the Ska complex colocalized with the coiled-coil region of the NDC80 complex, inside the Hec1 CH domain. Interestingly, most of the N- and C-termini of all Ska complex components also mapped near this region, suggesting that the bulk of the complex is not significantly extended along the NDC80 complex axis. The one exception is the C-terminus of Ska3, which mapped closely to, but still inside, the CH domain of Hec1, suggesting that the unstructured region of Ska3 may be somewhat elongated along the length of the NDC80 complex. These results are consistent with a recent study from Helgeson et al. (2018), in which the authors found a large number of contact points between Ska3 and the coiled-coil region of the NDC80 complex using cross-linking mass spectrometry.
The Hec1 loop domain has Ska complex–dependent and –independent functions in chromosome alignment
We report that mutation of the Hec1 loop domain prevents enhancement of NDC80 complex–microtubule binding by the Ska complex. This is possibly at odds with a recent study from Huis in’t Veld et al. (2019), which reported that removal of the loop domain from Hec1 did not affect the interaction between soluble NDC80 and Ska complexes. This difference could possibly reflect a requirement for the loop domain in the interaction of NDC80 and Ska complexes specifically on microtubules. Alternatively, since Ska3 phosphorylation by CDK1 increases the affinity of soluble Ska and NDC80 complexes for each other (Zhang et al., 2017; Huis in’t Veld et al., 2019), the phosphorylation state of Ska3 might impact the requirement of the Hec1 loop domain for the two complexes to associate. In such a scenario, dephosphorylated Ska complexes bind more weakly to NDC80 complexes in the presence of microtubules, and the loop domain is required for high-affinity interactions, specifically under these suboptimal binding conditions. Given our result that the loop domain is required in cells for microtubule-independent Ska complex loading to kinetochores, we do not favor the latter hypothesis.
Both chromosome alignment and kinetochore–microtubule attachments were severely perturbed in cells expressing Hec1 constructs containing a mutated loop domain—more so than in cells expressing 9D-Hec1 mutants or in cells depleted of Ska1 and Ska3. This was observed in previous studies (Zhang et al., 2012; Varma et al., 2012) and led us to ask whether the defects observed were entirely due to loss of Ska complex recruitment. To test this, we modified the loop mutant to include a nonphosphorylatable N-terminal tail domain, since we demonstrated that expression of the 9A-Hec1 mutant abrogated the need for Ska1 and Ska3 to form stable kinetochore–microtubule attachments. Cells expressing the 9A/ML-Hec1 mutant showed no improvement in either chromosome alignment or formation of stable kinetochore–microtubule attachments compared with those expressing ML-Hec1. Thus, we conclude that in addition to contributing to efficient Ska complex recruitment to kinetochores, the Hec1 loop has an additional, non-Ska complex–dependent role in forming stable attachments. We found that mutating short stretches of the loop sequence that contain at least two charged residues phenocopied expression of the full ML-Hec1 construct. Thus it is possible that the loop domain recruits additional factors, such as Cdt1, that are required for generation of stable, end-on kinetochore–microtubule attachments (Varma et al., 2012). Alternatively, the loop region could be critical for adoption of a conformation of NDC80 that is required for maintaining proper, end-on attachments to microtubules.
Overall, our results support a model in which the Hec1 tail domain, while not explicitly required for forming end-on kinetochore–microtubule attachments, is important in regulating the force-generating attachments between kinetochores and microtubule plus ends. They also suggest that the Ska complex is loaded to the central coiled-coil region of the NDC80 complex during mitosis to ensure proper force coupling at the kinetochore–microtubule interface, which may be particularly important at kinetochores containing NDC80 complexes with lower microtubule binding capacity (e.g., with highly phosphorylated tail domains). How tail domain phosphorylation and dephosphorylation are coordinated with Ska complex loading to kinetochores to ensure proper regulation of kinetochore–microtubule attachments is an important issue that requires future investigation.
MATERIALS AND METHODS
Cell culture
HeLa Kyoto cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic/antimycotic solution. RPE1 (American Type Culture Collection) cells were cultured in 1:1 Ham’s F12:DMEM supplemented with 10% FBS and 1% antibiotic/antimycotic solution. All cell lines were maintained at 37°C in 5% CO2.
