Abstract
Warfarin is a narrow therapeutic index anticoagulant drug and its use is associated with infrequent but significant adverse bleeding events. The international normalized ratio (INR) is the most commonly used biomarker to monitor and titrate warfarin therapy. However, INR is derived from a functional assay, which determines clotting efficiency at the time of measurement and is susceptible to technical variability. Protein induced by vitamin K antagonist‐II (PIVKA‐II) has been suggested as a biomarker of long‐term vitamin K status, providing mechanistic insights about variation in the functional assay. However, the currently available antibody‐based PIVKA‐II assay does not inform on the position and number of des‐carboxylation sites in prothrombin. The assay presented in this paper provides simultaneous quantification of carboxy and des‐carboxy prothrombin that are essential for monitoring early changes in INR and, thus, serves as the superior tool for managing warfarin therapy. Additionally, this assay permits the quantification of total prothrombin level, which is affected by warfarin treatment. Prothrombin recovery from plasma was 95% and the liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) assay was linear (r 2 = 0.98) with a dynamic range of 1–100 µg/mL. The assay interday precision was within 20%. A des‐carboxy peptide of prothrombin (GNLER) was negatively correlated with active prothrombin (Pearson r = 0.99, P < 0.0001), whereas its association was positively linked with INR values (Pearson r = 0.75, P < 0.015). This novel LC‐MS/MS assay for active and inactive prothrombin quantification can be applied to titrate anticoagulant therapy and to monitor the impact of diseases, such as hepatocellular carcinoma on clotting physiology.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Protein induced by vitamin K antagonist (PIVKA‐II) has been adopted as a long‐term marker of prothrombin activity. The conventional enzyme‐linked immunosorbent assay (ELISA) assay for PIVKA‐II quantification uses an antibody that is not validated to distinguish different prothrombin γ‐carboxylation states, some of which have varying degrees of biological activity. Moreover, the ELISA assay does not provide quantification of total and fully active (10 Gla residues) prothrombin.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ This study investigated the effect of warfarin on the des‐carboxylation of prothrombin and defined the des‐carboxylation sites that correlate well with international normalized ratio value.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ This study provides a high throughput and sensitive liquid chromatography‐tandem mass spectrometry assay for the quantification of des‐carboxylated, fully carboxylated, and total prothrombin using only 10 µL plasma.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ The test we developed provides a more comprehensive assessment of hepatic vitamin K status and post‐translational modification of factor II, which may improve the safety and efficacy of long‐term warfarin therapy.
Biologically active prothrombin (factor II) contains 10 γ‐carboxylated glutamic acid (Gla) residues, which are formed post‐translationally by vitamin K‐dependent hepatic γ‐glutamyl carboxylase during prothrombin biosynthesis in the liver.1 Warfarin exhibits its anticoagulant effect by inhibiting the recycling of vitamin K epoxide to the reduced form of vitamin K, a cofactor used by γ‐glutamyl carboxylase,2, 3 and thereby slowing the synthesis of active prothrombin, extending blood clotting time, and reducing the chance of a systemic blood clot in at‐risk individuals. The pharmacodynamic response to a fixed starting dose of warfarin varies substantially between individuals, necessitating an adjustment of dose to achieve a common, safe degree of anticoagulation. This interindividual variability in warfarin pharmacodynamic response can be explained partly by demographic and clinical factors, as well as polymorphisms in genes encoding the: (i) warfarin metabolizing enzyme (i.e., cytochrome P450 2C9 (CYP2C9)),4, 5 (ii) warfarin target enzyme, vitamin K epoxide reductase complex subunit 1,6, 7 and (iii) vitamin K catabolizing enzyme, CYP4F2.8 Achieving a safe therapeutic response can be challenging due to warfarin’s narrow therapeutic index and high interindividual variability in the dose requirement. International normalized ratio (INR) of prothrombin time is used as a biomarker of warfarin efficacy9 in order to avoid adverse events associated with both under and over anticoagulation. Prothrombin time represents the time needed for clot formation in a plasma sample under a defined chemical stimulus. Interindividual and intra‐individual variability in warfarin response can be due to drug interactions affecting warfarin metabolism, liver dysfunction, and alteration in dietary vitamin K intake.10, 11, 12, 13, 14 An INR value > 4 is associated with an increased risk of bleeding,15 and the risk of intracranial hemorrhage increases approximately twofold for every one unit rise in INR above 4.16
INR is a functional assay of pharmacodynamic response to anticoagulation therapy that measures prothrombin time under prespecified conditions, normalized for laboratory variation. Readouts of the assay can be affected by methodological variables, such as blood collection procedure and sample handling temperature and duration.17, 18 Moreover, depending on the half‐life of vitamin K‐dependent coagulation factors, there is a time‐lag between warfarin dose and the initiation of the therapeutic response. For example, during the initial days of warfarin therapy, prolongation of INR correlates with decline in factor VII (half‐life 4–6 hours), whereas the effect of altered prothrombin on INR appears later (half‐life 60–70 hours).9, 19 To address limitations of the INR assay, quantitation of plasma under‐ γ‐carboxylated forms of prothrombin, also referred to as protein induced by vitamin K antagonist (PIVKA‐II) has been adopted as it is considered to be a more specific and long‐term marker of prothrombin activity. PIVKA‐II in plasma exists as a mixture of proteoforms with a variable number (1–9) of non‐carboxylated glutamate residues in the prothrombin Gla domain in warfarin‐treated patients. Vitamin K deficiency, warfarin treatment and liver dysfunction (e.g., hepatic carcinoma) lead to PIVKA‐II build up in the blood.13, 20 Conventionally, an enzyme‐linked immunosorbent assay (ELISA) assay is used for plasma PIVKA‐II quantification. Although the ELISA method is sensitive, the antibody is not validated