Abstract
Behaviors emerge from activity throughout the brain, but non-invasive optical access in adult vertebrate brains is limited. We show that three-photon (3P) imaging through the head of intact adult zebrafish allows structural and functional imaging at cellular resolution throughout the telencephalon and deep into the cerebellum and optic tectum. With 3P imaging, considerable portions of the brain become non-invasively accessible from embryo to sexually mature adult in a vertebrate model.
Vertebrate behavior emerges from interactions of neurons spread widely throughout the brain, but non-invasive monitoring of neuronal activity at single cell resolution is limited. Translucent larval zebrafish allowed for the first non-invasive calcium imaging of vertebrate neurons, setting the stage for the burgeoning use of zebrafish for functional studies1–7. In contrast, opaque adult zebrafish (Fig. 1A) are less tractable for deep noninvasive approaches, although important imaging has been done of more accessible regions, mostly in brain explants 8,9 and, in one case, initial structural 3P imaging in the telencephalon of intact adult fish10. Here we evaluate non-invasive long wavelength 3P imaging in adult fish and image through the telencephalon and deep into the optic tectal and cerebellar regions, three widely-studied brain regions.
Fig. 1: Imaging deep into zebrafish and Danionella brains.
a. Adult zebrafish. b. Dorsal view of a zebrafish brain outline. Rostral is to the left. Tel: Telencephalon, OT: Optic tectum, CB: Cerebellum, HB: Hindbrain, SC: Spinal cord. c. CT of a zebrafish head. Red lines: sections through the head in CT cross sections at right. Arrows: brain regions, labeled as in B. White dot in HB: muscle. d. Signal intensity vs. depth from dye-labeled blood vessels to determine scattering lengths in labeled brain regions. Colors represent laser power, which increased with depth. e. Left side: 3D view of 3P imaging into cerebellum of an intact fish with nuclear localized GCaMP6s (Tg(elavl3:H2B-GCaMP6s)) (region shown in inset). GCaMP6s alone (green, on left) and overlain with THG (magenta on right). Arrowhead: skull. Long arrow: neuronal nuclei. Right: single slices containing larger, presumptive Purkinje cell nuclei (top), and smaller likely granule cell nuclei (bottom). f. 3D reconstruction from optic tectum of a fish with DSRed labeled glutamatergic neurons (Tg(vglut2a:loxP-DsRed-loxP-GFP)). Arrowhead: skull surface. DSRed labeled neurons are green for visibility. Left panel arrows: processes of glutamatergic neurons (upper arrow) and a deeper cell layer. Middle panel: THG channel (magenta). Right: overlay of THG fibers and DSRed neurons. g. Optical sections from a montage of Z stacks through one entire half of the telencephalon of an intact fish. Neuronal nuclei: green (Tg(elavl3:H2B-GCaMP6s)), THG:magenta. Rostral is to right. Arrowhead top left: skull. Dotted line, lower left: midline of the telencephalon, with an arrow marking neurons along the midline. h. Left and middle panels: NanoCT scan of an adult Danionella dracula head marking major brain regions. Left panel: sagittal view, with rostral to the left. Middle panel: cross section through the region imaged with 3P (red box) prior to the CT. Right: 3P imaging of blood vessels filled with fluorescein imaged at 1300 nm (green) and THG signal in red.
We imaged brains in 3–7 month old adults (N=41) in the Casper strain to minimize pigment interference11 (Fig. 1A,B). Using custom-built 3P imaging systems12,13, structural and functional imaging was mainly performed at 1300 nm, where green light emitting calcium indicators are optimized, with additional structural imaging at 1700 nm with red light emitting fluorophores. Third harmonic generation signal (THG) was simultaneously collected to allow visualization of surface structures, fiber tracts, blood vessels and blood flow (to assess viability)14 (Supp. Figs. 1, 2; Supp. video 1). Small to medium sized adults (mean standard length, tip of the head to base of the tail, 20.0 mm; range, 14.1–24.2 mm) were used to maximize optical access to the brain. CT scan images showed that brain regions vary in depth and the thickness of the overlying skull (about 50–115 μm ) and muscle (Fig. 1B,C), all of which may influence imaging success.
To assess in vivo optical properties of intact living fish using 3P, we imaged blood vessels labeled by retro-orbital injection of fluorescein or Texas red dextran to determine the decay of signal with depth (scattering length) at 1300 or 1700 nm in telencephalon, optic tectum, and cerebellum. Figure 1D shows a representative example of the results, as summarized in Supplementary Table 1. The telencephalon showed the longest scattering length (lowest decay with depth), consistent with its more translucent appearance in dissected brains. Shorter scattering lengths were measured in optic tectum and cerebellum. Signal-to-background ratio for labeled blood vessels and GCaMP6s labeled nuclei at different depths in the three brain regions studied is shown in Supplementary Fig 3. The upper bound for the axial resolution was ~ 2.9 μm (Supp. Fig. 4) at ~ 200 μm depth.
