Abstract
Natural language processing (NLP) technologies have been successfully applied to cancer research by enabling automated phenotypic information extraction from narratives in electronic health records (EHRs) such as pathology reports; however, developing customized NLP solutions requires substantial effort. To facilitate the adoption of NLP in cancer research, we have developed a set of customizable modules for extracting comprehensive types of cancer-related information in pathology reports (e.g., tumor size, tumor stage, and biomarkers), by leveraging the existing CLAMP system, which provides user-friendly interfaces for building customized NLP solutions for individual needs. Evaluation using annotated data at Vanderbilt University Medical Center showed that CLAMP-Cancer could extract diverse types of cancer information with good F-measures (0.80-0.98). We then applied CLAMP-Cancer to an information extraction task at Mayo Clinic and showed that we can quickly build a customized NLP system with comparable performance with an existing system at Mayo Clinic. CLAMP-Cancer is freely available for academic use.
Keywords: Natural Language Processing, Information Storage and Retrieval, Electronic Health Records
Introduction
Cancer is a highly heterogeneous group of related diseases with a growing body of evidence that cancer initiation, progression, metastasis, and response to treatment are directly associated with variability at the molecular level [1]. Technological advances have enabled us to identify an increasing number of molecular biomarkers, leading towards personalized oncology [2]. Personalized cancer research relies heavily on large clinical series of well annotated and high-quality data. One emerging research direction is to leverage large, real-world practice data stored in electronic health records (EHRs) to facilitate clinical and translational research [3]. However, much of this information (e.g., tumor characteristics) can only be found in narrative or semi-structured text in EHRs, such as pathology reports. Despite the move towards synoptic reporting by the pathology community [4], uptake of this formatting is not complete and synoptic reports are usually only issued for definitive resections, as mandated by the American College of Surgeon’s Commission on Cancer Standard 2.1. Natural language processing (NLP) technologies that can automatically extract and structure information from narrative documents have been extensively investigated in the medical domain and cancer sub-domain [5, 6]. In particular, many studies have focused on named entity recognition (NER), a fundamental NLP task that locates and classifies named entities to predefined categories (e.g., diseases, drugs, and lab tests).
Several NLP systems have been developed to process pathology reports, which contain rich information about tumor specimen characteristics [7]. An earlier study by Xu et al. extended the existing MedLEE system to extract information from pathology reports to support cancer studies [8]. Crowley et al. reported the Cancer Tissue Information Extraction System (caTIES), which focuses on de-identification of cancer specimens for research purposes, along with information retrieval and concept coding functionalities [9]. MedKAT (MedTAS/P) is a rule-based system, which aims to extract cancer characteristics from pathology reports including anatomic site, histology, and grade [10]. Recently, Savova et al reported another rule-based system named DeepPhe, which can extract summary of cancer phenotypes including morphology, topology, procedure, and staging information from clinical documents [11].
Despite the success of reported use cases on cancer information extraction from clinical text, NLP has not been widely used for routine EHR-based cancer research, likely due to implementation barriers. Because of the diversity and complexity of clinical documentation, significant effort is required to develop custom clinical NLP systems or to extend existing systems for individual applications. In a study that aimed to locally adopt a smoking module in the cTAKES system [12], users had to write extensive code to re-train modules and re-define rules [13], which could be challenging for programmers without much NLP expertise. Additionally, completeness and the accuracy of the information remain as major obstacles in computational processing of clinical texts. CLAMP (Clinical Language Annotation, Modeling, and Processing) [14] is a general clinical NLP system that provides not only high-performance NLP components but also user-friendly interfaces for building customized NLP pipelines for individual needs. In this study, we describe our work on developing a library of information extraction modules for important tumor informiton in pathology reports within CLAMP. Using these built-in modules and user-friendly interfaces of CLAMP, users can quickly build customized NLP solutions to extract cancer information from their local pathology reports, with minimal programming effort. We believe such a tool would greatly benefit cancer researchers to leverage clinical text for their studies.
Methods
CLAMP follows the pipeline-based architecture as defined by the Apache UIMA™ (Unstructured Information Management Applications) framework, where an NLP system consists of multiple components in a specific order. The development of CLAMP Cancer Modules involved building cancer-specific NLP components and extending CLAMP interface that allows end users to customize and assemble different components into an NLP pipeline for their specific needs.