to distinguish different prothrombin γ‐carboxylation states. The activity of the prothrombin is defined by its carboxylation states. Fully active prothrombin contains 10 carboxy glutamic acid residues (Glu). Carboxylation of prothrombin Glu residues is thought to occur stepwise and in a specific order described by Uehara et al.21 The absence of two carboxylation modifications (GC‐10 and GC‐9) leads to a 50% reduction in prothrombin activity, whereas the absence of three or more carboxylation modifications results in nearly complete loss of activity.22, 23 The ELISA assay does not provide quantification of total, fully active (10 Gla residues) prothrombin, which may bring added mechanistic value. An liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) assay that quantifies a peptide, ERECVEETCSY, representing one form of PIVKA‐II (des‐carboxylated at 7, 3, 6, and 5 Glu positions in the Gla domain) has been reported,24, 25 but again it does not test for fully active prothrombin nor carboxylation at specific glutamate residues. Thus, this peptide might not reflect early changes in coagulation status, as initial increases in INR likely reflect, in part, the absence of carboxylation at the last three glutamic acid residues (8–10) to be carboxylated (i.e., ANTFLEEVRK and GNLER). The assay also involves immuno‐enrichment of prothrombin prior to protein digestion, which is time‐consuming and expensive. To systematically address these limitations, we developed and validated an LC‐MS/MS assay to quantify multiple forms of des‐carboxylated that are shown to be critical for the activity (GC‐10 through GC‐7),22, 23 fully carboxylated, and total prothrombin levels using 10 µL of plasma and a high‐throughput prothrombin enrichment procedure. This novel, targeted proteomic method was applied to quantify des‐carboxy and fully carboxy prothrombin in plasma samples of warfarin‐treated and untreated individuals.
Targeted proteomics is a selective and quantitative methodology that utilizes stable isotope labeled surrogate peptides for the quantification of peptides or proteins in complex biological specimens in a high‐throughput manner. Moreover, targeted proteomics generally does not require immuno‐enrichment and protein fractionation to achieve sensitive quantification (down to low nanomolar levels). We believe that the method presented in this paper is superior to the ELISA assay in multiple ways: (i) it provides biochemical and mechanistic insight into the des‐carboxylation of glutamic acid residues that drive changes in prothrombin activity and blood clotting potential; (ii) it allows simultaneous quantification of multiple forms of prothrombin—fully active, partially active and inactive; and (iii) it allows quantification of total prothrombin levels that are also affected by warfarin or vitamin K deficiency. Moreover, this assay complements our previously developed LC‐MS/MS method for quantification of vitamin K metabolites in urine.26 The novel proteomics‐based assay is potentially applicable for (i) titrating anticoagulant therapy, (ii) investigating the impact of variable dietary vitamin K and genetic variation on prothrombin activity in the absence and presence of anticoagulation therapy, and (iii) for monitoring the impact of diseases, such as hepatocellular carcinoma on clotting physiology.
Methods
Chemicals and reagents
LC‐MS grade acetonitrile, methanol, chloroform, and formic acid were purchased from Fisher Scientific (Fair Lawn, NJ). Standard human prothrombin, trichloroacetic acid (TCA), and trifluoroacetic acid (TFA) were purchased from Sigma‐Aldrich (St. Louis, MO). Isopropanol (IPA) was purchased from VWR DBH Chemicals. The protein quantification bicinchoninic acid assay kit was purchased from Pierce Biotechnology (Rockford, IL). Ammonium bicarbonate buffer (98% purity) was purchased from Acros Organics (Geel, Belgium). Human serum albumin, a surrogate protein matrix, was obtained from Calbiochem (Billerica, MA). Iodoacetamide, dithiothreitol, MS grade trypsin, and surrogate heavy peptides representing carboxy‐forms and des‐carboxy‐forms of prothrombin were purchased from Thermo Fisher Scientific (Rockford, IL).
Sample procurement
The plasma samples were collected from two groups. Group 1 consisted of 12 individuals receiving antithrombotic warfarin therapy at the University of Washington Medical Center and group 2 were 12 healthy adults (controls) from University of Washington Medical Center. The samples from warfarin‐treated patients were archived, and anonymized specimens were obtained from University of Washington Laboratory Medicine. The group of adult controls were part of a University of Washington Institutional Review Board‐approved protocol for collecting anonymized blood samples to develop coagulation tests.
Sample preparation
Serum albumin was depleted from plasma (10 µL) using a published IPA‐TCA extraction methodology27 with few modifications. Briefly, 10 volumes of 0.1% TFA in IPA was vortex mixed with plasma for 2 minutes and centrifuged at 1,500 × g, 5 minutes at 4°C. The resulting supernatant that was primarily enriched with albumin was discarded. The protein pellet was washed with 0.5 mL ice cold methanol and re‐dissolved in a mixture of ammonium bicarbonate buffer (100 mM) and 4% sodium dodecyl sulfate (1:1, v/v, 100 µL). Extracted proteins were digested as described previously28 with slight modification. A detailed protein digestion protocol is described in the Supplemental Data.
LC‐MS/MS assay development and validation for quantification of carboxy and des‐carboxy prothrombin
Two peptides from the Gla region of prothrombin (carboxy/des‐carboxy forms) and three peptides from a non‐Gla region were selected as surrogates for active/inactive and total prothrombin, respectively (Figure 1). LC and multiple reaction monitoring (MRM) parameters for individual peptides were optimized using a digest of purified human prothrombin. The heavy labeled peptides were used as internal standards (Table S2). LC‐MS/MS analysis of carboxy and des‐carboxy peptides was performed on a SCIEX 6500 triple quadrupole mass spectrometer coupled to a Waters Acquity UPLC system. Chromatographic separation of peptides was achieved on a reversed phase HSS T3 C18 column (2.1 × 100 mm, 1.8 μ particle size; Figure S1). The mobile phase consisted of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) was used. The gradient conditions and MS parameters are reported in Table S1. Data were acquired by Analyst 1.6 software and analyzed by Skyline software 4.1 (MacCoss Lab, University of Washington, Seattle, WA).