To determine safe imaging parameters, we explored both nonlinear (at the focal point) and linear (away from the focal point, “heat”) damage. To assess nonlinear damage, we focused on one shallow depth in the telencephalon or optic tectum, gradually raising the power until tissue damage was apparent both during imaging and in subsequent examination of resulting 3D volumes near the imaging plane. Three adults were tested at 1300 nm by imaging the telencephalon within 200–300 μm of the skull surface at 4 Hz with a laser repetition rate of 333 kHz, similar to parameters described below for calcium imaging. Tissue damage within the field of view began at 11.3 mW (276 μm depth), 8.9 mW (180 μm depth), and 12 mW (267 μm depth) (Supp. video 2). The fish retained blood flow (on THG channel) in regions outside the targeted area in all cases. In our structural and functional imaging experiments below, we used much lower powers of 2–5 mW in superficial imaging. Power was then carefully increased with imaging depth to compensate for laser light attenuation by the tissue.
We tested linear damage at 1300 nm by first locating a shallow region in the optic tectum with obvious blood flow and then scanning the laser beam on a plane 800+ μm below that region while exposing the brain to increasingly higher laser powers for 10 minutes. No change in surface blood flow was seen after exposure at 148 mW and 162 mW (N=1 each). Our actual maximal powers (up to 92 mW) for deep imaging at 1300 nm were well below this, indicating that linear damage from heating by the unfocused light is unlikely to occur at power levels useful for imaging.
We non-invasively imaged genetically labeled neurons in all three brain regions, focusing primarily on the ability to image nuclear localized GCaMP6s under the elavl3 promoter, which produces sparse, widespread labeling in adults that assists regional identification 4,6,8,15. We also successfully imaged fish with glutamatergic neurons labeled with DSRed (Tg(vglut2a:loxP-DsRed-loxP-GFP)16), tdTomato (Tg(vglut2a:mtdtomato) and GFP (Tg(vglut2a:EGFP17).
In the cerebellar region, we imaged THG signal at about 800 μm below the surface of the head and GCaMP6s at 700–750 μm below. An example cerebellar reconstruction from GCaMP6s imaging is shown in Figure 1E. Individual optical sections also revealed larger nuclei more superficially and deeper tightly packed smaller ones, consistent with the disposition and size differences of Purkinje and granule cells (Fig. 1E) 17.
Imaging of glutamatergic neurons in the optic tectum revealed the known layering of fibers and cell bodies there18, clearly evident at both 1700 nm (Fig. 1F, Tg(vglut2a:loxP-DsRed-loxP-GFP) 16) and 1300 nm (Tg(vglut2a:EGFP) 17; not shown). In superficial layers of the optic tectum where cell bodies are sparse and axons and dendrites predominate, fiber layers were visible on the THG channel (Supp. Fig. 1, 2) as were some fluorescently labeled cell bodies. In the deepest tectal imaging (approx. 750 μm below the surface of the head), we were able to reach the largest tectal cellular layer, the stratum periventriculare, which contains densely packed neurons18.
The telencephalon, where some 3P imaging of structure has been previously achieved 10, was the most favorable region for imaging, consistent with its longer scattering length. Figure 1G and supplementary videos 3 and 4 show imaging through the depth of the telencephalon (to 1045 μm below the surface of the head) based on a montage of one complete side of the forebrain from a series of Z stacks containing neurons labeled with elavl3:GCaMP6s. The 3P approach thus allows for noninvasive access to the entire forebrain of an intact adult vertebrate model system19.
We also used blood vessel labeling to test the depth of 3P imaging of adult Danionella dracula (n=3), a member of a closely related genus containing emergent translucent fish models with smaller sized adults than zebrafish20. We imaged through the head, the deepest parts of the brain, and beyond, as shown in the paired CT and 3P images of the same fish in Figure 1H.
Deep, noninvasive functional studies have proven challenging in adult zebrafish, in contrast to the extensive imaging possible in young larval fish1,3,4,6. We set out to overcome this barrier. Figure 2 shows 3P functional imaging at different depths in the cerebellum, optic tectum and telencephalon. We imaged spontaneous calcium transients deep into these regions. To demonstrate the signal-to-noise ratio in our imaging, raw photon counts of the data in Figure 2 are presented in Supplementary Figures 5, 6. We were able to monitor calcium signals in over two hundred neurons within a 200 μm by 200 μm field-of-view in a densely labeled region of optic tectum (Supp. video 5). The deepest imaging was in the telencephalon where we could detect calcium transients at 750 μm into the head, near the ventral extent of this region. There was still considerable calcium signal (over two fold increases) even in the deepest regions.