Cell treatments and transfections
For all fixed cell experiments, cells were grown on sterile, acid-washed coverslips in six-well plates. All nucleic acid transfections were done in Optimem (Life Technologies). siRNA duplexes were transfected as follows: Ska3 (5′-AGACAAACAUGAACAUUAA-3′; Gaitanos et al., 2009) at 80 nM, Ska1 (5′-CCCGCTTAACCTATAATCAAA-3′; Hanisch et al., 2006) at 80 nM, and Hec1 (5′-CCCUGGGUCGUGUCAGGAA-3′; DeLuca et al., 2011) at 160 nM. All siRNA duplexes were transfected into HeLa cells using Oligofectamine (Thermo Fisher Scientific) according to the manufacturer’s protocol. For all siRNA transfections, cells were processed for immunofluorescence 48 h after addition of siRNA. Plasmids encoding Hec1-GFP and mCherry-tubulin were transfected into HeLa Kyoto cells with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocol. RPE1 cells were transfected using a nucleofector (Lonza) and using Lonza Kit L according to the manufacturer’s protocol with program X-001. Cells were processed for immunofluorescence 24 h following DNA transfection. For silence-rescue experiments of Hec1 and expression of Hec1 mutants in Ska-depleted cells, cells were transfected with siRNA using Oligofectamine and 24 h later were transfected with DNA using Lipofectamine 2000. At 24 h after DNA transfection (48 h post–siRNA addition), cells were processed for immunofluorescence. For end-on attachment experiments, transfection media was replaced with ice-cold DMEM 24 h post–DNA transfection and cells were incubated on ice for 12 min prior to fixation. For attachment-independent analysis of Ska3 localization, Hec1-GFP–transfected cells were arrested at the G2/M transition by inhibiting CDK1/Cyclin-B with 9 µM RO-3306 (Sigma-Aldrich) for 16 h and then extensively washed out with warm DMEM and placed into DMEM supplemented with 10 µM nocodazole (Tocris Bioscience), where they were incubated at 37°C for 1 h before being processed for immunofluorescence. For analysis of asynchronous, attachment-independent Ska3 levels (Supplemental Figure S2), cycling HeLa cell populations were treated with 10 µM nocodazole for 1 h without any synchronization and then were processed for immunofluorescence. For live cell experiments, cells were synchronized with thymidine the day after being seeded into glass-bottom live cell dishes (35 mm), washed out of the first thymidine treatment 16 h later, and subsequently transfected with plasmids encoding Hec1-GFP and mCherry-tubulin. At eight hours post-transfection, cells were subjected to a second thymidine treatment. The following day (16 h after the second thymidine treatment), cells were washed out into warm DMEM and incubated for nine hours prior to live cell imaging.
Western blotting
Hec1-GFP transfected cells were harvested 24 h post-transfection and stored at –20°C until use, at which time they were thawed on ice and resuspended in lysis buffer (4 mM dithiothreitol [DTT] in phosphate-buffered saline with protease inhibitors). Cells were lysed by sonication and subjected to Western blot analysis. Hec1 expression was detected using mouse anti–Hec1 9G3 (Novus Biologicals) at 1:500 and donkey anti-mouse horseradish peroxidase (Jackson ImmunoResearch) at 1:10,000. Hec1-GFP band intensity was quantified using MetaMorph software with ponceau-stained blots as a loading control. All mutant Hec1-GFP band intensities were normalized to WT-Hec1-GFP.