Figure 1.
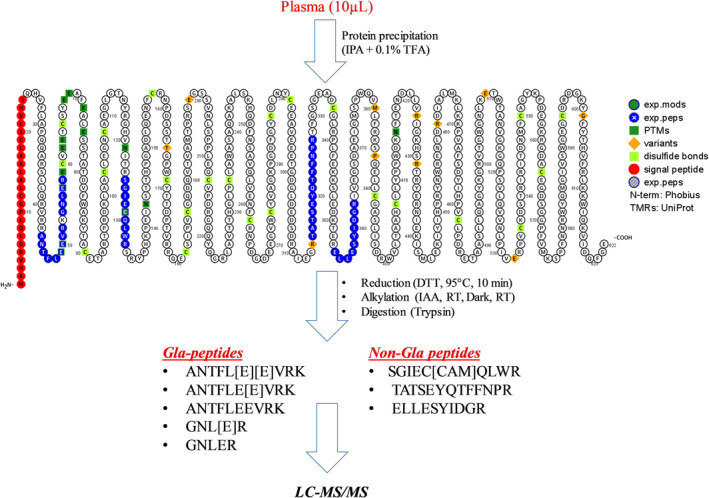
Schematic representation of sample preparation and analysis protocols. Proteins from plasma samples (10 µL) were precipitated with 0.1% trifluoroacetic acid (TFA) in isopropanol (IPA). Extracted proteins were reduced with dithiothreitol (DTT), alkylated with Iodoacetamide and digested with trypsin. Finally, digested samples were analyzed by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS). Prothrombin surrogate peptides from both glutamic acid (Gla)‐region and non‐Gla region (highlighted by blue color) were selected for the analysis.
The LC‐MS/MS assay was validated for linearity, precision, accuracy, and recovery. Linearity of the assay was assessed by spiking different concentrations of the standard prothrombin (1–100 µg/mL) into serum albumin. Accuracy and extraction recovery were assessed using the standard addition method by spiking standard prothrombin at two concentrations (40 and 80 µg/mL) into pooled human plasma from control adults. Interday precision of the assay was performed with the plasma samples processed on three different days. Prothrombin time for INR determination was measured using a STA‐Compact coagulation analyzer (Diagnostica Stago, Parsippany, NJ).
Data analysis
Peptide peaks were identified by matching the retention time with isotope labeled internal standards and alignment of selected precursor ion to daughter ion fragments. Data were normalized to internal standard and then with endogenous serum albumin to reduce the technical variability in the sample preparation. To address interindividual differences due to variable prothrombin concentration, active and inactive (Gla region peptides) prothrombin levels were normalized by non‐Gla peptide signals. Absolute or relative protein abundance (pmol/g) across healthy control and warfarin treated adults (group 1) and control adults (group 2) were then compared by the Kruskal‐Wallis followed by a Mann‐Whitney test. Associations between different peptide/protein measurements and INR were evaluated by linear regression. The protein abundance data are presented as mean ± SD.
Results
Selection of surrogate peptide and optimization of MRM transitions for quantification
We identified two surrogate tryptic peptides (ANTFLEEVRK and GLNER) using Skyline28 that represent Gla region of prothrombin covering the initial des‐carboxylation sites (GC‐10 through GC‐7 states). To quantify total prothrombin, we also identified three surrogate peptides from non‐Gla region of prothrombin (i.e., SGIECQLWR, TATSEYQTFFNPR, and ELLESTIDGR). For ANTFLEEVRK, four different possible carboxylation modifications on two glutamic acid residues (E) were possible (i.e., (i) carboxylation of both E residues, (ii) des‐carboxylation of both E residues, (iii) carboxylation of first E residue, and (iv) carboxylation of second E residue; Figure 1). Similarly, for GNLER, two different possible isoforms were predicted (Figure 1). y‐Ions and b‐ions for both Gla and non‐Gla peptides were selected for MRM quantification. However, Gla peptides did not show any signals for predicted MRM transitions. Therefore, to optimize the MRM transitions for carboxy‐peptides, we performed a pilot MS/MS experiment using custom synthesized carboxy peptide standards. We found that the neutral loss of CO2 molecule(s) from the y‐ion of the mono‐carboxylated or di‐carboxylated ANTFLEEVRK peptides resulted in identical fragmentation patterns of des‐carboxy and carboxy peptides (Figure 2). To selectively quantify individual prothrombin proteoforms, the LC method was optimized to separate the carboxy and des‐carboxy proteoforms of individual peptides, which resulted in good resolution between fully carboxylated ANTFL[E][E]VRK, mono‐carboxylated ANTFLE[E]VRK, and fully des‐carboxylated ANTFLEEVRK with retention times of 14.3, 12.5, and 13.0 minutes, respectively, confirmed by standard synthetic peptides (Figure S1) and fragmentation patterns (Figure 2). Des‐carboxy GNLER representing the absence of carboxylation at seventh glutamic acid residue (Figure S1), was detected at 6.0 minutes, whereas carboxy GNL[E]R was not observed in the reversed phase LC, perhaps due to (i) high‐polarity and early elution and/or (ii) poor ionization. The optimized LC and MRM transitions with y‐CO2 loss fragments are provided in Tables S1 and S2. Capitalizing on the CO2 loss during in‐source fragmentation of γ‐carboxylated peptides and construction of MRM transitions for selected peptide quantification is the major new technical finding of the present work that has broad applicability.
