Abstract
Mikania micrantha is a noxious invasive plant causing enormous economic losses and ecological damage. Soil microbiome plays an important role in the invasion process of M. micrantha, while little is known about its rhizosphere microbiome composition and function. In this study, we identified the distinct rhizosphere microbial communities of M. micrantha, by comparing them with those of two coexisting native plants (Polygonum chinense and Paederia scandens) and the bulk soils, using metagenomics data from field sampling and pot experiment. As a result, the enrichment of phosphorus-solubilizing bacteria Pseudomonas and Enterobacter was consistent with the increased soil available phosphorus in M. micrantha rhizosphere. Furthermore, the pathogens of Fusarium oxysporum and Ralstonia solanacearum and pathogenic genes of type III secretion system (T3SS) were observed to be less abundant in M. micrantha rhizosphere, which might be attributed to the enrichment of biocontrol bacteria Catenulispora, Pseudomonas, and Candidatus Entotheonella and polyketide synthase (PKS) genes involved in synthesizing antibiotics and polyketides to inhibit pathogens. These findings collectively suggested that the enrichment of microbes involved in nutrient acquisition and pathogen suppression in the rhizosphere of M. micrantha largely enhances its adaptation and invasion to various environments.
Keywords: Rizosphere bacteria, Mikania micrantha, beneficial microbes, nutrition, pathogen
Introduction
The rhizosphere is the interface where the complex interactions among soil, microbes, and the host plant are maintained (Philippot et al., 2013). Plants selectively harbor specific microbes through root exudates that contain carbohydrates, amino acids, and organic acid ions, which act as carbon source and nutrients for microbial growth (Reinhold-Hurek et al., 2015). Rhizosphere microbes play pivotal roles in plant growth, nutrient uptakes, and disease suppression (Bulgarelli et al., 2015; Edwards et al., 2015).
Invasive alien species (IAS) could reduce the richness and abundance of native species in the invaded regions, or even dramatically change the local ecological system (Pyšek and Richardson, 2008). Compared with native plants, invasive plants generally performed higher value of inherent traits on physiology, leaf-area allocation, shoot allocation, and growth rate (Van Kleunen et al., 2010) and also exhibit higher abilities to capture and utilize resources at both above- and below-ground processes, such as photosynthesis and nutrition uptake (Van Der Heijden et al., 2008). The soil microbes play important roles in the establishment of invasive plants and might also act as drivers of plant invasions (Dawson and Schrama, 2016). Previous studies showed that invasive plants can recruit different soil microbes to promote their growth over native plants (Reinhart et al., 2003). The indigenous soil microbial communities are altered due to the exotic invading plants (Kourtev et al., 2002). For example, the Alnus trabeculosa increased the soil bacterial diversity in the invaded regions (Xueping et al., 2016). Another invasive plant Centaurea maculosa enhanced its competitive ability through enriching mycorrhizal fungi that changes soil nutrient availability (Marler et al., 1999). In addition, other studies also showed that many invasive plants have fewer pathogens in rhizosphere than native plants do, escaping from pathogenic agents in soil (Mitchell and Power, 2003). C. maculosa could reduce local soil pathogens in invaded regions, therefore investing less in unused defense and more into growth to increase competitiveness against natives (Callaway et al., 2004). An invasive plant may influence soil nutrient content through the soil microbial communities (Piper et al., 2015; Zhao et al., 2019). For example, the invasive tree staghorn sumac changed the structure of soil N2-fixing bacterial communities to enhance soil N availability (Wu et al., 2019). Solidago gigantea enhances phosphorus (P) turnover rates in soil (Chapuis-Lardy et al., 2006), and C. maculosa increases available P in soil (Thorpe et al., 2006). Invasive plants increased the availability of vital nutrients, thus gaining a competitive advantage, which might be an important contributor to invasion success (Castro-Díez et al., 2014).
Mikania micrantha (Asteraceae family), an extremely fast-growing vine, is one of the top 100 worst IAS in the world (Lowe et al., 2000), causing severe substantial damages to natural ecosystems (Day et al., 2016) and economic losses (Macanawai et al., 2012). Several mechanisms have been proposed to explain the success of M. micrantha invasion, such as rapid growth caused by high regeneration capacity of each vine node (Li X. et al., 2013) and extraordinary biological characteristics including high seed production and germination (Hu and But, 1994), the strong allelopathic effects on other native plant and soil microbes (Chen et al., 2009), and high nutrient (NPK) turnover rates in soil (Sun et al., 2019; Liu et al., 2020). Recently, we have published the genome of M. micrantha, as well as its rhizosphere metagenome, and also found out that the rhizosphere microbes of M. micrantha could increase the bioavailable nitrogen content to speed up the nitrogen cycle (Liu et al., 2020), which might contribute to its rapid growth as well as invasion. Enhancing the availability of soil P is also one of the major factors for the success of plant invasion. In recent studies on P acquisition of M. micrantha, it was shown that the contents of soil available P and plant tissues of M. micrantha were significantly higher than that of native plants. However, very few studies have explained the component and mechanism of P-solubilizing bacteria. We hypothesized that the enrichment of P-solubilizing microorganisms will contribute to the available P in M. micrantha rhizosphere. Except for the nutrient acquisition mechanism of plant invasion, the well-known enemy release mechanism that escapes from its natural enemies in its native ranges was also confirmed in other invasive plants. Some invasive plants were not only associated with higher ability of nutrients uptake but also harbored few known pathogens that were more abundant in the rhizosphere of native plants or accumulated pathogens in the soil that are harmful to natives. The research on the invasion mechanism of M. micrantha mainly focuses on inherent superiority, allelopathy, and nutrient acquisition, and there is a paucity of research on the influence of pathogenic microorganisms in the M. micrantha rhizosphere. We hypothesized that few known pathogens were harbored in M. micrantha rhizosphere because of the allelopathy of its leaves and roots. In this study, using these metagenomic data, we investigated the phosphorus solubilizing bacteria and pathogens in the rhizosphere of M. micrantha, to better understand the role of the rhizosphere microbiome in M. micrantha invasion.
Materials and Methods
Experimental Design and Sampling Collection
In order to test the contribution of rhizosphere bacteria to M. micrantha invasion, we conducted a pot experiment with M. micrantha and its two neighboring native species, namely, Polygonum chinense and Paederia scandens. These two plants are chosen as the control species because based on the investigation from the field sample, not only are these frequently and stably present in the invasive community of M. micrantha, but also the reproduction strategies of these two plants are very similar to those of M. micrantha (Sun et al., 2019). The seeds of three plants were germinated and grew to about 10 cm for transplanting. Seedlings of three plants, respectively, planted in the pot (20 cm diameter) filled with natural field soil were collected from the non-invasive area near the invader M. micrantha monoculture, which is located in the dry riverbed of Liuxi River, Guangzhou City, Guangdong Province, China. Four treatments (three plants plus a blank control) were replicated six times (two plants per pot with 7 kg fresh soil) and put in a greenhouse.