Fig. 2. Functional imaging of major brain regions in intact adult zebrafish.
Panels a-c: functional imaging in the Tg(elavl3:H2B-GCaMP6s) at different depths in cerebellum, optic tectum, and telencephalon. In each panel, the top row shows, from left to right, optical sections at increasing depths in that brain region. The images show raw fluorescence signal, averaged over the length of imaging with intensity mapped to the color bar at the right (red is the brightest pixel in each image and blue the dimmest). Middle row: Delta F/F traces from the numbered neurons in each frame. Bottom row: Raster plots of delta F/F intensity from neurons in the section. Lower right color bar maps the magnitude of the delta F/F signal. Note: the duration of imaging in the deepest telencephalic regions was shorter than the others, so that the time base is expanded.
In sum, we show deep 3P non-invasive imaging in several major, widely-studied brain regions of mature adult zebrafish, including access to the entire forebrain of a vertebrate model. Large portions of a vertebrate brain are now accessible for non-invasive structural and functional studies from embryo to adult.
Online Methods
Animal Preparation
Adult zebrafish between 3 to 7 months post-fertilization (Casper, Tg(elavl3::H2B-GCaMP6s)6; Tg(vglut2a:loxP-DsRed-loxP-GFP)16; Tg(vglut2a:EGFP)17; Tg(vglut2a: mtdtomato) were used. Fish have indeterminate growth with the rate varying with parameters such as feeding and fish density, so adults can vary widely in size. We primarily chose small to medium-sized adults to facilitate the percentage of brain structure accessible with deep imaging (mean size in our experiments = 20 mm standard length (tip of head to base of tail), 14.1–24.2 mm range). Animals were anesthetized in 0.2 mg/mL tricaine solution (pH 7.2) and then injected with 2 μL of pancuronium bromide (0.4 μg/μL in Hanks). The fish were stabilized by placing them in a “V” shaped groove cut in agar and held in place by a Kimwipe draped over the fish caudal to the head and glued onto the agar surface. A drop of bupivacaine was place on the surface of the head for anaesthetic. Fish were perfused through the mouth and over the gills at a rate of 2 mL/min with an ESI MP2 Peristaltic Pump (Elemental Scientific) with well-oxygenated temperature-controlled fish system water (4 L reservoir heated with a Finnex 800W heater controlled via a Heater Controller set to 80F).
Adult Danionella dracula were anesthetized in 0.03% benzocaine solution and their vasculature labeled via injection of a 10% solution of dextran fluorescein (10000 MW, Invitrogen) into blood vessels in a highly vascularized region caudal to the operculum and rostral to the heart. Fish were stabilized by positioning them on a cured Sylgard 184 Silicone Elastomer (Dow Consumer Solutions)-containing dish and kept upright with stainless steel supports embedded in the cured Sylgard. Fish were perfused through the mouth and over the gills at a rate of 1 mL/min with an ESI MP2 Peristaltic Pump (Elemental Scientific) with well-oxygenated temperature-controlled fish system water (4 L reservoir heated with a Top Fin Betta Aquarium Heater set to 77F (25C).
All procedures were in accord with the U.S. National Institutes of Health guidelines for animal use in experiments, and were approved by Cornell University’s Institutional Animal Care and Use Committee.
Computerized Tomography (CT) Scanning of Zebrafish and Danionella
Adult zebrafish were sacrificed in tricaine solution. Whole fish were fixed in 4% paraformaldehye at 4° C for 48 hours. They were then transferred to a solution containing 0.1% iodine metal (I2) and 0.2% potassium iodide (KI) in water until imaging 4 days later.
Zebrafish were scanned on a Zeiss Versa 520 CT scanner at 80 kV/7W. The instrument was set to yield a 4.9 μm resolution using a 4x objective. The CT image was reconstructed from 2001 x-ray fluoroscopy images with an exposure time of 3 s each, using the standard Zeiss reconstruction software. Skull thickness above telencephalon, optic tectum, and cerebellum was measured on the CT by averaging three dorsal locations above each brain region.
Adult Danionella were sacrificed in benzocaine solution. Whole fish were fixed in 4% paraformaldehyde at 4° C for 24 hours and then stained with 1% iodine metal and 2% potassium iodide. The stained fish was scanned at 120 kV/10W on the Zeiss Versa 520, using the 4x objective and a resolution of 2.7 μm. The machine took 2401 exposures of 0.7 seconds each, and the CT data was reconstructed using the standard Zeiss reconstruction software.