Figure 2.
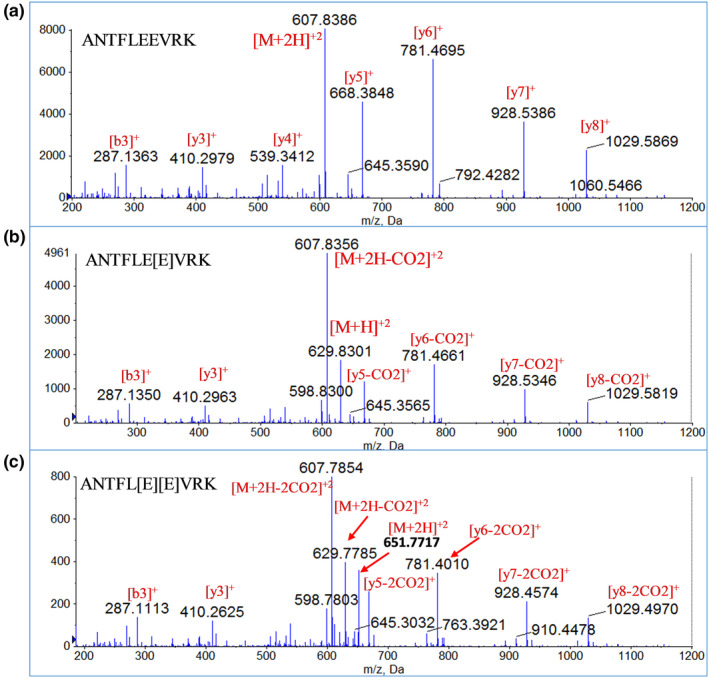
Tandem mass spectrometry (MS/MS) spectra of carboxy and des‐carboxy forms of a representative peptide, ANTFLEEVRK. (a) MS/MS spectrum of parent ion, m/z 607.8386 (ANTFLEEVRK – des‐carboxy peptide) showing y‐ions as dominant fragment, (b) MS/MS spectrum of parent ion, m/z 829.8301 (ANTFLE[E]VRK – mono–carboxy peptide) showing ions with one CO2 loss (y‐CO2) as dominant fragment and (c) MS/MS spectrum of parent ion, 651.7717 (ANTFL[E][E]VRK – di‐carboxy peptide) showing ions with two CO2 losses (y‐2CO2) as dominant fragments.
Optimized plasma sample preparation protocol
Plasma proteomics are often challenging due to ionization suppression and high background because of highly abundant proteins (e.g., albumin) in plasma. Here, we optimized a recently published cost‐effective albumin depletion method27 to enrich prothrombin, which utilizes IPA and TCA to remove albumin. For the modification, we replaced TCA by TFA, as the latter is a better solvent for dissolving serum albumin.29 High (> 95%) and reproducible recovery was observed with the standard addition method using optimized conditions (i.e., 0.1% TFA‐IPA and 10 µL plasma).
LC‐MS/MS assay validation
Analyte recovery with the LC‐MS/MS assay was calculated using three replicates of standard prothrombin addition to the pooled plasma samples, with two different prothrombin concentrations (40 and 80 µg/mL). At both concentrations, the extraction recovery was > 95% (Figure 3). The assay was linear (r 2 = 0.98; Figure 3) across a 100‐fold dynamic concentration range (1–100 µg/mL). All of the calibration curve points were analyzed in triplicate. Limit of detection and lower limit of quantification values were determined based on signal‐to‐noise ratios of 3 and 10, respectively. The assay was relatively sensitive, with a lower limit of quantification of 1 µg/mL of protein and a precision of ± 20%. The interday precision of the assay was also within ± 20%, as assessed by extracting and digesting prothrombin from plasma on three separate days (Table S3). Importantly, the use of isotope labeled internal standards in LC‐MS/MS assay provides high accuracy and precision in a complex matrix, unlike the ELISA assay. For example, incorporation of heavy internal standards into the method addressed analytical variabilities, including ion suppression/matrix effect in the MS/MS. Additionally, peptide correlation provides a quality check on the data. Peptides from the non‐Gla region correlates very well both in control and warfarin treated group (Figure S3 ,a–c). Although correlation between fully carboxy peptide from Gla region and peptide from non‐Gla region highlights lower levels of carboxy peptides in warfarin treated group (Figure S3 ,d–f). Similarly, correlation between des‐carboxy peptides and peptide from non‐Gla region highlights higher levels of des‐carboxy peptides in the warfarin‐treated group (Figure S3 ,g–i).
Figure 3.
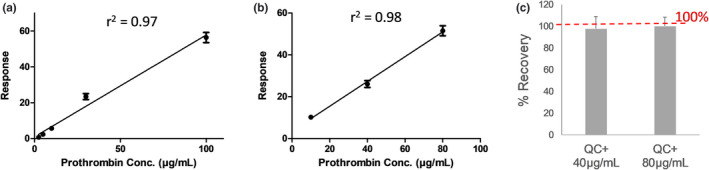
Assay performance. The linearity of the liquid chromatography‐tandem mass spectrometry assay was determined by spiking heavy peptide into human serum albumin (a‐ 2.5–100 µg/mL) and human serum matrix (b‐ 10, 40, and 80 µg/mL). Recovery was calculated by spiking the standard prothrombin at 40 and 80 µg/mL into pooled control serum samples (c).