Developing Cancer-Specific NLP components
As the primary goal is to extract important information related to cancer, we limited the scope to the “diagnosis” section of pathology reports and followed these steps to build CLAMP Cancer Modules:
Define an Information Model for Pathology Reports
We reviewed available models including the Cancer Disease Knowledge Representation Model (CDKR) used in the MedKAT [10] and the information model in caTIES [9]. As our goal is to support clinical research, we specified data elements for pathology reports as recommended by the College of American Pathologists (CAP) (Figure 1). We reviewed all available CAP cancer templates (available at http://www.cap.org/cancerprotocols) as of January 2017, including the biomarker templates, and defined a list of minimum elements and relations to be included in CLAMP-Cancer Modules.
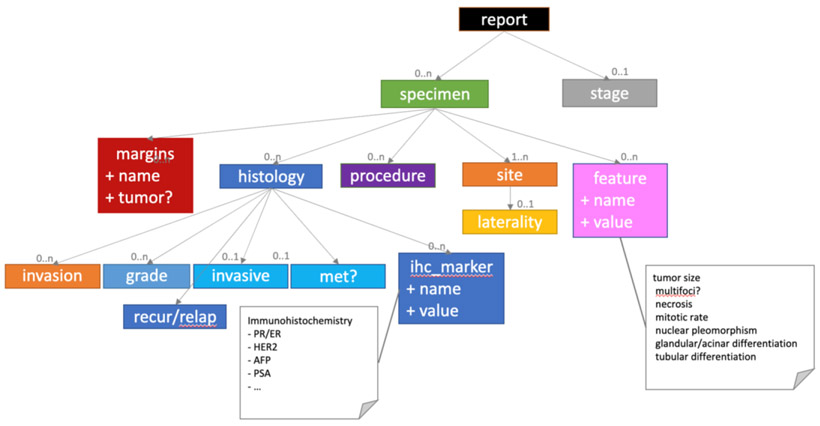
Figure 1.
CLAMP information model is based on data element suggestions of College of American Pathologists
We define Primary-site (site of the source of the material extracted from the body) as the root concept, with relations to Specimen (often numbered in the reports), Sub-site (detailed sub-anatomical sites related to the primary sites), Procedure (methods for specimen extraction from the body, e.g., biopsy), and Histology diagnosis (the morphology of cancer cells, e.g., “squamous cell carcinoma”). In addition, we also extract several important attributes defining histologic behavior and malignant potential, including Tumor Grade, Tumor Size, Invasiveness (invasion status and sites infiltrated by the tumor), Tumor Margin (tumor involvement in excisional margins), as well as Tumor Biomarkers and Values (e.g., estrogen and progesterone receptors).
Annotate a Corpus of Pathology Reports following the Information Model
Based on the information model, we developed an annotation guideline, which specifies the types of entities and relations for annotation, as well as different examples. We randomly selected 400 pathology reports from patients with a cancer ICD9 code at Vanderbilt University Medical Center (VUMC) in 2010. Four domain experts manually reviewed each pathology report and annotated all the entities and relations following the guideline, using CLAMP. Following a typical evaluation design for NLP studies, the annotated corpus was then divided into a training set (200 reports) and a test set (200 reports).
Develop Various Entity and Relation Extraction Components of CLAMP Cancer Modules
As CLAMP Cancer Modules extract a broad range of entities and their relations from pathology reports, different approaches were implemented to achieve optimal performance. For named entity recognition, we implemented regular expression-based, dictionary lookup-based, as well as machine learning-based approaches. For relation extraction, both rule-based and machine learning approaches were developed. Hybrid approaches that combine rules and machine learning were also used to optimize the performance. All the resources collected for developing these components (e.g., dictionaries, rules, and models) are available in CLAMP Cancer Modules and users can use them as-is or customize them to improve performance on their local data.
Evaluating CLAMP Cancer Modules
The evaluation of CLAMP Cancer Modules has two aspects. First, we developed the default CLAMP Cancer Modules using the training set from VUMC and evaluated them using the test dataset from VUMC. For each type of entity, we report precision, recall, and F-measure on recognizing entities only, using both exact and partial (relaxed) matching criteria [15]. We also report the system performance by considering both entity and relation information (e.g., a detected “sub_site” entity is correct only if both the entity and its relation to the linked “primary_site” are correct). Then, we apply CLAMP Cancer Modules to the same information extraction task as the MedKAT system [10]. Using CLAMP Cancer Modules, we assembled a customized NLP pipeline and compared it with MedKAT, using the same annotated corpus from Mayo Clinic.