3P system
The 3P imaging was done with a previously published custom-built system.12,13. A schematic description of the system is shown in Supplementary Fig. 7.
Experimental setup at 1300 nm
Excitation Source
The excitation source for 3PM at 1300 nm was a noncollinear optical parametric amplifier (NOPA, Spectra Physics) pumped by a regenerative amplifier (Spirit, Spectra Physics). The NOPA provided an average power of ~500 mW (1,250 nJ per pulse at 400 kHz repetition rate). A half-wave plate (HWP) and a polarization beamsplitter cube (PBS) were used for power control.
Pulse compression
A two-prism (SF11 glass) compressor was used to compensate for the normal dispersion of the optics of the light source and the microscope, including the objective. The pulse duration (measured by second-order interferometric autocorrelation) under the objective was ~60 fs after optimizing the prism compressor (Supp. Fig. 7).
Imaging setup
Images were taken with a custom-built microscope with a high-numerical aperture objective (Olympus XLPLN25XWMP2, 25X, NA 1.05). The signal was epi-collected through the objective and then reflected by a dichroic beam splitter (FF705-Di01–25 × 36, Semrock) to the detectors. The detection system had two channels: one for the green fluorescence signal emitted by the calcium indicator and the other for third harmonic generation (THG) signal at λ = 433 nm. A 488-nm dichroic beam splitter (Di02-R488–25×36, Semrock) was inserted in the signal path at 45° between the two PMTs to separate the THG and fluorescence. We used a photomultiplier tube (PMT) with GaAsP photocathode (H7422–40) for the fluorescence signal and an ultra bialkali PMT (R7600U–200) for the THG signal. The optical filters for the fluorescence and the THG channels were 520/60 and 420/40 bandpass filters (Semrock), respectively.
Experimental setup at 1700 nm
Excitation Source
The excitation source for 3PM at 1700 nm was an optical parametric amplifier (OPA, Opera-F, Coherent) pumped by an amplifier (Monaco, Coherent). The excitation wavelength was ~ 1,680 nm. The average power of the source was ~330 mW (1,000 nJ per pulse at 333 kHz repetition rate). A half-wave plate (HWP) and a polarization beamsplitter cube (PBS) were used for power control.
Pulse compression
A 4 mm thick silicon plate was placed in the optical beam path to compensate for the anomalous dispersion of the optical elements of the light source and the microscope, including the objective. In order to maximize transmission, the silicon plate was placed at the Brewster’s angle. The pulse duration (measured by second-order interferometric autocorrelation) under the objective was ~70 fs.
Imaging setup
Images were taken with a custom-built microscope with a high-numerical aperture objective (Olympus XLPLN25XWMP2, 25X, NA 1.05). The signal was epi-collected through the objective and then reflected by a dichroic beam splitter (FF705-Di01–25 × 36, Semrock) to the detectors. The detection system had two channels: one for the red fluorescence signal emitted by DSRed or tdTomato and the other for third harmonic generation (THG) signal at λ = 560 nm. A 573-nm dichroic beam splitter (Di02-R561–25×36, Semrock) was inserted in the signal path at 45° between the two PMTs to separate the THG and fluorescence. We used a photomultiplier tube (PMT) with GaAsP photocathode (H7422–40) for the fluorescence signal and an ultra bialkali PMT (R7600U–200) for the THG signal. The optical filters for the fluorescence and THG channels were 593/40 and 556/25 bandpass filters (Semrock), respectively.
For both systems, the fish was placed on a motorized stage (M-285, Sutter Instrument Company). A computer running the ScanImage 3.8 module on MATLAB (MathWorks) software was used to control the stage translation and image acquisition. The PMT current was converted to voltage and amplified using a transimpedance amplifier (c7319, Hamamatsu). Analog-to-digital conversion was performed by a data acquisition card (NI PCI-6115, National Instruments).
The field of view ranged from 200 to 650 um. The pixel rate for 3P imaging is limited by the repetition rate of the excitation source, which is lower than that in 2P imaging. The fast, 4.25Hz per frame scanning rate used for functional imaging corresponds to about one laser pulse per pixel. For activity imaging, however, signal from the entire nucleus was integrated to generate the activity trace. Depending on the exact size, a single neuron consists of tens to hundreds of pixels within the field of view. Therefore, the number of excitation pulses per neuron per frame is large, even at the limit of 1 excitation pulse per pixel, enabling a high frame rate at a relatively low repetition rate. For structural imaging, longer pixel dwell time and/or averaging over multiple frames eliminates any impact of the low repetition rate. More details of dwell times, averaging etc. for structural and functional imaging are below.
