Abstract
Quantitative analysis of next-generation sequencing (NGS) data requires discriminating duplicate reads generated by PCR from identical molecules that are of unique origin. Typically, PCR duplicates are identified as sequence reads that align to the same genomic coordinates using reference- based alignment. However, identical molecules can be independently generated during library preparation. Misidentif ¡cation of these molecules as PCR duplicates can introduce unforeseen biases during analyses. Here, we developed a cost-effective sequencing adapter design by modifying Illumina TruSeq adapters to incorporate a unique molecular identifier (UMI) while maintaining the capacity to undertake multiplexed, single-index sequencing. Incorporation of UMIs into TruSeq adapters (TrUMIseq adapters) enables identification of bona fide PCR duplicates as identically mapped reads with identical UMIs. Using TrUMIseq adapters, we show that accurate removal of PCR duplicates results in improved accuracy of both allele frequency (AF) estimation in heterogeneous populations using DNA sequencing and gene expression quantification using RNA-Seq.
Keywords: unique molecular identifier (UMI), RNA-Seq, TruSeq, PCR duplicates, TrUMIseq
Next-generation sequencing (NGS) enables a variety of quantitative assays, including frequency estimates of rare alleles from a population of cells and expression profiling using RNA-Seq. A critical technical issue associated with sequencing library preparation protocols for quantitative analyses is minimizing PCR duplicates originating from library amplification prior to cluster generation (1–3). PCR duplicates represent redundant information that can inflate perceived read depth of specific genome or transcriptome sequences and, therefore, introduce biases in detecting minor frequency alleles in heterogeneous populations (4) or result in over-estimation of fragments derived from specific mRNAs.
In practice, PCR duplicates are removed using bioinformatics tools that detect duplicates based on sequence identity and alignment information; however, one cannot know the rate of false-positive or false-negative duplicate detection using this approach because there is no independent means of assessing whether an identical sequence read is the result of PCR amplification or reflects an independently generated molecule that is identical by chance. To identify unique molecules in complex mixtures, stochastic labeling of DNA molecules with unique labels was introduced (5,6). Labeling biomolecules with random sequences has been adopted for NGS applications through the use of unique molecular identifiers (UMIs) (7–11). Identifying unique molecules using UMIs is essential for single-cell RNA-Seq protocols that rely on amplification from