Three months later, we randomly selected five replicates of each treatment and the rhizosphere soil of three plants and control soil were collected. Plants were removed carefully and shaken lightly; then, the soil remaining attached to the root surface was collected with sterile water. The separated soil solution was centrifuged at 8000 r/min for 10 min to collect soil samples. The collected soils were stored at −80°C until use for microbial community analysis. M. micrantha is an ecologically harmful weed in the natural environment. We chose the natural field of M. micrantha monoculture with the dominant two coexisting native plants (P. chinense and P. scandens) in the dry riverbed of Liuxi River in Guangzhou City. We separated five (5 m by 5 m) sampling plots by more than 200 m and used the same method of pot experiment to collect 15 rhizosphere samples of three plants and five control samples, which is in the uninvaded area near M. micrantha monoculture by more than 500 m, for a total of 20 samples.
DNA Extraction and Sequencing
A combination of bacteria cell lysis steps was applied before DNA extraction. The soil microbial cells were subjected to six freeze–thaw cycles (alternating vortex for 5 min, then liquid nitrogen for 5 min, and incubation at 65°C for 5 min). Next, DNA was extracted from all samples using the PowerSoil DNA isolation kit following the manufacturer’s protocol (MO BIO Laboratories, QIAGEN Inc., United States). The DNA quality and quantity were checked by the Nanodrop and Qubit device, and the DNA quantity of each sample was at least 1 μg. Then, DNA fragments (200–400 bp) were processed by ultrasonic instrument (Thermo Fisher Scientific, Covaris S220). The library was constructed using TruSeq DNA PCR-Free Library Prep Kit as per standard protocol (Illumina, United States) and then sequencing was performed on Illumina HiSeq 2500 with each sample having about 10 Gb sequencing data.
Metagenomic Analyses
The raw reads were cleaned by removing adaptor sequences, low-quality sequences, host sequences, and unpaired reads by in-house software clean_adapter, clean_lowqual, and filter_unpaired_reads.pl1, resulting in a clean and high-quality reads data with average error rate < 0.001. Then, the clean reads from each sample and pooled for four groups (M. micrantha, P. chinense, P. scandens, and control) were assembled by Megahit (v1.1.3). After filtering the contig length less than 500 bp, gene prediction was performed using MetaProdigal (v2.6.3), and then we filtered out the gene models with cds length less than 102 bp. The protein models of each sample and each group were also performed using MetaProdigal (v2.6.3). The non-redundant gene catalog was obtained using the genes predicted from each sample and each group by cd-hit-est (v4.6.6) with the criteria of identity > 95%, and overlap > 90% (parameter “−c 0.95 −n 10 −G 0 −aS 0.9”). The non-redundant protein catalog was obtained from the combination of protein files of each sample and each group by in-house software fishInWinter.pl2.
To generate the taxonomic information, the non-redundant protein sequences were aligned against the NCBI-NR database using DIAMOND (v0.8.28.90) software with the parameter “blastp –evalue 10 –max-target-seqs 250” (Buchfink et al., 2015). CARMA3 software (parameter “carma –classify-blast –type p –database p”) was used to assign the taxonomic annotation of the unigenes (Gerlach and Stoye, 2011). Thus, we obtained the non-redundant genes and their corresponding species classification. To obtain functional information for the gene set, the non-redundant protein sequences were searched (E value < 1e-5) against the KEGG protein database (release 79) using DIAMOND software (Kanehisa et al., 2004). To calculate the relative gene abundance, the clean reads from each sample were aligned against the non-redundant gene catalog by BWA-MEM (alignment length ≥ 50 bp and identity > 95%) (Li and Durbin, 2009). The alignments were parsed to produce the reads count abundance (Huang et al., 2018). Based on the taxonomic assignments using CARMA3, the relative abundance of each phylum, genus, species, and KO was calculated by summing the abundances of corresponding genes belonging to each category per sample. Similarly, the relative abundance profile of genes was also summarized into KEGG functional profiles for the functional analysis.
Functional Bacteria and Genes Collection
The bacteria and genes involved in soil microbial P-solubilizing and mineralization, pathogen, and defense were searched based on previous publications and are shown in Supplementary Tables S1–S6 (Weller et al., 2002; Garbeva et al., 2004; Beth Mudgett, 2005; Raaijmakers and Mazzola, 2012; Sharma et al., 2013; Raj et al., 2014; Alori et al., 2017; Han et al., 2018; Dai et al., 2019). The names, KOs, and functions of the genes associated with P solubilizing and mineralization, type III secretion/effector systems, and polyketide synthase (PKS) are shown in Supplementary Tables S2, S4, S6, respectively.
Microbial Composition Analysis
At the gene level, Shannon index was used to analyze microbial alpha diversity using the non-redundant genes of individual samples. The overall differences in the bacterial community structures were calculated by non-metric multidimensional scaling (NMDS) using non-redundant genes of individual samples based on Bray–Curtis dissimilarity values and implemented in in R package “Phyloseq.”
Statistical Analysis
Based on the relative abundance profiles at the phyla or genera level, the significantly differential abundances in the control soil and rhizospheres of M. micrantha, P. chinense, and P. scandens were determined using Kruskal–Wallis test with Dunn’s multiple comparison (BH methods for multiple tests adjustment). The relative abundance of microbial species and functional genes involved in P solubilization, pathogens, and defense in the control soil and rhizospheres of three plants is compared using Kruskal–Wallis test with Dunn’s multiple comparison (BH methods for multiple tests adjustment).