Results
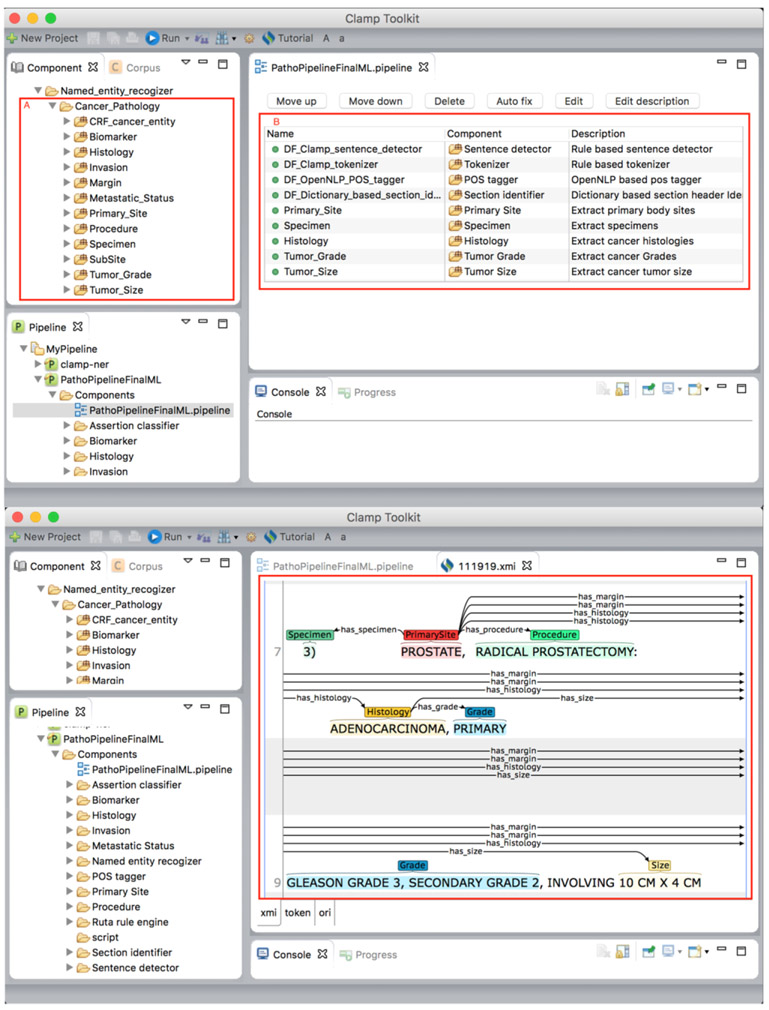
The developed CLAMP Cancer Modules consist of several components with machine learning models, dictionaries and rule sets to extract various types of cancer specific information from pathology reports following the CAP recomendations. Figure 2-a shows the cancer-specific modules in CLAMP and how they can be used to build customized NLP pipelines for local pathology reports. In Figure 2-b shows document annotation for cancer specific entities, and relationships among them.
Figure 2.
a. CLAMP Cancer module implements several cancer specific components (A), can be used to build customized pipelines (B). b. Pipeline annotates pathology reports to extract entities as well as their relationships.
Table 1 (a) shows the evaluation results of CLAMP Cancer Modules for different types of entities and relations. The default components of CLAMP Cancer Modules achieved good F-measures for entity recognition, with a range of 0.87 to 0.99, for exact matching criterion. A quick manual review shows that a fair number of relevant entities are missed by the system (e.g., a low recall in “invasion”), probably due to the complexity of expression patterns of these entities and the relatively limited size of the training corpus. When the relaxed matching criterion was used, CLAMP Cancer Modules achieved higher performance for some entities (e.g., Histology improved from 0.88 to 0.97), indicating some errors were about entity boundary only. When the relationships of entities were taken into consideration, performance dropped as expected (F-measures within 0.82-0.98), because of some known challenges for relation extraction (e.g., long-distance dependency). Table 1 (b) shows the results of the customized NLP pipeline built for Mayo Clinic’s pathology reports using CLAMP Cancer Modules, as well as the results from MedKAT. It took 12 hours for a developer to build the customized NLP pipeline for the specified task using CLAMP Cancer Modules, and it achieved comparable performance as MedKAT, indicating high effectiveness of CLAMP-Cancer for building customized NLP pipelines for pathology reports. Although MedKAT has slightly higher performance in certain entities, the production time and the adaptability remain as the major advantage of using CLAMP.