Correlation of INR with carboxy and des‐carboxy prothrombin
The mean total plasma prothrombin concentrations were 47.5 ± 6.7 and 30.1 ± 13.4 µg/mL for control and warfarin treated samples, respectively. Total prothrombin was significantly reduced (*P < 0.02; Figure 4 a) in the warfarin‐treated group compared with the control group. Further, no significant correlation was observed between INR and total prothrombin when testing within the warfarin‐treated group (Figure 5 a). In addition, fully carboxylated prothrombin, represented by the surrogate peptide with γ‐carboxylation at glutamic acid residues 9 and 10 (ANTFL[E][E]VRK), was quantified to monitor the effect of warfarin treatment on the final steps of prothrombin post‐translational modification (i.e., biological activation). To address interindividual variability in total prothrombin concentration and its effect on the quantification of fully carboxylated and des‐carboxy prothrombin proteoforms, the average non‐Gla peptide response was used to normalize the carboxy and des‐carboxy prothrombin levels.
Figure 4.
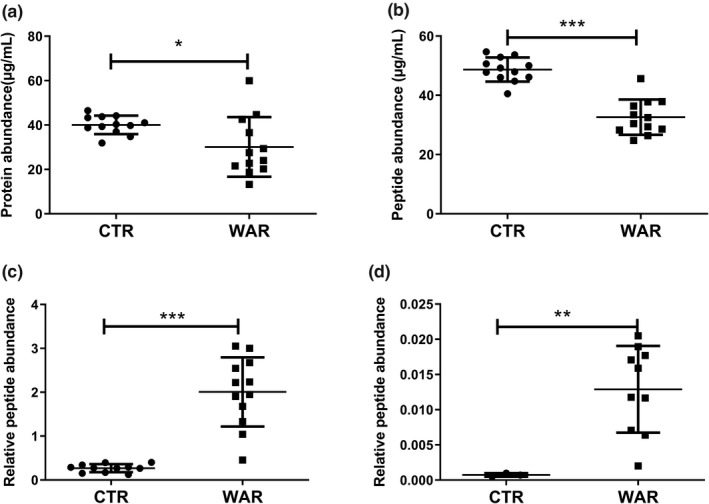
Comparison of abundance of total prothrombin, carboxy peptide (ANTFL[E][E]VRK) and des‐carboxy peptides (ANTFLEEVRK and GLNER) in serum samples from control and warfarin treated individuals. Each dot and square represent the individual subjects from control, and warfarin treated groups, respectively. Error bars indicate the standard deviation. Significant reduction in the levels of total prothrombin (P = 0.02) and carboxylated prothrombin (P < 0.0001) in warfarin treated subjects compared with control subjects was observed. Des‐carboxylated prothrombin was significantly (P < 0.0001) elevated in warfarin treated subjects. Des‐carboxy peptide ANTFLEEVRK is a low response peptide, which was quantifiable in first analysis only (n = 1). Mann‐Whitney test. *P value < 0.05, **P value < 0.01 and ***P value < 0.001.
Figure 5.
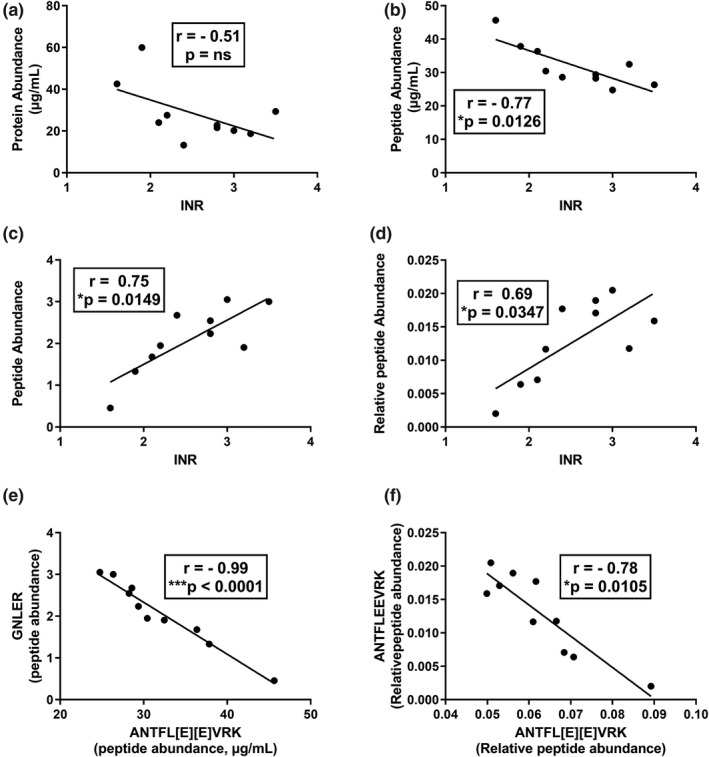
Correlation between international normalized ratio (INR) value with abundance of total prothrombin, carboxy peptide, and des‐carboxy peptides in serum samples from warfarin‐treated subjects. (a–d) represent correlations with INR in warfarin‐treated samples (group 1). (e,f) Represent the correlation between carboxy peptide ANTFL[E][E]VRK with des‐carboxy peptides GNLER and ANTFLEEVRK in the same sample set. INR values negatively correlated with carboxy peptides while positively correlated with des‐carboxy peptides. In addition, there was a strong negative correlation between carboxy peptide and des‐carboxy peptides.
The mean plasma concentrations of fully carboxylated prothrombin were 49.2 ± 1.2 and 32.6 ± 5.9 µg/mL for the control and warfarin treated samples, respectively (Figure 4 b). Fully carboxylated prothrombin levels were significantly reduced in the warfarin treated subgroup as compared with the control subgroup (Figure 4 b). Fully carboxylated prothrombin levels were reduced to ~ 90–50% of mean control and accompanied by an INR of 1.6–3.5. There was a significant negative correlation (Pearson r = −0.77, *P = 0.013) between fully carboxylated prothrombin concentration and the INR value (Figure 5 b).
