SUMMARY
A single mouse blastomere from until 8-cell embryo can generate an entire blastocyst. Whether laboratory cultured cells retain a similar generative capacity remains unknown. Starting from a single stem cell type, extended pluripotent stem (EPS) cells, we established a 3D differentiation system that enabled the generation of blastocyst-like structures (EPS-blastoids) through lineage segregation and self-organization. EPS-blastoids resembled blastocysts in morphology and cell lineage allocation and recapitulated key morphogenetic events during preimplantation and early postimplantation development in vitro. Upon transfer, some EPS-blastoids underwent implantation, induced decidualization, and generated live, albeit disorganized, tissues in utero. Single-cell and bulk RNA-sequencing analysis revealed that EPS-blastoids contained all three blastocyst cell lineages and shared transcriptional similarity with natural blastocysts. We also provide proof-of-concept that EPS-blastoids can be generated from adult cells via cellular reprogramming. EPS-blastoids provide a unique platform for studying early embryogenesis and pave the way to create viable synthetic embryos using cultured cells.
Keywords: Extended Pluripotent Stem (EPS) cells, totipotent, Blastoids, EPS-blastoid, blastocyst, implantation, reprogramming
Graphical Abstract
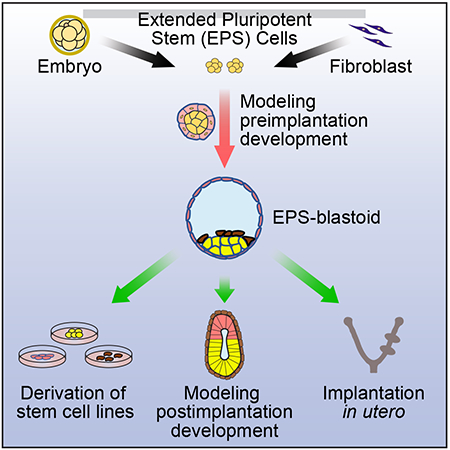
A blastocyst-like structure is generated from a single stem cell type as well as from somatic reprogrammed cells and these artificial embryo-like structures can implant, induce decidualization, and give rise to cells of the three embryonic founding tissues: epiblast, trophectoderm, and primitive endoderm.
INTRODUCTION
Mammalian embryogenesis starts with a fertilized egg, which divides to generate blastomeres. Mouse blastomeres are considered totipotent until the 8-cell stage (Papaioannou et al., 1989; TARKOWSKI, 1959). The blastomeres then compact, polarize and undergo the first two lineage segregations to generate three different cell types of a blastocyst. The first lineage segregation specifies an outer trophectoderm (TE) layer and an inner cell mass (ICM). ICM cells later segregate into epiblast (EPI) and primitive endoderm (PE) lineages. Stem cell lines from all three blastocyst lineages have been derived and stably maintained in vitro, namely embryonic stem cells (ESCs) (Evans and Kaufman, 1981; Martin, 1981), trophoblast stem cells (TSCs) (Tanaka et al., 1998), and extraembryonic endoderm stem cells (XENs) (Kunath, 2005). With all the necessary parts available, it may be possible to de novo assemble an artificial embryo using cultured stem cells.
Early attempts to generate embryo-like structures relied on the spontaneous differentiation of EPI-derived ESCs in 3D culture, thereby producing structurally disorganized cell aggregates, termed as embryoid bodies (Doetschman et al., 1985; Evans and Kaufman, 1981; Martin, 1981), which are a popular in vitro model for studying differentiation and pluripotency (Berge et al., 2008; Fuchs et al., 2012). Organized embryo-like structures can also be generated from ESCs in 2D or 3D differentiation cultures. For example, under inductive cues, mouse and human ESCs could establish spatial patterns resembling a gastrulating embryo when cultured in a soft fibrin matrix and micropatterned condition, respectively (Poh et al., 2014; Warmflash et al., 2014). In addition, pulsing mouse ESC aggregates undergoing differentiation with a WNT/β-catenin pathway activator in a shaking 3D culture gave rise to elongated gastruloids (Beccari et al., 2018). Others have used multiple stem cell types to assemble embryo-like structures. Aggregation of ESCs, TSCs, and/or XENs generated embryo-like structures that recapitulated several key morphogenetic events characteristic of early postimplantation development, including lumenogenesis, epithelialization, and symmetry breaking to specify mesoderm and primordial germ cell-like cells (Harrison et al., 2017; Sozen et al., 2018; Zhang et al., 2019).
These successes have stimulated the interest in recreating a preimplantation blastocyst from cultured stem cells. Recently, Rivron et al. reported the early success of generating mouse blastocyst-like structures, termed blastoids, by the sequential aggregation of ESCs and TSCs in microwells (Rivron et al., 2018). These blastoids (hereafter referred to as ETS-blastoids) morphologically resemble blastocysts and can implant and induce the formation of decidua upon embryo transfer. Notwithstanding the success, however, generation of ETS-blastoids involves the use of two peri-implantation stem cell lines and is unable to model preimplantation development. In addition, ETS-blastoids poorly developed into postimplantation embryo-like structures in vitro (Vrij et al., 2019) and only generated trophoblast cell types in vivo (Rivron et al., 2018).
Blastocysts can be generated from a single totipotent blastomere at the 2-, 4-, and 8-cell stage (Papaioannou et al., 1989; TARKOWSKI, 1959; TARKOWSKI and Wroblewska, 1967). In recent years, there have been significant advances in identifying and stabilizing cultured stem cells with totipotent-like features. It was first observed that mouse ESCs could transiently cycle in and out of a 2-cell embryo-like state (Macfarlan et al., 2012; Morgani et al., 2013). More recently, we and others have developed culture conditions that can stabilize totipotent-like features in cultured extended pluripotent stem cells or expanded potential stem cells (hereafter referred to as EPS cells). EPS cells can efficiently contribute to both embryonic (Em) and extraembryonic (ExEm) lineages in vitro and in vivo (Yang et al., 2017a; 2017b). Based on these results, we hypothesize that EPS cells alone can differentiate and self-organize into blastoids that contain all three founder tissues of a blastocyst: EPI, TE, and PE. Here, we report the generation of blastoids from mouse EPS cells using a newly developed 3D differentiation system. From these blastoids (herein referred to as EPS-blastoids), we were able to derive ESCs, TSCs, and XENs. When further cultured, EPS-blastoids generated structures characteristic of an E5.0—E5.5 postimplantation embryo. In utero EPS-blastoids were capable of implantation, triggering decidualization, and gave rise to structures containing live tissues of EPI, TE, and PE origins. Bulk and single-cell RNA-sequencing (RNA-Seq) analysis revealed that EPS-blastoids were transcriptionally similar to natural E3.5 blastocysts and contained all three blastocyst cell lineages. Also, we were able to generate EPS-blastoids from mouse fibroblasts, providing proof-of-principle that embryo-like structures can also be generated from somatic cells
RESULTS
A 3D Differentiation System for Generating Blastocyst-like Structures from EPS Cells
The ability of a single EPS cell to contribute to all three blastocyst lineages suggests that, under certain condition, EPS cells can differentiate and self-organize into blastocyst-like structures (Figure 1A). To test this idea, we first seeded dissociated mouse EPS cells into microwells (~ 5 cells/microwell) to form small aggregates. During early preimplantation development, TE cells are specified before ICM cells (Posfai et al., 2017). Thus, we first focused on optimizing conditions that bias the differentiation of EPS cells toward trophoblasts. We tested a commonly used TSC culture medium (Tanaka et al., 1998) and found that although cell aggregates grew well in TSC medium alone, they failed to form any sphere-like structures (Figure 1B). Interestingly, mixing an embryo culture medium (KSOM) with TSC medium at a 1:1 ratio induced cavity formation in a small number of cell aggregates (Figure 1B). The effect of the KSOM could be replicated with another embryo culture medium M16 (Figure S1A). Later we switched from KSOM: TSC medium to KSOM: ETS medium (a 2:1:1 mixture of KSOM, N2B27 basal, and TSC basal medium), which supports the growth of both ESC and TSC aggregates (Harrison et al., 2017).
Figure 1. A 3D Differentiation System for Generating Blastocyst-like Structures from EPS Cells.
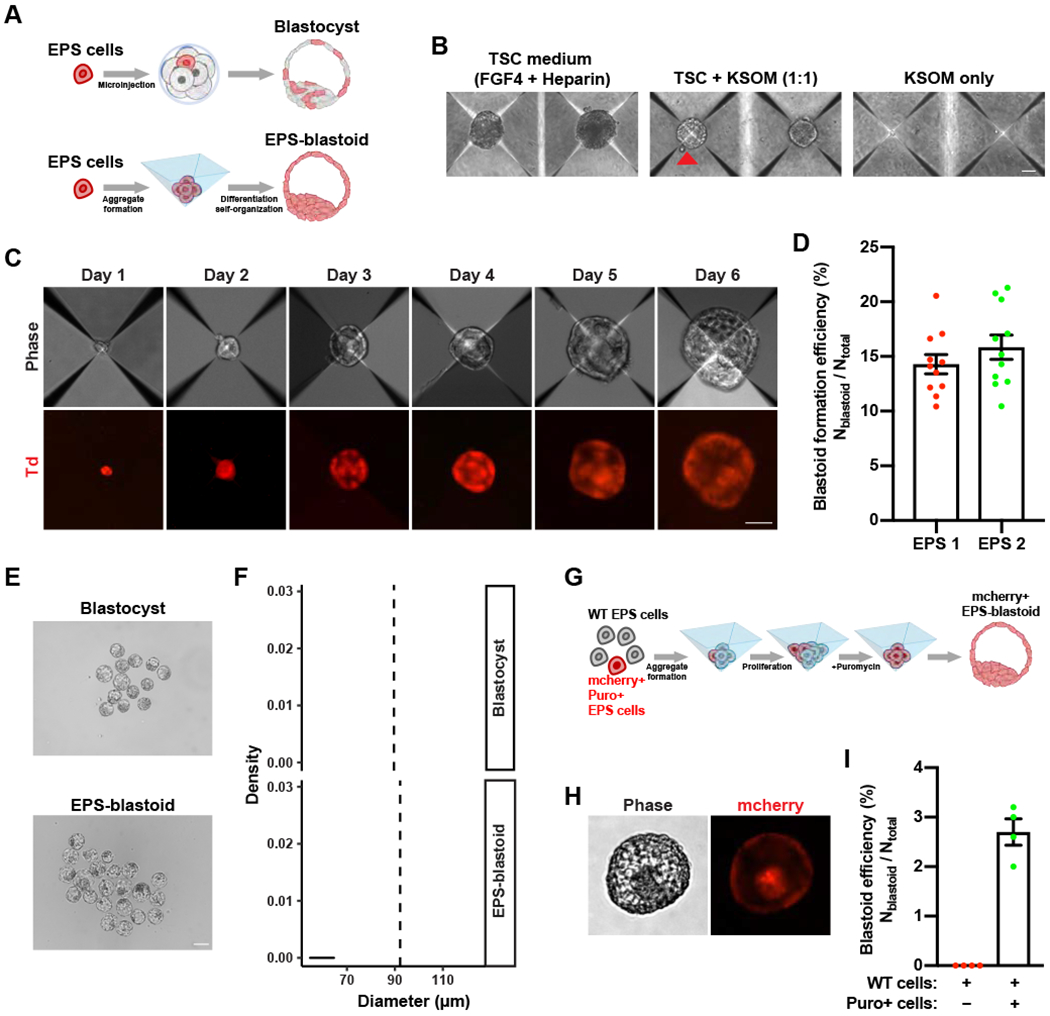
(A) Top panel: A diagram showing that a single EPS cell can contribute to both embryonic (Em) and extraembryonic (ExEm) lineages in the blastocyst after injection into an 8-cell embryo. Bottom panel: A diagram showing that EPS cells differentiate and self-organize into an EPS-blastoid.
(B) Phase contrast images of EPS cell aggregates cultured in the indicated medium conditions for four days. The red triangle indicates an EPS-blastoid.
(C) Representative phase contrast (upper panel) and fluorescence images (lower panel) of EPS cell aggregates at the indicated time point showing the formation of EPS-blastoids. Phase, phase contrast; Td, tdTomato.
(D) Quantification of EPS-blastoids formation efficiency. n = 11 independent assays for each EPS cell line.
(E) Phase contrast images of E3.5 blastocysts (upper panel) and EPS-blastoids (lower panel).
(F) Histograms showing the distribution of the diameters of E3.5 blastocysts (upper panel) and EPS-blastoids (lower panel). n = 55 blastocysts and n = 95 EPS-blastoids. The vertical dotted line denotes the mean of the group.
(G) A diagram showing the strategy for a single EPS cell to generate a clonal EPS-blastoid.
(H) Phase contrast (left) and fluorescent (right) images of an EPS-blastoid generated using the strategy shown in (G).
(I) Quantification of EPS-blastoids formation efficiency in the assay described in (G). n = 4 independent assays.
Data are represented as mean ± SEM. Scale bars, 50 μm (B, C), 100 μm (E), 20 μm (H).
To further improve the differentiation condition, we screened for different growth factors, cytokines, and small molecules to identify condition(s) that enable the generation of structures mimicking blastocysts (EPS-blastoids), which contain a cavity and an inner cell mass. We found that a combination of FGF4, Heparin, BMP4, CHIR99021, and A83-01 resulted in the highest efficiency of EPS-blastoid formation. We also treated EPS cells with Y-27632, a Rho kinase (ROCK) inhibitor, on the day of seeding to enhance cell survival. Using this optimized condition, we consistently observed that EPS-blastoids emerged three days after cell seeding (Figure 1C). EPS-blastoids continued to enlarge, reaching an early blastocyst-like size at around day five or day six. To quantify the efficiency of EPS-blastoid formation, we counted the total number of blastocyst-like structures and divided it by the total number of cell aggregates. We tested two independent EPS cell lines and found ~15% of the cell aggregates exhibited typical blastocyst-like morphology at day five or day six (Figure 1D; Table S1). The average diameter of these EPS-blastoids was comparable to that of E3.5 blastocysts (Figure 1E and 1F).
WNT/β-catenin signaling pathway is dispensable for blastocyst formation (Figure S1B and S1C) (Biechele et al., 2013; Haegel et al., 1995). However, WNT3a can transiently up-regulate CDX2 (a trophoblast transcription factor) in mouse ESCs (He et al., 2008), and canonical WNT pathway activation was necessary for the generation of ETS-blastoids (Rivron et al., 2018). Similarly, we found that the WNT antagonists, XAV939 or IWR-1-endo, significantly inhibited EPS-blastoids formation (Figure S1D and S1E; Table S1).
Next, we tested whether a single EPS cell could give rise to an entire EPS-blastoid. Surprisingly, we found individual EPS cell did not survive in the differentiation system. To overcome this problem, we mixed puromycin resistant and mCherry+ EPS cell with helper wild type EPS cells at the ratio of 1:10 for the initial plating. Low concentration of puromycin (0.25 μg/ml) was added to the differentiation medium 24 hours later to eliminate helper cells gradually (Figure 1G). Using this strategy, we could generate clonal EPS-blastoid from a single EPS cell with an efficiency of ~2.7% (Figure 1H and 1I; Table S1). These results demonstrate that a single EPS cell retains the capacity to form an entire EPS-blastoid.
We also examined whether extended pluripotency was required for generating blastoids. To this end, we compared blastoid formation efficiencies between an ES cell line (Tanimoto et al., 2008) and EPS cells converted from the same ES cell line. We found while ~8% of ES-converted EPS cell aggregates formed EPS-blastoids, little to no (~ 0.2%) ES cell aggregates generated blastocyst-like structures (ES-blastoids) (Figure S1F and S1G; Table S1).
Besides EPS cells cultured in LCDM condition (Yang et al., 2017b), EPS cells generated and cultured under a different culture condition developed by Pengtao Liu’s group (referred to as Liu-EPS) (Yang et al., 2017a) also generated EPS-blastoids using our 3D differentiation system in ~11% of cell aggregates (Figure S1H and S1I; Table S1). We found that Liu-EPS-blastoids were of similar size as E3.5 blastocysts (Figure S1J).