small amounts of starting material (12). Studies have shown that the use of UMIs improves the accuracy of NGS assays, and various protocols incorporating UMIs have been developed using modifications of the SMART protocol (7), amplicon library preparation (4), and tagmentation (13). However, no method for counting unique molecules currently exists that is compatible with TruSeq library workflows— the most commonly employed method for constructing Illumina sequencing libraries.
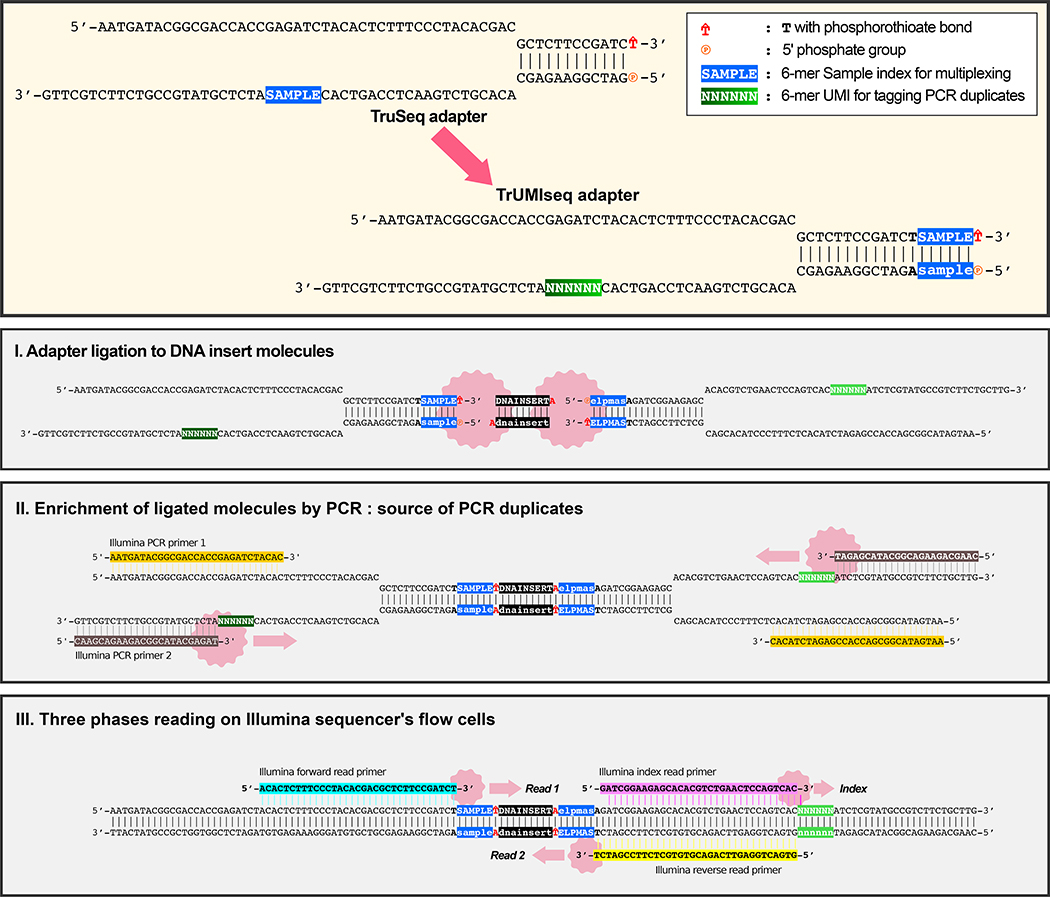
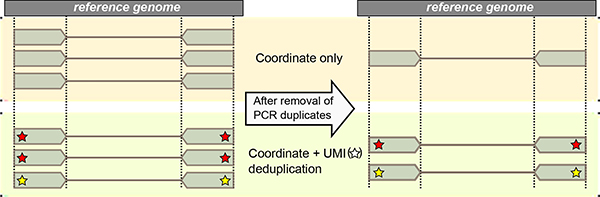
We developed a novel, cost-effective sequencing adapter design that enables identification of true PCR duplicates while maintaining the ability to perform sample multiplexing through modification of the widely used single-index TruSeq adapter. We moved the multiplexing sample index to the 5’ end of the adapter proximate to the ligation site of the DNA insert and placed a 6-bp UMI, generated by random incorporation of bases during oligonucleotide synthesis, at the position that typically contains the sample index (Figure 1A). Whereas uniquely formed molecules may contain identical insert sequences or identical UMIs, the chance of both occurring is exceedingly rare; therefore, reads with both identical mapping coordinates and identical UMI sequences are defined as true PCR duplicates (Figure 1B).
Figure 1. Accurate detection of PCR duplicates using TrUMIseq adapters.
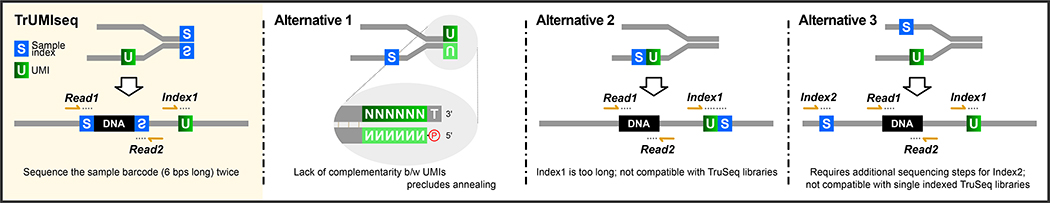
(A) TrUMIseq adapters are based on TruSeq adapters, with relocation of the sample index and addition of a unique molecular identifier (UMI). Libraries are generated and sequenced with TrUMIseq adapters using the identical ligation, PCR, and sequencing primers and protocols currently used for TruSeq adapters in either paired-end (PE) or single-end (SE) sequencing mode. After Step II, the two complementary strands of a double-stranded cDNA molecule will be barcoded with two different UMIs and sequenced as independent reads. When using a strand-specific RNA-Seq protocol, one of the cDNA strands is destroyed prior to PCR amplification. (B) Removal of PCR duplicates using TrUMIseq adapters. Whereas coordinate-based deduplication depends on mapping information only, the use of UMIs enables distinction between true PCR duplicates that have identical UMIs (red star) from independently generated molecules that have different UMIs (yellow star). (C) Comparison between TrUMIseq adapters and possible alternative configurations of UMIs and sample indices potentially compatible with single-index TruSeq workflows. TrUMIseq adapters can be easily incorporated into any single-index TruSeq protocol without requiring either specialized methods for preparing adapters or specialized sequencing steps.