Navigation around the head and brain of the fish
Navigation under the 3P imaging conditions initially proved challenging, as surface features under magnification, and using the THG channel, were not intuitive without familiarity. In order to ease navigation to known areas for imaging, we implemented a wider field of view light objective and white light imaging using a camera system to establish a relative coordinate system, defining reference points based on skull sutures and other anatomical features of the head. With experience, we were eventually able to more easily identify regions in single slices based on the THG signal from the surface structures, fiber patterns and the disposition of neurons (see Supp. Figs. 1, 2).
Decay Length Measurements
Decay lengths were quantified by examining signal intensities at different depths by imaging fluorescent dye injected into the bloodstream of wild-type unlabeled Casper fish. Approximately 1 μL of the dye, (20% concentration, 500 kDa MW fluorescein dextran or 70 kDa MW Texas Red dextran in Hanks solution for 1300 nm and 1700 nm excitation, respectively) was injected by hand retro-orbitally on the morning of the experiment. Positively labeled fish were screened by imaging the tail with a fluorescent dissecting microscope (Leica M205 FA) immediately after injection to look for clearly labeled blood vessels. For each region to be measured, stacks were taken at 5 to 10 μm steps starting deepest and moving shallower, and power was adjusted in order to avoid saturation of the PMTs and potential damage to the fish.
For analysis, after background subtraction and averaging, regions containing blood vessels were segmented from the image for each slice using a custom Matlab script to extract pixels 2–3 standard deviations above the mean intensity. An average signal intensity was produced for each slice by averaging the segmented pixels. Linear decay curves were fit to log(avg. signal / power^3) and reported as (3/slope) of the fit, which gives the length in microns through which the signal intensity will decay by a factor of 1/e3. Alternatively, the top 0.1% of pixels was also identified as over 3.29 standard deviation above background. The results were similar with both approaches.
Nonlinear and linear damage thresholds
Nonlinear damage was assessed in the telencephalon or optic tectum by imaging at 4.25 Hz (the rate used for our functional data) or 8.5 Hz for 4 minutes at a depth between 200 to 300 μm (including skin and skull) in the telencephalic or optic tectal regions with a repetition rate of 333 kHz in animals expressing GCaMP6s (Tg(elavl3:H2B-GCaMP6s)). When power was sufficiently high to start damaging tissue, damage was apparent during live imaging by sustained increase in brightness of GCaMP signals of neurons followed by a development of absence in the signal that appears as a black area with a whiter halo, where the tissue damage was likely most severe (Supp. video 2).
Linear damage was assessed by examining the presence or absence of blood flow on the THG channel in living animals near the surface of the optic tectum after sustained exposure for 10 minutes to a beam of systematically varying power focused at a depth of 800 μm below the surface. Animals were considered heat-damaged if blood flow occurred pre-exposure in a region but not post-exposure to deep imaging at the tested intensity. The overall viability of the animal was evident by assessing blood flow outside the exposed area.
Structural Imaging
For structural imaging using both 1300 and 1700 nm excitation, the maximum power was limited to about 90 mW at the deepest depths imaged in order to ensure a lack of damage to the tissue. As this appeared to be well below the damage limit, it is possible that additional power usage could result in good signal-to-noise ratios at even greater depths than reported here. Surface imaging required approximately 3 mW power. As we imaged deeper into the brain, power and the number of averages per slice were increased. Structural images were collected at exposures of 6 microseconds per pixel and frame rates of 0.53 Hz, with averaging of 3–10 images depending on depth. The longest structural imaging, for the entire telencephalon lobe, took over 4 hours, about 3 of which was actual scanning to collect the stacks.
For structural reconstructions, image stacks were assembled using a Matlab program autoAssembleStacks.m and Imaris (Bitplane). The program averaged .tif files and placed them according to the z-coordinates present in the header of the ScanImage.tif files. To obtain a concatenated stack from substacks collected at increasing power with depth, each substack was normalized such that the 99th percentile of pixels was at 2^15 (half the range of a 16bit number). Overlaps between stacks were averaged.
For more complex multi-column assemblies, such as that of the entire telencephalon on one side of the brain, each averaged substack was normalized to a single shallow slice with many labeled cells. The intensities in the reference slice were converted into the range [0, 2^15] and then the imhist function in Matlab was used to compute a histogram. Histogram equalization of the other substacks was accomplished by using the histeq contrast matching function. The larger assembly was then produced by using image coordinates from the microscope motor and the program XUVstitch. Finally, the normalize layers function in Imaris was used, followed by median smoothing (3×3×1) to produce the final images.