EPS-blastoid Formation Recapitulates Key Preimplantation Developmental Processes
Next, we investigated whether key cellular and molecular events characteristic of early preimplantation development could be recapitulated during EPS-blastoid formation. Beginning at the 8-cell stage, blastomeres undergo compaction, which is characterized by the formation of intercellular junctions (Rossant and Tam, 2009; Wang et al., 2008). To determine whether a similar process occurs during EPS-blastoid formation, we monitored the dynamics of EPS cell aggregation during the first 18 hours. Four hours after seeding, cells were found loosely connected. At ~18 hours, cells started to form compact aggregates (Figure 2A), with the cell adhesion protein, E-cadherin, and the tight junction protein, ZO1, beginning to accumulate at the cell-cell junctions, reminiscent of a compacted 8-cell embryo (Figure 2B, 2C, S2A, and S2B). The fast kinetics of EPS cell aggregate formation was likely due to the high levels of the Cdhl and Tjpl (genes encoding E-cadherin and ZO1, respectively) mRNA and ZO1 protein in cultured EPS cells (Figure S2C and S2D).
Figure 2. EPS-blastoid Formation Recapitulates Key Preimplantation Developmental Processes.
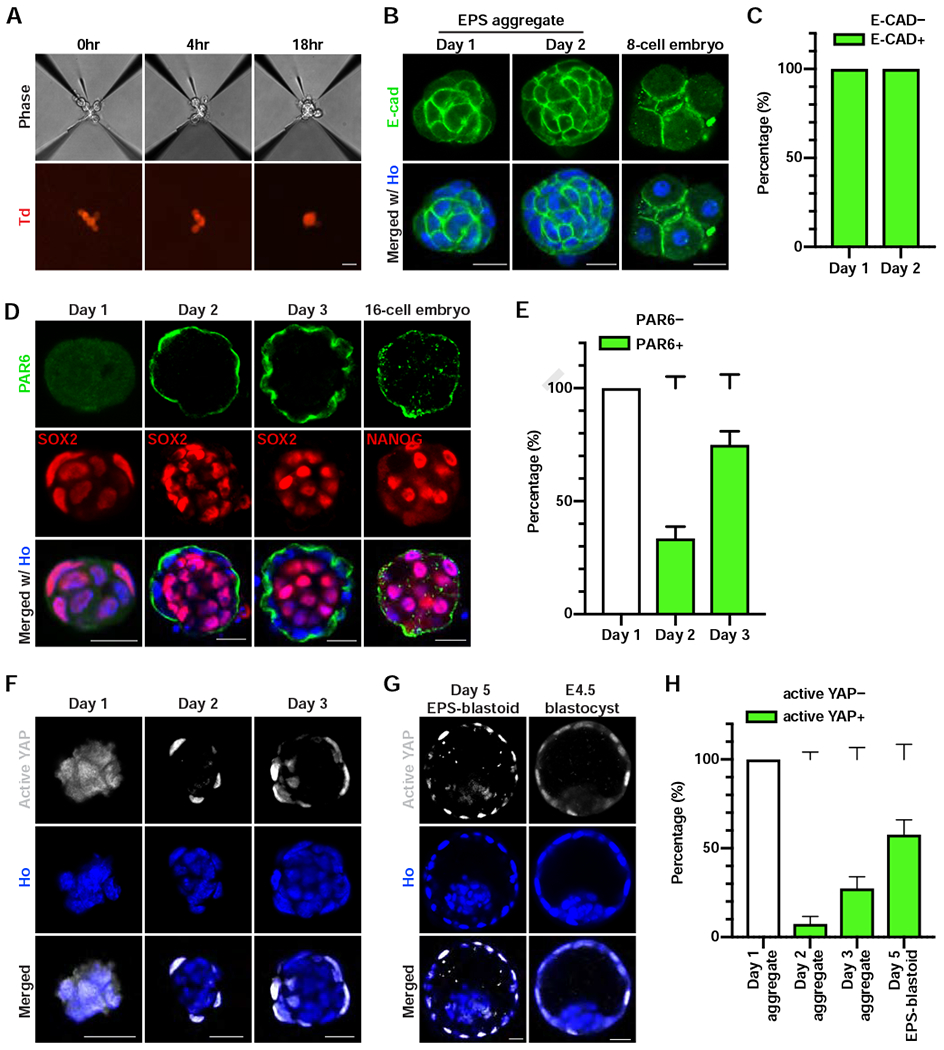
(A) Phase contrast (upper panel) and fluorescent (lower panel) images of EPS cells at the indicated time after cell seeding. Td, tdTomato.
(B) Immunofluorescence staining of an EPS aggregate at day 1 (left), day 2 (middle), and a compacted 8-cell embryo (right) for E-cadherin (E-CAD).
(C) Quantification of the percentage of cell aggregates showing E-CAD+ staining at the indicated day. n = 3 biological replicates for each time point.
(D) Immunofluorescence staining of EPS aggregates at the indicated time points and 16-cell embryos for PAR6 and SOX2 or NANOG.
(E) Quantification of the percentage of cell aggregates showing a PAR6 ring at the indicated time points. n = 3 biological replicates for each time point.
(F and G) Immunofluorescence staining for active YAP in EPS aggregates at the indicated time point (F), an EPS-blastoid (G), and an E4.5 blastocyst (G).
(H) Quantification of the percentage of structures showing active YAP+ at the indicated time points. n = 3 independent assays for day 1, day 2 aggregates, and day 5 EPS-blastoids; n = 5 independent assays for day 3 aggregates.
Data are represented as mean ± SEM. Scale bars, 20 μm. Ho, Hoechst.
Following blastomere compaction, polarization begins. Apical domain proteins, such as atypical protein kinase C (aPKC) and the Par complex proteins (PAR3 and PAR6), start to accumulate at the apical side of the blastomeres (Chazaud and Yamanaka, 2016; Rossant and Tam, 2009). Next, we examined whether EPS cell aggregates also underwent polarization. At day 1, PAR6 protein was absent from all cell aggregates examined, which homogenously expressed the pluripotency factor SOX2 (Figure 2D). At day 2, a fraction of cell aggregates (~33%) began to show PAR6 enrichment on the apical surface, reminiscent of a 16-cell embryo (Figure 2D and 2E). At day 3, the majority of cell aggregates (~75%) exhibited a polarized pattern of PAR6 enrichment towards the apical side (Figure 2D and 2E). These data support the claim that EPS-blastoid formation recapitulates the process of polarization characteristic of early preimplantation development.
During early embryogenesis, the Hippo/YAP signaling pathway plays an essential role in specifying the TE and ICM lineages (Kaneko and DePamphilis, 2013; Nishioka et al., 2009; Posfai et al., 2017; Rayon et al., 2014; Yagi et al., 2007). To determine whether the formation of EPS-blastoids depends on YAP activity, we monitored YAP localization during EPS-blastoid induction. YAP is predominantly localized in the cytoplasm in cultured EPS cells (Figure S2E). At day 2, YAP could be found in the nucleus of some outside cells of the aggregate (Figure 2F). This YAP nuclear localization was observed in ~ 7% of aggregates (Figure 2H). At day 3, ~27% of aggregates contained outside cells with nuclear localization of YAP (Figure 2F and 2H). In the EPS-blastoids collected at day 5, nuclear YAP localization was evident in most outside cells, whereas inside cells still exhibited cytoplasmic YAP localization (Figure 2G). This YAP localization pattern mirrored that in the blastocysts and was found in about 60% of EPS-blastoids (Figure 2G and 2H). Furthermore, inhibiting the interaction between YAP and TEAD4 via a small molecule inhibitor prevented cavity formation in mouse embryos and EPS aggregates (Figure S2F, S2G, S2H, and S2I; Table S1). Therefore, similar to blastocyst, EPS-blastoid formation also requires an intact Hippo/YAP signaling pathway.
In an early female blastocyst, while both X-chromosomes are active in the ICM, the paternally-inherited X chromosome is preferentially inactivated in TE (Takagi and Sasaki, 1975; West et al., 1977). To check the X-chromosome status in different lineages of EPS-blastoids, we obtained an epiblast stem cell (EpiSC) line, which contained a green fluorescent protein (GFP) transgene in the paternal X-chromosome (X-GFP). X-GFP EpiSCs were FACS-sorted to obtain a pure GFP negative population (Paternally X inactivated, or Xpi-GFP) (Bao et al., 2009). We converted Xpi-GFP EpiSCs to EPS cells by culture adaptation. Over passaging, we observed the percentage of GFP+ cells increased, indicating gradual reactivation of the inactivated paternal X chromosome during conversion. Next, we FACS-sorted out GFP+ EPS cells and used them to generate EPS-blastoids. We stained these blastoids with NANOG and CDX2 and found that a large fraction of them (79%, 11/14) contained GFP+ cells only in the ICM-like compartment (Figure S2J and S2K). These data suggest that upon EPS cell differentiation, paternal X-chromosome was preferentially silenced in TE-like cells while most ICM-like cells contained two active X chromosomes.
Collectively, our results demonstrate EPS-blastoid formation recapitulate key molecular and cellular processes characteristic of early preimplantation development.
EPS-blastoids Possess Three Lineages of Blastocysts
E3.5 mouse blastocysts have two cell lineages, the external TE layer and the internal ICM. We checked whether EPS-blastoids also had these two early blastocyst lineages. Immunofluorescence analysis revealed that cells in the outer layer of EPS-blastoids expressed the TE transcription factors CDX2 and EOMES (Figure 3A and S3A). The outer layer of cells also expressed the trophoblast-specific cytokeratin KRT8 (Figure 3B). Within the ICM-like compartment of EPS-blastoids, we detected the expression of pluripotency factors SOX2, NANOG, and OCT4 (Figure 3C, 3D, and 3E). Co-staining of CDX2 and NANOG confirmed the presence of both TE- and ICM-like lineages within the same EPS-blastoid (Figure S3B). Of the 140 EPS-blastoids examined, 74.2% had correctly allocated TE- (CDX2+) and ICM-like (SOX2+) lineages, 15% had only the TE-like lineage, 1.4% had only the ICM-like lineage, and 9.3% exhibited mislocalization of the TE- and/or ICM-like lineages (Figure 3F). We also counted the number of cells within both the TE- and ICM-like compartments in EPS-blastoids collected on day 4, 5, and 6. Day 4 and 5 EPS-blastoids had fewer cells in both lineages than E3.5 blastocysts (Figure 3G). At day 6, cell numbers in both lineages were comparable to E3.5 blastocysts (Figure 3G). These results demonstrate that the cellular composition of EPS-blastoids resembles early blastocysts.
Figure 3. EPS-blastoids Possess Three Lineages of Blastocysts.
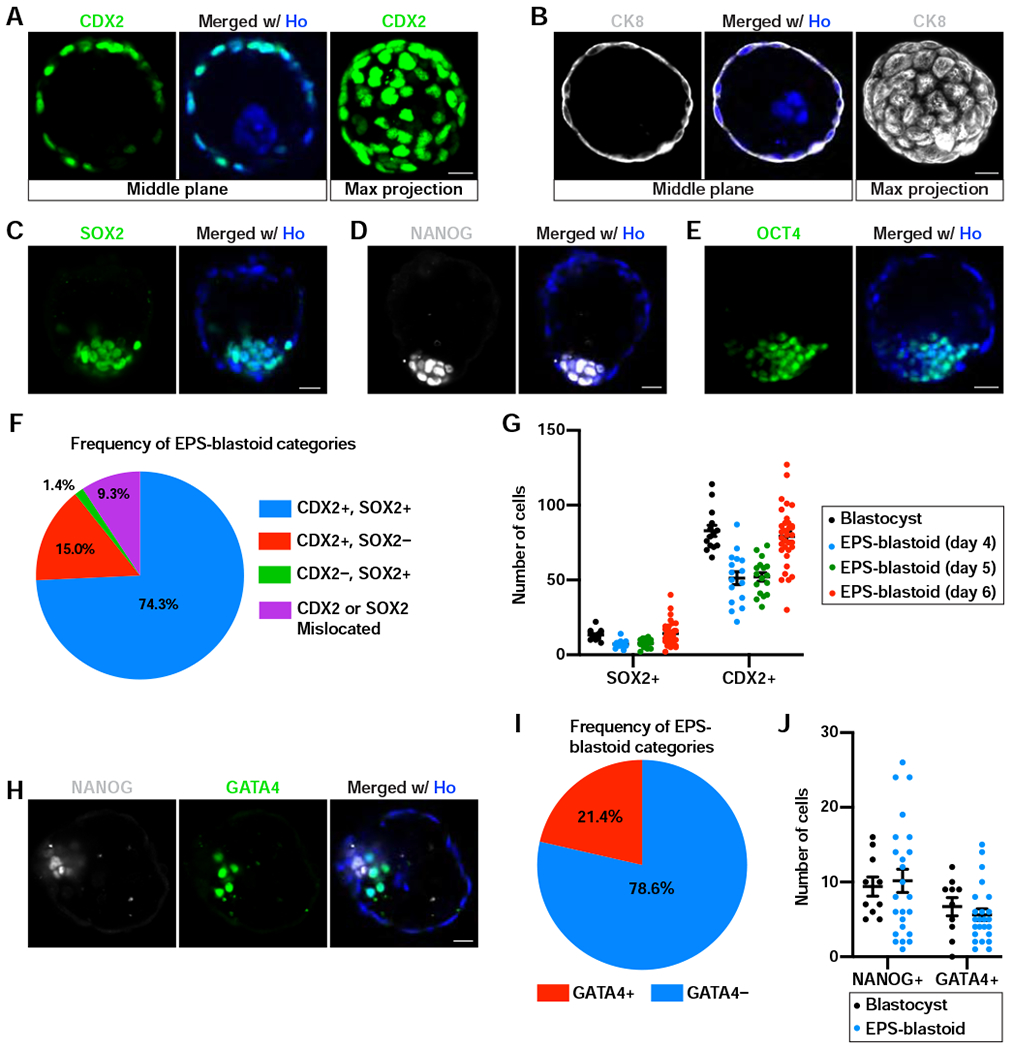
(A and B) Immunofluorescence staining of EPS-blastoids for CDX2 (A) and CK8 (B). The rightmost panel is the maximum intensity projection of z-stack images of the indicated protein.
(C-E) Immunofluorescence staining of EPS-blastoids for SOX2 (C), NANOG (D), and OCT4 (E).
(F) Quantification of the frequency of different EPS-blastoid categories based on CDX2 and SOX2. n = 140 EPS-blastoids.
(G) Quantification of the number of cells with SOX2+ or CDX2+ staining in the ICM or TE compartment, respectively, of the indicated samples. n =14 blastocysts, 16 EPS-blastoids at day 4, 17 EPS-blastoids at day 5, and 34 EPS-blastoids at day 6.
(H) Immunofluorescence staining of an EPS-blastoid for NANOG and GATA4.
(I) Quantification of the frequency of EPS-blastoids with or without GATA4+ PE-like cells. n = 112 EPS-blastoids.
(J) Quantification of the number of cells with NANOG+ or GATA4+ staining in the EPI- or PE-like compartment, respectively, of blastocysts or EPS-blastoids. n = 15 blastocysts and 24 EPS-blastoids.
Data are represented as mean ± SEM. Scale bars, 20 μm. Ho, Hoechst.
See also Figure S3.
We examined when the segregation of TE and ICM lineage occurred during EPS-blastoid formation and how the cells of the two lineages were spatially distributed in the aggregates. To this end, we analyzed the expression of CDX2 and SOX2 in EPS cell aggregates collected from day 1 to day 5. At day 1, we found that ~55% of the aggregates exhibited a composition of mixed CDX2+ and SOX2+ cell. This percentage continued to increase from day 1 to day 3 (Figure S3C and S3D). Non-blastoid EPS cell aggregates at day 4 or day 5 were also mostly composed of CDX2+ and SOX2+ cells (Figure S3C and S3D). However, CDX2+ cells were randomly distributed, and the number of CDX2+ cells varied among aggregates.