To fully address the effect of warfarin on prothrombin carboxylation, additional prothrombin proteoforms were quantified; des‐carboxy tryptic peptide GNLER representing the absence of carboxylation at Glu residues 7, 8, 9, and 10 (GC‐6 or lower, assuming ordered γ‐carboxylation) and ANTFLEEVRK representing the absence of carboxylation at glutamic acid residues 9 and 10 (GC‐8 or lower). Plasma levels of both des‐carboxy peptides were significantly increased (***P < 0.0001) in the warfarin‐treated group, as compared with its control group (Figure 4 c,d). Additionally, the levels of the two des‐carboxy peptides (GNLER and ANTFLEEVRK) were positively correlated (Pearson r = 0.75 and 0.69) with the INR values (Figure 5 c,d). Interestingly, the increase in GNLER and ANTFLEEVRK was sevenfold and eightfold, respectively, whereas INR values ranged from 1.6–3.5. There was a strong negative correlation (Pearson r = −0.99 and −0.78) between peptides corresponding to fully carboxylated prothrombin (ANTFL[E][E]VRK) and des‐carboxy prothrombin peptides (GNLER and ANTFLEEVRK, respectively) representing GC‐6 or less and GC‐8 or less proteoform states, respectively (Figure 5 e).
Discussion
Active prothrombin contains ten γ‐carboxylated glutamic acid residues (Gla region). Carboxylation of glutamate residues in the Gla region reportedly takes place in a specific order resulting in different prothrombin proteoforms (GC‐1 to GC‐10), as shown in Figure S2.21 During warfarin therapy or vitamin K deficiency, PIVKA‐II is detected in blood as unspecified (i.e., with regard to site of des‐carboxylation) des‐carboxylated prothrombin. When tested in vitro, the absence of two carboxylation modifications (GC‐10 and GC‐9) leads to a 50% reduction in prothrombin activity, whereas the absence of three or more carboxylation modifications results in nearly complete loss of activity.22, 23 Accordingly, we posited that quantification of individual des‐carboxy peptides, representing glutamic acid residues that are the last four to be carboxylated (i.e., GC‐10, GC‐9, GC‐8, and GC‐7; Figure S2), will more accurately represent the status of prothrombin under‐carboxylation in blood during warfarin therapy. Two tryptic peptides ANTFLEEVRK and GNLER, that cover the GC‐10 through GC‐7, were selected for precisely quantifying the extent of warfarin effect during anticoagulation therapy.
While quantifying the fully carboxylated peptides, the predicted MRM transitions through Skyline for Gla peptides did not work. This was not surprising, as the analysis of Gla region peptides by MS is challenging and hindered by (i) presence of multiple carboxy modifications that impacts ionization efficiency of Gla peptides in ESI positive mode, (ii) in‐source fragmentation of the γ‐carboxy group, and (iii) an unpredictable and unstable fragmentation pattern due to γ‐carboxylation of multiple glutamic acid residues. Methylation of carboxy groups can be used to generate stable fragments30; however, an unpredictable number of methylated carboxy sites complicates the data analysis using this strategy. Identification of neutral loss of CO2 molecule(s) from the y‐ion of the carboxylated ANTFLEEVRK peptides confirmed by MS/MS analysis was the key finding for the optimizing the MRM transition for the carboxylated peptides.
Plasma proteomics often encountered with higher matrix effect due to high abundant proteins. To address this, a number of approaches can be used, such as protein fractionation,31, 32 albumin depletion by immunoaffinity column,33, 34 and immunoaffinity purification of target proteins.14, 20 Prothrombin proteoforms have been analyzed from plasma by multiple sample preparation protocols, such as immune‐enrichment, using specific prothrombin antibody or depleting this highly abundant protein using resin‐based columns.25, 35 However, these methods are low throughput, expensive, and also prone to variability due to nonspecific binding of the antibody. The optimized protocol provides prothrombin enrichment and a reduction in ion suppression because of the removal of highly abundant proteins, such as albumin. The optimized TFA‐IPA method is a high throughput strategy that can be automated, similar to ELISA, for prothrombin analysis. Moreover, this approach can be extended to enrich other γ‐carboxylated proteins and small molecules, where ionization is hindered due to the presence of highly abundant plasma proteins.
Warfarin treatment affects γ‐carboxylation of prothrombin leading to decreased levels of total prothrombin. This could be due to reduced stability of under‐carboxylated prothrombin or a reduction in prothrombin synthesis. Fully carboxylated prothrombin represents the most active form of the protein, whereas mono‐descarboxy prothrombin (ANTFLE[E]VRK) and di‐descarboxy prothrombin (ANTFLEEVRK) correspond to progressively less active carboxy prothrombin, whereas lower des‐carboxy prothrombin states represented by the GNLER peptide are reported to be inactive. Our results were consistent with the literature that warfarin treatment significantly reduces the levels of full carboxylated prothrombin whereas accumulation of des‐carboxy proreform (PIVKA‐II) in the plasma were positively correlated with increasing INR value. More precisely, des‐carboxy prothrombin representing completely inactive proteoform (GNLER) shows strong negative correlation with fully carboxylated proteoform also validate our results. PIVKA‐II has been identified in a biomarker in multiple pathophysiological conditions. Apart from its role in warfarin treatment, the PIVKA‐II assay is also useful for the diagnosis and monitoring of hepatocellular carcinoma (HCC), particularly in patients with HCC with low alpha fetoprotein levels.20, 36, 37, 38
Vitamin K deficiency is also associated with an increase in the plasma PIVKA‐II levels,39 thus it has importance in vitamin K‐related disorders. High PIVKA‐II values also have been reported for liver cirrhosis, hepatitis, benign nodule, and hepatic adipose infiltration. Among these conditions, HCC presents with significantly higher PIVKA‐II levels, as compared with others that exhibit small or no statistically significant changes.13, 24 However, the standard ELISA‐based assay may not distinguish between the differentially under‐carboxylated forms of prothrombin, which our novel method can accomplish. Such discrimination may provide added diagnostic specificity.