Our design enables straightforward incorporation into existing single-index TruSeq workflows, and sequencing adapters can easily be prepared using oligonucleotide synthesis and annealing. Alternative designs based on TruSeq adapters either preclude duplex formation by annealing or require additional sequencing cycles or custom sequencing steps that make them incompatible with existing single-index TruSeq workflows (Figure 1C).
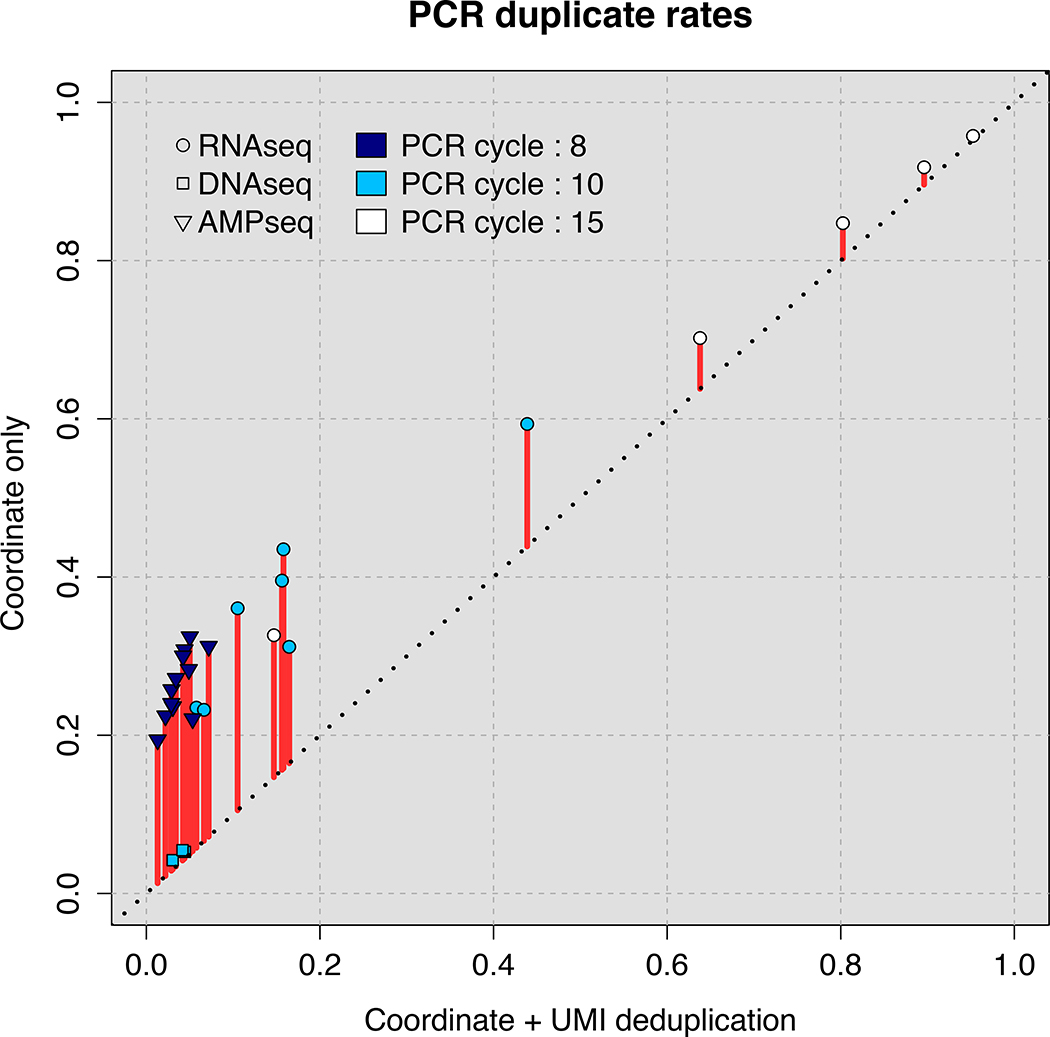
We compared rates of PCR duplicate detection using conventional genome coordinate-based methods versus UMI-based PCR duplicate detection. Mapping information and UMIs for three different sequencing protocols using TrUMIseq adapters and samples derived from budding yeast {Saccharomyces cerevisiae) were determined: (i) allele frequency (AF) estimation from whole-genome DNA sequencing (DNA-Seq); (ii) AF estimation from targeted sequencing of amplicons (AMP-Seq); and (iii) strand- specific RNA-Seq. The PCR duplicate rate is proportional to the number of PCR cycles used during library preparation and differs depending on the method of detection (Figure 2A). When considering only mapping information, the duplicate rate ranges 20%−40% for libraries prepared using <10 PCR cycles and up to 90% for libraries amplified using 15 cycles. By contrast, when incorporating UMI information to identify bona fide PCR duplicates, the duplication rate decreases to <10% for libraries constructed using <10 PCR cycles. Thus, up to 20% more unique sequencing reads can be recovered using TrUMIseq adapters that would otherwise be incorrectly discarded without the use of UMIs.