Immunofluorescence microscopy
Cells were rinsed in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 4 mM MgCl2, pH 7.0) and permeabilized in lysis buffer (PHEM + 1.0% Triton X–100) for 5 min at 37°C. Post-lysis, cells were quickly washed in PHEM and subsequently fixed in freshly made fixative solution (4% paraformaldehyde in PHEM) for 20 min at 37°C. After fixation, cells were subjected to three 5-min washes in PHEM-T (PHEM + 0.1% Triton X–100), quickly rinsed in PHEM, and blocked in 10% boiled donkey serum (BDS) in PHEM for 1 h at room temperature. Following blocking, primary antibodies diluted in 5% BDS in PHEM were added to cells and incubated for 1 h at room temperature followed by 16 h at 4°C. Primary antibodies were used as follows: human anti-centromere antibody (ACA) at 1:300 (Antibodies, Inc.), mouse anti-tubulin (DM1α) at 1:600 (Sigma-Aldrich), mouse anti-Hec1 (9G3) at 1:3000 (Novus Biologicals), rabbit anti-phosphorylated Hec1-pSer69 (pS69) at 1:3000 (DeLuca et al., 2018), mouse anti-Ska3 at 1:500 (Santa Cruz Biotechnology), rabbit anti-Ska3 at 1:300 (GeneTex), and rabbit anti-Astrin at 1:1000 (Sigma-Aldrich). After primary antibody incubation, unbound antibody was washed off using three 5-min PHEM-T rinses, followed by a quick wash in PHEM. Secondary antibodies (conjugated to Alexa 488, Cy3 dye, or Alexa 647; Jackson ImmunoResearch) were diluted 1:1000 in 5% BDS except mouse Ska3 and rabbit Ska3, for which secondary antibodies were diluted 1:500 and 1:300, respectively. Cells were incubated in secondary antibody for 45 min at room temperature, and unbound antibody was washed off with 3 × 5 min PHEM-T washes followed by a quick rinse in PHEM. Cells were then incubated in a 2 ng/ml 4’,6-diamidino-2-phenylindole (DAPI) solution (diluted in PHEM) for 30 s, subjected to two 5-min PHEM-T washes, quickly rinsed in PHEM, and mounted onto glass slides using an antifade solution (90% glycerol + 0.5% N-propyl gallate). Following mounting, coverslip edges were sealed with nail polish and slides were stored at 4°C.
Fixed cell imaging
All fixed cell images were acquired using a DeltaVision Personal DV Imaging system (GE Healthcare) on an IX71 inverted microscope (Olympus) using SoftWoRx software (GE Healthcare). All fixed cell experiments were imaged using a 60 × 1.42 NA differential interference contrast Plan Achromat oil immersion lens (Olympus). Images were acquired using a CoolSNAP HQ2 camera (Photometrics/Roper Technologies) for a final magnification of 107 nm/pixel. For two-color distance measurements, a 1.6 magnification lens was inserted in the light path, providing a final magnification of 67 nm/pixel at the camera sensor.
Live cell imaging
For live cell imaging experiments, cells were seeded into custom-built glass-bottom 35 mm dishes and imaged in Leibovitz’s L-15 medium (Invitrogen) supplemented with 10% FBS, 7 mM HEPES, and 4.5 g/l glucose, pH 7.0. HeLa Kyoto cells were double thymidine synchronized and transfected with plasmids encoding Hec1-GFP and mCherry-tubulin as described above. After incubation for 9 h after the second thymidine washout, cells were imaged at 37°C on a Nikon Ti-E microscope equipped with a Piezo Z-control (Physik Instrumente), stage top incubation system (Okolab), and spinning disk confocal scanner unit (CSUX1; Yokogawa), using a 0.6 NA 40X objective and an iXon DU888 EM-CCD camera (Andor). Z-stacks were acquired taking seven planes at 1 µm steps using 488 and 594 nm lasers to excite GFP and mCherry, respectively. For each experiment, 20 fields were imaged at 5-min intervals for 12 h.