As early blastocysts further develop, ICM segregates into two lineages: EPI and PE. Therefore, we examined whether EPS-blastoids could develop into a late blastocyst-like structure comprised of all three blastocyst lineages: TE, EPI, and PE. Indeed, in some EPS-blastoids, we detected GATA4+ PE-like cells enclosing the NANOG+ EPI-like compartment, reminiscent of a peri-implantation E4.5 blastocyst (Figure 3H). This pattern was observed in 24 out of 112 EPS-blastoids (21.4%) collected on day 5 or 6 (Figure 3I). The number of EPI- and PE-like cells in these GATA4+ EPS-blastoids were comparable to that in E4.5 blastocysts, although EPS-blastoids exhibited higher variations (Figure 3J).
We also checked the expression of the lineage markers in EPS-blastoids generated from ES-converted EPS, Liu-EPS, as well as a single EPS cell. Consistently, all of these EPS-blastoids exhibited blastocyst-like allocation of cell lineages, as revealed by CDX2, SOX2/NANOG, and GATA4 staining (Figure S3E–S3H).
Transcriptome Analysis of EPS-blastoids
We performed RNA-Seq analysis using individual EPS-blastoids and compared their transcriptomes to E3.5 early blastocysts and E3.0 morulae from published datasets (Sampath Kumar et al., 2017; Wang et al., 2018). Principle component analysis revealed that EPS-blastoids were closer to blastocysts than morulae on both PC1 and PC2 axis (Figure 4A). Unsupervised correlation clustering also showed that EPS-blastoids clustered closer to blastocysts than to morulae (Figure S4A). These RNA-seq data also revealed differentially expressed genes (DEGs) between EPS-blastoids, blastocysts, and morulae (Table S2). We identified 928 DEGs (including novel genes from a de novo transcriptome assembly; 1.8% of all genes) between EPS-blastoids and blastocysts (Figure S4B and S4D). In contrast, 4707 DEGs (8.9% of all genes) were found between EPS-blastoids and morulae (Figure S4C). DEGs between EPS-blastoids and blastocysts were enriched in pathways related to metabolism (Figure S4E). Thus, at the bulk RNA-Seq level, EPS-blastoids more resembled blastocysts than morulae.
Figure 4. Transcriptome Analysis of EPS-blastoids.
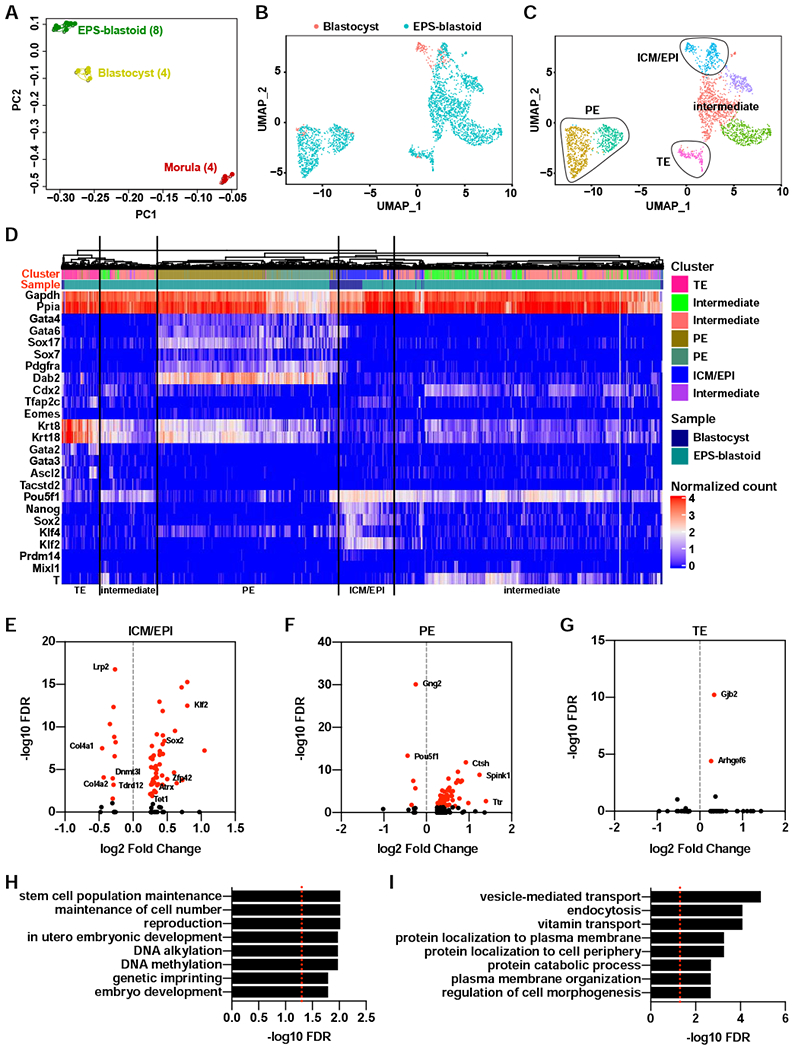
(A) Principle component analysis (PCA) of bulk RNA-Seq data from individual EPS-blastoid, blastocyst, and morula. The number of biological replicates in each group was shown inside the parenthesis.
(B) A Umap plot of 2702 cells from blastocysts and EPS-blastoids after alignment using the Seurat package.
(C) A Umap plot showing the clustering of all cells. The identities of each cluster were determined based on the expression of the lineage markers.
(D) Unsupervised clustering analysis showing the cells of similar lineage identities cluster to each other regardless of sample type. The cluster row indicates the subpopulation defined in (C). The sample row indicates the sample type.
(E to G) Dot plots showing the differentially expressed genes (DEGs) in the ICM/EPI lineage (E), PE lineage (F), and TE lineage (G) between blastocysts and EPS-blastoids. Genes with FDR exceeding the statistical significance cutoff (FDR < 0.05) are labeled with red color.
(H and I) Gene ontology analysis of biological functions for DEGs in the ICM/EPI lineage (H) and PE lineage (I) between blastocyst and EPS-blastoids. Red dotted line indicates the cutoff (FDR < 0.05).
To gain insights into the similarities and the differences within each lineage between EPS-blastoids and blastocysts, we performed single-cell RNA-Seq and profiled the transcriptomes of over 2700 single cells collected from EPS-blastoids and blastocysts. Integrated analysis using SEURAT revealed that the cells from EPS-blastoids and blastocysts largely overlapped with each other (Figure 4B). Clustering analysis divided all cells into 7 clusters, with 4 of them shared by both EPS-blastoids and blastocysts (Figure 4C and S4F). We examined the expression of a panel of 15 lineage markers and determined the lineages associated with each cluster (Figure S4G). Based on the expression pattern of marker genes, we identified one cluster as ICM/EPI, one cluster as TE, and two clusters as PE. The remaining three clusters, which came mostly from EPS-blastoids, showed mixed expression of both ICM/EPI and TE markers and may represent intermediate and/or uncommitted cell types (Figure 4C). In addition, we performed unsupervised clustering analysis and confirmed the clustering of analogous lineages from both samples (Figure 4D). Overall our results confirm that EPS-blastoids contain all three blastocyst cell lineages.
To uncover differences within each lineage between EPS-blastoids and blastocysts, we performed functional annotation of DEGs. 53 DEGs were identified for the ICM/EPI lineage between the two samples (Figure 4E; Table S3), which were enriched with functional terms related to stem cell maintenance, reproduction, and DNA methylation (Figure 4H; Table S3). Two pluripotency transcription factors Sox2 and Klf2 were expressed at lower levels (by ~28% and ~43% respectively), and Tet1 and Dnmt3L, two DNA methylation related enzyme genes, showed ~18% decreased level in EPS-blastoids (Figure 4E). For the PE lineage, 67 DEGs were identified between EPS-blastoids and blastocysts, which were mostly enriched in terms related to vesicle transport and endocytosis (Figure 4F and 4I; Table S3). For the TE lineage, only two DEGs (Gjb2 and Arhgel6) were identified to be significantly different between the two samples (Figure 4G; Table S3).
In Vitro Developmental Potential of EPS-blastoids
ESCs, TSCs, and XEN cells, which are considered the in vitro counterparts of EPI, TE, and PE lineages, respectively, could all be derived directly from blastocysts. We asked whether EPS-blastoids could also give rise to these three stem cell lines. Using the ground state culture condition (Ying et al., 2008), ESC lines were successfully established from four out of five EPS-blastoids. These ESCs formed colonies with similar morphologies to those generated from natural blastocysts and expressed the pluripotency factors OCT4, NANOG, and SOX2, but not the trophoblast marker CDX2 (Figure 5A and S5A). Injection of EPS-blastoid-derived ESCs into natural blastocysts resulted in adult chimeras (Figure 5B). We also derived eight TSC lines from 17 EPS-blastoids. These TSCs morphologically resembled those generated from natural blastocysts and expressed the TE transcription factors CDX2 and EOMES, but not OCT4 or NANOG (Figure 5C and S5B). Injection of EPS-blastoid-derived TSCs into blastocyst followed by embryo transfer generated chimeric placental tissues that contained CK8+ EPS-blastoid TSC-derivatives (Figure 5D). Finally, two XEN cell lines were also established from six EPS-blastoids (Figure 5E). These EPS-blastoid-derived XEN cells expressed the PE transcription factors GATA4 and GATA6 (Figure S5C and S5D) and could chimerize host yolk sac in utero (Figure 5F).
Figure 5. In Vitro Developmental Potential of EPS-blastoids.
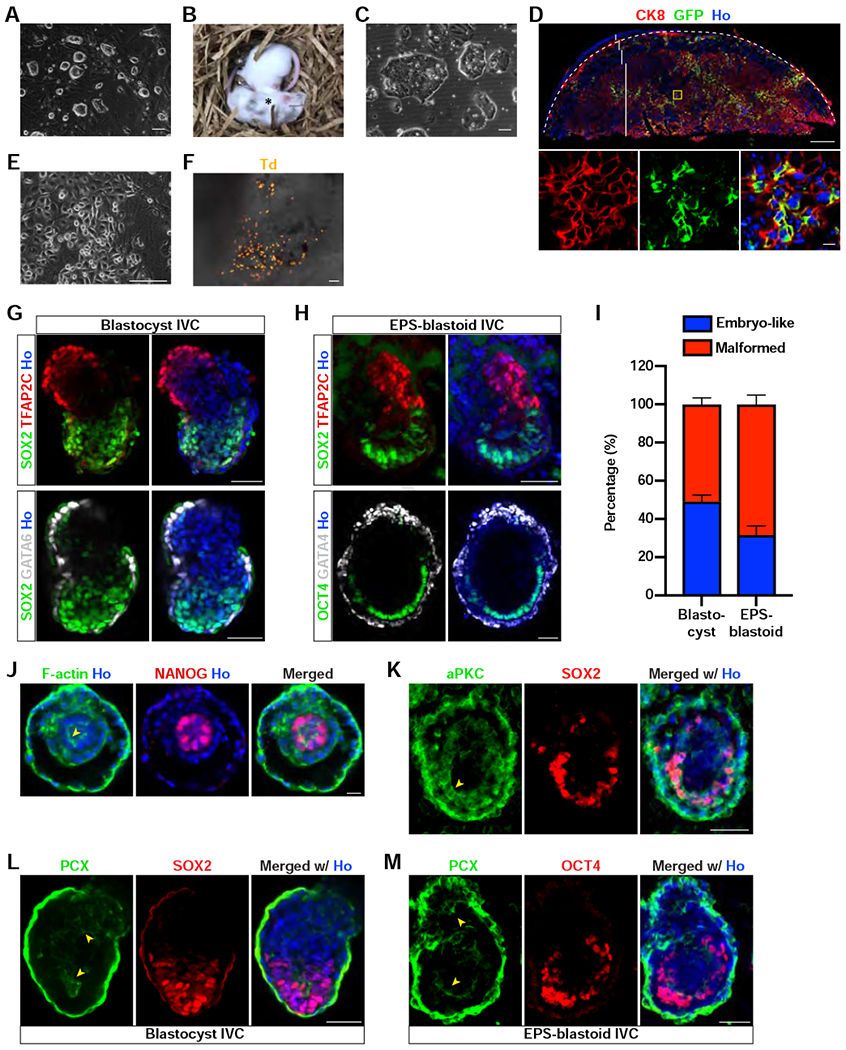
(A) Phase contrast image of de novo derived ES cell lines from EPS-blastoids.
(B) Brightfield image of two littermates generated from blastocyst injected with EPS-blastoid-derived ES cells showing that these cells can contribute to chimeric mice. The star symbol denotes a chimeric mouse.
(C) Phase contrast image of de novo derived TS cell lines from EPS-blastoids.
(D) Immunofluorescence staining of a placental section for CK8 and GFP. The panels below are the enlargement of the yellow boxed region. The placenta was delineated by a dotted line; dec, decidua layer; gc, giant cell layer; sp, spongiotrophoblast layer; laby, labyrinth layer.
(E) Phase contrast image of de novo derived XEN cell lines from EPS-blastoids.
(F) Brightfield image of a yolk sac overlaid with tdTomato epifluorescence image showing EPS-blastoid-derived XEN cells can contribute to the developing yolk sac. Td, tdTomato.
(G) Immunofluorescence staining of blastocyst-derived postimplantation embryo-like structures for TFAP2C and SOX2 (upper panel) or GATA6 and SOX2 (lower panel).
(H) Immunofluorescence staining of EPS-blastoid-derived postimplantation embryo-like structures for TFAP2C and SOX2 (upper panel) or GATA4 and OCT4 (lower panel).
(I) Quantification of the percentage of postimplantation embryo-like and malformed structures formed after in vitro culture of blastocysts and EPS-blastoids. n = 3 and 4 independent assays for blastocysts and EPS-blastoids, respectively.
(J) Immunofluorescence staining of an EPS-blastoid-derived peri-implantation embryo-like structure for F-actin and NANOG showing the formation of rosette EPI-like structure.
(K) Immunofluorescence staining of an EPS-blastoid-derived postimplantation embryo-like structure for aPKC and SOX2. Yellow arrowhead denotes the apical domain.
(L and M) Immunofluorescence staining of a blastocyst- (L) or an EPS-blastoid- (M) derived postimplantation embryo-like structure for PCX and SOX2 (L) or PCX and OCT4 (M). Yellow arrowheads denote the enrichment of PCX protein around the lumens in both the EPI and ExE-like structure. PCX, podocalyxin.
Data are represented as mean ± SEM. Scale bar, 500 μm (D, upper), 100 μm (A, C, E, and F), 50 μm (G, H, K, L, and M), 20 μm (D, bottom; and J). Ho, Hoechst.
See also Figure S5.
A recently developed in vitro culture (IVC) system enabled the development of mouse and human blastocysts beyond the implantation stages in vitro, and has also been successfully used to assemble postimplantation embryo-like structures (Bedzhov and Zernicka-Goetz, 2014; Bedzhov et al., 2014a; Harrison et al., 2017; Sozen et al., 2018). We tested whether the IVC condition could support the development of EPS-blastoids beyond the implantation stage. In agreement with the reports from the Zernicka-Goetz group (Bedzhov and Zernicka-Goetz, 2014), in vitro culture of blastocysts generated an egg cylinder structure with extraembryonic ectoderm (ExE, marked by TFAP2C) and EPI (marked by SOX2) as two hemispheres enclosed by the visceral endoderm (VE, marked by GATA6) (Figure 5G). Similar structures formed when EPS-blastoids were cultured in the IVC media (Figure 5H). Approximately 49% of cultured blastocysts and 32% cultured EPS-blastoids generated an organized egg cylinder structure (Figure 5I).