Results
Microbial Structure of the Rhizosphere Microbiome
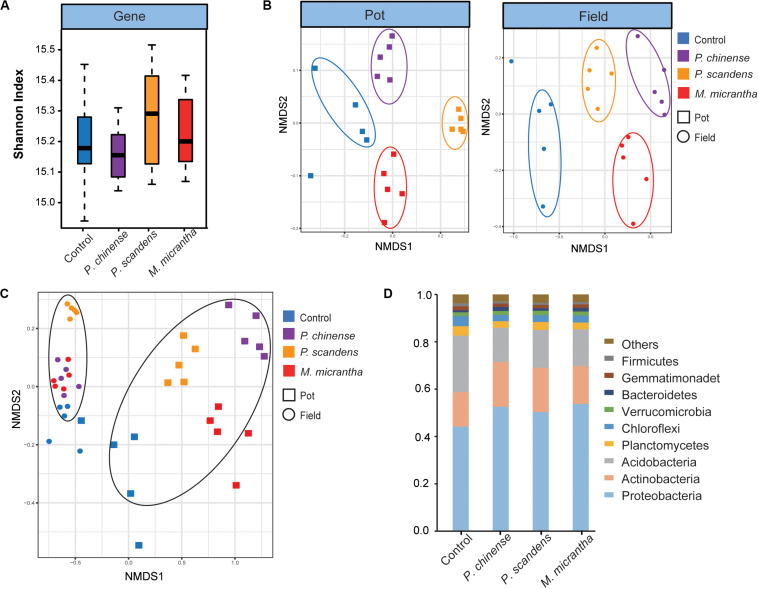
Using the metagenomic data and non-redundant gene set of M. micrantha genome project, deposited in NCBI (SRR8936416–SRR8936475) and AGIS ftp-site3, we investigated the microbial structure of the rhizosphere of M. micrantha and two native plants (P. chinense and P. scandens). The microbial alpha diversity (Shannon index) at the gene level showed no significant difference between the control soil (bulk soil) and rhizospheres (P > 0.05) (Figure 1A). However, at the gene level, the NMDS analysis revealed the distinct microbial community differences among the rhizospheres of M. micrantha, two native plants, and control soil at both pot experiment and invaded site (Figure 1B). Moreover, the NMDS plots showed that there was a clear separation between the pot experiment and field invaded site, indicating that rhizosphere microbial community was largely influenced by environmental conditions (Figure 1C). The dominant prokaryotic phyla found in the control and rhizosphere community included Proteobacteria, Actinobacteria, Acidobacteria, Planctomycetes, and Chloroflexi (Figure 1D), which was consistent with previous studies (Lu-Irving et al., 2019). The community differences between the control soil and rhizospheres of M. micrantha and native plants were also explored. Proteobacteria and Actinobacteria occupied higher percentages than in the control soil, whereas Acidobacteria has lower percentages (P < 0.05, Dunn test) in the rhizospheres (Figure 1D). This suggests that some bacteria from bulk soil are selected to inhabit in the rhizospheres.
FIGURE 1.
Microbial structure in the rhizosphere of Mikania micrantha (M. micrantha), Polygonum chinense (P. chinense), and Paederia scandens (P. scandens), and control (bulk) soil. (A) Comparison of the microbial diversity at the gene level. (B) The non-metric multidimensional scaling (NMDS) plot of microbial communities at both pot experiment and invaded site. The analysis was based on Bray–Curtis dissimilarities at the gene level. (C) The NMDS plot of microbial communities in all samples, based on Bray–Curtis dissimilarities at the gene level. (D) Relative abundances at the phylum level.
Distinctive Enrichment of Plant Microbes
The microbial compositions of the rhizosphere of M. micrantha and the two native plants were analyzed at the genus level, at both pot experiment and invaded site. From the metagenomic data, genes could be classified to the genus level by CARMA3 software. The relative abundance of genus in each sample was calculated according to reads count at the genus level. In total, the top 69 genera (relative abundance > 0.01%) accounted for 94.8% of the total relative abundance of classified genera, and 45 of them were enriched (P < 0.05, Dunn test) in rhizospheres compared to the bulk soil, most of which belong to Proteobacteria and Actinobacteria (Figure 2). Moreover, 30 genera were all enriched in rhizospheres of three plants, including Bradyrhizobium, Mesorhizobium, Rhizobium, Burkholderia, Paraburkholderia, Methylobacterium, Novosphingobium, Pseudomonas, Enterobacter, Bacillus, Nocardioides, and Streptomyces, many species of which were known as plant beneficial microbes that can facilitate nutrition acquisition, improve resistance to abiotic stress, and control phytopathogens (Figure 2) (Ahemad and Kibret, 2014; Cordovez et al., 2018; Vives-Peris et al., 2018). The enrichment of these plant microbes might facilitate plant assembling beneficial endosphere bacteria from the rhizosphere soil.
FIGURE 2.
Comparison of average relative abundance at the genus level in rhizospheres of M. micrantha and two native plants. The relative abundance of each genus was colored according to the row z score ((value - row mean)/row standard deviation). The comparisons of microbes in plant rhizospheres and control soil were compared by the Kruskal–Wallis test with Dunn’s multiple comparison test (*P < 0.05 and **P < 0.01).
Previous studies have shown that the plant species and varieties could influence the composition of their rhizosphere (Philippot et al., 2013; Zhang et al., 2019). In our study, the microbe enrichment in rhizospheres of M. micrantha and native plants was also distinctive. Enterobacter, Pseudomonas, Cupriavidus, and Phenylobacterium relatively occupied higher percentages in M. micrantha rhizosphere compared to P. chinense and P. scandens rhizospheres (Figure 2). Many species belonging to Enterobacter and Pseudomonas (Meena et al., 2017; Zheng et al., 2019) are well known plant-beneficial microbes, and Cupriavidus and Phenylobacterium species were reported to participate in the mineralization of soil organic P and degrade organic material (De La Cruz-Barrón et al., 2017). In comparison, P. chinense rhizosphere was enriched with Variovorax, Bosea, and Acinetobacter, and some species of which could inhibit pathogens and supply N for plant growth (Rilling et al., 2018; Bruisson et al., 2019), and P. scandens rhizosphere was enriched with Anaeromyxobacter, Frankia, Streptacidiphilus, and Amycolatopsis (Figure 2), with nitrogen-fixing (Chaia et al., 2010) and antimicrobial activity (Buszewski et al., 2018). In summary, although many bacteria are shared among three plant species, each plant still recruits distinctive microbes in rhizosphere, possibly due to their different root exudates.
Enrichment of Pseudomonas and Enterobacter to Enhanced Phosphorus Solubilization
Phosphorus (P), is an essential element for plant growth and development (Sharma et al., 2013), playing important roles in many metabolic processes of plant, including photosynthesis, signal transduction, energy transfer, respiration, macromolecular biosynthesis (Khan et al., 2010), and nitrogen fixation (Kouas et al., 2005). Microorganisms are major members of the soil P cycle, improving available P to plants (Khan et al., 2009). We have previously reported that the available P content in M. micrantha rhizosphere was significantly higher than that in the rhizosphere of two native plants (Liu et al., 2020).