Protein expression and purification
Glutathione-S-transferase (GST)-NDC80Bonsai (Ciferri et al., 2008) was a generous gift from Andrea Musacchio (Max Planck Institute of Molecular Physiology, Dortmund, Germany). GST-NDC80Bronsai constructs were generated from GST-NDC80Bonsai (parent vector backbone = pGEX6P1-2RBS). Specifically, Nuf21-348/Spc24122-197, Hec11-506/Spc25118-224 fragments were obtained by PCR from parent vectors of each protein while creating 20–base pair overhangs for Gibson reaction for cloning back into BamH1/Age1-digested GST-Bonsai plasmid. Cloning of ∆80-NDC80Bronsai was carried out using the same fragments, except the Hec1 PCR used a forward primer with amino acid 81 immediately following the start codon. ML-NDC80Bronsai was generated by producing a PCR fragment of Hec11-461 with the mutant loop sequence from the cell expression vector used in this study and annealing it into the NDC80Bronsai vector digested with Sac1/Afl2. NDC80Bonsai and NDC80Bronsai constructs were expressed and purified using the following scheme: BL21-DE3 cells were transformed with NDC80 constructs, and cultures were grown to the appropriate OD600 before induction overnight (16 h) at 18°C with 400 µM isopropyl β-d-1-thiogalactopyranoside. All steps after induction were carried out at 4°C. The next morning, cells were pelleted by centrifugation and resuspended in lysis buffer (25 mM Tris, pH 7.6, 300 mM NaCl, 1 mM EDTA) supplemented with protease inhibitors (Pierce Protease Inhibitor tablets; Thermo Scientific) and 1 mM DTT (Gold Bio). Resuspended cells were lysed using a microfluidic chamber at 80 psi. The resulting lysed mixture was cleared of cell debris by centrifugation at 40,000 rpm for 45 min in a Beckman L8-70M ultracentrifuge using a TY70-TI rotor. Supernatant was applied to glutathione-agarose resin (Pierce resin; Thermo Scientific) (preequilibrated in lysis buffer), and the mixture was rocked gently for 1 h. Following binding, unbound protein was washed from the resin with lysis buffer, and resin-bound protein was eluted by GST-tag cleavage overnight with human rhinovirus 3C protease (HRV3C protease, expressed and purified in-house). Elutions were pooled, concentrated, and run on a GE Superdex 200 HiLoad 16/60 sizing column in lysis buffer supplemented with 5% glycerol and 1 mM DTT. Protein fractions were pooled and concentrated, and glycerol was added to 20% final volume before small aliquots were snap-frozen in liquid nitrogen and stored at –80°C.
Purification of recombinant human Ska complex (SkaC) was carried out as described previously (Abad et al., 2016). Briefly, BL21-Gold Escherichia coli cells were cotransformed with equal amounts of the individual Ska1, GST-Ska2, and Ska3 plasmids. Cells were grown to the appropriate OD600 before induction overnight (16 h) at 18°C with 400 µM isopropyl β-d-1-thiogalactopyranoside.. All steps after induction were carried out at 4°C. The next morning, cells were pelleted by centrifugation and resuspended in SkaC lysis buffer (20 mM Tris, pH 8.0, 500 mM NaCl) supplemented with protease inhibitors and 5 mM DTT. Resuspended cells were lysed by microfluidics as specified in NDC80 complex purifications. The resulting lysed mixture was cleared of cell debris by centrifugation at 40,000 rpm for 45 min as noted for NDC80 complex purifications. The supernatant from lysed cells was applied to glutathione-agarose resin (preequilibrated in SkaC lysis buffer) and rocked gently for 3 h. Following binding, unbound protein was washed away with lysis buffer, and resin was further washed with chaperone buffer (20 mM Tris, pH 8.0, 1 M NaCl, 50 mM KCl, 10 mM MgCl2, 2 mM ATP, 5 mM DTT) to remove associated protein chaperones. Resin-bound protein was then eluted using three sequential elutions for 1 h each in elution buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 50 mM glutathione, 5 mM DTT). Elutions were pooled and dialyzed overnight into column buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 5 mM DTT), and tags were simultaneously cleaved with tobacco etch virus (TEV) protease overnight while rocking. Cleaved SkaC was further purified by gel filtration on a Superose 6 Increase 10/300 in column buffer. Protein-containing fractions were collected and concentrated, and the protein was snap-frozen in liquid nitrogen and stored at –80°C.