We also examined the cellular and molecular events that contributed to the generation of the egg cylinder structures from EPS-blastoids. During the peri-implantation stage, EPI cells become polarized and form a rosette-like structure with apical domains clustered in the center. In cultured EPS-blastoids, we observed that F-actin was enriched in the center of the EPI-like compartment and the cells adopted a rosette-like configuration, reminiscent of a peri-implantation embryo at ~E4.5—E4.75 stage (Figure 5J). The polarity protein aPKC lined the cells that formed the cavity within the EPI-like compartment (Figure 5K), characteristic of an E5.25—E5.5 embryo. Lumenogenesis in the postimplantation embryos depends on membrane repulsion mediated by podocalyxin (PCX) (Bedzhov and Zernicka-Goetz, 2014). As seen in natural embryos, PCX levels were highest on the apical side of the cells enclosing the lumen in both the EPI- and ExE-like compartments of EPS-blastoid-derived structures (Figure 5L and 5M). Lumenogenesis also depends on signaling from the VE-derived basement membrane, and we found that both the EPI- and ExE-like compartments were surrounded by a laminin-containing basement membrane adjacent to the GATA4+ VE-like cells (Figure S5E and S5F).
In sum, EPS-blastoids could give rise to functional ESCs, TSCs, and XENs, and upon further cultivation, could develop into postimplantation embryo-like structures.
In Vivo Developmental Potential of EPS-blastoids
A more stringent functional test for blastoids is to determine whether they can develop into fetuses in utero. To this end, we transferred EPS-blastoids into pseudopregnant mice at 2.5 days post coitum (dpc) and analyzed their in vivo developmental potential. At 7.5 dpc, decidua formed in the uteri of both control mice and surrogates that had been transferred with EPS-blastoids (Figure 6A and S6A). We confirmed vascular permeability of EPS-blastoid-derived implantation sites by Evans blue dye (Figure 6B and S6B). Transfer experiments were performed using EPS-blastoids generated from several different lines and overall ~7% of transferred EPS-blastoids implanted and induced decidualization (Figure S6C; Table S4). This efficiency is comparable to that reported for ETS-blastoids (Rivron et al., 2018). Although control decidua appeared uniform in size, the size of EPS-blastoid-induced decidua varied, with some similar to the control and others much smaller (Figure S6D). Genomic PCR analysis using a primer pair specific for the tdTomato gene showed that the decidua tissue contained tdTomato+ cells derived from EPS-blastoids (Figure 6C). Immunohistochemistry analysis of decidua sections also confirmed the presence of tdTomato+ cells (Figure 6D). In addition, an embryonic axis was established in deciduae induced by both a control blastocyst and an EPS-blastoid (Figure 6E). We observed that some structures were formed within the EPS-blastoid-derived deciduae at 6.5, 7.5, and 8.5 dpc, respectively (Table S5), which all appeared retarded or malformed when compared with control E6.5—E8.5 embryos (Figure 6F, 6G, S6E, and S6F. Nonetheless, we were able to detect the presence of OCT4+, EOMES+, and GATA4+ cells in sections prepared from these structures (Figure 6H–6J, S6G–S6I). These results suggest that EPS-blastoids can implant, trigger decidualization, and continue to grow inside the uterus.
Figure 6. In Vivo Developmental Potential of EPS-blastoids.
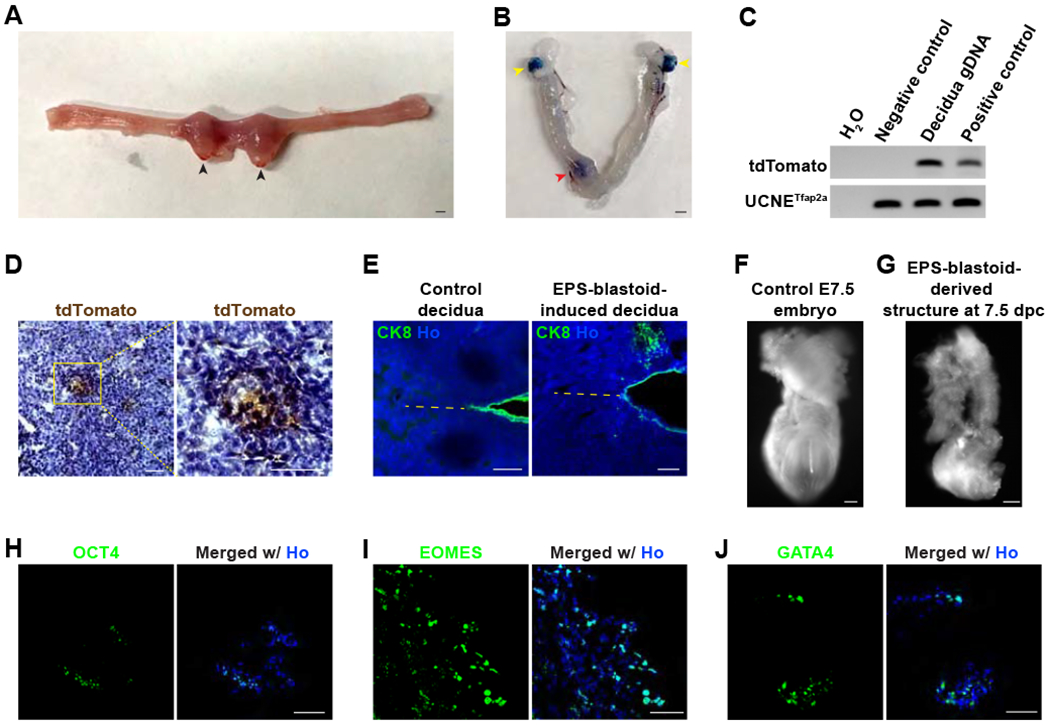
(A) Brightfield image showing the formation of decidua in the mouse uterus 5 days after EPS-blastoids transfer at 2.5 dpc. Black arrowheads indicate deciduae.
(B) Brightfield image of a mouse uterus 5 days after EPS-blastoids transfer at 2.5 dpc with Evan’s blue staining. The red arrowhead indicates a decidua. Yellow arrowheads denote the ovaries.
(C) PCR analysis of genomic DNA for the tdTomato gene reveals the presence of EPS-blastoid-derived cells in the decidua tissue. UCNETfap2a was used as an internal loading control.
(D) Immunohistochemistry analysis of decidua sections showing the decidua contains EPS-blastoid-derived tdTomato+ cells. The image on the right is the enlargement of the yellow box region.
(E) Immunofluorescence staining of a section from control decidua (left) or EPS-blastoid-induced decidua (right) for CK8. The dotted line indicates the embryonic axis. AM, antimesometrial pole; M, mesometrial pole.
(F and G) Brightfield images of a control E7.5 embryo (F) or an in vivo EPS-blastoid-derived structure recovered from decidua at 7.5 dpc (5 days after EPS-blastoids transfer) (G).
(H - J) Immunofluorescence staining of sections from an in vivo EPS-blastoid-derived structure recovered from decidua at 7.5 dpc (5 days after EPS-blastoids transfer) for OCT4 (H), EOMES (I), and GATA4 (J).
Scale bar, 1 mm (A, and B), 100 μm (D, E, F, and G), and 50 μm (H, I, and J). Ho, Hoechst.
Generation of EPS-blastoids from Somatic Cells
As a proof-of-concept that EPS-blastoids can be generated without using any cells of embryonic origin, we next tested generating EPS-blastoids from somatic cells. Through somatic cell reprogramming, we established EPS cells from mouse ear fibroblasts (induced EPS cells, or iEPS cells), which were subsequently used for blastoid formation. We successfully generated blastoids from iEPS cells (referred to as iEPS-blastoids) in ~15% of aggregates (Figure 7A and 7B; Table S1). Similar to EPS-blastoids, iEPS-blastoids also morphologically resembled natural blastocysts and were of similar size as E3.5 blastocysts (Figure 7A and 7C). In addition, the process of the induction of iEPS-blastoids recapitulated the compaction, polarization, and changes in subcellular YAP localization (Figure 7D, 7E, and 7F). iEPS-blastoids displayed the correct spatial expression of markers for both embryonic and extraembryonic lineages (Figure 7G and S7A). In addition, further culture of iEPS-blastoids in IVC media generated egg-cylinder structures containing ExE-, EPI-, and VE-like compartments (marked by TFAP2C, SOX2/OCT4, and GATA4, respectively) (Figure 7H, S7B, and S7C). Lastly, iEPS-blastoids were also able to implant into the uterus and induced the formation of decidua (Figure 7I). Collectively, these data demonstrated that iEPS-blastoids generated from adult somatic cells are similar to those from embryo-derived stem cells.
Figure 7. Generation of EPS-blastoids from Somatic Cells.
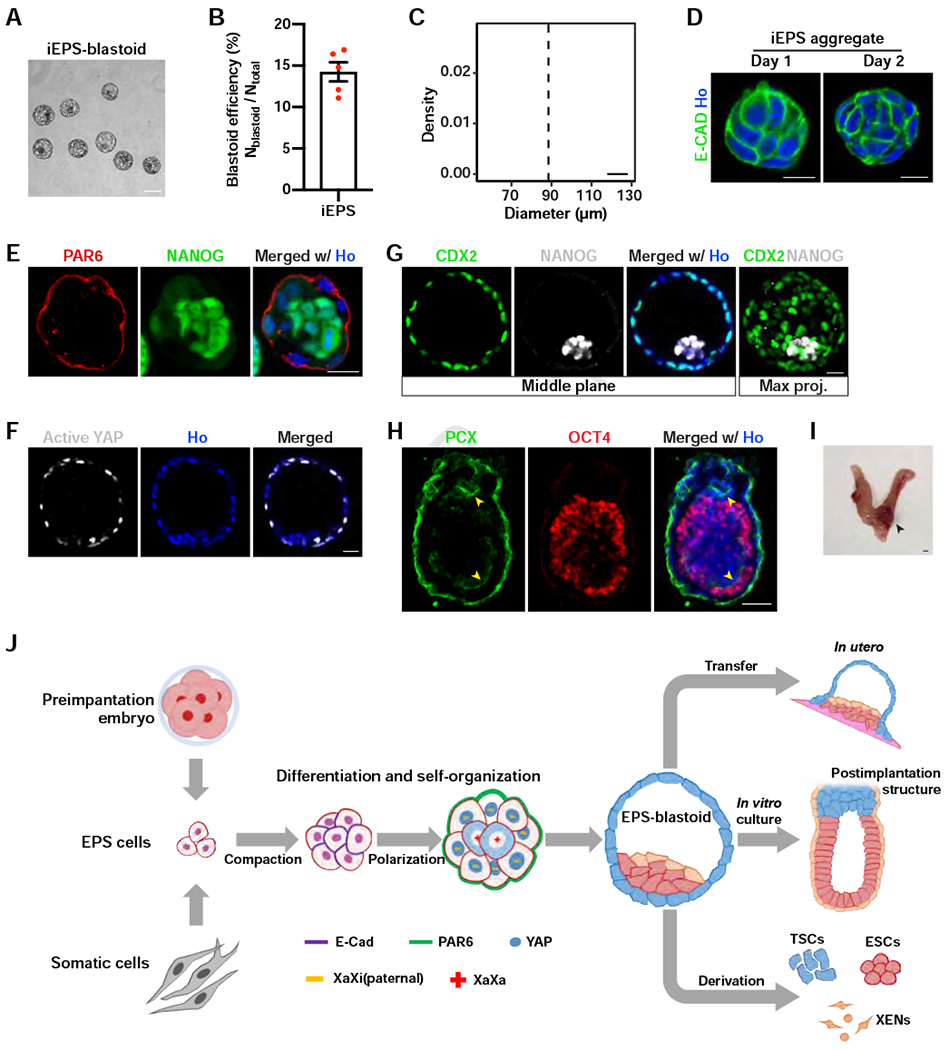
(A) A phase contrast image of iEPS-blastoids.
(B) Quantification of iEPS-blastoids formation efficiency. n = 5 independent assays.
(C) Histogram showing the distribution of diameters of iEPS-blastoids. n = 23 iEPS-blastoids.
(D) Immunofluorescence staining of iEPS aggregates at the indicated day of blastoid induction for E-cadherin.
(E) Immunofluorescence staining of an iEPS aggregate at day 2 of blastoid induction for PAR6 and NANOG.
(F) Immunofluorescence staining of an iEPS-blastoid for active YAP.
(G) Immunofluorescence staining of an iEPS-blastoid for CDX2 and NANOG. The rightmost panel shows the maximum intensity projection of z-stack images of the indicated protein.
(H) Immunofluorescence staining of a postimplantation embryo-like structure from in vitro culture of iEPS-blastoids for PCX and OCT4. The yellow arrowheads indicate the lumens lined with PCX.
(I) Brightfield image showing the formation of decidua in the mouse uterus 5 days after iEPS-blastoids transfer at 2.5 dpc. Black arrowhead indicates decidua.
(J) A diagram summarizing the major findings of this study.
Data are represented as mean ± SEM. Scale bar, 1 mm (I), 100 μm (A), 50 μm (H), and 20 μm (D, E, F, and G). Ho, Hoechst.
DISCUSSION
It has recently been demonstrated that EPS cells that maintain bi-developmental potential towards both Em and ExEm lineages can be derived and stably maintained in culture. Here we showed that EPS cells alone could differentiate and self-organize to generate blastocyst-like structures that share several cellular, molecular, and functional features with natural blastocysts (Figure 7J). Our method provides a unique and highly malleable in vitro system for studying early preimplantation development, and with further optimization, may potentially be harnessed to generate fully functional synthetic embryos in vitro.
EPS-blastoid formation recapitulates several key early preimplantation developmental processes, including compaction, polarization, changes in subcellular YAP localization, and paternal XCI in TE. To our best knowledge, this represents the first time that these events are modeled in an artificial embryo, which indicates that EPS-blastoid formation shares and follows certain similar developmental paths with those that generate a mouse blastocyst. More analyses of other molecular and cellular processes are warranted to provide a better understanding of the similarities and differences between the blastocyst and EPS-blastoid formation. This, in turn, may help us to better understand the evolutionarily distinct molecular signals that underlie the different forms and patterns that arise during embryogenesis.
In addition, our EPS-only blastoid approach has several additional advantages. First, unlike ETS-blastoids, which are generated by sequential aggregation using multiple cells from two stem cell types (ESCs and TSCs), all cells within EPS-blastoids come from a single cell type (even from a single cell). This enables a more straightforward interpretation of the readouts after introducing genetic or epigenetic changes. Second, EPS-blastoids can be further cultured in vitro and recapitulate several key morphogenetic processes of early postimplantation development, forming an egg cylinder embryo-like structure. Interestingly, Rivron’s group recently reported that ETS-blastoids developed poorly into postimplantation structures in vitro. An additional PE induction step was devised to promote the generation of postimplantation-like structures from ETS-blastoids (Vrij et al., 2019). We believe one key difference could be due to EPS cells’ superior ability to generate PE cells than ESCs.
Bulk RNA-Sseq analysis of individual EPS-blastoids revealed that they were more similar to blastocysts than morulae. Single-cell RNA-Seq analysis confirmed that EPS-blastoids contained all three cell lineages of blastocysts. We also identified several DEGs for each lineage (EPI/ICM, TE, and PE) between EPS-blastoids and blastocysts. For PE, DEGs seem to be enriched in terms related to vesicle transport and endocytosis. For ICM/EPI, a group of DNA methylation- and genomic imprinting-related genes, namely Tet1, Dnmt3L, Zfp42, Atrx, and Tdrd12, were expressed at lower levels in EPS-blastoids than blastocysts. These data suggest that epigenetic abnormalities, well-known defects for embryonic development (Barton et al., 1991; Surani et al., 1990), likely play a negative role in EPS-blastoids development in utero. Another notable observation from single-cell RNA-Seq analysis is that there are several subpopulations between EPI/ICM and TE. These cells likely represent uncommitted or improperly differentiated cells, likely as a result of suboptimal differentiation condition. Further optimization of differentiation culture and protocol is needed to overcome these deficiencies.
Since our EPS-blastoid formation protocol includes small molecules and growth factors that promote differentiation and growth of the TE lineage, it raises the concern that pluripotency of the ICM-like compartment may be compromised. But this is unlikely because 1) we derived chimera competent ESC lines from EPS-blastoids; 2) the ICM-like cells further segregated into EPI and PE; 3) following IVC culture, the EPI-like compartment was polarized, adopted a rosette-like morphology, and underwent lumenogenesis; and 4) OCT4-expressing cells could be detected in tissues isolated from EPS-blastoid deciduae in vivo. These results collectively indicate that the ICM-like compartment of EPS-blastoids remains functional, at least until lumenogenesis during early postimplantation development.