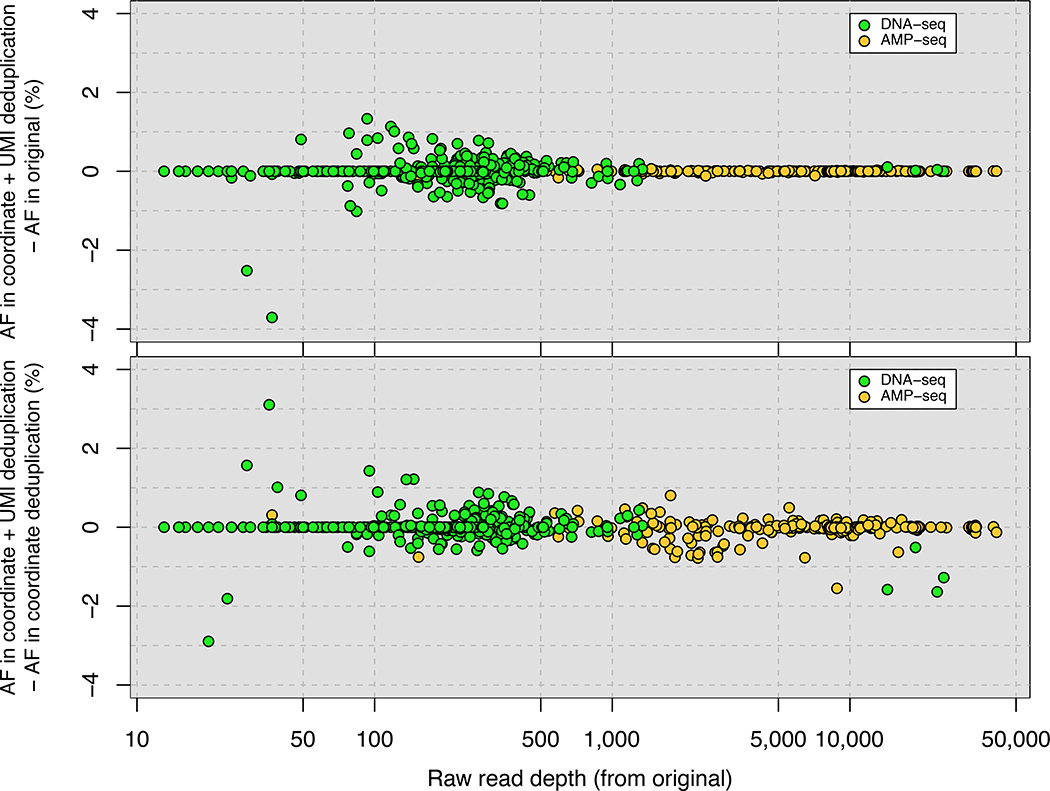
Figure 2. Accurate removal of PCR duplicates improves quantitative sequencing assays.
