Abstract
Summary
The software pipeline SHOGUN profiles known taxonomic and gene abundances of short-read shotgun metagenomics sequencing data. The pipeline is scalable, modular and flexible. Data analysis and transformation steps can be run individually or together in an automated workflow. Users can easily create new reference databases and can select one of three DNA alignment tools, ranging from ultra-fast low-RAM k-mer-based database search to fully exhaustive gapped DNA alignment, to best fit their analysis needs and computational resources. The pipeline includes an implementation of a published method for taxonomy assignment disambiguation with empirical Bayesian redistribution. The software is installable via the conda resource management framework, has plugins for the QIIME2 and QIITA packages and produces both taxonomy and gene abundance profile tables with a single command, thus promoting convenient and reproducible metagenomics research.
Availability and implementation
1 Introduction
Next-generation sequencing technology has led to a massive influx in the amount of metagenomic data, creating the potential to discover the causal roles microbes play in the many complex ecosystems they influence (Buermans and den Dunnen, 2014). The quantification of taxonomic and gene abundance profiles from metagenomic data are often carried out using custom, in-house workflows leading to redundant implementations of software and the inability to reproduce results across labs and studies (da Veiga Leprevost et al., 2017). To tackle these challenges, we propose the SHOGUN pipeline that assembles current-best practices in the field into a single, easy to use, and flexible framework to carry out known taxonomic and gene abundance profiling of metagenomic whole-genome shotgun (WGS) data (Hillmann and Knights, 2017).
2 Shogun pipeline
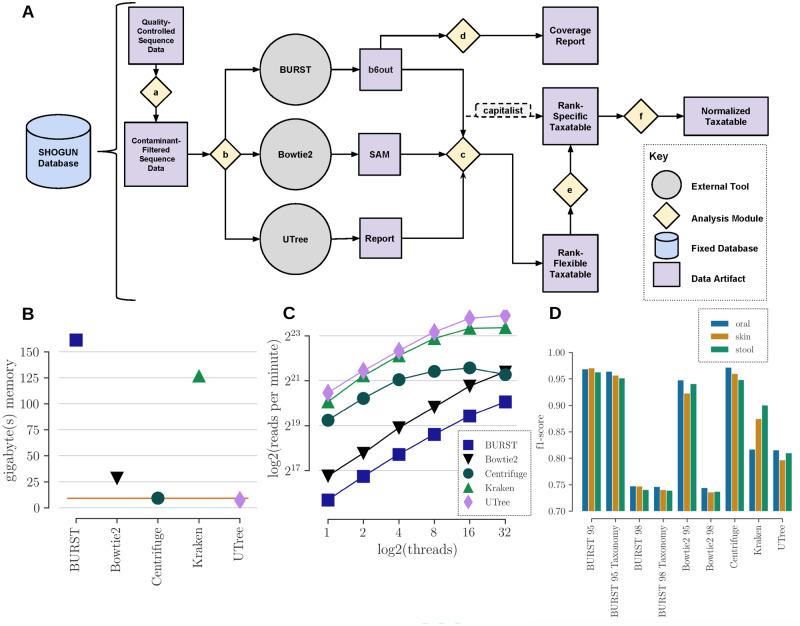
SHOGUN is a command-line tool that can be installed via the Anaconda (Anaconda Software Distribution, 2017) resource management framework with a single command, is open source and freely available via the GNU Affero General Public License and is well-documented and easy to use. SHOGUN is also available as a plugin for the QIIME2 (Bolyen et al., 2018) and QIITA (Gonzalez et al., 2018) packages. SHOGUN includes contaminate read filtering, relative abundance profiling and coverage with a full description of the methods in Figure 1. The command line interface was designed using a modular subcommand framework, so that a user can run the whole pipeline at once or each step individually. The codebase is unit-tested and every version of the pipeline receives a unique version tag that can rolled-back to for complete analysis reproducibility.
Fig. 1.