Although mouse EPS cells exhibit bi-directional developmental potency (in vivo chimera formation (Yang et al., 2017b) and in vitro blastoid generation (this study)) as well as some molecular features of early preimplantation embryos, they are clearly not equivalent to totipotent blastomeres. Nonetheless, and despite that EPS-blastoids were generated in an artificial milieu vastly different from that present in the in vivo microenvironment, the fact that several evolutionary conserved developmental processes were faithfully recapitulated highlights the plastic yet remarkably regulatory nature of early mammalian embryos. The transcriptional differences between laboratory created and naturally evolved blastocysts uncovered in our study, may reflect distinct molecular trajectories that, nonetheless, lead to the generation of similarly patterned structures. A deeper understanding of these differences may serve towards the development of a protocol to generate fully functional blastoids from cultured stem cells, as well as to help discover new intrinsic and extrinsic factors that safeguard normal mammalian embryogenesis.
In summary, we have established a 3D differentiation system for generating blastoids from cultured EPS cells derived from embryonic and adult sources. Our studies can serve as a framework for advancing the development of fully functional synthetic blastocysts, not only in mice but also in other mammalian species, including humans. As such this system could be used as an in vitro model for studying fundamental questions in both preimplantation and early postimplantation mammalian embryogenesis, modeling diseases related to early pregnancy, high-throughput pharmacological and toxicological screens, and possibly bioengineered embryogenesis.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to the Lead Contact, Juan Carlos Izpisua Belmonte (belmonte@salk.edu). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All procedures related to animals were performed following the ethical guidelines of the Salk Institute for Biological Studies. Animal protocols were reviewed and approved by the Salk Institute Institutional Animal Care and Use Committee (IACUC) before any experiments were performed. C57BL/6J (Stock No: 000664 | Black 6), ICR mice (Stock No: 009122), and C57BL/6-Tg(CAG-EGFP)1Osb/J (Stock No: 003291) were obtained from The Jackson Laboratory. To prepare pseudopregnant surrogates, ICR female mice (8 - 12 weeks old) in the estrus were mated with vasectomized ICR male mice. Mice were housed in a 12 hr light/12 hr dark cycle in a temperature-controlled facility with free access to water andfood.
Culture of Mouse Embryos
Mouse 2-cell embryo or blastocysts were flushed out of the uterus of pregnant female C57BL/6J or ICR and cultured in drops of KSOM medium (homemade or Millipore, MR-020P-5D) covered by a layer of mineral oil (Sigma-Aldrich, M8410) in a humidified incubator under 5% CO2 at 37°C. Homemade KSOM was prepared according to a previously published recipe (Wu et al., 2017). The KSOM medium contains: NaCl (95 mM), KCl (2.5 mM), KH2PO4 (0.35 mM), MgSO4 (0.2 mM), NaHCO3 (25 mM), CaCl2 (1.71 mM), Na2-EDTA (0.01 mM), L-glutamine (1.0 mM), Na lactate (10 mM), Na pyruvate (0.2 mM), glucose (5.56 mM), essential amino acid (EAA; 10.0 ml/l), nonessential amino acid (NEAA; 5.0 ml/l), and BSA (4 g/l). In some experiments, KSOM-HEPES medium was used. KSOM-HEPES medium was prepared using the same amount of chemicals as KSOM with the following changes: using lower amount of NaHCO3 (5 mM), the addition of HEPES-Na (20 mM), without EAA or NEAA, and substitution of BSA by PVA (0.1 g/l). All reagents were from Sigma-Aldrich except for NEAA and EAA, which were from Thermo Fisher Scientific. The sex of the mouse embryos was not determined. Both male and female embryos were used in all experiments.
To culture blastocysts beyond the implantation stage, we used a protocol developed by the Zernicka-Goetz group (Bedzhov and Zernicka-Goetz, 2014; Bedzhov et al., 2014b). Blastocysts were first treated in drops of Tyrode’s Solution, Acidic (Sigma-Aldrich) for one to two minutes to digest the zona pellucida. The zona-free blastocysts were washed in drops of KSOM medium and transferred into u-Slide 8 well (ibidi, 80826) containing pre-equilibrated IVC-1 medium (Cell Guidance Systems, M11-25). We plated 20 - 25 blastocysts into each well of u-Slide. Within 2 to 3 days, blastocysts attached to the plate and medium was replaced with pre-equilibrated IVC-2 (Cell Guidance Systems, M12-25). The culture continued for an additional 4 - 6 days and was fixed with 4% PFA for 15 min at room temperature for immunofluorescence analysis.
Culture of Mouse Stem Cells
All stem cell lines were cultured on a layer of irradiated CF1 mouse embryonic fibroblasts (MEF) under 20% O2 and 5% CO2 at 37°C. The chimera-competent naïve ES cell line (B6N-22; male) was a gift from Fumihiro Sugiyama (Tanimoto et al., 2008). ES cells were cultured in N2B27-based medium. N2B27 basal medium was composed of 1:1 mixture of DMEM/F-12 (11330-032) and Neurobasal (21103-049) supplemented with 0.5X N2 (17502-048), 0.5X B27 (17504-044), 1X NEAA (11140-050), 1X GlutaMAX (35050-061), 0.1 mM 2-mercaptoethanol (21985-023), and 0.1% BSA (15260-037, optional) or 5% KnockOut Serum Replacement (10828-028, optional) (all from Thermo Fisher Scientific). Mouse ESCs were maintained in N2B27 medium supplemented with 10 ng/ml hLIF (Peprotech, 300-05), 3 μM CHIR99021 (Reagents Direct, 27-H76), and 1 μM PD0325901 (Selleck Chemicals, S1036) (hereinafter referred to as N2B272iL) (Ying et al., 2008) on a layer of irradiated MEF and passaged every two to three days using TrypLE (Thermo Fisher Scientific, 12604-013). The B6 GFP+ naïve ES cell line was derived from C57BL/6-Tg(CAG-EGFP)1Osb/J blastocyst using the 2iL protocol. Blastocysts were collected from timed-pregnant mice and transferred onto a MEF feeder layer in a 96-well plate and cultured in N2B272iL medium. The cell outgrowth was dissociated and replated on new MEF feeder cells. Cell lines were established by dissociating individual colony using 0.05% trypsin-EDTA and replating into a new well.
The two EPS cell lines (EPS 1 and EPS 2, tdTomato+; both were male) derived from 8-cell embryos were obtained from the Hongkui Deng’s lab. These two cell lines EPS cells were cultured on irradiated MEF cells in N2B27 basal medium supplemented with 10 ng/ml LIF (Peprotech, 300-05), 3 μM CHIR99021 (Reagents Direct, 27-H76), 2 μM (S)-(+)-Dimethindene maleate (Tocris, 1425), and 2 μM minocycline hydrochloride (Santa Cruz Biotechnology, sc-203339) (hereinafter referred to as N2B27LCDM). In some experiments, the EPSC culture protocol developed by Pentao Liu’s lab was used (Yang et al., 2017b). The Liu-EPS culture medium was CDF12 basal medium supplemented with 10 ng/ml hLIF (Peprotech, 300-05), 3 μM CHIR99021 (Reagents Direct, 27-H76), 1 μm PD0325901 (Selleck Chemicals, S1036), 4 μM JNK Inhibitor VIII (Millipore, 420135), 10 μM SB203580 (Tocris, 1402), 0.3 μM A-419259 (Tocris, 3914), and 5 μM XAV939 (Sigma-Aldrich, X3004). EPS cells were routinely passaged every two days at a ratio of 1:10 to 1:20. CDF12 basal medium was composed of DMEM/F-12 (11330-032) supplemented with 20% KnockOut Serum Replacement (10828-028), 1X NEAA (11140050), 1X GlutaMAX (35050-061), and 0.1 mM 2-mercaptoethanol (21985-023) (all from Thermo Fisher Scientific). The female X-GFP mEpiSC (female) was a gift from Dr. Azim Surani (Bao et al., 2009). EpiSCs were cultured in CDF12 medium supplemented with 12.5 ng/ml bFGF (Peprotech, 100-18B). To convert naïve ESCs or EpiSCs into EPS cells, naïve ESC or EpiSCs were first seeded on MEF feeder cells with ESC or EpiSC medium, respectively. After 24 hr, the medium was removed and replaced with EPS medium. The conversion process usually completed after five passages in the EPS conditions.
TSCs were maintained in basal TSC medium supplemented with 25 ng/ml rhFGF4 (R&D, 235F4025) and 1 μg/ml Heparin (Sigma-Aldrich, H3149) on a layer of irradiated MEF (Tanaka et al., 1998). TSC basal medium was composed of RPMI 1640 (11875-093) supplemented with 20% Fetal Bovine Serum (FBS) (16000-044), 1X GlutaMAX (35050061), 1X Sodium pyruvate (11360-070), and 0.1 mM 2-mercaptoethanol (21985-023) (all from Thermo Fisher Scientific). TSCs were passaged every five to seven days at 1:5-1:10 using 0.05% trypsin (Thermo Fisher Scientific, 25300-054). In addition, XEN cells were cultured in the TSC basal medium and passaged every 4 to 5 days using TrypLE.
Culture of Mouse Ear Fibroblasts
Mouse ear fibroblasts were derived by plating minced ear tissue in DMEM (11950-040) supplemented with 10% FBS (16000-044), 1X NEAA (11140-050), and 1X GlutaMAX (35050-061) (all from Thermo Fisher Scientific) under 20% O2 and 5% CO2 at 37°C. When confluent, ear fibroblasts were split using TrypLE (Thermo Fisher Scientific, 12604-013) at 1:5.
METHOD DETAILS
Reprogramming of Mouse Ear Fibroblasts
Retrovirus containing the four Yamanaka factors (Takahashi and Yamanaka, 2006) were packaged in 293 cells by transfection of pMXs-c-Myc, pMXs-Klf4, pMXs-Sox2, pMXs-Oct3/4 (all from Addgene). Mouse ear fibroblasts at passage 1 to 3 were used for reprogramming into iPS cells by incubation with the mixed retrovirus for two days. Then medium was replaced with CDF12 supplemented with 10ng/ml LIF (CDF12LIF). iPS colonies were picked up, dissociated with TrypLE, and replated into a new MEF well for establishing individual iPS cell line. iPS cells were routinely cultured in either CDF12LIF or N2B272iL iPS cells were converted into EPS cells by culturing in N2B27LCDM for at least five passages.
Lentiviral Transduction of EPS Cells
Lentiviral particle encoding the puromycin resistant gene and the mCherry gene (Lenti-EF1a-puromycin-mcherry) (Liao et al., 2015) were packaged in 293 cells via transfection. Medium supernatant containing the lentiviral particles was collected 48 hr after transfection and concentrated by ultracentrifugation at 25,000 r.p.m (82,700g) at 4°C for 2 hr using a Beckman ultracentrifuge (Beckman Coulter) (Kutner et al., 2009). The lentivirus pellet was resuspended with cold DPBS. EPS cells were transduced by incubating with lentivirus-containing N2B27LCDM for 48 hr. Upon passaging, puromycin (1 μg/ml; InvivoGen, ant-pr-1) was supplemented in the medium to eliminate untransduced cells.
Generation of EPS-blastoids
EPS colonies were dissociated into single cells by incubation with TrypLE (Thermo Fisher Scientific, 12604-013). Cell resuspension was transferred into a 0.1% gelatin-coated plate and incubated at 37°C for 30 min to allow irradiated MEF cells attach to the plate. The supernatant containing the EPS cells were collected, filtered through a 40 pm cell strainer, and counted using the TC-10 counter (Bio-Rad, 1450001). AggreWell 400 (STEMCELL Technologies, 34415) was prepared following the manufacturer’s instructions. EPS-blastoid basal medium is composed of 25% TSC basal medium (see above), 25% N2B27 basal medium (see above), and 50% KSOM (see above). In some experiments, M16 (Sigma-Aldrich, M7292) was used to replace KSOM. Approximately 6,000 cells (five cells per microwell for 1200 microwells) were resuspended in EPS-blastoid basal medium supplemented with 2 μM ROCK inhibitor Y-27632 (Reagents Direct, 53-B80-50), 12.5 ng/ml rhFGF4 (R&D, 235F4025), 0.5 μg/ml Heparin (Sigma-Aldrich, H3149), 3 μM GSK3 inhibitor CHIR99021 (Reagents Direct, 27-H76), 5 ng/ml BMP4 (Proteintech, HZ-1040), and 0.5 μM A83-01 (Axon Medchem, 1421) and seeded into one well of the 24-well AggreWell plate. The plate was centrifuged at 300g for one minute and transferred into an incubator. The day of cell seeding was counted as day 0 of the process. Medium was removed 24h later (day 1) and replaced with fresh medium without Y-27632. Additional medium change is optional for the rest of the EPS-blastoid formation process. Starting from day 4, blastoids were manually picked up using a mouth pipette (Sigma-Aldrich, A5177) under a stereomicroscope for analysis or downstream experiments. For testing of the effect of antagonists or inhibitors on EPS-blastoid induction, chemicals were added to the medium at day 1. XAV939 (5 μM; Tocris, 3748) or IWR-1-endo (10 μM; STEMCELL Technologies, 72562) was used to inhibit Wnt signaling. Verteporfin (2 μM; Tocris, 5305) was used to inhibit YAP/TEAD interaction. For testing whether a single EPS cell can form a blastoid, WT cells and Puromycin+/mcherry+ cells were mixed at a ratio of 10:1 and seeded onto Aggrewell 400 as stated above. Puromycin (0.25 μg/ml; InvivoGen, ant-pr-1) was added to the medium 24 hr later to eliminate helper cells gradually.
Derivation of Three Types of Stem Cells from EPS-blastoids
To derive ES and TS cells, individual EPS-blastoid was transferred onto a MEF feeder layer in a 96-well plate and cultured with ES culture medium (N2B272iL) or TS culture medium, respectively. Within 2-3 days, EPS-blastoids attached to the plate and outgrowth was observed. Outgrowth was dissociated with 0.05% trypsin and plated into a new plate with MEF feeders. Individual colony was manually picked, dissociated, and seeded onto a new 96-cell MEF feeders for cell line derivation. The derivation of XEN cell line was performed following an established protocol (Rugg-Gunn, 2017) with modifications. EPS-blastoids were plated individually in a 24-well plate pre-coated with MEF feeders in XEN derivation medium (TSC basal medium supplemented with 25 ng/ml rhFGF4 and 1 μg/ml Heparin). The outgrowth was formed around day 3. And medium was changed every 3 days until the XEN cells are around 80% confluent. The XEN cells were dissociated into single cells using TrypLE Express for 5 min at 37°C with gentle pipetting. Dissociated cells were transferred into a 15 ml falcon tube containing 5x volume of digestion mixture and collected by centrifugation at 300g for 5 min. After removal of the supernatant, the XEN cells were resuspended and seeded into a 6-well plate pre-coated with MEF feeders. After 2-3 passages, the medium was switched to XEN culture medium.
Chimeric assays of ES and TS cells from EPS-blastoids
ICR female mice were superovulated by intraperitoneal (i.p.) injection of 7.5 Unit of pregnant mares’ serum gonadotrophin (PMSG; Prospec-Tany Technogene, HOR-272) and 46-48 hr later 7.5 Unit of human chorionic gonadotrophin (HCG; Sigma-Aldrich, CG10-1VL), and then mated with ICR male mice immediately. The blastocysts were flushed from female mice 3.5 days after detection of the vaginal plug, and cultured in KSOM under 37°C with an atmosphere of 5% CO2 in the air. The blastoid-derived ES or TS cells were dissociated into single cells and placed into the working medium before blastocyst injection. For each chimeric blastocyst, 12-15 ES or TS cells were injected into the cavity of blastocyst assisted with a PIEZO impact drive (Primetech, Ibaraki, Japan). After injection, the chimeric blastocysts were rinsed three times and cultured in KSOM at 37°C with an atmosphere of 5% CO2 in the air. Fifteen to twenty chimeric blastocysts, which re-expanded after injection, were transferred into the uterine horn of 2.5 dpc pseudopregnant ICR female mice. For the ES chimeric group, full-term pups were delivery naturally from the pregnant mice at 17.5 days after embryo transfer; for TS chimeric group, the E14.5 fetus was dissected from uterine of pregnant mice 12.5 days after embryo transfer. Placenta was fixed with 4% PFA overnight and embedded in OCT. Frozen sections (10 μm thick) were cut using a microtome cryostat (Leica, model# CM1900-3-1).