TIRF microscopy
Immediately prior to the microtubule binding assays, the protein was flash-thawed and centrifuged at 90,000 × g to remove large aggregates. Supernatant was collected and concentration measured by Bradford assay. TIRF microscopy (TIRFM) binding assays were performed as described previously (Ecklund et al., 2017). Briefly, flow chambers were constructed by adhering plasma cleaned, silanized coverslips (22 × 30 mm) to glass slides with double-sided tape. Silanized coverslips were incubated with a rat anti-tubulin antibody (8 μg/ml, YL1/2; Accurate Chemical & Scientific Corporation) for 5 min, and then blocked with 1% Pluronic F-127 solution (Fisher Scientific) for 5 min. Taxol-stabilized, Alexa647-labeled microtubules (made by mixing fluorescently labeled and unlabeled porcine tubulin at a 1:12.5 ratio) diluted in BRB80 (80 mM PIPES, 1 mM EGTA, 1 mM MgCl2) supplemented with 20 µM taxol were flowed into the chamber and incubated for 5–10 min, and then unbound microtubules were washed out with one chamber volume of SN (“Ska-NDC80”) buffer (20 mM Tris, pH 7.0, 50 mM NaCl, 6 mg/ml bovine serum albumin [BSA], 4 mM DTT, 20 µM taxol). GFP-NDC80 complex (either alone or supplemented with 10 nM unlabeled Ska complex) diluted to the appropriate concentration in SN buffer was introduced to the chamber, and the binding reaction was incubated for 2 min. Two more additions of NDC80 complex (or NDC80 complex + Ska complex) were subsequently perfused into the chamber to allow the binding reaction to reach equilibrium. Two minutes after the third addition (after binding reaction had reached equilibrium as determined by time-lapse imaging), TIRF images were collected from 10 individual fields. For analysis in BRB80 and BRB20 (Supplemental Figure S4), all steps were performed as above, except protein (either GFP-NDC80 alone or supplemented with SkaC, or SkaC-GFP) was diluted into either BRB80 or BRB20 supplemented with 6 mg/ml BSA, 4 mM DTT, and 20 µM taxol. All TIRFM images were collected at room temperature using a 1.49 NA 100 X Plan Apo TIRF oil immersion lens on a Nikon Ti-E inverted microscope equipped with an iXon3 DU897 EM-CCD camera (Andor) for a final pixel size of 160 nm/pixel.
Data analysis
For all mutant expression studies, cells were initially scored for kinetochore GFP fluorescence intensity as “low,” “intermediate,” or “high.” Cells scored as having low Hec1-GFP fluorescence were discarded from analysis. Subsequently, cells were scored for Hec1-pS69 staining, and only cells with undetectable Hec1-pS69 were analyzed where appropriate (for mutants lacking intact Ser-69). For experiments in which kinetochore fluorescence intensity was quantitatively measured, only cells within a defined range of Hec1-GFP kinetochore fluorescence intensity were analyzed to ensure that expression levels were equal for all mutants (see Supplemental Figure S1C). Measurement of kinetochore fluorescence intensity in fixed cells was measured from non-deconvolved, non-compressed images using a custom program in MatLab (Mathworks) courtesy of X. Wan (University of North Carolina at Chapel Hill; Wan et al., 2009). For analysis of Ska3 levels at attached kinetochores (Figures 1, A–C, and 4, G and H, and Supplemental Figure S1), cells from mutants lacking Hec1 Ser-69 with greater than 25% Hec1-pS69 levels measured in WT-Hec1-GFP cells were discarded from analysis to reduce effects of endogenous, non-mutant kinetochore Hec1. Measurements of end-on attachment and interkinetochore distances in cold-treated cells were performed in SoftWoRx Explorer software. End-on attachment was analyzed by selecting random kinetochores in the kinetochore channel and then subsequently overlaying the tubulin channel and scoring whether spindle microtubules terminated at the preselected kinetochores (lateral attachments were not quantified). For analysis of end-on attachment in Ska-depleted cells, only kinetochores between the spindle poles were analyzed, as polar chromosomes remained unattached in all conditions (Figure 2E). Interkinetochore distances were analyzed by measuring the distance between Hec1-GFP signals from two kinetochores in a sister pair in the same z-plane. For chromosome alignment analysis, bipolar Hec1-GFP expressing cells post–nuclear envelope breakdown were scored as either aligned (metaphase plate with <5 chromosomes off the plate) or unaligned (no metaphase plate, or metaphase plate with five or more chromosomes off the plate). For analysis of multipolarity, Hec1-GFP expressing cells were stained with anti-tubulin antibodies and assessed for the number of spindle poles.
For two-color distance measurements, analyses were performed on kinetochore pairs in which their maximum fluorescence intensity centroids were not separated by more than one focal plane of 0.2 μm. Centroids of each test antibody signal were calculated using the custom program SpeckleTracker in MatLab (MathWorks), which was provided by X. Wan and T. Salmon (University of North Carolina, Chapel Hill). Distances were also calculated in the SpeckleTracker program. A detailed description of the protocol can be found in Wan et al. (2009).