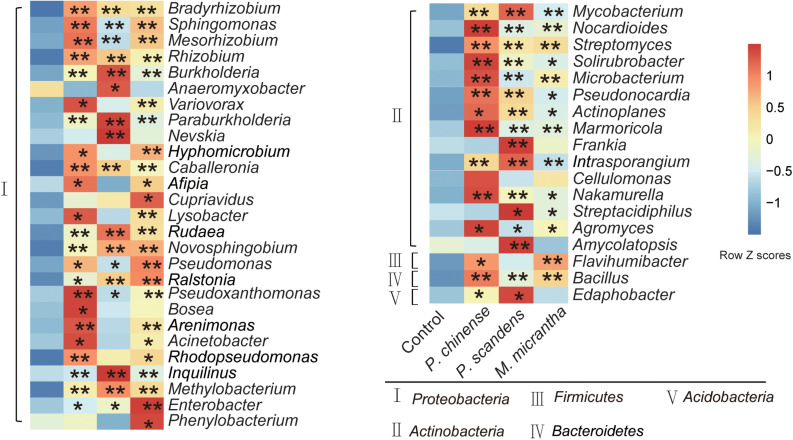
Phosphorus solubilizing microorganisms (PSMs), such as Pseudomonas, Bacillus, Enterobacter, and Burkholderia (Babalola and Glick, 2012), can increase soil available P via solubilization and mineralization of unavailable P in organic matter and minerals. These PSMs were enriched in the rhizospheres of M. micrantha, P. chinense, and P. scandens (Figure 3A); however, the relative abundance of PSMs is different. Enterobacter was most highly enriched in M. micrantha rhizosphere, with its average relative abundance 7-fold and 100-fold higher than that in P. chinense and P. scandens rhizosphere, respectively (Figure 3A). Similarly, the average relative abundance of Pseudomonas was also 1.5-fold and 13-fold higher than those in P. chinense and P. scandens rhizosphere, respectively (Figure 3A). In the invaded field site, the plant-growth promotion bacteria Pseudomonas putida (Możejko-Ciesielska and Serafim, 2019) and Enterobacter asburiae (Teng et al., 2019) were more abundant in M. micrantha rhizosphere (Figure 3B). On the other hand, Bacillus and Burkholderia were more enriched in rhizosphere of P. chinense and P. scandens (1.2- and 2.5-fold that in M. micrantha rhizosphere), which might also contribute to the solubilization of soil unavailable phosphorus. Taken together, the recruitment of these PSM would help to increase the available P content in M. micrantha rhizosphere.
FIGURE 3.
Enhanced soil-borne available P in the rhizosphere of M. micrantha and two native plants. (A) The relative abundance of phosphate bacteria. (B) The relative abundance of Enterobacter asburiae and Pseudomonas putida. On each boxplot, the central mark indicates the median, the bottom and top edges of the box indicate the interquartile range (IQR), and the whiskers represent the maximum and minimum data points. (C) The relative abundance of genes coding for P solubilization and P mineralization. PQQGDH, quinoprotein glucose dehydrogenase; InP, inorganic pyrophosphatase; CPLS, C-P lyase subunit; ExP, exopolyphosphatase; GDP, glycerophosphoryl diester phosphodiesterase; ALP, alkaline phosphatase; PhH, phosphonoacetate hydrolase; 2APT, 2-aminoethylphosphonate-pyruvate transaminase; PeH, phosphonoacetaldehyde hydrolase; PhT, phosphotriesterase; ACP, acid phosphatase. The C-P lyase subunit was calculated as the total abundances of gene phnF, phnG, phnH, phnI, phnJ, phnK, phnL, phnM, phnN, phnO, and phnP. Error bars indicate average value ± SEM of indicated replicates. The pairwise comparisons of rhizosphere in each plant and control soil were used by the Kruskal–Wallis test with Dunn’s multiple comparison test (*P < 0.05 and **P < 0.01).
Next, the genes generally contained in PSM were analyzed, including those coding for P mineralizing and solubilizing enzymes (Dai et al., 2019). The genes coding for organic P mineralization, such as C-P lyase, phosphonoacetate hydrolase, and phytase, as well as the genes coding for inorganic pyrophosphatase responsible for the inorganic P solubilization, were all enriched in rhizosphere of M. micrantha and two native plants (P < 0.05) (Figure 3C). The genes involved in alkaline phosphatase phoD were more abundant in rhizosphere of M. micrantha (P = 0.045) and P. chinense (P = 0.07), whereas phoA showed no significant difference (P > 0.05) (Figure 3C). The relative abundance of other genes showed no significant difference (P > 0.05) (Figure 3C). These results indicated that the rhizosphere microbes in M. micrantha and P. chinense may contribute to available P through the similar P mineralization mechanism in terms of alkaline phosphatase.
Fewer Pathogens in M. micrantha Rhizosphere Microbiota
The plant-associated microbiome, as the second genome of the plant, has great influence on plant growth and health (Berendsen et al., 2012). To suppress the pathogen attack, plants could be able to recruit protective microorganisms in the rhizosphere, as the complement of the plant innate immune system (Mendes et al., 2014).
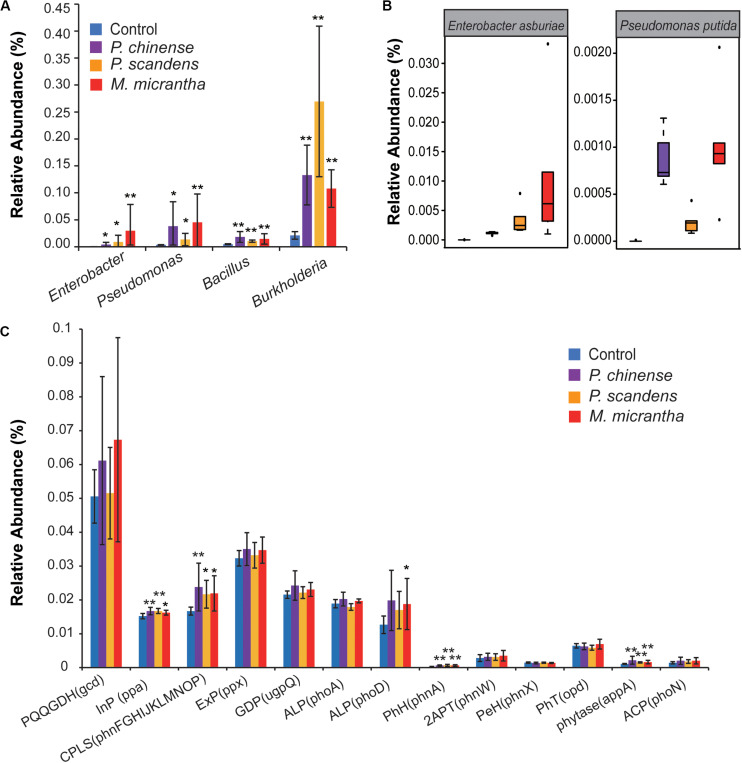
The aggressive soil-borne pathogens were analyzed (Supplementary Table S3). Although many pathogens could not be detected in our data, we found that the pathogens of Fusarium oxysporum (Srinivas et al., 2019) and Ralstonia solanacearum (Genin and Denny, 2012) were enriched in P. chinense (sevenfold and twofold) and P. scandens (sixfold and fourfold) rhizosphere compared to M. micrantha rhizosphere (Figure 4A). Besides, the genes involved in the host–pathogen interactions (Supplementary Table S4) [type III secretion system (T3SS)] were more abundant in the rhizosphere of P. scandens (P = 0.04) than in M. micrantha rhizosphere (Figure 4B). Plants could inhibit pathogen attack by secreting antimicrobials or recruiting the biocontrol microbes that have the relevant antimicrobial gene cluster (Berendsen et al., 2012). The biocontrol microbes (Supplementary Table S5), such as Pseudomonas, Catenulispora, and Candidatus Entotheonella, were more abundant in rhizosphere of M. micrantha than that in two native plants (Figures 3A, 4C). It is known that some species belonging to Catenulispora, Pseudomonas, and Candidatus Entotheonella could suppress pathogen by producing antibiotics and polyketides (Zettler et al., 2014; Kurnia et al., 2017). In our results, type II PKS genes (Supplementary Table S6) that were involved in synthesizing aromatic polyketides that could control plant disease (Han et al., 2018) were also enriched in M. micrantha rhizosphere (P = 0.002) (Figure 4D), whereas type III PKS genes were not different among three plants. These results indicated that the biocontrol bacteria might contribute to the less pathogens by antimicrobial aromatic polyketides in M. micrantha rhizosphere.