Chimeric assay of XEN cells derived from EPS-blastoids
ICR female mice in natural estrous cycles were mated with same strain males, and blastocysts were collected from uterine at 3.5 dpc in KSOM-HEPES. They were cultured in the KSOM under 37°C and 5% CO2 until microinjection of XEN cells. The EPS-blastoid derived XEN cells were dissociated into single cells and placed into KSOM on ice before blastocyst injection. Fifteen cells were introduced into the blastocoel assisted with a PIEZO impact drive. Microinjected blastocysts were cultured in KSOM until embryo transfer to the surrogates. Microinjected blastocysts were surgically transferred to the uterine horn of 2.5 dpc pseudopregnant ICR females. 15-18 blastocysts were transferred to each surrogate. E11.5 fetuses were dissected from uterine for analysis.
In Vitro Culture of EPS-blastoids Beyond Implantation
EPS-blastoids were cultured beyond the implantation stage using a protocol as for blastocyst (see above). EPS-blastoids were manually picked up using a mouth pipette, washed twice with pre-equilibrated IVC-1 medium (Cell Guidance Systems, M11), and transferred into a μ-Slide 8-well (ibidi, 80826) containing the IVC-1 medium. Around 20 - 30 EPS-blastoids were plated in one well of the μ-Slide. Within one or two days, EPS-blastoids attached to the plate. Once the EPS-blastoids attached, the medium was switched to IVC-2 medium (Cell Guidance Systems, M12). In two to four days, postimplantation embryo-like structures emerged and were fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature for immunofluorescence staining analysis.
EPS-blastoid Transfer
EPS-blastoids were manually picked up under a stereomicroscope and transferred into KSOM droplets using a mouth pipette. The surrogate at 2.5 days post coitum (dpc) was anesthetized with ketamine (Putney) and xylazine (Akorn) and the uterine horn was exposed surgically. After three washes in KSOM, EPS-blastoids were loaded to the pipette with air bubble and transferred to the uterine horn, which was previously punctured with a 27G needle. Around 20 EPS-blastoids were transferred into each uterine horn. The process of transfer was typically performed within 20-30 min per surrogate. A C-section was performed at 6.5, 7.5, or 8.5 dpc, and the uterus was dissected out. For staining with Evans blue, the surrogate mice received a tail vein injection of 0.5% Evans Blue (MP Biomedicals, 151108) 15 min before the C-section. Deciduae were dissected out of the uterus, and embryo-like structures were dissected out of the deciduae. Tissue samples were fixed with 4% PFA overnight and embedded in OCT. Frozen sections (10 μm thick) were cut using a microtome cryostat (Leica, model# CM1900-3-1).
Immunofluorescence staining
Immunofluorescence for 2D cell culture, 3D cell aggregates, EPS-blastoid, early mouse embryos, and postimplantation embryo-like structures was performed following a previously established protocol (Gu et al., 2018) with small modifications. The samples were fixed with freshly prepared 4% PFA in PBS for 15 min at room temperature and permeabilized with 0.2% Triton X-100 in PBS for 15 min. Samples were then blocked with blocking buffer (PBS containing 5% normal donkey serum (NDS), 2% BSA, and 0.1% Tween 20) at room temperature for two hours or overnight (O/N) at 4°C. Primary antibodies diluted in blocking buffer were applied to samples and incubated for two hours at room temperature or O/N at 4°C. Samples were washed for three times with PBS containing 0.1% Tween 20 followed by the incubation with fluorescence-conjugated secondary antibodies diluted in blocking buffer (2 - 5 pg/ml) for 1 hr (2D culture) or 2 hr (3D structures and postimplantation embryo-like structures) at room temperature. Samples were washed for three times with PBS containing 0.1% Tween 20. Nuclei were counterstained with Hoechst 33342 at 1 pg/ml. In some experiments for staining with membrane-associated protein (E-cadherin, ZO1, and PAR6), Saponin (0.1%; MP Biomedicals, 102855) was used for permeabilization and wash to replace Triton X-100 and Tween20. For staining of tissue cryosections, an additional step of antigen retrieval between permeabilization and blocking was performed to incubate the sections in 1X HistoVT One (Nacalai Tesque, 06380-05) at 70°C for 20 min. When using mouse antibody on mouse tissue, Mouse on Mouse Basic Kit (Vector Laboratories, Cat# BMK-2202) was used after blocking with normal serum and BSA. Also, to reduce background, 1X TruBlack Lipofuscin Autofluorescence Quencher (Biotium, 23007) was applied to sections as the last step of the staining process. Image acquisition was performed using a Zeiss LSM 710 or 880 confocal microscope. Images were processed using Fiji (ImageJ, v2.0.0) (Rueden et al., 2017)(Schindelin et al., 2012) or Zen (Zeiss). 3D cell counting was performed using the Imaris software (Oxford Instruments). The primary antibodies and dilutions used were: anti-CDX2 (1:100; Biogenex, MU392A), anti-KRT8 (1:5; Developmental Studies Hybridoma Bank, TROMA-1), anti-EOMES (1:200; Abcam, Ab23345), anti-ECAD (1:200; Dako, M3612), anti-ZO1 (1:150, Invitrogen, 61-7300), Rabbit anti-OCT4 (1:200; Abcam, ab19857), Rabbit anti-GATA6 (1:100; Cell Signaling Technology, 5851), Mouse anti-laminin gamma 1 (1:5; DSHB, 2E8), Goat anti-GATA6 (1:200; R and D Systems, AF1700), anti-PAR6 (1:50, Santa Cruz Biotechnology, sc-166405), anti-YAP (1:100, Santa Cruz Biotechnology, sc101199), anti-active YAP (1:100, Abcam, ab205270), anti-GFP (1:1000, MBL, M048-3), anti-TFAP2C (1:50; Santa Cruz Biotechnology, sc-12762), anti-OCT4 (1:100; Santa Cruz Biotechnology, sc-5279), anti-NANOG (1:100; Invitrogen, 14-5761-80), anti-SOX2 (1:100; R&D, AF2018), anti-GATA4 (1:100; Santa Cruz Biotechnology, sc-1237), anti-aPKC (1:50; Santa Cruz Biotechnology, sc-17781), and anti-Podocalyxin (PCX) (1:200; R&D, MAB1556). Secondary antibodies used were: Alexa Fluor 488 Donkey anti-Rat IgG (H+L) (Thermo Fisher Scientific, A-21208), Alexa Fluor 488-AffiniPure Donkey Anti-Goat IgG (H+L) (Jackson ImmunoResearch Labs, 705-545-147), Alexa Fluor 488-AffiniPure Donkey Anti-Mouse IgG (H+L) (Jackson ImmunoResearch Labs, 715-545-151), Alexa Fluor 488-AffiniPure Donkey Anti-Rabbit IgG (H+L) (Jackson ImmunoResearch Labs, 711545-152), Alexa Fluor 555 Donkey Anti-Mouse IgG (H+L) (Thermo Fisher Scientific, A-31570), Alexa Fluor 555 Donkey anti-Rat IgG (H+L) (Abcam, ab150154), Alexa Fluor 647 Donkey anti-Rat IgG (H+L) (Abcam, ab150155), Alexa Fluor 647-AffiniPure Donkey Anti-Goat IgG (H+L) (Jackson ImmunoResearch Labs, 705-605-147), Alexa Fluor 647-AffiniPure Donkey Anti-Mouse IgG (H+L) (Jackson ImmunoResearch Labs, 715-605-151), Alexa Fluor 647-AffiniPure Donkey Anti-Rabbit IgG (H+L) (Jackson ImmunoResearch Labs, 711-605-152), DyLight 550 Donkey anti-Goat IgG (H+L) (Thermo Fisher Scientific, SA5-10087), DyLight 550 Donkey anti-Rabbit IgG (H+L) (Thermo Fisher Scientific, SA5-10039). F-actin was directly stained with Phalloidin CruzFluor 488 Conjugated antibody (1:1000; Santa Cruz Biotechnology, sc-363791) along with other secondary antibodies in blocking buffer.
Immunohistochemistry
Immunohistochemistry (IHC) was performed using the ImmPRESS™ HRP Anti-Rabbit IgG (Peroxidase) Polymer Detection Kit (Vector laboratories, MP-7401) according to the manufacturer’s instructions with minor modifications. The frozen sections were firstly treated with citrate-based (Vector laboratories, H-3300) for antigen unmasking. Blocking was done with 2.5% normal horse blocking serum included in the kit for 1 hr at room temperature. The primary antibody for tdTomato (1:200; Rockland, 600-401-379) was applied to the sections and incubated overnight at 4°C. All washes were performed with 0.1% Tween-20 (PBS) for 5 min. The color was developed using ImmPACT DAB Peroxidase (HRP) Substrate (Vector laboratories, SK-4105) according to the manufacturer’s instructions. Lastly, the sections were counterstained with hematoxylin and went through a series of ethanol-based dehydration and xylene-based clearing and mounted with mounting medium.
Genomic DNA PCR
Decidua tissue sample was minced and resuspended in TE buffer. Tissue was digested by treating with 0.3 mg/ml proteinase K (Thermo Fisher Scientific, AM2546) at 55°C overnight. Genomic DNA preparations were incubated at 95°C for 5min to inactivate proteinase K before used for PCR. An ultraconserved noncoding element (UNCE) overlapping with the Tfap2a locus was used as an internal control (Cohen et al., 2016). The tdTomato gene was amplified by a nested PCR. The first round of PCR was done with an external primer set: 5’-GGC GAG GAG GTC ATC AAA GAG T-3’, 5’-ATG GTG TAG TCC TCG TTG TGG G-3’. PCR product of the first PCR was diluted at 1:200 and 1pl of the diluted sample was used as the template for the second round nested PCR with the following primers: 5’-ACA TCC CCG ATT ACA AGA AGC-3’, 5’-TTG TAG ATC AGC GTG CCG TC-3’. All PCR reactions were performed with PrimeSTAR GXL DNA Polymerase (Clontech, R050B). PCR products were resolved in a 2% agarose gel with TBE buffer. Images were acquired using a Bio-Rad Gel Doc XR+ system with Image Lab software.
Transcriptome Analysis
Total RNA was isolated from eight individual EPS-blastoids collected at day five using the TRIzol (Thermo Fisher Scientific, 15596026) method. RNA-Seq libraries were constructed using the Illumina Smart-Seq2 (Picelli et al., 2013) using Nextera XT DNA sample preparation kit (Illumina, FC-131-1096) and Nextera XT 24-index kit (Illumina, FC-131-1001), and 2x150 bp pair-end sequencing was performed on an Illumina HiSeq Xten. Sequencing reads were filtered and mapped to the mouse genome build mm10 using the HISAT2 alignment program (Kim et al., 2019). De novo transcriptome assembly and transcript and gene abundance calculations were performed using the StringTie assembler (Pertea et al., 2015). The expression values of each gene were normalized using FPKM. RNA-Seq data of morula stage and E3.5 early blastocyst stage embryos were obtained from published datasets (GSE98150 and GSE87504, respectively) (Sampath Kumar et al., 2017; Wang et al., 2018). Raw read data were downloaded and processed using the same pipeline as that used for EPS-blastoid data. Differentially expressed genes (DEGs) were calculated using the R package ballgown (Frazee et al., 2015). DEGs were deemed significant if they passed the following cutoff parameters: FPKM > 1, absolute value of log2 ratio > 1, and Q-value (adjusted p-value) < 0.05. Gene ontology (GO) and KEGG pathway analyses were performed using Fisher’s exact test, and the false discovery rate (FDR) was controlled by the BH method. Principle components analysis was performed using the R package ade4 (Dray and Dufour, 2007). Cluster analysis was performed using the R package pvclust (Suzuki and Shimodaira, 2006). Heatmaps were generated using the R package pheatmap (Kolde, 2012).
Single-cell RNA-Seq library generation
EPS-blastoids were manually picked up using mouth pipette and washed three times in PBS containing 0.04% BSA. Around 500 EPS-blastoids were harvested and dissociated with a homemade enzyme mix composed of 0.5X versene (Lonza, 17711E), 0.5X Acumax (Innovative Cell Tech, AM105), and 0.05X Dnase (STEMCELL Technologies, 07900) at 37°C for 30min with agitation. Dissociated cells were spun down and wash with PBS + 0.04%BSA for three times and resuspended in the same buffer. Cell density was determined by a TC10 cell counter (Bio-Rad, 1450001). Blastocysts were dissociated using the same protocol. Dissociated cells (~4800 cells for EPS-blastoids and ~1000 cells for blastocysts) were loaded into the Chromium Single Cell B Chip (10X Genomics, PN-120262) and processed in the Chromium single cell controller (10X Genomics) to generate single-cell gel beads in the emulsion according to the manufacturer’s protocol. The library was generated using the Chromium Single Cell 3’ Reagent Kits v3 (10X Genomics, PN-1000092) and Chromium i7 Multiplex Kit (10X Genomics, PN-120262) according to the manufacturer’s manual. The two libraries were pooled and sequenced using Nextseq 500 (150 cycles, high output).
Single-cell RNA-Seq data analysis
We used STAR v2.5.1b1 (Dobin et al., 2013) to align reads to the 10x Genomics prebuilt mm10 reference genome and utilized the CellRanger v3.0.2 (10X Genomics) software for blastocysts (288 cells) and EPS-blastoids (3528 cells) datasets with the default setting for de-multiplexing to generate feature-barcode matrix. The R package Seurat v3.0.12 (Stuart et al., 2019) was used to read and analyze feature-barcode matrix following the steps: First, we filtered the cells that have unique feature counts over 5000 according to quality control matrix plots (184 and 2518 cells in the blastocysts and EPS-blastoids group passed the filter, respectively); Then UMI counts were normalized with NormalizeData function using the default settings. Seurat’s RunUMAP function was used to perform a non-linear dimension reduction and clustering with resolution setting at 0.2. Differentially expressed genes within the clusters between blastocysts and EPS-blastoids were determined by the FindMarkers function using a bimodal likelihood ratio test. For the differentially expressed genes, we tested whether each had enriched GO terms in biological process and molecular functions using the ToppGene Suite3 (Chen et al., 2009). Unsupervised clustering analysis (UCA) was performed using the R package ComplexHeatmap v2.1.0 (Gu et al., 2016) with clustering_distance_columns = “manhattan”.
QUANTIFICATION AND STATISTICAL ANALYSIS
The sample size was not predetermined using any statistical methods or packages before experimentation. Quantification details on the number of biological replicates (n value) and data presentation were included in figure legends. Values were shown as the mean and error bars represented SEM unless otherwise indicated. Statistical analysis details were described in figure legends or method details. No method was used to determine whether the data met assumptions of the statistical approach. Differences were considered to be significant when the P (or adjusted P) values were smaller than 0.05. Graphs were generated using Prism or R package ggplot2 (Wickham, 2016) or other R packages described in the method details.
DATA AND CODE AVAILABILITY
R scripts used for the single-cell RNA-Seq analysis are available upon request. The sequencing data have been deposited at the NCBI Gene Expression Omnibus under the following accession number: GSE135289 (bulk RNA-Seq) and GSE135701 (single-cell RNA-Seq).
Supplementary Material
Figure S1. Additional Data on the Establishment of a 3D Differentiation System to Induce Blastoids from EPS Cells, Related to Figure 1
(A) Phase contrast images of multicellular structures in microwells after five days in the blastoid induction medium containing with either KSOM (left) or M16 (right) embryo culture medium. The red triangles indicate EPS-blastoids. Scale bar, 100 μm.