Adult female Danionella dracula (3 fish total) with a range in standard length from 15.7–16.7 mm were imaged. We used an injection of fluorescein caudal to the operculum to fill blood vessels in the fish, which we then imaged intact and alive. One to three image stacks ranging in z depth from 400–1000 μm were taken per fish at the midline near caudal midbrain and rostral cerebellum, where the brain is thickest in z. For structural imaging using 1300 nm excitation, maximum imaging power was 13.6 mW at the deepest depths imaged. Surface imaging required at most 3 mW power after the objective. As we imaged deeper into the brain, power was increased. Structural images were collected at exposures of 6 and 13 microseconds(μs) per pixel and frame rates of 0.47 and 0.23 Hz, with averaging of 3–10 images, depending on depth. Figure 1H (right panel) shows an example imaging at 6 us/pixel with a frame rate of 0.47 Hz. Images were assembled and viewed as for zebrafish above. While our imaging was 3P, Danionella dracula are so transparent that even 2P imaging may make much of their brain accessible.
Ca++ Imaging
Ca++ imaging was performed in healthy animals as assessed by blood flow visible on the THG channel. Images were acquired at a 3 μs exposure per pixel and a frame rate of 4.25 Hz, at minimal power needed to visualize the label. As expected with the differences in decay length, the ability to functionally image deep into the zebrafish adult brain varied across the major brain regions. The changes in neuronal calcium could be observed in telencephalon greater than 600 μm (see Fig. 2, 656 μm). The deepest functional imaging in the optic tectum was at 561 μm, and the deepest calcium imaging in the cerebellum was at 372 μm. The powers used were well below those that led to linear or nonlinear damage in our testing of damage thresholds. Though most functional imaging sessions were about 4–5 minutes long at a given plane, we performed several longer experiments, including one calcium imaging experiment where we imaged continuously for 1 hour at 4.25 Hz.
After acquisition of calcium imaging data, a correction was made for small lateral movements of the imaged region and background was subtracted. Cell segmentation was accomplished using Imaris (Bitplane). For each time point, a neuron’s intensity was calculated as the mean of its segmented pixels. Traces were further processed by averaging subsequent frames (halving the frequency to 2.125 Hz), smoothed using a moving average of 5 frames and passed through a low-pass Butterworth filter with cutoff at 0.43 Hz. ΔF/F was calculated by using the median of a 60 s window around each point as the baseline.
For photon counts, instrument background was estimated from calcium imaging videos using the presence of shuttered pixels on the first frame of each video (approximately the first two lines). Subsequently, the mean-background-subtracted image intensity per neuron was multiplied by the number of pixels segmented for that neuron and divided by a constant factor 880 which was determined experimentally12,13 as the scaling factor for photon counts of the instrumentation. Finally, flux was computed as a rate (photon counts per second per neuron) by multiplying the counts by the image acquisition frequency (typically 4.25 Hz).
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code Availability
The software codes used for the analyses are available from the corresponding author upon reasonable request.
Extended Data
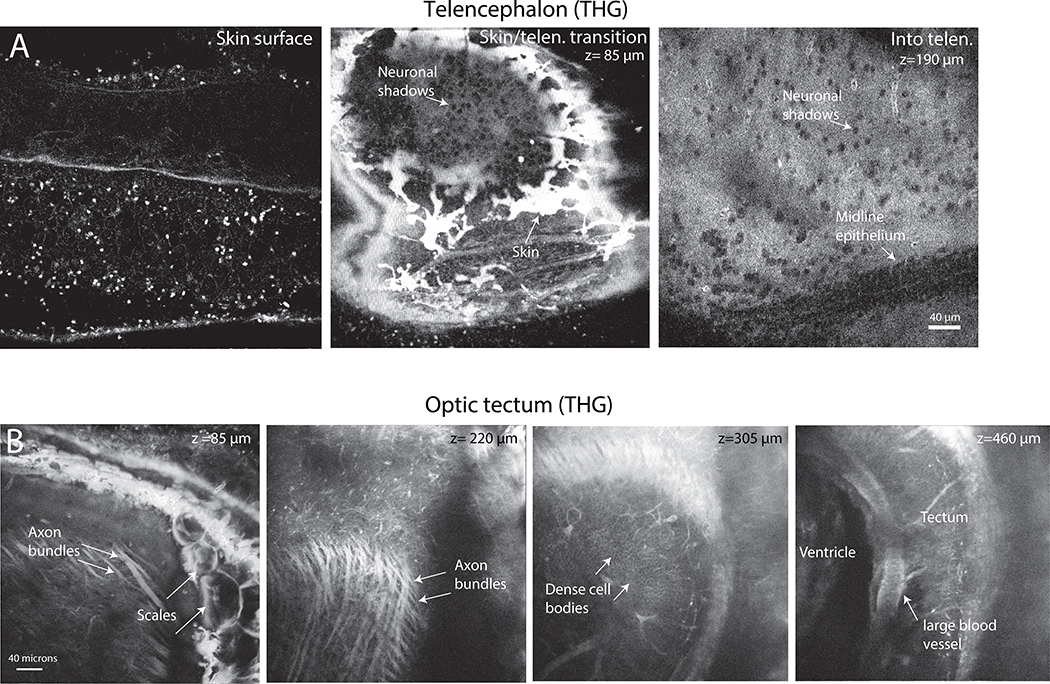
Extended Data Fig. 1. Structural features visible in optical sections with THG that facilitate navigation through the brain.