Table 1 –
Evaluation results of CLAMP Cancer Modules
| Type of information |
# of ent.* |
Entity only (Exact/Relaxed Matching) |
Entity and Relation | ||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F-measure | Precision | Recall | F-measure | ||
| Specimen | 310 | 0.99/0.99 | 0.99/0.99 | 0.99/0.99 | 0.97 | 0.98 | 0.98 |
| Primary-site | 351 | 0.98/0.99 | 0.98/0.99 | 0.98/0.99 | 0.98 | 0.98 | 0.98 |
| Sub-Site | 187 | 0.96/0.98 | 0.82/0.83 | 0.89/0.90 | 0.88 | 0.78 | 0.83 |
| Procedure | 339 | 0.98/0.99 | 0.98/0.99 | 0.98/0.99 | 0.97 | 0.97 | 0.97 |
| Histology | 553 | 0.91/1.00 | 0.85/0.93 | 0.88/0.97 | 0.90 | 0.85 | 0.86 |
| Tumor Grade | 92 | 0.96/1.00 | 0.88/0.91 | 0.92/0.96 | 0.91 | 0.83 | 0.86 |
| Tumor Size | 60 | 0.96/0.96 | 0.90/0.90 | 0.93/0.93 | 0.88 | 0.83 | 0.85 |
| Tumor Margin | 93 | 0.92/0.99 | 0.91/0.98 | 0.92/0.98 | 0.80 | 0.79 | 0.80 |
| Invasion | 71 | 0.92/1.00 | 0.83/0.90 | 0.87/0.95 | 0.86 | 0.78 | 0.82 |
| Biomarker | 107 | 0.95/0.99 | 0.90/0.94 | 0.92/0.96 | 0.88 | 0.84 | 0.86 |
|
(a) Performance of CLAMP-Cancer components on the VUMC test corpus * The number of each type of entities in the test corpus of 200 notes. | |||||||
| CLAMP-Cancer | MedKAT | ||||||
| Precision | Recall | F-1 | Precision | Recall | F-measure | ||
| Tumor size | 1.00 | 0.99 | 0.99 | 1.00 | 1.00 | 1.00 | |
| Dimension Extend | 1.00 | 0.99 | 0.99 | 0.99 | 1.00 | 0.99 | |
| Dimension Unit | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Tumor Site | 0.94 | 0.89 | 0.92 | 0.96 | 0.95 | 0.96 | |
| Histology | 0.91 | 0.92 | 0.92 | 0.96 | 0.98 | 0.97 | |
| Grade | 1.00 | 0.88 | 0.94 | 0.93 | 0.97 | 0.99 | |
| Date | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| (b) Performance comparison of CLAMP Cancer Modules and MedKAT on the same information extraction task from 302 pathology reports at Mayo Clinic | |||||||
The number of each type of entities in the test corpus of 200 notes.
Discussion and Conclusions
The contribution of CLAMP Cancer Modules is two-fold. First, it is a new NLP system that extracts comprehensive cancer information from pathology reports, including some that are not included by previous systems, e.g., biomarkers. Moreover, CLAMP Cancer Modules allow users to quickly build customized NLP pipelines by leveraging existing components and user-friendly interfaces. To the best of our knowledge, this is the first attempt to build cancer-focused NLP system that integrates into a drag and drop graphical user interface (GUI) for building customized NLP solutions for individualized needs. As pathology reports contain important cancer information, we believe that CLAMP Cancer will facilitate wide adoption of NLP in cancer research.
We acknowledge that cancer knowledge is constantly evolving and new standards, such as the AJCC 8th Edition, may require re-annotation for additional concepts. As a full-featured NLP system, CLAMP Cancer Modules ease such tasks. To further accelerate EHR-based cancer research, we plan to extend CLAMP Cancer Modules in several directions. First, we will develop an encoding function to further map extracted cancer information to existing terminologies, e.g., ICD-O-3, or to standard value sets. We will then expand CLAMP Cancer Modules to include other types of clinical reports containing rich cancer information, e.g., radiology reports. We also plan to conduct formal usability studies of CLAMP Cancer Modules and iteratively improve the user interface based on users’ feedback.