(B) Phase contrast images of mouse embryos 48hrs after treating with either vehicle (left) or XAV939 (5 μM) (right) at the 4-cell stage. Scale bar, 100 μm.
(C) Quantification of the cavity area in the mouse embryos shown in (B). Data are represented as mean ± SEM; n = 6 embryos in each group.
(D) Phase contrast images of multicellular structures in microwells after five days in blastoid induction medium supplemented with vehicle (left), XAV939 (middle), or IWR-1-endo (right). The red triangles indicate EPS-blastoids. Scale bar, 100 μm.
(E) Quantification of EPS-blastoids formation efficiency with the indicated treatment. Data are represented as mean ± SEM; n = 5 independent assays for each group.
(F) Phase contrast images of multicellular structures in microwells after five days of blastoid induction from ES-converted EPS (left) or ES (right) cells. The red triangles indicate EPS-blastoids. Scale bar, 100 μm.
(G) Quantification of EPS-blastoids formation efficiency for ES-converted EPS or ES cells. Data are represented as mean ± SEM; n = 3 independent assays for each group.
(H) Phase contrast image of blastoids generated from Liu-EPS cells. Scale bar, 100 μm.
(I) Quantification of EPS-blastoids formation efficiency from Liu-EPS cells. Data are represented as mean ± SEM; n = 4 independent assays.
(J) Quantification of the diameter of blastocyst or Liu-EPS-blastoids. n = 55 blastocysts; n = 25 Liu-EPS-blastoids.
Figure S2. Additional Data on the Characterization of Preimplantation Developmental Processes during EPS-blastoids Formation, Related to Figure 2
(A) Immunofluorescence staining of an EPS aggregate at day 1 (left) and a compacted 8-cell embryo (right) for ZO1. Ho, Hoechst. Scale bars, 20 μm.
(B) Quantification of the percentage of cell aggregates showing ZO1+ staining at day 1 or day 2. Data are represented as mean ± SEM; n = 3 biological replicates for each time point.
(C) A heatmap showing the FPKM values of the indicated genes in two EPS and ES cell lines. FPKM, Fragments Per Kilobase of transcript per Million mapped reads.
(D and E) Immunofluorescence staining of 2D EPS cells for ZO1 and OCT4 (D) or YAP (E). Ho, Hoechst. Scale bar, 50 μm.
(F) Phase contrast images of mouse embryos 48hrs after treating with either vehicle (left) or VP (right) at the 4-cell stage. Scale bar, 100 μm. VP, verteporfin.
(G) Quantification of the cavity area in the mouse embryos shown in (F). Data are represented as mean ± SEM; n = 6 embryos in each group.
(H) Phase contrast images of multicellular structures in microwells after five days of blastoid induction in medium supplemented with vehicle (left) or VP (right). The red triangles indicate EPS-blastoids. Scale bar, 100 μm. VP, verteporfin.
(I) Quantification of EPS-blastoids formation efficiency with the indicated treatment. Data are represented as mean ± SEM; n = 4 independent assays for each group.
(J) Immunostaining of an EPS-blastoid from a paternal X-GFP cell line for CDX2, NANOG, and X-GFP. Ho, Hoechst. Scale bar, 20 μm.
(K) Quantification of the frequency of different EPS-blastoid categories based on paternal X-GFP expression pattern. n = 14 X-GFP EPS-blastoids.
Figure S3. Additional Data on the Characterization of the Three Cell Lineages in the EPS-blastoids, Related to Figure 3
(A and B) Immunofluorescence staining of EPS-blastoids for EOMES and OCT4 (A) or CDX2 and NANOG (B). Ho, Hoechst. Scale bars, 20 μm.
(C) Immunofluorescence staining of EPS aggregates at the indicated day for SOX2 and CDX2 expression. Ho, Hoechst. Scale bars, 10 μm.
(D) Quantification of different patterns of SOX2 and CDX2 expression in EPS cell aggregates at the indicated day. n = 47, 47, 36, 27, and 40 for EPS cell aggregates at day 1, 2, 3, 4, and 5, respectively.
(E and F) Immunofluorescence staining of ES-converted EPS-blastoids for CDX2 and SOX2 (E), or GATA4 and NANOG (F). Ho, Hoechst. The rightmost panel in E is the maximum intensity projection of z-stack images of the indicated protein. Scale bars, 20 μm.
(G) Immunofluorescence staining of a Liu-EPS-blastoid for CDX2 and SOX2. Ho, Hoechst. The rightmost panel shows the maximum intensity projection of z-stack images of the indicated protein. Scale bars, 20 μm.
(H) Immunofluorescence staining of an EPS-blastoid generated from a single EPS cell for CDX2, SOX2, and mCherry. Ho, Hoechst. Scale bars, 20 μm.
Figure S4. Additional Data on Transcriptome Analysis of EPS-blastoids at the Bulk and Single Cell Level, Related to Figure 4
(A) Unsupervised average clustering analysis of RNA-Seq data from individual EPS-blastoid, blastocyst, and morula.
(B) A summary of differential gene expression analysis between EPS-blastoids and blastocysts.
(C) A summary of differential gene expression analysis between EPS-blastoids and morulae.
(D) A volcano plot showing the differentially expressed genes (DEGs) between EPS-blastoids and blastocysts.
(E) Pathways enrichment analysis of DEGs between EPS-blastoids and blastocysts.
(F) A Umap plot showing the clustering of cells from blastocysts (left) or EPS-blastoids (right), respectively.
(G) The expression of lineage-specific genes shown in Umap plots.
Figure S5. Additional data on Functional Characterization of EPS-blastoids in vitro, Related to Figure 5
(A) Immunofluorescence staining of ES cells derived from EPS-blastoids for OCT4, NANOG, SOX2, and CDX2. Scale bar, 50 μm.
(B) Immunofluorescence staining of TS cells derived from EPS-blastoids for EOMES, CDX2, OCT4, and NANOG. Scale bar, 100 μm.
(C and D) Immunofluorescence staining of EPS-blastoids-derived XEN cells for GATA6 (C) and GATA4 (D). Ho, Hoechst. Scale bar, 100 μm.
(E and F) Immunofluorescence staining of postimplantation embryo-like structures from in vitro culture of EPS-blastoids for Laminin and SOX2 (E), or Laminin and GATA4 (F). Scale bar, 50 μm.
Figure S6. Additional Data on the Functional Characterization of EPS-blastoids in vivo, Related to Figure 6
(A) Brightfield image of the uterus of a control mouse at 7.5 dpc. Black triangles indicate deciduae. Scale bars, 1 mm.
(B) Brightfield image of a decidua (circled with dotted line) after removing the uterus wall showing blood infiltration (indicated by black arrowheads). Scale bar, 1 mm.
(C) Quantification of the efficiency of decidua formation per EPS-blastoid transferred into the mouse uterus. Data are represented as mean ± SEM; n = 10 independent assays.
(D) Brightfield images of deciduae recovered from control 7.5 dpc mice (left) or surrogate mice at 7.5 dpc with EPS-blastoids transfer at 2.5 dpc. Scale bar, 1 mm.
(E) Brightfield images of control embryos at the indicated embryonic days. BF, brightfield. Scale bar, 100 μm.
(F) Brightfield (left) and fluorescent (right) images of EPS-blastoid-derived structures recovered from decidua at the indicated time points. EPS-blastoids were transferred at 2.5 dpc. BF, brightfield. Td, tdTomato. Scale bar, 100 μm.
(G - I) Immunofluorescence staining for OCT4 (H), EOMES (I), and GATA4 (J) in tissue sections of an in vivo EPS-blastoid-derived structure recovered from decidua at 6.5 dpc (4 days after EPS-blastoids transfer). Ho, Hoechst. Scale bar, 50 μm.
Figure S7. Additional Data on the Characterization of EPS-blastoids Generated from Adult Fibroblast, Related to Figure 7
(A) Immunofluorescence staining of iEPS-blastoids for NANOG and GATA4. Ho, Hoechst. Scale bar, 20 μm.
(B-C) Immunofluorescence staining of postimplantation embryo-like structure from in vitro culture of iEPS-blastoids for TFAP2C and SOX2 (B), or F-actin, GATA4, and OCT4 (C). Ho, Hoechst. Scale bar, 50 μm.
Table S1. Summary of EPS-blastoid Efficiency, Related to Figure 1, Figure 2, and Figure 7
Table S1-1. Summary of EPS-blastoid efficiency from different EPS cell lines, Related to Figure 1 and Figure 7. Table S1-2. Summary of EPS-blastoid efficiency with treatment of Wnt antagonists, Related to Figure 1. Table S1-3. Summary of EPS-blastoid Efficiency from a single EPS cell, Related to Figure 1. Table S1-4. Summary of EPS-blastoid Efficiency from ES and ES-converted EPS, Related to Figure 1. Table S1-5. Summary of EPS-blastoid efficiency with treatment of YAP-TEAD inhibitor, related to Figure 2.
Table S2. List of DEGs Identified from Analysis of Bulk RNA-Seq Data from Individual EPS-blastoid, Blastocyst, and Morula, Related to Figure 4
Table S2-1. List of DEGs Between EPS-blastoids and Blastocysts, Related to Figure 4.
Table S2-2. List of DEGs Between EPS-blastoids and Morulae, Related to Figure 4.
Table S3. Summary of GO Term Analysis of DEGs for Each Lineage Between Blastocysts and EPS-blastoids, Related to Figure 4
Table S4. Summary of Decidualization Efficiency of EPS-blastoids Transfer Experiments, Related to Figure 6
Table S5. Summary of the Efficiency of Recovered Embryo/Embryo-like Structures from Deciduae, Related to Figure 6
Highlights.
A method that enables the generation of blastocyst-like structures from EPS cells
EPS-blastoids resemble blastocyst in morphology and cell lineage allocation
EPS-blastoid formation recapitulates early developmental events in vitro
EPS-blastoids are able to implant in utero
ACKNOWLEDGMENTS
We thank Travis Berggren for his continuous advice and guidance. We thank Dr. Fumihiro for the B6N-22 mESC line. We are thankful to Dr. Azim Surani for the female X-GFP mEpiSC. We are grateful for technical assistance from C. O’Connor and C. Fitzpatrick from Salk flow cytometry core for FACS sorting, U. Manor and T. Zhang from the Waitt Advanced Biophotonics Core for confocal imaging, N. Hah and G. Chou from the Salk Sequencing Core for single-cell RNA-Seq experiments. We are also thankful for all the GEL-B lab members for helpful discussion and constructive criticism, and M. Schwarz and P. Schwarz for administrative assistance. We would also like to express our gratitude to Dave O’Keefe for his contribution toward manuscript preparation. C.Z. was supported by the Larry L. Hillblom Foundation, Paul F. Glenn Foundation, and Salk Women & Science Special Award. This work was also supported by the National Key R&D Program of China (2016YFC1000601 to Y.Y.). L.Y. was partially supported by a trainee fellowship from the Hamon Center for Regenerative Science & Medicine. J.W. was a Virginia Murchison Linthicum Scholar in Medical Research and funded by Cancer Prevention & Research Institute of Texas (CPRIT). This project was supported by the G. Harold and Leila Y. Mathers Charitable Foundation, The Moxie Foundation, The Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002), The Hewitt Foundation, NIH (5 DPI DK113616) and Universidad Catolica San Antonio de Murcia.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
None
REFERENCES
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, and Surani MA (2009). Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 2013 500:7462 461, 1292–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton SC, Ferguson-Smith AC, Fundele R, and Surani MA (1991). Influence of paternally imprinted genes on development. Development 113, 679–687. [DOI] [PubMed] [Google Scholar]
- Beccari L, Moris N, Girgin M, Turner DA, Baillie-Johnson P, Cossy A-C, Lutolf MP, Duboule D, and Arias AM (2018). Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 2013 500:7462 562, 272–276. [DOI] [PubMed] [Google Scholar]
- Bedzhov I, and Zernicka-Goetz M (2014). Self-Organizing Properties of Mouse Pluripotent Cells Initiate Morphogenesis upon Implantation. Cell 156, 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedzhov I, Leung CY, Bialecka M, and Zernicka-Goetz M (2014a). In vitro culture of mouse blastocysts beyond the implantation stages. Nat Protoc 9, 2732–2739. [DOI] [PubMed] [Google Scholar]
- Bedzhov I, Leung CY, Bialecka M, and Zernicka-Goetz M (2014b). In vitro culture of mouse blastocysts beyond the implantation stages. Nat Protoc 9, 2732–2739. [DOI] [PubMed] [Google Scholar]
- Berge ten, D., Koole W, Fuerer C, Fish M, Eroglu E, and Nusse R (2008). Wnt Signaling Mediates Self-Organization and Axis Formation in Embryoid Bodies. Cell Stem Cell 3, 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biechele S, Cockburn K, Lanner F, Cox BJ, and Rossant J (2013). Porcn-dependent Wnt signaling is not required prior to mouse gastrulation. Development 140, 2961–2971. [DOI] [PubMed] [Google Scholar]
- Chazaud C, and Yamanaka Y (2016). Lineage specification in the mouse preimplantation embryo. Development 143, 1063–1074. [DOI] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, and Jegga AG (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MA, Wert KJ, Goldmann J, Markoulaki S, Buganim Y, Fu D, and Jaenisch R (2016). Human neural crest cells contribute to coat pigmentation in interspecies chimeras after in utero injection into mouse embryos. Proc Natl Acad Sci USA 113, 1570–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman TC, Eistetter H, Katz M, Schmidt W, and Kemler R (1985). The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol 87, 27–45. [PubMed] [Google Scholar]
- Dray S, and Dufour A-B (2007). The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software 22. [Google Scholar]
- Evans MJ, and Kaufman MH (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 2013 500:7462 292, 154–156. [DOI] [PubMed] [Google Scholar]
- Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, and Leek JT (2015). Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol 33, 243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs C, Scheinast M, Pasteiner W, Lagger S, Hofner M, Hoellrigl A, Schultheis M, and Weitzer G (2012). Self-Organization Phenomena in Embryonic Stem Cell-Derived Embryoid Bodies: Axis Formation and Breaking of Symmetry during Cardiomyogenesis. Cells Tissues Organs 195, 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Posfai E, and Rossant J (2018). Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat. Biotechnol 36, 632–637. [DOI] [PubMed] [Google Scholar]
- Gu Z, Eils R, and Schlesner M (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, and Kemler R (1995). Lack of beta-catenin affects mouse development at gastrulation. Development 121, 3529–3537. [DOI] [PubMed] [Google Scholar]
- Harrison SE, Sozen B, Christodoulou N, Kyprianou C, and Zernicka-Goetz M (2017). Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 356, eaal1810. [DOI] [PubMed] [Google Scholar]
- He S, Pant D, Schiffmacher A, Meece A, and Keefer CL (2008). Lymphoid Enhancer Factor 1-Mediated Wnt Signaling Promotes the Initiation of Trophoblast Lineage Differentiation in Mouse Embryonic Stem Cells. Stem Cells 26, 842–849. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ, and DePamphilis ML (2013). TEAD4 establishes the energy homeostasis essential for blastocoel formation. Development 140, 3680–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, and Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol 37, 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R (2012). Pheatmap: pretty heatmaps. R Package Version 61, 915. [Google Scholar]
- Kunath T (2005). Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 132, 1649–1661. [DOI] [PubMed] [Google Scholar]
- Kutner RH, Zhang X-Y, and Reiser J (2009). Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc 4, 495–505. [DOI] [PubMed] [Google Scholar]
- Liao H-K, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, Suzuki K, Xu R, Hishida T, Chang C-J, et al. (2015). Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun 6, 6413. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, and Pfaff SL (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 2013 500:7462 487, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.a 78, 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani SM, Canham MA, Nichols J, Sharov AA, Migueles RP, Ko MSH, and Brickman JM (2013). Totipotent Embryonic Stem Cells Arise in Ground-State Culture Conditions. Cell Rep 3, 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N, Inoue K-I, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. (2009). The Hippo Signaling Pathway Components Lats and Yap Pattern Tead4 Activity to Distinguish Mouse Trophectoderm from Inner Cell Mass. Developmental Cell 16, 398–410. [DOI] [PubMed] [Google Scholar]