A. Telencephalic region, showing from left to right: the skin surface features; the transition into the brain as shadows of neurons become visible; and deeper in the telencephalon where the shadows of the neurons clustered at the midline are evident, as are scattered neurons throughout the section. B. Optic Tectum, showing from left to right: axon bundles on the surface of the tectum just below the skin and skull, with scales on the skin visible; deeper axon bundles in fiber layers of the tectum; densely packed shadows of cell bodies deep in the tectum; a major blood vessel lying adjacent to the ventricle deep in tectum. OT N= 16, Tel N=12.
Extended Data Fig. 2. Third harmonic generation images at different depths in the cerebellar and optic tectal areas.
Squares on brain diagrams on the right show approximate areas of imaging, and dashed lines represent approximate brain midlines. A. Cerebellar region from shallow to deep imaging. At 60 μm, some skin is visible (red asterisk) due to the curvature of the brain, as well as several blood vessels (red triangles) under the skin and above the skull. At 120 μm, skull features are evident. Deeper into the brain, fiber tracts predominate (red arrows). B. Optic tectal region from shallow to deep. At 147 μm, some skull is evident (green square) as well as some brain. Tectal fiber tracts are evident (green asterisk) at multiple depths (see 147, 245 and 378 μm) with a thick commissural fiber tract present (green triangle) at 490 μm. At deeper layers fiber tracts predominate (green arrows). CB N=10, OT N=16.
Extended Data Fig. 3. Signal to Background ratio.
Signal-to-background ratio (SBR) in shallow and deep regions in three areas of the brain. (A-C) Fluorescence intensity profiles along the lines across GCaMP6s-labeled neuronal nuclei (N=1 each). (D-F) Fluorescence intensity profiles along the lines across blood vessels labelled with fluorescein (N=2 each). To produce each plot, a five-pixel-wide line was drawn across the blood vessel or nucleus. The 5 pixel values were averaged to get an intensity value for the corresponding position along the line. The x axis corresponds to the lines shown on the images to the right. Scale bar 50 μm.
Extended Data Fig. 4. 3P Axial resolution measurement.
A. 3P image of a small fluorescein-labeled blood vessel (blue arrowhead) imaged about 200 μm deep into the optic tectum. Image stack was taken with 0.3 μm axial steps at 3 μs pixel time (512×512 pixels/frame) and 4 times averaging. B. Intensity profile along the axial direction fitted with a Gaussian curve to characterize the upper bound of axial resolution. The full width at half maximum of the Gaussian fit is 2.9 μm. N=1.
Extended Data Fig. 5. Example photon counts from telencephalic neurons.
Rows A-C show neurons at different depths. In each panel, the plots in magenta and green show photon counts from two neurons at that depth, with the monitored neurons marked with m and g in the optical section shown at the right. Tel functional imaging N= 9.
Extended Data Fig. 6. Raster plot of photon counts for all of the neurons shown in main Fig. 2.
Neurons were monitored at different depths in: A. telencephalon, B. optic tectum and C. cerebellum. Functional imaging: Tel N = 9, OT N = 6, CB N = 2.