Regarding limitations of this study, the method we developed does not provide quantification of all possible des‐carboxy prothrombin proteoforms (e.g., detection of des‐carboxylation at GC positions 8 or 1–6) because we used trypsin as proteolytic enzyme and it generates a single large poorly ionization peptide containing those glutamic acid residues. An alternative digestion method (e.g., chymotrypsin) might address this limitation.
To our understanding, this is the first comprehensive LC‐MS/MS method for the simultaneous quantification of fully carboxy (active) prothrombin, different des‐carboxy prothrombin proteoforms, and total prothrombin. This method is highly sensitive, requiring only 10 µL of plasma, unlike the ELISA and INR assays, which require 100–200 µL of plasma. Further, this method has potential application for dry‐blood spot analysis, due to its low sample volume requirement. This may facilitate the analysis when venipuncture is not feasible (for patients living in remote settings), or less accepted by the study population. The LC‐MS/MS quantitative proteomics approach developed for quantification of prothrombin proteoforms may also be applicable to the quantification of other vitamin K‐dependent proteins, such as coagulation factor VII, IX, X, osteocalcin, matrix Gla protein, protein C, protein S, and protein Z.
Funding
This work was supported in part by grants from the National Institutes of Health, P01 GM116691 and T32 GM007750.
Conflict of Interest
The authors declared no competing interests for this work.
Author Contribution
A.B., B.P., A.E.R., and K.T. wrote the manuscript. A.B., B.P., A.E.R., and K.T. designed the research. A.B., J.K.E., D.E.S., N.A., and D.V. performed the research. A.B. and B.P. analyzed the data.
Supporting information
Supplemental Data. Tables S1–S3, Figures S1–S3.
References
- 1. Tie, J.K. et al Characterization of vitamin K‐dependent carboxylase mutations that cause bleeding and nonbleeding disorders. Blood 127, 1847–1855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agarwal, S. , Hachamovitch, R. & Menon, V. Current trial‐associated outcomes with warfarin in prevention of stroke in patients with nonvalvular atrial fibrillation: a meta‐analysis. Arch. Intern. Med. 172, 623–631; discussion 631–623 (2012). [DOI] [PubMed] [Google Scholar]
- 3. Cove, C.L. & Hylek, E.M. An updated review of target‐specific oral anticoagulants used in stroke prevention in atrial fibrillation, venous thromboembolic disease, and acute coronary syndromes. J. Am. Heart Assoc. 2, e000136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daly, A.K. , Rettie, A.E. , Fowler, D.M. & Miners, J.O. Pharmacogenomics of CYP2C9: functional and clinical considerations. J. Pers. Med. 8, (2018). 10.3390/jpm8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Flora, D.R. , Rettie, A.E. , Brundage, R.C. & Tracy, T.S. CYP2C9 genotype‐dependent warfarin pharmacokinetics: impact of CYP2C9 genotype on R‐ and S‐warfarin and their oxidative metabolites. J. Clin. Pharmacol. 57, 382–393 (2017). [DOI] [PubMed] [Google Scholar]
- 6. Fohner, A.E. et al Variation in genes controlling warfarin disposition and response in American Indian and Alaska Native people: CYP2C9, VKORC1, CYP4F2, CYP4F11, GGCX. Pharmacogenet. Genomics 25, 343–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henderson, L.M. et al VKORC1 and novel CYP2C9 variation predict warfarin response in Alaska native and American Indian people. Clin. Transl. Sci. 12, 312–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang, J.E. et al Effect of genetic variability in the CYP4F2, CYP4F11, and CYP4F12 genes on liver mRNA levels and warfarin response. Front. Pharmacol. 8, 323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Costa, I.M. et al Therapeutic monitoring of warfarin: the appropriate response marker. J. Pharm. Pharmacol. 52, 1405–1410 (2000). [DOI] [PubMed] [Google Scholar]
- 10. Cropp, J.S. & Bussey, H.I. A review of enzyme induction of warfarin metabolism with recommendations for patient management. Pharmacotherapy 17, 917–928 (1997). [PubMed] [Google Scholar]
- 11. Holbrook, A.M. et al Systematic overview of warfarin and its drug and food interactions. Arch. Intern. Med. 165, 1095–1106 (2005). [DOI] [PubMed] [Google Scholar]
- 12. Rohde, L.E. , de Assis, M.C. & Rabelo, E.R. Dietary vitamin K intake and anticoagulation in elderly patients. Curr. Opin. Clin. Nutr. Metab. Care 10, 1–5 (2007). [DOI] [PubMed] [Google Scholar]
- 13. Yu, R. , Tan, Z. , Xiang, X. , Dan, Y. & Deng, G. Effectiveness of PIVKA‐II in the detection of hepatocellular carcinoma based on real‐world clinical data. BMC Cancer 17, 608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lamerz, R. , Runge, M. , Stieber, P. & Meissner, E. Use of serum PIVKA‐II (DCP) determination for differentiation between benign and malignant liver diseases. Anticancer Res. 19, 2489–2493 (1999). [PubMed] [Google Scholar]
- 15. Makris, M. , van Veen, J.J. & Maclean, R. Warfarin anticoagulation reversal: management of the asymptomatic and bleeding patient. J. Thromb. Thrombolysis 29, 171–181 (2010). [DOI] [PubMed] [Google Scholar]
- 16. Hylek, E.M. & Singer, D.E. Risk factors for intracranial hemorrhage in outpatients taking warfarin. Ann. Intern. Med. 120, 897–902 (1994). [DOI] [PubMed] [Google Scholar]