For analysis of TIRFM microtubule binding assays, GFP-NDC80 complex–microtubule binding was quantitated using ImageJ software (National Institutes of Health). The NDC80 fluorescent signal was measured along the microtubule axis (as determined from the Alexa647-tubulin signal), and the “background” signal was measured using the same mask (created along the microtubule’s length) in a region immediately adjacent to the microtubule. Corrected signal intensity was measured by subtracting the background signal from the GFP signal on the microtubule. Raw GFP-NDC80 fluorescence intensity at each concentration (averaged across all three replicates) was plotted, and curves were fitted using a Specific binding model with a Hill fit in Prism (GraphPad). All statistical analyses were carried out using Prism (GraphPad).
Supplementary Material
Acknowledgments
We thank members of the DeLuca laboratory for helpful discussions, Ted Salmon and Xiaohu Wan for providing MatLab code to carry out kinetochore fluorescence intensity analysis and two-color fluorescence mapping experiments, and Gary Gorbsky (Oklahoma Medical Research Foundation) for the Ska1-GFP expression plasmid and for insightful discussions. This work was supported by National Institutes of Health grants R01GM088371 and R35GM130365 (to J.G.D.).
Abbreviations used:
- CH
calponin homology
- GFP
green fluorescent protein
- TIRF
total internal reflection fluorescence.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-05-0286) on May 13, 2020.
REFERENCES
- Abad MA, Medina B, Santamaria A, Zou J, Plasberg-Hill C, Madhumalar A, Jayachandran U, Redli PM, Rappsilber J, Nigg EA, Jeyaprakash AA. (2014). Structural basis for microtubule recognition by the human kinetochore Ska complex. Nature Commun 2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abad MA, Zou J, Medina-Pritchard B, Nigg EA, Rappsilber J, Santamaria A, Jeyaprakash AA. (2016). Ska3 Ensures timely mitotic progression by interacting directly with microtubules and Ska1 microtubule binding domain. Sci Rep 34042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Musinipally V, Matson D, Tooley J, Stukenberg PT, Nogales E. (2012). Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat Struct Mol Biol , 1161–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. (2010). The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature , 805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auckland P, Clarke NI, Royle SJ, McAinsh AD. (2017). Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment. J Cell Biol , 1623–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA. (2009). Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol , 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. (1999). The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev , 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Wheelock M, Funabiki H, Earnshaw WC. (2012). The Chromosomal Passenger Complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol , 789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. (2012). Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol , 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur DK, Gassmann R, Cook B, Oegema K, Desai A. (2013). Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science , 1239–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur DK, Prevo B, Hattersley N, Lewellyn L, Corbett KD, Oegema K, Desai A. (2017). Dephosphorylation of the Ndc80 tail stabilizes kinetochore-microtubule attachments via the Ska complex. Dev Cell , 424–437.e.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. (2006). The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell , 983–997. [DOI] [PubMed] [Google Scholar]
- Ciferri C, DeLuca JG, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A. (2005). Architecture of the human Ndc80-Hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem , 29088–29095. [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, DeLuca JG, De Wulf P, et al. (2008). Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell , 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Wan X, Hirel CB, Salmon ED. (2006). Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol , 1711–1718. [DOI] [PubMed] [Google Scholar]
- Conti D, Gul P, Asifa I, Martín-Durán JM, Pickersgill RW, Draviam VM. (2019). Kinetochores attached to microtubule-ends are stabilized by Astrin bound PP1 to ensure proper chromosome segregation. eLife, e49325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum JR, Potapova TA, Sivakumar S, Daniel JJ, Flynn JN, Rankin S, Gorbsky GJ. (2011). Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr Biol , 1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum JR, Wren JD, Daniel JJ, Sivakumar S, Mcavoy JN, Potapova TA, Gorbsky GJ. (2009). Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr Biol , 1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. (2006). Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell , 969–982. [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Musacchio A. (2012). Structural organization of the kinetochore-microtubule interface. Curr Opin Cell Biol , 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca KF, Lens SM, DeLuca JG. (2011). Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci , 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca KF, Meppelink A, Broad AJ, Mick JE, Peersen OB, Petkas S, Lens SMA, DeLuca JG. (2018). Aurora A kinase phosphorylates Hec1 to regulate metaphase kinetochore-microtubule dynamics. J Cell Biol , 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirel PB, Keyes BE, Chatterjee M, Remington CE, Burke DJ. (2012). A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces cerevisiae. Genetics , 753–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecklund KH, Morisaki T, Lammers LG, Marzo MG, Stasevich TJ, Markus SM. (2017). She1 affects dynein through direct interactions with the microtubule and the dynein microtubule-binding domain. Nat Commun , 2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad B, Kuijt TE, Kops GJ. (2015). Kinetochore-microtubule attachment is sufficient to satisfy the human spindle assembly checkpoint. Nat Commun , 8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad B, Vertesy A, Kuijt TEF, Sacristan C, van Oudenaarden A, Kops GJPL. (2019). Spindle checkpoint silencing at kinetochores with submaximal microtubule occupancy. J Cell Sci , 321589. [DOI] [PubMed] [Google Scholar]
- Gaiatnos TN, Santamaria A, Jeyaprakash AA, Wang G, Conti E, Nigg EA. (2009). Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J , 1375–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes GJ, DeLuca JG. (2009). Connecting with Ska, a key complex at the kinetochore-microtubule interface. EMBO J , 1442–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, DeLuca JG. (2008). Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol , 1778–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Sillje HHW, Nigg EA. (2006). Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J , 5504–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeson LA, Zelter A, Riffle M, Maccoss MJ, Asbury CL, Davis TN. (2018). Human Ska complex and Ndc80 complex interact to form a load-bearing assembly that strengthens kinetochore-microtubule attachments. Proc Natl Acad Sci USA , 2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis in’t Veld PJ, Volkov VA, Stender I, Musacchio A, Dogterom M. (2019). Molecular determinants of the Ska-Ndc80 interaction and their influence on microtubule tracking and force-coupling. eLife , e49539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janczyk P, Skorupka KA, Tooley JG, Matson DR, Kestner CA, West T, Pornillos O, Stukenberg PT. (2017). Mechanism of Ska recruitment by Ndc80 complexes to kinetochores. Dev Cell , 438–449.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash AA, Santamaria A, Jayachandran U, Chan YW, Benda C, Nigg EA, Conti E. (2012). Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol Cell , 274–286. [DOI] [PubMed] [Google Scholar]
- Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, Lechner J. (2009). Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signaling. EMBO J , 1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber S. (2011). Quantitative phosphoproteomics identifies Substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal , rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn V, Musacchio A. (2015). The Aurora B kinase in chromosome bi-orientation and spindle checkpoint signaling. Front Oncol , 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J, Dumont S. (2019). Mammalian kinetochores count attached microtubules in a sensitive and switch-like manner. J Cell Biol , 3583–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F, Hornung P, Westermann S. (2010). The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol , 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. (2013). Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J Cell Biol , 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AF, Udy DB, Dumont S. (2017). Hec1 tail phosphorylation differentially regulates mammalian kinetochore coupling to polymerizing and depolymerizing microtubules. Curr Bio , 1692–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Logarinho E. (2014). Mitotic spindle multipolarity without centrosome amplification. Nat Cell Biol , 386–394. [DOI] [PubMed] [Google Scholar]
- Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. (2007). Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol Cell Proteomics , 2200–2211 [DOI] [PubMed] [Google Scholar]
- Manning AL, Compton DA. (2007). Mechanisms of spindle-pole organization are influenced by kinetochore activity in mammalian cells. Curr Biol , 260–255. [DOI] [PubMed] [Google Scholar]
- Mattiuzzo M, Vargiu G, Totta P, Fiore M, Ciferri C, Musacchio A, Degrassi F. (2011). Abnormal kinetochore-generated pulling forces from expressing a N-terminally modified Hec1. PLoS One , e16307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Johnson ML, Stukenberg PT. (2008). Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80Hec1. Curr Biol , 1785–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen M, Sillje H, Sauer G, Nigg EA, Ko R. (2006). Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA , 5391–5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH. (2009). RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci , 2436–2445. [DOI] [PubMed] [Google Scholar]