FIGURE 4.
Fewer pathogens and more biocontrol bacteria in M. micrantha rhizosphere. (A) The relative abundance of pathogens of Fusarium oxysporum and Ralstonia solanacearum. (B) The relative abundance of pathogenic genes of type III secretion systems (T3SS). (C) The relative abundance of biocontrol bacteria of Catenulispora and Candidatus Entotheonella. (D) The relative abundance of genes coding for type II polyketide synthase (PKS). On each boxplot, the central line indicates the median, the bottom and top edges of the box indicate the interquartile range (IQR), and the whiskers represent the maximum and minimum data points.
Discussion
The success of plant invasion depends on enemy release, enhanced nutrient acquisition, and adaptations to the physical environment (Dawkins and Esiobu, 2016). Recently, increased attention has been paid to the interactions between soil microbes and plant invasions (Dawkins and Esiobu, 2018). In this study, we investigated the role of soil microbes in plant invasions by comparing the taxonomic and functional difference of rhizosphere community between the invasive plant M. micrantha and two native plants (P. chinense and P. scandens) at invaded field site and pot experiment. Since the pot experiment lasted only 3 months, and each plant grew independently without competition, obvious microbial differences between pot experiment and invaded site were observed (Figure 1C). However, many plant-associated microbes were enriched in rhizospheres both in the pot experiment and the invaded site, and these genera were generally higher in the invaded field than those in the pot experiment, indicating their important roles in the natural environment. The interactions between an invasive plant and associated soil communities changed across the invaded range (Nunes et al., 2019). In our study, we found that there is a difference of M. micrantha rhizosphere between the pot experiment and the field site. As a plant killer, more field samples of the rhizosphere microbes of M. micrantha across latitudinal gradients in its invaded range should be analyzed to understand the interactions between its performance and soil microbes. This could provide an important basis for controlling its spread. By comparing the microbes in the rhizospheres and in the control soil, we found that the relative abundance of Proteobacteria and Actinobacteria was higher in the rhizosphere than in the control soil, whereas Acidobacteria was more abundant in control soil (Figure 1C). The distinct enrichment may be attributed to the abundant nutrients in rhizosphere, which promote the copiotrophic microorganisms (Ling et al., 2017) and the inhibited growth of oligotrophic microorganisms (Fierer et al., 2007).
The competition of invasive species with native species depends largely on the abilities of accession in resources (Seabloom et al., 2003). P is an essential macronutrient for plant growth and development (Lidbury et al., 2016), and microorganisms play an important role in soil P cycling and in regulating P availability (Dai et al., 2019). In this study, we found that Enterobacter and Pseudomonas might contribute to the increased soil available P content, and helped M. micrantha to outcompete native species and ultimately facilitate plant invasion (Figure 3A). Although the gene of gcd was not significantly different when the field and potted samples were analyzed together (Figure 3C), it was found significantly enriched in rhizosphere of M. micrantha (P = 0.0008) and P. chinense (P = 0.02) (Supplementary Figure S1) in the invaded site. The relative abundance of the gcd gene in M. micrantha rhizosphere was 1.2-fold of that in P. chinense rhizosphere and 1.5-fold of that in P. scandens rhizosphere in the field. Besides, even genes coding for alkaline phosphatase were at a similar level in the rhizosphere of M. micrantha and P. chinense, and the highly elongated, deep, and extensive root system of M. micrantha may still promote the efficient uptake of the released available P in soil.
Invasive plants may benefit from introduction to new regions where they can escape pathogens on the native ranges (Lu-Irving et al., 2017). Recently, Ramirez et al. (2019) found that the range-expanding plants harbored fewer pathogens compared to native species in the new range, through the analysis of the microbiome of European continental range-expanding plant species along a latitudinal gradient. This result was consistent with our study, which revealed that the pathogens and pathogenic genes, including the soil-borne pathogen F. oxysporum and R. solanacearum, as well as T3SS, were depleted in M. micrantha rhizosphere compared to the native plants (Figures 4A,B). Correspondingly, many biocontrol bacteria such as Catenulispora, Pseudomonas, and Candidatus Entotheonella, which release antibiotics and polyketides to inhibit pathogens (Kurnia et al., 2017; Mori et al., 2018), were enriched in M. micrantha rhizosphere. In addition, Mikania sesquiterpene lactones (STLs) have allelopathic effects on native plants and antibacterial activities (Li Y. et al., 2013), which may also contribute to the fewer pathogens in M. micrantha rhizosphere. In summary, the fewer pathogens and more protective microorganisms that inhabit the M. micrantha rhizosphere potentially benefit root growth and nutrient uptake, thus possibly enabling the successful invasion. However, there is a lack of difference in the soil microbes in M. micrantha between the origin and invaded one. Evidences for the resource availability and pathogen release in soil of invasive plants would require combined tests in the native and invaded ranges. Hence, in order to comprehensively understand the role of soil microorganisms in M. micrantha invasion, the metagenome of M. micrantha rhizosphere in the native range and the differences to their introduced range would need to be studied in the future. Although we showed the differences of microbial community and functional genes among the rhizosphere of three plants, the observed changes would require further experimental study.
Conclusion
Mikania micrantha rhizosphere has a distinct bacteria community structure that is clearly separated from the native plants and the bulk soil. Although some common microbes are observed in the rhizosphere of both M. micrantha and two native plants, M. micrantha rhizosphere specifically recruited Cupriavidus, Enterobacter, Pseudomonas, and Phenylobacterium, which played important roles in resource acquisition, plant hormone regulation, and pathogen suppression. On the other hand, the rhizosphere of native plants P. chinense and P. scandens recruited some other distinctive plant microbes. According to our analysis, the previously found higher soil available P content in M. micrantha rhizosphere was possibly contributed by the enrichment of P-solubilizing bacteria Enterobacter and Pseudomonas. Moreover, pathogens including F. oxysporum and R. solanacearum and pathogenic genes of T3SS were less abundant in M. micrantha rhizosphere compared to the two native plants. In contrast, the biocontrol bacteria such as Catenulispora, Pseudomonas, and Candidatus Entotheonella, as well as the PKS genes were enriched in M. micrantha rhizosphere to develop antibacterial activities. Taken together, these findings deepen our understanding of the microbial composition and function in M. micrantha rhizosphere, as well as the two native plants, and thus provide useful information that would help develop efficient technologies to control the invasion of M. micrantha.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found in the NCBI under the accession numbers SRR8936416–SRR8936475.