(A) Schematic overview of the computational pipeline SHOGUN. For every step in the SHOGUN pipeline, the user must supply the pre-formatted SHOGUN database folder. To run every step shown here in a single command, the user can select the pipeline subcommand. Otherwise, the analysis modules can be run independently. (a) Filter–The input quality-controlled reads are aligned against the contamination database using BURST to filter out all reads that match human associated genome content. (b) Align–The contamination-free reads are aligned against the reference genome database. The user has the option to select one or all of the three alignment tools BURST, Bowtie2, or UTree. (c) Assign-taxonomy–Given the data artifacts from a SHOGUN alignment tool, output a Biological Observation Matrix (BIOM) format profile with the rows being rank-flexible taxonomies, the columns are samples and the entries are counts for each given taxonomy per sample. The alignment tool BURST has two run modes, taxonomy and capitalist. If the capitalist mode is enabled, a rank-specific BIOM file is output instead. (d) Coverage–The output from BURST can be utilized to analyze the genome coverage of each taxonomy across all samples in the alignment file. This can useful for reducing the number of false positive alignments by removing taxonomies below a minimum coverage score. (e) Redistribute–The rank-flexible profile is summarized into a rank-specific profile. (f) Normalize–Each sample in the profile is normalized to the median depth of all the samples for count-based analysis tools that use BIOM tables. (B) RAM memory usage in gigabytes of each of the aligners using the Kraken timing dataset. The horizontal red line depicts the size of the Rep82 database. (C) The scaling and efficiency in reads per minute of each of the aligners across many threads per process. Tools were selected based on an open-source codebase, ability to make a custom reference database and an output file containing mappings per sequence. The fastest tools are the alignment-free methods Kraken and UTree. Each of the tools scale efficiently across many threads per process. (D) The F1-score for each aligners’ per read taxonomic profiles on the simulated stool, oral and skin communities. For the alignment methods, two different thresholds at 95% and 98% for alignment identification were tested to account for recall bias in highly-divergent reads
3 Taxonomic abundance profiling
The profiling algorithm of known taxonomies within SHOGUN is broken up into three primary steps: sequence alignment, taxonomic assignment and rank-specific relative abundance estimation.
3.1 Sequence alignment
SHOGUN implements application wrappers for three distinct alignment algorithms: Bowtie2 (Langmead and Salzberg, 2012), a Burrows–Wheeler alignment algorithm, BURST (Al-Ghalith and Knights, 2017), an optimal, exhaustive Needleman–Wunsch alignment algorithm and UTree (Al-Ghalith and Hillmann, 2017), a k-mer-based alignment algorithm.
3.2 Taxonomic assignment
Due to shared regions of the genome across microbes, a query sequence often aligns to multiple reference genomes equally well. When a query sequence matches multiple genomes, SHOGUN uses a confidence weighted last-common ancestor algorithm to assign a single taxonomic match to each sequence. The confidence for a taxonomic clade is calculated by the matches within that clade divided by the sum of all sequence matches. The most specific taxonomic clade above a confidence threshold is selected as the single match. This read-disambiguation scheme, known in SHOGUN as taxonomy mode, results in a rank-flexible taxonomy assignment, where taxonomic abundance profiles contain a mixture of taxonomic levels; i.e. some queries are classified at the kingdom level, some at the phylum level and others at the species level. If all annotations are at the same level, such as the species level and the taxonomic profile is known as being rank-specific. The BURST aligner can be run in the SHOGUN taxonomy mode, or can optionally perform its own read-disambiguation scheme that returns rank-specific relative profiling known as capitalist (Al-Ghalith and Knights, 2017).
3.3 Rank-specific relative abundance estimation
SHOGUN implements empirical Bayesian redistribution of reads in a similar fashion to the Bracken algorithm, where higher levels of the taxonomic tree are redistributed to lower levels according to each genome’s uniqueness, number of hits in a profile and length (Lu et al., 2017). SHOGUN implements the Bayesian redistribution using BURST taxonomy profiling instead of the Kraken profiling.
4 Gene abundance profiling
Gene abundance profiles are obtained in a similar manner to taxonomic profiles and only returns genes labeled within the reference database. The three steps for gene abundance profiling are sequence alignment to an annotated nucleotide gene database, gene assignment and relative abundance estimation. When a query sequence matches multiple reference genes, we are unable to leverage taxonomical associations between genes for disambiguation and therefore are only able to leverage the capitalist reference gene disambiguation scheme.