- 17. van Geest‐Daalderop, J.H. , Mulder, A.B. , Boonman‐de Winter, L.J. & Hoekstra, M.M. , & van den Besselaar, A.M.H.P . Preanalytical variables and off‐site blood collection: influences on the results of the prothrombin time/international normalized ratio test and implications for monitoring of oral anticoagulant therapy. Clin. Chem. 51, 561–568 (2005). [DOI] [PubMed] [Google Scholar]
- 18. Mohammed Saghir, S.A. , Al‐Hassan, F.M. , Alsalahi, O.S. , Abdul Manaf, F.S. & Baqir, H.S. Optimization of the storage conditions for coagulation screening tests. J. Coll. Physicians Surg. Pak. 22, 294–297 (2012). [PubMed] [Google Scholar]
- 19. Kuruvilla, M. & Gurk‐Turner, C. A review of warfarin dosing and monitoring. Proc. (Bayl. Univ. Med. Cent.) 14, 305–306 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu, R. et al Performance of protein induced by vitamin K absence or antagonist‐II (PIVKA‐II) for hepatocellular carcinoma screening in Chinese population. Hepat. Mon. 15, e28806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uehara, S. , Gotoh, K. , Handa, H. , Honjo, K. & Hirayama, A. Process of carboxylation of glutamic acid residues in the Gla domain of human des‐gamma‐carboxyprothrombin. Clin. Chim. Acta 289, 33–44 (1999). [DOI] [PubMed] [Google Scholar]
- 22. Malhotra, O.P. , Nesheim, M.E. & Mann, K.G. The kinetics of activation of normal and gamma‐carboxyglutamic acid‐deficient prothrombins. J. Biol. Chem. 260, 279–287 (1985). [PubMed] [Google Scholar]
- 23. Ratcliffe, J.V. , Furie, B. & Furie, B.C. The importance of specific gamma‐carboxyglutamic acid residues in prothrombin. Evaluation by site‐specific mutagenesis. J. Biol. Chem. 268, 24339–24345 (1993). [PubMed] [Google Scholar]
- 24. Sohn, A. , Kim, H. , Yu, S.J. , Yoon, J.H. & Kim, Y. A quantitative analytical method for PIVKA‐II using multiple reaction monitoring‐mass spectrometry for early diagnosis of hepatocellular carcinoma. Anal. Bioanal. Chem. 409, 2829–2838 (2017). [DOI] [PubMed] [Google Scholar]
- 25. Sohn, A. et al Fully validated SRM‐MS‐based method for absolute quantification of PIVKA‐II in human serum: Clinical applications for patients with HCC. J. Pharm. Biomed. Anal. 156, 142–146 (2018). [DOI] [PubMed] [Google Scholar]
- 26. McDonald, M.G. et al A new LC‐MS assay for the quantitative analysis of vitamin K metabolites in human urine. J. Lipid. Res. 60, 892–899 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu, G. et al A novel and cost effective method of removing excess albumin from plasma/serum samples and its impacts on LC‐MS/MS bioanalysis of therapeutic proteins. Anal. Chem. 86, 8336–8343 (2014). [DOI] [PubMed] [Google Scholar]
- 28. Bhatt, D.K. & Prasad, B. Critical issues and optimized practices in quantification of protein abundance level to determine interindividual variability in DMET proteins by LC‐MS/MS proteomics. Clin. Pharmacol. Ther. 103, 619–630 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katz, J.J. Anhydrous trifluoroacetic acid as a solvent for proteins. Nature 174, 509–509 (1954). [DOI] [PubMed] [Google Scholar]
- 30. Hallgren, K.W. , Zhang, D. , Kinter, M. , Willard, B. & Berkner, K.L. Methylation of gamma‐carboxylated Glu (Gla) allows detection by liquid chromatography‐mass spectrometry and the identification of Gla residues in the gamma‐glutamyl carboxylase. J. Proteome Res. 12, 2365–2374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keshishian, H. et al Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Mol. Cell. Proteomics 14, 2375–2393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee, S.E. et al Plasma proteome biomarkers of inflammation in school aged children in Nepal. PLoS One 10, e0144279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tu, C. et al Depletion of abundant plasma proteins and limitations of plasma proteomics. J. Proteome Res. 9, 4982–4991 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bellei, E. et al High‐abundance proteins depletion for serum proteomic analysis: concomitant removal of non‐targeted proteins. Amino Acids 40, 145–156 (2011). [DOI] [PubMed] [Google Scholar]
- 35. Toyoda, H. et al Novel method to measure serum levels of des‐gamma‐carboxy prothrombin for hepatocellular carcinoma in patients taking warfarin: a preliminary report. Cancer Sci. 103, 921–925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fujiyama, S. , Morishita, T. , Shibata, J. & Sato, T. Clinical usefulness of plasma PIVKA‐II assay and its limitations in patients with hepatocellular carcinoma. Gan To Kagaku Ryoho. 16, 1129–1138 (1989). [PubMed] [Google Scholar]
- 37. Zakhary, N.I. , Khodeer, S.M. , Shafik, H.E. & Abdel Malak, C.A. Impact of PIVKA‐II in diagnosis of hepatocellular carcinoma. J. Adv. Res. 4, 539–546 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marrero, J.A. et al Des‐gamma carboxyprothrombin can differentiate hepatocellular carcinoma from nonmalignant chronic liver disease in American patients. Hepatology 37, 1114–1121 (2003). [DOI] [PubMed] [Google Scholar]
- 39. Dituri, F. et al PIVKA‐II plasma levels as markers of subclinical vitamin K deficiency in term infants. J. Matern. Fetal Neonatal Med. 25, 1660–1663 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data. Tables S1–S3, Figures S1–S3.