Acknowledgements
This study is partially supported by grants from NCI U24 CA194215, NIGMS R01 GM103859, NIGMS R01 GM102282, R01 LM011829, CPRIT R1307, and UTHealth Innovation for Cancer Prevention Research Training Program Pre-doctoral Fellowship (CPRIT # RP160015).
Footnotes
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the Cancer Prevention and Research Institute of Texas.
Disclosure
Dr. Soysal, Mr. Wang, Dr. Xu, and UTHealth have research-related financial interests in Melax Technologies, Inc.
References
- [1].Goossens N, Nakagawa S, Sun X, and Hoshida Y, "Cancer biomarker discovery and validation," (in eng), Transl Cancer Res, vol. 4, no. 3, pp. 256–269, June 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kalia M, "Biomarkers for personalized oncology: recent advances and future challenges," (in eng), Metabolism, vol. 64, no. 3 Suppl 1, pp. S16–21, March 2015. [DOI] [PubMed] [Google Scholar]
- [3].Yu P, Artz D, and Warner J, "Electronic health records (EHRs): supporting ASCO’s vision of cancer care," (in eng), Am Soc Clin Oncol Educ Book, pp. 225–31, 2014. [DOI] [PubMed] [Google Scholar]
- [4].Srigley JR et al. , "Standardized synoptic cancer pathology reporting: a population-based approach," (in eng), J Surg Oncol, vol. 99, no. 8, pp. 517–24, June 15 2009. [DOI] [PubMed] [Google Scholar]
- [5].Yim WW, Yetisgen M, Harris WP, and Kwan SW, "Natural Language Processing in Oncology: A Review," (in eng), JAMA Oncol, vol. 2, no. 6, pp. 797–804, June 01 2016. [DOI] [PubMed] [Google Scholar]
- [6].Spasić I, Livsey J, Keane JA, and Nenadić G, "Text mining of cancer-related information: Review of current status and future directions," International Journal of Medical Informatics, vol. 83, no. 9, pp. 605–623, September 2014. 2014. [DOI] [PubMed] [Google Scholar]
- [7].Burger G, Abu-Hanna A, Keizer N. d., and Cornet R, "Natural language processing in pathology: a scoping review," (in en), Journal of Clinical Pathology, vol. 69, no. 11, pp. 949–955, 2016-July-22 2016. [DOI] [PubMed] [Google Scholar]
- [8].Xu H, Anderson K, Grann VR, and Friedman C, "Facilitating cancer research using natural language processing of pathology reports," Studies in health technology and informatics, vol. 107, no. Pt 1, pp. 565–572, 2003. [PubMed] [Google Scholar]
- [9].Crowley RS, Castine M, Mitchell K, Chavan G, McSherry T, and Feldman M, "caTIES: a grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research," J Am Med Inform Assoc, vol. 17, no. 3, pp. 253–64, May-Jun 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Coden A et al. , "Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model," Journal of Biomedical Informatics, vol. 42, no. 5, pp. 937–949, October 2009. 2009. [DOI] [PubMed] [Google Scholar]
- [11].Savova GK et al. , "DeepPhe: A Natural Language Processing System for Extracting Cancer Phenotypes from Clinical Records," Cancer Research, vol. 77, no. 21, pp. e115–e118, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Savova GK, Ogren PV, Duffy PH, Buntrock JD, and Chute CG, "Mayo clinic NLP system for patient smoking status identification," (in eng), J Am Med Inform Assoc, vol. 15, no. 1, pp. 25–8, 2008. Jan-Feb 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liu M et al. , "A Study of Transportability of an Existing Smoking Status Detection Module across Institutions," presented at the AMIA Annu Symp Proc, 2012. Available: http://dx.doi.org/ [PMC free article] [PubMed] [Google Scholar]
- [14].Soysal E et al. , "CLAMP - a toolkit for efficiently building customized clinical natural language processing pipelines," (in eng), J Am Med Inform Assoc, November 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Uzuner Ö, South BR, Shen S, and DuVall SL, "2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text," (in eng), J Am Med Inform Assoc, vol. 18, no. 5, pp. 552–6, 2011. Sep-Oct 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]