5 Materials and methods
To validate the performance of the pipeline, we selected representative genomes from bacteria, archea and viruses from the publicly available RefSeq nucleotide database version number 82 (Rep82) (Tatusova et al., 2014). We identified genes using UniProt (Bateman et al., 2017) annotations obtained by running Prokka (Seemann, 2014) on all the bacterial genomes and mapping them to Kyoto Encyclopedia of Genes and Genomes (Kanehisa et al., 2012) annotations. To test each internal alignment engine’s accuracy of relative abundance estimation of metagenomic communities, we created a simulated community with known species level taxonomy. The data were simulated according to abundances obtained from Human Microbiome Project using the top 100 most abundant species from each general body habitat according to the original study’s results (Turnbaugh et al., 2007). Reads were simulated from a strain of those species according to the average proportion of that taxonomy in their respective group using the tool dwgsim (Homer, 2017) with default settings. The two tools outside of the SHOGUN framework utilized were Kraken and Centrifuge (Kim et al., 2016); for a more complete benchmark of accuracy please compare the relative accuracies to the Lindgreen et al. (2016) evaluation. The alignment methods were evaluated using F1-scores (the average of precision and recall) of the known species assignment versus the identified species. Each of the taxonomic assignment methods was also evaluated for speed and memory usage using the query sequences from the Kraken timing dataset and the Rep82 reference database. The results of profiling are summarized in Figure 1B–D, demonstrating comparable performance of alignment-based methods over a range of runtime efficiencies.
The following benchmarked tools were installed from the Anaconda channel ‘knights-lab’ using versions SHOGUN = 1.0.5, Utree = 2.0 rf and BURST = 0.99.7f. The rest of the tools were installed from the Anaconda channel ‘bioconda’ with versions Bowtie2==2.3.4.1–0, Kraken = 1.1–1, Centrifuge = 1.0.3–2 and dwgsim = 1.1.11-5.
Funding
This work was supported by the University of Minnesota Masonic Cancer Center (to D.K.), National Institutes of Health grant R01AI121383 (to D.K.), grant P01DK078669 and in-kind support from National Science Foundation 1565057 and the Alfred P. Sloan Foundation 2017–9838 (to R.K., QIIME and QIITA) and by the Masonic Cancer Center at the University of Minnesota.
Conflict of Interest: D.K. serves as CEO of CoreBiome, a company involved in the commercialization of microbiome analysis. CoreBiome is now a wholly owned subsidiary of OraSure. These interests have been reviewed and managed by the University of Minnesota in accordance with its conflict-of-interest policies.
References
- Anaconda Software Distribution. Anaconda, Inc, 2017, conda.io. (1 June 2017, date last accessed).
- Al-Ghalith G., Hillmann B. (2017) Knights-lab/UTree: UTree 2.0 SigNature edition: SHOGUN release. Technical report, Zenodo.
- Al-Ghalith G., Knights D. (2017) Knights-lab/BURST: BURST v0.99.4a. Technical report, Zenodo.
- Bateman A. et al. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res., 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E. et al. (2018) QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. Technical report e27295v2, PeerJ Inc.
- Buermans H.P.J., den Dunnen J.T. (2014) Next generation sequencing technology: advances and applications. Biochim. Biophys. Acta, 1842, 1932–1941. [DOI] [PubMed] [Google Scholar]
- da Veiga Leprevost F. et al. (2017) BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics (Oxford, England), 33, 2580–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A. et al. (2018) Qiita: rapid, web-enabled microbiome meta-analysis. Nat. Methods, 15, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmann B., Knights D. (2017) Knights-lab/SHOGUN: Release 1.0.0. Technical report, Zenodo.
- Homer N. (2017) DWGSIM: whole genome simulator for next-generation sequencing. Original-date: 2011-10-19T00:19:14Z.https://github. com/nh13/DWGSIM. (1 June 2017, date last accessed).
- Kanehisa M. et al. (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res., 40, D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. (2016) Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res., 26, 1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgreen S. et al. (2016) An evaluation of the accuracy and speed of metagenome analysis tools. Sci. Rep., 6, 19233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. (2017) Bracken: estimating species abundance in metagenomics data. Peer J. Comp. Sci., 3, e104. [Google Scholar]
- Seemann T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics (Oxford, England), 30, 2068–2069. [DOI] [PubMed] [Google Scholar]
- Tatusova T. et al. (2014) RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res., 42, D553–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J. et al. (2007) The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature, 449, 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]