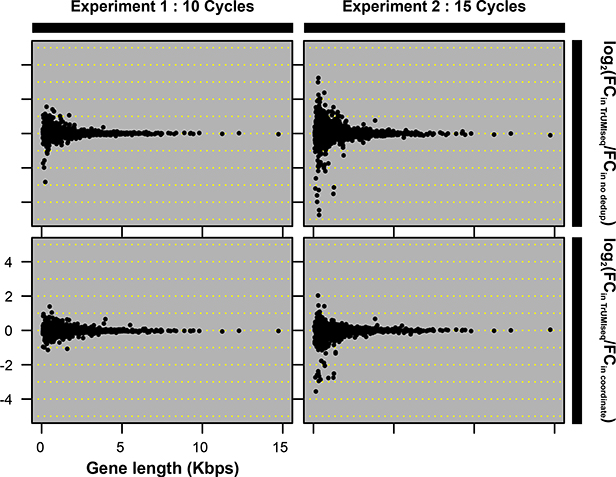
(A) Comparison of PCR duplicate detection rates using mapping coordinates only and mapping coordinates plus unique molecular identifiers (UMIs). (B) Differences in allele frequency (AF) estimates using deduplication based on mapping coordinates in conjunction with UMIs compared with no deduplication (upper panel) or deduplication based on mapping coordinates only (lower panel). A total of 482 (DNA-Seq) and 276 (Amp-Seq) SNPs were studied. (C) The effect of the number of PCR cycles on estimates of differential expression (DE) levels for all mRNAs using directional RNA-Seq. In each experiment, 2 biological replicates were prepared using either 10 cycles (Experiment 1) or 15 cycles (Experiment 2) of PCR. The fold change in Experiments 1 and 2 compared with 3 biological replicates of a reference sample generated using 10 PCR cycles was determined. The y-axis is the log2-transformed ratio between the fold change determined using UMI- and coordinate-based deduplication and either no deduplication (upper panel) or coordinate-only deduplication (lower panel).
We found that each sequencing protocol differs in the estimated PCR duplicate rate. AMP-Seq and RNA-Seq data have very different estimated duplication rates using the two deduplication methods (represented as triangular or circular data points in Figure 2A). Interestingly, our DNA-Seq data showed very low rates of PCR duplicates, <5%, regardless of the duplication detection method (rectangular data points in Figure 2A). This is likely due to two factors: (i) libraries obtained from whole genomes are more complex than libraries prepared from genome subsets (i.e., the transcriptome or targeted loci), and (ii) a larger quantity of starting material was used in our DNA-Seq library preparations.
Quantitative sequencing enables both detection and estimation of AFs of variants in heterogeneous samples, such as human tumors or microbial populations. We investigated the impact of a reduced rate of false-positive PCR duplicate detection when quantifying rare AFs in genome sequencing data from heterogeneous populations of yeast cells evolving under selection. We compared differences in AF estimates of SNPs identified in DNA-Seq and AMP-Seq libraries following removal of PCR duplicates (Figure 2B). The majority of SNPs show <1% difference in their AF estimation using any deduplication method. However, the difference in estimated AF increases to up to 4% as read depth decreases for both types of samples, demonstrating that correctly identifying PCR duplicates is critical for quantifying minor frequency alleles from sequencing data. AFs in heterogeneous populations are typically estimated from sequencing data with genome coverage ranging 50-fold to 300-fold. Therefore, it is notable that we find that accurate deduplication has the greatest impact on the accuracy of AF estimation at sequence read depths <500-fold.
Next, we tested the impact of the deduplication approaches on RNA-Seq quantification of gene expression. In general, deduplication of RNA-Seq data using UMIs improves the accuracy of estimation of differential gene expression, although this improvement is modest, which is consistent with previous reports (14). Importantly, we find that deduplication based on UMIs and mapping information improves differential expression analysis when applied to data generated using a higher number of PCR cycles (Figure 2C). Shorter transcripts appear to be most susceptible to misidentification of PCR duplicates (Supplementary Figure S1). Thus, the use of UMIs for deduplication of RNA-Seq data is of particular importance when making libraries from small amounts of starting material.
One potential technical issue associated with TrUMIseq adapters is that the presence of the sample index at the beginning of Read 1 results in low nucleotide diversity across a flow cell in the first 7 nucleotides of all reads. However, we have found that multiplexing libraries with different sample indices (Supplementary Table S1) in a single sequencing lane mitigates reduction of data quality associated with low base complexity using different Illumina sequencing platforms and a variety of quantities of PhiX spike-in (Supplementary Table S2). We analyzed the frequency of homopolymeric sequences in UMIs from deduplicated data generated on a NextSeq instrument (Illumina) and found an aberrantly high abundance of the GGGGGG UMI (Supplementary Table S3). The occurrence of poly-G sequences with high-quality base calls is a known problem with Illumina two-color chemistry, as G is detected on the basis of an absence of signal in both channels. UMIs that occur at frequencies much greater than expected should be excluded from downstream analyses. TrUMIseq adapters do not enable distinction of the two complementary strands that originate from a single DNA fragment in DNA-Seq or AMP-Seq (Step III In Figure 1A). However, the use of a strand-specific RNA-Seq protocol, as in our study, means that only a single UMI is associated with each cDNA molecule.