Author Contributions
BL, WF, and WQ conceived the study. LY, HeW, YZ, FJ, and SW collected the samples and analyzed the data. YR, CL, HL, WQ, HaW, and FW provided suggestions and helped in the checking. YZ, SW, BL, WQ, and WF helped to revise the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the Shenzhen Science and Technology Program (JCYJ20170303154245825), the Associated Fund of Dapeng District (PT20170310 and PT20170309), and the research program of Urban Management Bureau of Shenzhen Municipality (No. 201914), as well as the Agricultural Science and Technology Innovation Program and The Elite Young Scientists Program of CAAS, Fundamental Research Funds for Central Non-Profit Scientific Institution (No. Y2017JC01), the Agricultural Science and Technology Innovation Program Cooperation and Innovation Mission (CAAS-XTCX2016), and Fund of Key Laboratory of Shenzhen (ZDSYS20141118170111640). Projects subsidized by special funds for science technology innovation and industrial development of Shenzhen Dapeng New District (Grant No. KJYF202001-03).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01462/full#supplementary-material
References
- Ahemad M., Kibret M. (2014). Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J. King Saud Univ. Sci. 26 1–20. 10.1016/j.jksus.2013.05.001 [DOI] [Google Scholar]
- Alori E. T., Glick B. R., Babalola O. O. (2017). Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 8:971. 10.3389/fmicb.2017.00971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babalola O. O., Glick B. R. (2012). The use of microbial inoculants in African agriculture: current practice and future prospects. J. Food Agricult. Environ. 10 540–549. [Google Scholar]
- Berendsen R. L., Pieterse C. M., Bakker P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17 478–486. 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Beth Mudgett M. (2005). New insights to the function of phytopathogenic bacterial type III effectors in plants. Ann. Rev. Plant Biol. 56 509–531. 10.1146/annurev.arplant.56.032604.144218 [DOI] [PubMed] [Google Scholar]
- Bruisson S., Zufferey M., L’haridon F., Trutmann E., Anand A., Dutartre A., et al. (2019). Endophytes and epiphytes from the grapevine leaf microbiome as potential biocontrol agents against phytopathogens. Front. Microbiol. 10:2726. 10.3389/fmicb.2019.02726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12:59. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., Garrido-Oter R., Münch P. C., Weiman A., Dröge J., Pan Y., et al. (2015). Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17 392–403. 10.1016/j.chom.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszewski B., Railean-Plugaru V., Pomastowski P., Rafińska K., Szultka-Mlynska M., Golinska P., et al. (2018). Antimicrobial activity of biosilver nanoparticles produced by a novel Streptacidiphilus durhamensis strain. J. Microbiol. Immunol. Infect. 51 45–54. 10.1016/j.jmii.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Callaway R. M., Thelen G. C., Rodriguez A., Holben W. E. (2004). Soil biota and exotic plant invasion. Nature 427 731–733. 10.1038/nature02322 [DOI] [PubMed] [Google Scholar]
- Castro-Díez P., Godoy O., Alonso A., Gallardo A., Saldaña A. (2014). What explains variation in the impacts of exotic plant invasions on the nitrogen cycle? A meta-analysis. Ecol. Lett. 17 1–12. 10.1111/ele.12197 [DOI] [PubMed] [Google Scholar]
- Chaia E. E., Wall L. G., Huss-Danell K. (2010). Life in soil by the actinorhizal root nodule endophyte Frankia. A review. Symbiosis 51 201–226. 10.1007/s13199-010-0086-y [DOI] [Google Scholar]
- Chapuis-Lardy L., Vanderhoeven S., Dassonville N., Koutika L.-S., Meerts P. (2006). Effect of the exotic invasive plant Solidago gigantea on soil phosphorus status. Biol. Fertil. Soils 42 481–489. 10.1007/s00374-005-0039-4 [DOI] [Google Scholar]
- Chen B.-M., Peng S.-L., Ni G.-Y. (2009). Effects of the invasive plant Mikania micrantha HBK on soil nitrogen availability through allelopathy in South China. Biol. Invas. 11 1291–1299. 10.1007/s10530-008-9336-9 [DOI] [Google Scholar]
- Cordovez V., Schop S., Hordijk K., De Boulois H. D., Coppens F., Hanssen I., et al. (2018). Priming of plant growth promotion by volatiles of root-associated Microbacterium spp. Appl. Environ. Microbiol. 84 e1865–e1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Liu G., Chen H., Chen C., Wang J., Ai S., et al. (2019). Long-term nutrient inputs shift soil microbial functional profiles of phosphorus cycling in diverse agroecosystems. ISME J. 14 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins K., Esiobu N. (2016). Emerging insights on Brazilian pepper tree (Schinus terebinthifolius) invasion: the potential role of soil microorganisms. Front. Plant Sci. 7:712. 10.3389/fpls.2016.00712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins K., Esiobu N. (2018). The invasive Brazilian Pepper Tree (Schinus terebinthifolius) is colonized by a root microbiome enriched with alphaproteobacteria and unclassified spartobacteria. Front. Microbiol. 9:876. 10.3389/fmicb.2018.00876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson W., Schrama M. (2016). Identifying the role of soil microbes in plant invasions. J. Ecol. 104 1211–1218. 10.1111/1365-2745.12619 [DOI] [Google Scholar]
- Day M. D., Clements D. R., Gile C., Senaratne W. K. A. D., Shen S. C., Weston L. A., et al. (2016). Biology and Impacts of Pacific Islands Invasive Species. 13. Mikania micrantha Kunth (Asteraceae). Pacific Sci. 70 257–285. 10.2984/70.3.1 [DOI] [Google Scholar]
- De La Cruz-Barrón M., Cruz-Mendoza A., Navarro-Noya Y. E., Ruiz-Valdiviezo V. M., Ortíz-Gutiérrez D., Ramírez-Villanueva D. A., et al. (2017). The bacterial community structure and dynamics of carbon and nitrogen when maize (Zea mays L.) and its neutral detergent fibre were added to soil from Zimbabwe with contrasting management practices. Microb. Ecol. 73 135–152. 10.1007/s00248-016-0807-8 [DOI] [PubMed] [Google Scholar]
- Edwards J., Johnson C., Santos-Medellín C., Lurie E., Podishetty N. K., Bhatnagar S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. 112 E911–E920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Bradford M. A., Jackson R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88 1354–1364. 10.1890/05-1839 [DOI] [PubMed] [Google Scholar]