- Roscioli E, Germanova TE, Smith CA, Embacher PA, Erent M, Thompson AI, Burroughs NJ, McAinsh AD. (2019). Ensemble-level organization of human kinetochores and evidence for distinct tension and attachment sensors. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Arthanari H, Boeszoermenyi A, Natalia M, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, et al. (2012). The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell , 968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Kiyomitsu T, Hori T, Backer CB, Fukagawa T, Cheeseman IM. (2010). Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. J Cell Biol , 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Daum JR, Tipton AR, Rankin S, Gorbsky GJ. (2014). The spindle and kinetochore-associated (Ska) complex enhances binding of the anaphase-promoting complex/cyclosome (APC/C) to chromosomes and promotes mitotic exit. Mol Biol Cell , 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Janczyk PŁ, Qu Q, Brautigam CA, Stukenberg PT, Yu H, Gorbsky GJ. (2016). The human SKA complex drives the metaphase-anaphase cell cycle transition by recruiting protein phosphatase 1 to kinetochores. eLife , e12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D, Gassman R, Oegema K, Desai A. (2011). Uncoordinated loss of chromatid cohesion is a common outcome of extended metaphase arrest. PLoS One , e22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin LJR, Guimaraes GJ, DeLuca JG. (2011). The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol Biol Cell , 759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Badger BL, Haase J, Ohashi T, Erickson HP, Salmon ED, Bloom K. (2016). How the kinetochore couples microtubule force and centromere stretch to move chromosomes. Nat Cell Biol , 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Long SK, Salmon ED. (2018). An optimized method for 3D fluorescence co-localization applied to human kinetochore protein architecture. eLife , e32418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJR, Nasmyth K. (2002). Evidence that the Ipl1-Sli5 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell , 317–329. [DOI] [PubMed] [Google Scholar]
- Tauchman EC, Boehm FJ, DeLuca JG. (2015). Stable kinetochore-microtubule attachment is sufficient to silence the spindle assembly checkpoint in human cells. Nat Commun , 10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis M, Slabicki M, Junqueira M, Paszkowski M, Sontheimer J, Kittler R, Heninger A, Glatter T, Kruusmaa K, Poser I, et al. (2009). Comparative profiling identifies C13orf as a component of the Ska complex required for mammalian cell division. EMBO J , 1453–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. (2010). Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol , 713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley JG, Miller SA, Stukenberg PT, Drubin DG. (2011). The Ndc80 complex uses a tripartite attachment point to couple microtubule depolymerization to chromosome movement. Mol Biol Cell , 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit NT, Gestaut DR, Tien JF, Vollmar BS, Gonen T, Asbury CL. (2012). The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc Natl Acad Sci USA , 16113–16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Chandrasekaran S, Sundin LJR, Reidy KT, Wan X, Chasse DAD, Nevis KR, DeLuca JG, Salmon ED, Cook JG. (2012). Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat Cell Biol , 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Salmon ED. (2012). The KMN protein network—chief conductors of the kinetochore orchestra. J Cell Sci , 5927–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. (2009). Protein architecture of the human kinetochore microtubule attachment site. Cell , 672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. (2007). The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol , 54–59. [DOI] [PubMed] [Google Scholar]
- Welburn JPI, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, III, Cheeseman IM. (2009). The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell , 374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. (2008). Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol , 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimbish RT, DeLuca JG. (2020). Hec1/Ndc80 tail domain function at the kinetochore-microtubule interface. Front Cell Dev Biol , 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Wei R, Cheng E, Ngo B, Lee WH. (2009). Hec1 contributes to mitotic centrosomal microtubule growth for proper spindle assembly through interaction with Hice1. Mol Biol Cell , 4686–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo TY, Choi J, Conway W, Yu C, Pappu RV, Needleman DJ. (2018). Measuring NDC80 binding reveals the molecular basis of tension-dependent kinetochore-microtubule attachments. Elife , 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev AB, Mick JE, Maslennikov E, Nikashin B, DeLuca JG, Grishchuk EL. (2015). Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol Biol Cell , 1829–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev AV, Sundin LJR, DeLuca KF, Grishchuk EL, DeLuca JG. (2014). Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J Cell Biol , 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG. (1995). Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol , 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Kelstrup CD, Hu X, Hansen MJK, Singleton MR, Olsen JV, Nilsson J. (2012). The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci , 3243–3253. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Chen Y, Yang L, Liu H. (2018). Multitasking Ska in chromosome segregation: its distinct pools might specify various functions. BioEssays , 1700176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Sivakumar S, Chen Y, Gao H, Yang L, Yuan Z, Yu H, Liu H. (2017). Ska3 phosphorylated by Cdk1 binds Ndc80 and recruits Ska to kinetochores to promote mitotic progression. Curr Biol , 1477–1484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.