Our results illustrate the utility of TrUMIseq adapters for distinguishing true PCR duplicates from randomly generated identical molecules. The procedure for library preparation and sequencing using TrUMIseq adapters uses existing single-index TruSeq protocols and primers and, therefore, is readily implemented with or alongside existing TruSeq-based workflows. TrUMIseq adapters are highly cost-effective, as oligonucleotide synthesis and purification costs ~$150. A simple annealing reaction can be used to make a stock of 20 mM adapter, which allows construction of hundreds to thousands of libraries. Because TrUMIseq adapters are completely compatible with existing TruSeq workflows, including the routine use of PhiX for increasing base diversity, there are no additional costs associated with their implementation. Thus, the use of TrUMIseq adapters for accurate detection of PCR duplicates provides an inexpensive and straightforward means of improving quantitative data quality for any sequencing application that currently uses TruSeq adapters.
Supplementary Material
METHOD SUMMARY.
TrUMIseq adapters incorporate unique molecular identifiers (UMIs) in TruSeq adapters while maintaining the capacity to multiplex sequencing libraries using existing single-index workflows. The use of UMIs increases the accuracy of quantitative sequencing assays, including allele frequency (AF) estimation and RNA-Seq, by enabling accurate detection of PCR duplicates.
Acknowledgments
We thank Tara Rock and the Genomics Core Facility at the New York University Center for Genomics and Systems Biology for assistance in implementing TrUMIseq, and members of the Gresham lab for helpful discussions. This work was funded by the National Institute of Health (R01GM107466) and the National Science Foundation (MCB1244219). This paper is subject to the NIH Public Access Policy.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary material for this article is available at www.BioTechniques.com/article/114608.
References
- 1.Aird D, Ross MG, Chen W-S, Danielsson M, Fennell T, Russ C, Jaffe DB, Nusbaum C, and Gnirke A 2011. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 12:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoeijmakers WAM, Bartfai R, Françoijs K-J, and Stunnenberg HG 2011. Linear amplification for deep sequencing. Nat. Protoc 6:1026–1036. [DOI] [PubMed] [Google Scholar]
- 3.Dabney J and Meyer M 2012. Length and GC-biases during sequencing library amplification: a comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. Biotechniques 52:87–94. [DOI] [PubMed] [Google Scholar]
- 4.Smith EN, Jepsen K, Khosroheidari M, Rassenti LZ, D’Antonio M, Ghia EM, Carson DA, Jamieson CH, et al. 2014. Biased estimates of clonal evolution and subclonal heterogeneity can arise from PCR duplicates in deep sequencing experiments. Genome Biol. 75:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hug H and Schuler R 2003. Measurement of the number of molecules of a single mRNA species in a complex mRNA preparation. J. Theor. Biol 227:615–624. [DOI] [PubMed] [Google Scholar]
- 6.Fu GK, Hu J, Wang P-H, and Fodor SPA 2011. Counting individual DNA molecules by the stochastic attachment of diverse labels. Proc. Natl. Acad. Sci. USA 703:9026–9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kivioja T, Vàhàrautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, and Taipale J 2011. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 2:72–74. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, Prindle MJ, Kuong KJ, et al. 2014. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat. Protoc. 9:2586–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu GK, Xu W, Wilhelmy J, Mindrinos MN, Davis RW, Xiao W, and Fodor SPA 2014. Molecular indexing enables quantitative targeted RNA sequencing and reveals poor efficiencies in standard library preparations. Proc. Natl. Acad. Sci. USA 777:1891–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, and Linnarsson S 2014. Quantitative single-cell RNA-Seq with unique molecular identifiers. Nat. Methods 77:163–166. [DOI] [PubMed] [Google Scholar]
- 11.Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, et al. 2017. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol. Cell 65:631–643.e4. [DOI] [PubMed] [Google Scholar]
- 12.Stegle O, Teichmann SA, and Marioni JC 2015. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet 76:133–145. [DOI] [PubMed] [Google Scholar]
- 13.Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, et al. 2017. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parekh S, Ziegenhain C, Vieth B, Enard W, and Hellmann I 2016. The impact of amplification on differential expression analyses by RNA-Seq. Sci. Rep 6:25533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.