- Garbeva P. V., Van Veen J., Van Elsas J. (2004). Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Ann. Rev. Phytopathol. 42 243–270. 10.1146/annurev.phyto.42.012604.135455 [DOI] [PubMed] [Google Scholar]
- Genin S., Denny T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Ann. Rev. Phytopathol. 50 67–89. [DOI] [PubMed] [Google Scholar]
- Gerlach W., Stoye J. (2011). Taxonomic classification of metagenomic shotgun sequences with CARMA3. Nucleic Acids Res. 39 e91–e91. 10.1093/nar/gkr225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. W., Choi G. J., Kim B. S. (2018). Antimicrobial aromatic polyketides: a review of their antimicrobial properties and potential use in plant disease control. World J. Microbiol. Biotechnol. 34 163. [DOI] [PubMed] [Google Scholar]
- Hu Y., But P. (1994). A study on life cycle and response to herbicides of Mikania micrantha. ACTA Scientiar. Nat. Univ. SunYatSeni 33 88–95. [Google Scholar]
- Huang P., Zhang Y., Xiao K., Jiang F., Wang H., Tang D., et al. (2018). The chicken gut metagenome and the modulatory effects of plant-derived benzylisoquinoline alkaloids. Microbiome 6 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32 D277–D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. A., Jilani G., Akhtar M. S., Naqvi S. M. S., Rasheed M. (2009). Phosphorus solubilizing bacteria: occurrence, mechanisms and their role in crop production. J. Agricult. Biol. Sci. 1 48–58. [Google Scholar]
- Khan M. S., Zaidi A., Ahemad M., Oves M., Wani P. A. (2010). Plant growth promotion by phosphate solubilizing fungi–current perspective. Arch. Agron. Soil Sci. 56 73–98. 10.1080/03650340902806469 [DOI] [Google Scholar]
- Kouas S., Labidi N., Debez A., Abdelly C. (2005). Effect of P on nodule formation and N fixation in bean. Agron. Sustain. Dev. 25 389–393. 10.1051/agro:2005034 [DOI] [Google Scholar]
- Kourtev P. S., Ehrenfeld J. G., Häggblom M. (2002). Exotic plant species alter the microbial community structure and function in the soil. Ecology 83 3152–3166. 10.1890/0012-9658(2002)083[3152:epsatm]2.0.co;2 [DOI] [Google Scholar]
- Kurnia N. M., Uria A. R., Kusnadi Y., Dinawati L., Zilda D. S., Hadi T. A., et al. (2017). Metagenomic survey of potential symbiotic bacteria and polyketide synthase genes in an indonesian marine sponge. HAYATI J. Biosci. 24 6–15. 10.1016/j.hjb.2017.04.004 [DOI] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Shen Y., Huang Q., Fan Z., Huang D. (2013). Regeneration capacity of small clonal fragments of the invasive Mikania micrantha HBK: effects of burial depth and stolon internode length. PLoS One 8:e84657. 10.1371/journal.pone.0084657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li J., Li Y., Wang X.-X., Cao A.-C. (2013). Antimicrobial Constituents of the Leaves of Mikania micrantha HB K. PLoS One 8:e76725. 10.1371/journal.pone.0076725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidbury I. D., Murphy A. R., Scanlan D. J., Bending G. D., Jones A. M., Moore J. D., et al. (2016). Comparative genomic, proteomic and exoproteomic analyses of three Pseudomonas strains reveals novel insights into the phosphorus scavenging capabilities of soil bacteria. Environ. Microbiol. 18 3535–3549. 10.1111/1462-2920.13390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling N., Chen D., Guo H., Wei J., Bai Y., Shen Q., et al. (2017). Differential responses of soil bacterial communities to long-term N and P inputs in a semi-arid steppe. Geoderma 292 25–33. 10.1016/j.geoderma.2017.01.013 [DOI] [Google Scholar]
- Liu B., Yan J., Li W., Yin L., Li P., Yu H., et al. (2020). Mikania micrantha genome provides insights into the molecular mechanism of rapid growth. Nat. Commun. 11 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe S., Browne M., Boudjelas S., De Poorter M. (2000). 100 of the World’s Worst Invasive Alien Species: A Selection From the Global Invasive Species Database. Auckland: Invasive Species Specialist Group Auckland. [Google Scholar]
- Lu-Irving P., Harenčár J., Sounart H., Welles S. R., Swope S. M., Baltrus D. A., et al. (2017). Escape from bacterial diversity: potential enemy release in invading yellow starthistle (Centaurea solstitialis) microbiomes. bioRxiv [Preprint] 10.1101/119917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu-Irving P., Harenčár J. G., Sounart H., Welles S. R., Swope S. M., Baltrus D. A., et al. (2019). Native and invading yellow starthistle (Centaurea solstitialis) microbiomes differ in composition and diversity of bacteria. mSphere 4 e88–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macanawai A., Day M., Tumaneng-Diete T., Adkins S. (2012). Impact of Mikania micrantha on crop production systems in Viti Levu. Fiji. Pak. J. Weed Sci. Res. 18 357–365. [Google Scholar]
- Marler M. J., Zabinski C. A., Callaway R. M. (1999). Mycorrhizae indirectly enhance competitive effects of an invasive forb on a native bunchgrass. Ecology 80 1180–1186. 10.1890/0012-9658(1999)080[1180:mieceo]2.0.co;2 [DOI] [Google Scholar]
- Meena V. S., Meena S. K., Verma J. P., Kumar A., Aeron A., Mishra P. K., et al. (2017). Plant beneficial rhizospheric microorganism (PBRM) strategies to improve nutrients use efficiency: a review. Ecol. Eng. 107 8–32. 10.1016/j.ecoleng.2017.06.058 [DOI] [Google Scholar]
- Mendes L. W., Kuramae E. E., Navarrete A. A., Van Veen J. A., Tsai S. M. (2014). Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 8:1577. 10.1038/ismej.2014.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C. E., Power A. G. (2003). Release of invasive plants from fungal and viral pathogens. Nature 421 625–627. 10.1038/nature01317 [DOI] [PubMed] [Google Scholar]
- Mori T., Cahn J. K., Wilson M. C., Meoded R. A., Wiebach V., Martinez A. F. C., et al. (2018). Single-bacterial genomics validates rich and varied specialized metabolism of uncultivated Entotheonella sponge symbionts. Proc. Natl. Acad. Sci. 115 1718–1723. 10.1073/pnas.1715496115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Możejko-Ciesielska J., Serafim L. S. (2019). Proteomic response of Pseudomonas putida KT2440 to dual carbon-phosphorus limitation during mcl-PHAs synthesis. Biomolecules 9:796. 10.3390/biom9120796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes K. A., Fitzpatrick C. R., Kotanen P. M. (2019). Soil biota composition and the performance of a noxious weed across its invaded range. Ecography 42 1671–1681. 10.1111/ecog.04562 [DOI] [Google Scholar]