- Papaioannou VE, Mkandawire J, and Biggers JD (1989). Development and phenotypic variability of genetically identical half mouse embryos. Development 106, 817–827. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, and Sandberg R (2013). Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098. [DOI] [PubMed] [Google Scholar]
- Poh Y-C, Chen J, Hong Y, Yi H, Zhang S, Chen J, Wu DC, Wang L, Jia Q, Singh R, et al. (2014). Generation of organized germ layers from a single mouse embryonic stem cell. Nat Commun 5, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai E, Petropoulos S, de Barros FRO, Schell JP, Jurisica I, Sandberg R, Lanner F, and Rossant J (2017). Position- and Hippo signaling-dependent plasticity during lineage segregation in the early mouse embryo. Elife 6, 2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, Cañon S, Sasaki H, Hadjantonakis A-K, la Pompa JL, et al. (2014). Notch and Hippo Converge on Cdx2 to Specify the Trophectoderm Lineage in the Mouse Blastocyst. Developmental Cell 30, 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivron NC, Frias-Aldeguer J, Vrij EJ, Boisset J-C, Korving J, Vivié J, Truckenmüller RK, van Oudenaarden A, van Blitterswijk CA, and Geijsen N (2018). Blastocyst-like structures generated solely from stem cells. Nature 2013 500:7462 557, 106–111. [DOI] [PubMed] [Google Scholar]
- Rossant J, and Tam PPL (2009). Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136, 701–713. [DOI] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, and Eliceiri KW (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugg-Gunn P (2017). Derivation and Culture of Extra-Embryonic Endoderm Stem Cell Lines. Cold Spring Harb Protoc 2017, pdb.prot093963. [DOI] [PubMed] [Google Scholar]
- Sampath Kumar A, Seah MKY, Ling KY, Wang Y, Tan JHL, Nitsch S, Lim SL, Lorthongpanich C, Wollmann H, Low DHP, et al. (2017). Loss of maternal Trim28causes male-predominant early embryonic lethality. Genes Dev. 31, 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozen B, Amadei G, Cox A, Wang R, Na E, Czukiewska S, Chappell L, Voet T, Michel G, Jing N, et al. (2018). Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol 20, 979–989. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Kothary R, Allen ND, Singh PB, Fundele R, Ferguson-Smith AC, and Barton SC (1990). Genome imprinting and development in the mouse. Dev. Suppl 89–98. [PubMed] [Google Scholar]
- Suzuki R, and Shimodaira H (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542. [DOI] [PubMed] [Google Scholar]
- Takagi N, and Sasaki M (1975). Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 2013 500:7462 256, 640–642. [DOI] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, and Rossant J (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075. [DOI] [PubMed] [Google Scholar]
- Tanimoto Y, Iijima S, Hasegawa Y, Suzuki Y, Daitoku Y, Mizuno S, Ishige T, Kudo T, Takahashi S, Kunita S, et al. (2008). Embryonic stem cells derived from C57BL/6J and C57BL/6N mice. Comp. Med 55, 347–352. [PMC free article] [PubMed] [Google Scholar]
- TARKOWSKI AK (1959). Experiments on the development of isolated blastomers of mouse eggs. Nature 2013 500:7462 184, 1286–1287. [DOI] [PubMed] [Google Scholar]
- TARKOWSKI AK, and Wróblewska J (1967). Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol 18, 155–180. [PubMed] [Google Scholar]
- Vrij EJ, Scholte op Reimer YS, Frias-Aldeguer J, Misteli Guerreiro I, Kind J, Koo B-K, van Blitterswijk CA, and Rivron NC (2019). Chemically-defined induction of a primitive endoderm and epiblast-like niche supports post-implantation progression from blastoids. 120, 173 10.1101/510396 [DOI] [Google Scholar]
- Wang C, Liu X, Gao Y, Yang L, Li C, Liu W, Chen C, Kou X, Zhao Y, Chen J, et al. (2018). Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol 20, 620–631. [DOI] [PubMed] [Google Scholar]
- Wang H, Ding T, Brown N, Yamamoto Y, Prince LS, Reese J, and Paria BC (2008). Zonula occludens-1 (ZO-1) is involved in morula to blastocyst transformation in the mouse. Developmental Biology 318, 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmflash A, Sorre B, Etoc F, Siggia ED, and Brivanlou AH (2014). A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 11, 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JD, Frels WI, Chapman VM, and Papaioannou VE (1977). Preferential expression of the maternally derived X chromosome in the mouse yolk sac. Cell 12, 873–882. [DOI] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: elegant graphics for data analysis. Springer. [Google Scholar]
- Wu J, Platero-Luengo A, Sakurai M, Sugawara A, Gil MA, Yamauchi T, Suzuki K, Bogliotti YS, Cuello C, Morales Valencia M, et al. (2017). Interspecies Chimerism with Mammalian Pluripotent Stem Cells. Cell 168, 473–486. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, and Buonanno A (2007). Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134, 3827–3836. [DOI] [PubMed] [Google Scholar]
- Yang J, Ryan DJ, Wang W, Tsang JC-H, Lan G, Masaki H, Gao X, Antunes L, Yu Y, Zhu Z, et al. (2017a). Establishment of mouse expanded potential stem cells. Nature 2013 500:7462 550, 393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu B, Xu J, Wang J, Wu J, Shi C, Xu Y, Dong J, Wang C, Lai W, et al. (2017b). Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell 169, 243–257. e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q-L, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, and Smith A (2008). The ground state of embryonic stem cell self-renewal. Nature 2013 500:7462 453, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen T, Chen N, Gao D, Shi B, Kong S, West RC, Yuan Y, Zhi M, Wei Q, et al. (2019). Implantation initiation of self-assembled embryo-like structures generated using three types of mouse blastocyst-derived stem cells. Nat Commun 10, 1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Additional Data on the Establishment of a 3D Differentiation System to Induce Blastoids from EPS Cells, Related to Figure 1
(A) Phase contrast images of multicellular structures in microwells after five days in the blastoid induction medium containing with either KSOM (left) or M16 (right) embryo culture medium. The red triangles indicate EPS-blastoids. Scale bar, 100 μm.
(B) Phase contrast images of mouse embryos 48hrs after treating with either vehicle (left) or XAV939 (5 μM) (right) at the 4-cell stage. Scale bar, 100 μm.
(C) Quantification of the cavity area in the mouse embryos shown in (B). Data are represented as mean ± SEM; n = 6 embryos in each group.
(D) Phase contrast images of multicellular structures in microwells after five days in blastoid induction medium supplemented with vehicle (left), XAV939 (middle), or IWR-1-endo (right). The red triangles indicate EPS-blastoids. Scale bar, 100 μm.
(E) Quantification of EPS-blastoids formation efficiency with the indicated treatment. Data are represented as mean ± SEM; n = 5 independent assays for each group.
(F) Phase contrast images of multicellular structures in microwells after five days of blastoid induction from ES-converted EPS (left) or ES (right) cells. The red triangles indicate EPS-blastoids. Scale bar, 100 μm.
(G) Quantification of EPS-blastoids formation efficiency for ES-converted EPS or ES cells. Data are represented as mean ± SEM; n = 3 independent assays for each group.
(H) Phase contrast image of blastoids generated from Liu-EPS cells. Scale bar, 100 μm.
(I) Quantification of EPS-blastoids formation efficiency from Liu-EPS cells. Data are represented as mean ± SEM; n = 4 independent assays.
(J) Quantification of the diameter of blastocyst or Liu-EPS-blastoids. n = 55 blastocysts; n = 25 Liu-EPS-blastoids.
Figure S2. Additional Data on the Characterization of Preimplantation Developmental Processes during EPS-blastoids Formation, Related to Figure 2
(A) Immunofluorescence staining of an EPS aggregate at day 1 (left) and a compacted 8-cell embryo (right) for ZO1. Ho, Hoechst. Scale bars, 20 μm.
(B) Quantification of the percentage of cell aggregates showing ZO1+ staining at day 1 or day 2. Data are represented as mean ± SEM; n = 3 biological replicates for each time point.
(C) A heatmap showing the FPKM values of the indicated genes in two EPS and ES cell lines. FPKM, Fragments Per Kilobase of transcript per Million mapped reads.
(D and E) Immunofluorescence staining of 2D EPS cells for ZO1 and OCT4 (D) or YAP (E). Ho, Hoechst. Scale bar, 50 μm.
(F) Phase contrast images of mouse embryos 48hrs after treating with either vehicle (left) or VP (right) at the 4-cell stage. Scale bar, 100 μm. VP, verteporfin.
(G) Quantification of the cavity area in the mouse embryos shown in (F). Data are represented as mean ± SEM; n = 6 embryos in each group.
(H) Phase contrast images of multicellular structures in microwells after five days of blastoid induction in medium supplemented with vehicle (left) or VP (right). The red triangles indicate EPS-blastoids. Scale bar, 100 μm. VP, verteporfin.
(I) Quantification of EPS-blastoids formation efficiency with the indicated treatment. Data are represented as mean ± SEM; n = 4 independent assays for each group.
(J) Immunostaining of an EPS-blastoid from a paternal X-GFP cell line for CDX2, NANOG, and X-GFP. Ho, Hoechst. Scale bar, 20 μm.
(K) Quantification of the frequency of different EPS-blastoid categories based on paternal X-GFP expression pattern. n = 14 X-GFP EPS-blastoids.
Figure S3. Additional Data on the Characterization of the Three Cell Lineages in the EPS-blastoids, Related to Figure 3
(A and B) Immunofluorescence staining of EPS-blastoids for EOMES and OCT4 (A) or CDX2 and NANOG (B). Ho, Hoechst. Scale bars, 20 μm.
(C) Immunofluorescence staining of EPS aggregates at the indicated day for SOX2 and CDX2 expression. Ho, Hoechst. Scale bars, 10 μm.
(D) Quantification of different patterns of SOX2 and CDX2 expression in EPS cell aggregates at the indicated day. n = 47, 47, 36, 27, and 40 for EPS cell aggregates at day 1, 2, 3, 4, and 5, respectively.
(E and F) Immunofluorescence staining of ES-converted EPS-blastoids for CDX2 and SOX2 (E), or GATA4 and NANOG (F). Ho, Hoechst. The rightmost panel in E is the maximum intensity projection of z-stack images of the indicated protein. Scale bars, 20 μm.
(G) Immunofluorescence staining of a Liu-EPS-blastoid for CDX2 and SOX2. Ho, Hoechst. The rightmost panel shows the maximum intensity projection of z-stack images of the indicated protein. Scale bars, 20 μm.
(H) Immunofluorescence staining of an EPS-blastoid generated from a single EPS cell for CDX2, SOX2, and mCherry. Ho, Hoechst. Scale bars, 20 μm.
Figure S4. Additional Data on Transcriptome Analysis of EPS-blastoids at the Bulk and Single Cell Level, Related to Figure 4
(A) Unsupervised average clustering analysis of RNA-Seq data from individual EPS-blastoid, blastocyst, and morula.
(B) A summary of differential gene expression analysis between EPS-blastoids and blastocysts.
(C) A summary of differential gene expression analysis between EPS-blastoids and morulae.
(D) A volcano plot showing the differentially expressed genes (DEGs) between EPS-blastoids and blastocysts.
(E) Pathways enrichment analysis of DEGs between EPS-blastoids and blastocysts.
(F) A Umap plot showing the clustering of cells from blastocysts (left) or EPS-blastoids (right), respectively.
(G) The expression of lineage-specific genes shown in Umap plots.
Figure S5. Additional data on Functional Characterization of EPS-blastoids in vitro, Related to Figure 5
(A) Immunofluorescence staining of ES cells derived from EPS-blastoids for OCT4, NANOG, SOX2, and CDX2. Scale bar, 50 μm.
(B) Immunofluorescence staining of TS cells derived from EPS-blastoids for EOMES, CDX2, OCT4, and NANOG. Scale bar, 100 μm.
(C and D) Immunofluorescence staining of EPS-blastoids-derived XEN cells for GATA6 (C) and GATA4 (D). Ho, Hoechst. Scale bar, 100 μm.
(E and F) Immunofluorescence staining of postimplantation embryo-like structures from in vitro culture of EPS-blastoids for Laminin and SOX2 (E), or Laminin and GATA4 (F). Scale bar, 50 μm.
Figure S6. Additional Data on the Functional Characterization of EPS-blastoids in vivo, Related to Figure 6
(A) Brightfield image of the uterus of a control mouse at 7.5 dpc. Black triangles indicate deciduae. Scale bars, 1 mm.
(B) Brightfield image of a decidua (circled with dotted line) after removing the uterus wall showing blood infiltration (indicated by black arrowheads). Scale bar, 1 mm.
(C) Quantification of the efficiency of decidua formation per EPS-blastoid transferred into the mouse uterus. Data are represented as mean ± SEM; n = 10 independent assays.
(D) Brightfield images of deciduae recovered from control 7.5 dpc mice (left) or surrogate mice at 7.5 dpc with EPS-blastoids transfer at 2.5 dpc. Scale bar, 1 mm.
(E) Brightfield images of control embryos at the indicated embryonic days. BF, brightfield. Scale bar, 100 μm.
(F) Brightfield (left) and fluorescent (right) images of EPS-blastoid-derived structures recovered from decidua at the indicated time points. EPS-blastoids were transferred at 2.5 dpc. BF, brightfield. Td, tdTomato. Scale bar, 100 μm.
(G - I) Immunofluorescence staining for OCT4 (H), EOMES (I), and GATA4 (J) in tissue sections of an in vivo EPS-blastoid-derived structure recovered from decidua at 6.5 dpc (4 days after EPS-blastoids transfer). Ho, Hoechst. Scale bar, 50 μm.
Figure S7. Additional Data on the Characterization of EPS-blastoids Generated from Adult Fibroblast, Related to Figure 7
(A) Immunofluorescence staining of iEPS-blastoids for NANOG and GATA4. Ho, Hoechst. Scale bar, 20 μm.
(B-C) Immunofluorescence staining of postimplantation embryo-like structure from in vitro culture of iEPS-blastoids for TFAP2C and SOX2 (B), or F-actin, GATA4, and OCT4 (C). Ho, Hoechst. Scale bar, 50 μm.
Table S1. Summary of EPS-blastoid Efficiency, Related to Figure 1, Figure 2, and Figure 7
Table S1-1. Summary of EPS-blastoid efficiency from different EPS cell lines, Related to Figure 1 and Figure 7. Table S1-2. Summary of EPS-blastoid efficiency with treatment of Wnt antagonists, Related to Figure 1. Table S1-3. Summary of EPS-blastoid Efficiency from a single EPS cell, Related to Figure 1. Table S1-4. Summary of EPS-blastoid Efficiency from ES and ES-converted EPS, Related to Figure 1. Table S1-5. Summary of EPS-blastoid efficiency with treatment of YAP-TEAD inhibitor, related to Figure 2.
Table S2. List of DEGs Identified from Analysis of Bulk RNA-Seq Data from Individual EPS-blastoid, Blastocyst, and Morula, Related to Figure 4
Table S2-1. List of DEGs Between EPS-blastoids and Blastocysts, Related to Figure 4.
Table S2-2. List of DEGs Between EPS-blastoids and Morulae, Related to Figure 4.
Table S3. Summary of GO Term Analysis of DEGs for Each Lineage Between Blastocysts and EPS-blastoids, Related to Figure 4
Table S4. Summary of Decidualization Efficiency of EPS-blastoids Transfer Experiments, Related to Figure 6
Table S5. Summary of the Efficiency of Recovered Embryo/Embryo-like Structures from Deciduae, Related to Figure 6
Data Availability Statement
R scripts used for the single-cell RNA-Seq analysis are available upon request. The sequencing data have been deposited at the NCBI Gene Expression Omnibus under the following accession number: GSE135289 (bulk RNA-Seq) and GSE135701 (single-cell RNA-Seq).