- Philippot L., Raaijmakers J. M., Lemanceau P., Van D. P., Wim H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11 789–799. 10.1038/nrmicro3109 [DOI] [PubMed] [Google Scholar]
- Piper C. L., Lamb E. G., Siciliano S. D. (2015). Smooth brome changes gross soil nitrogen cycling processes during invasion of a rough fescue grassland. Plant Ecol. 216 235–246. 10.1007/s11258-014-0431-y [DOI] [Google Scholar]
- Pyšek P., Richardson D. M. (2008). “Traits associated with invasiveness in alien plants: where do we stand?,” in Biological Invasions. Ecological Studies (Analysis and Synthesis),W. Nentwig (Heidelberg: Springer; ), 97–125. 10.1007/978-3-540-36920-2_7 [DOI] [Google Scholar]
- Raaijmakers J. M., Mazzola M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Ann. Rev. Phytopathol. 50 403–424. 10.1146/annurev-phyto-081211-172908 [DOI] [PubMed] [Google Scholar]
- Raj R., Paul D., Babyson R. S. (2014). Molecular characterization of phosphate solubilizing bacteria (PSB) and plant growth promoting rhizobacteria (PGPR) from pristine soils. Int. J. Innovat. Sci. Eng. Technol. 1 317–324. [Google Scholar]
- Ramirez K. S., Snoek L. B., Koorem K., Geisen S., Bloem L. J., ten Hooven F., et al. (2019). Range-expansion effects on the belowground plant microbiome. Nat. Ecol. Evol. 3 604–611. 10.1038/s41559-019-0828-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart K. O., Packer A., Van Der Putten W. H., Clay K. (2003). Plant–soil biota interactions and spatial distribution of black cherry in its native and invasive ranges. Ecol. Lett. 6 1046–1050. 10.1046/j.1461-0248.2003.00539.x [DOI] [Google Scholar]
- Reinhold-Hurek B., Bünger W., Burbano C. S., Sabale M., Hurek T. (2015). Roots shaping their microbiome: global hotspots for microbial activity. Ann. Rev. Phytopathol. 53 403–424. 10.1146/annurev-phyto-082712-102342 [DOI] [PubMed] [Google Scholar]
- Rilling J. I., Acuña J. J., Sadowsky M. J., Jorquera M. A. (2018). Putative nitrogen-fixing bacteria associated with the rhizosphere and root endosphere of wheat plants grown in an andisol from southern chile. Front. Microbiol. 9:2710. 10.3389/fmicb.2018.02710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabloom E. W., Harpole W. S., Reichman O., Tilman D. (2003). Invasion, competitive dominance, and resource use by exotic and native California grassland species. Proc. Natl. Acad. Sci. 100 13384–13389. 10.1073/pnas.1835728100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. B., Sayyed R. Z., Trivedi M. H., Gobi T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas C., Devi D. N., Murthy K. N., Mohan C. D., Lakshmeesha T., Singh B., et al. (2019). Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: biology to diversity–A review. Saudi J. Biol. Sci. 26 1315–1324. 10.1016/j.sjbs.2019.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Ou Q., Yu H., Li N., Peng C. (2019). The invasive plant Mikania micrantha affects the soil foodweb and plant-soil nutrient contents in orchards. Soil Biol. Biochem. 139:107630 10.1016/j.soilbio.2019.107630 [DOI] [Google Scholar]
- Teng Z., Chen Z., Zhang Q., Yao Y., Song M., Li M. (2019). Isolation and characterization of phosphate solubilizing bacteria from rhizosphere soils of the Yeyahu Wetland in Beijing. China. Environ. Sci. Pollut. Res. 26 33976–33987. 10.1007/s11356-018-2955-5 [DOI] [PubMed] [Google Scholar]
- Thorpe A. S., Archer V., Deluca T. H. (2006). The invasive forb, Centaurea maculosa, increases phosphorus availability in Montana grasslands. Appl. Soil Ecol. 32 118–122. 10.1016/j.apsoil.2005.02.018 [DOI] [Google Scholar]
- Van Der Heijden M. G., Bardgett R. D., Van Straalen N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11 296–310. 10.1111/j.1461-0248.2007.01139.x [DOI] [PubMed] [Google Scholar]
- Van Kleunen M., Weber E., Fischer M. (2010). A meta-analysis of trait differences between invasive and non-invasive plant species. Ecol. Lett. 13 235–245. 10.1111/j.1461-0248.2009.01418.x [DOI] [PubMed] [Google Scholar]
- Vives-Peris V., Gómez-Cadenas A., Pérez-Clemente R. M. (2018). Salt stress alleviation in citrus plants by plant growth-promoting rhizobacteria Pseudomonas putida and Novosphingobium sp. Plant Cell Rep. 37 1557–1569. 10.1007/s00299-018-2328-z [DOI] [PubMed] [Google Scholar]
- Weller D. M., Raaijmakers J. M., Gardener B. B. M., Thomashow L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Ann. Rev. Phytopathol. 40 309–348. [DOI] [PubMed] [Google Scholar]
- Wu B., Wang S., Wei M., Zhou J., Jiang K., Du D., et al. (2019). The invasive tree staghorn sumac affects soil N2-fixing bacterial communities in north China. Plant Biol. 21 951–960. 10.1111/plb.13003 [DOI] [PubMed] [Google Scholar]
- Xueping C., Zhang X., Xi’e Z., Zhang H., Liang X., Yanrui L., et al. (2016). Exotic plant Alnus trabeculosa alters the composition and diversity of native rhizosphere bacterial communities of Phragmites australis. Pedosphere 26 108–119. 10.1016/s1002-0160(15)60027-3 [DOI] [Google Scholar]
- Zettler J., Xia H., Burkard N., Kulik A., Grond S., Heide L., et al. (2014). New aminocoumarins from the rare actinomycete Catenulispora acidiphila DSM 44928: identification, structure elucidation, and heterologous production. Chembiochem 15 612–621. 10.1002/cbic.201300712 [DOI] [PubMed] [Google Scholar]
- Zhang J., Liu Y., Zhang N., Hu B., Jin T., Xu H., et al. (2019). NRT1. 1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 37 676–684. 10.1038/s41587-019-0104-4 [DOI] [PubMed] [Google Scholar]
- Zhao M., Lu X., Zhao H., Yang Y., Hale L., Gao Q., et al. (2019). Ageratina adenophora invasions are associated with microbially mediated differences in biogeochemical cycles. Sci. Total Environ. 677 47–56. 10.1016/j.scitotenv.2019.04.330 [DOI] [PubMed] [Google Scholar]
- Zheng M. M., Wang C., Li W. X., Song W. F., Shen R. F. (2019). Soil nutrients drive function and composition of phoC-Harboring bacterial community in acidic soils of Southern China. Front. Microbiol. 10:2654. 10.3389/fmicb.2019.02654 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found in the NCBI under the accession numbers SRR8936416–SRR8936475.