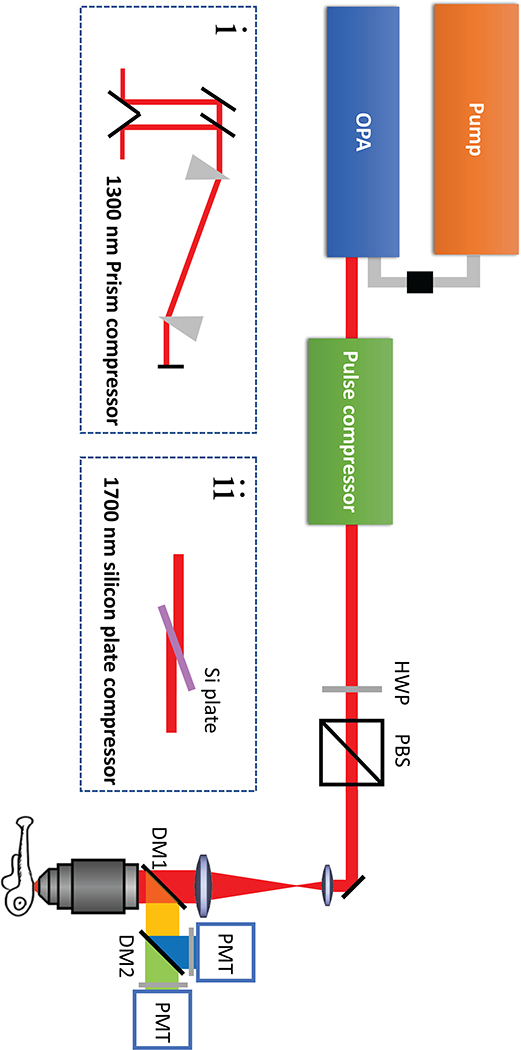
Extended Data Fig. 7. Experimental setup.
(a) schematic drawing of the excitation sources and the optical path of the imaging setup. OPA – optical parametric oscillator, HWP – Half wave plate, PBS – Polarizing Beam splitter, DM – Dichroic mirror, PMT – Photo-multiplier. inset(i) - Optical path for the prism compressor for the 1300 nm setup. inset(ii) - Optical path for the silicon plate compressor for the 1700 nm setup.
Supplementary Material
Video of nonlinear damage induced by excessive laser intensity
Blood flow imaged with THG at 8.5 Hz
Functional imaging with GCaMP6s 300 μm deep into the optic tectum for approximately 6 minutes in a region with densely packed neurons from the same optical section and data shown in Fig. 2B.
Video from a montage of Z stacks through the depth of telencephalon on one entire side of the brain from superficial to deep
3D video of one half of the telencephalon made from the Z stacks shown in Supp. video 3.
Acknowledgements
Supported by: NSF DBI-1707312 and NIH 1U01NS103516 and NSF IOS-1457108. We thank C. Walcott for photographic assistance, D. Mclean for his drawing skill, M. Farrar for the mTdTomato construct, and T. Porri for performing the CT scans.
Footnotes
Ethics Declaration:
All procedures were in accord with the U.S. National Institutes of Health guidelines for animal use in experiments, and were approved by Cornell University’s Institutional Animal Care and Use Committee.
Competing interest declaration.
The authors declare no competing interests.
References
- 1.Fetcho JR & O’Malley DM Visualization of Active Neural Circuitry in the Spinal-Cord of Intact Zebrafish. J Neurophysiol 73, 399–406 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Cox KJ & Fetcho JR Labeling blastomeres with a calcium indicator: a non-invasive method of visualizing neuronal activity in zebrafish. J Neurosci Methods 68, 185–191, doi:0165–0270(96)00067–2 [pii] (1996). [DOI] [PubMed] [Google Scholar]
- 3.O’Malley DM, Kao Y-H & Fetcho JR Imaging the Functional Organization of Zebrafish Hindbrain Segments during Escape Behaviors. Neuron 17, 1145–1155, doi: 10.1016/S0896-6273(00)80246-9 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Higashijima S, Masino MA, Mandel G & Fetcho JR Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. J Neurophysiol 90, 3986–3997, doi: 10.1152/jn.00576.200300576.2003 [pii] (2003). [DOI] [PubMed] [Google Scholar]
- 5.Ahrens MB et al. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 485, 471–477, doi: 10.1038/nature11057 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vladimirov N et al. Light-sheet functional imaging in fictively behaving zebrafish. Nat. Methods 11, 883–884, doi: 10.1038/nmeth.3040 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Andalman AS et al. Neuronal Dynamics Regulating Brain and Behavioral State Transitions. Cell, doi: 10.1016/j.cell.2019.02.037 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J et al. Early development of functional spatial maps in the zebrafish olfactory bulb. J Neurosci 25, 5784–5795 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobson GA, Rupprecht P & Friedrich RW Experience-Dependent Plasticity of Odor Representations in the Telencephalon of Zebrafish. Curr Biol 28, 1–14 e13, doi: 10.1016/j.cub.2017.11.007 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Guesmi K et al. Dual-color deep-tissue three-photon microscopy with a multiband infrared laser. Light: Science & Applications 7, doi: 10.1038/s41377-018-0012-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White RM et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189, doi: 10.1016/j.stem.2007.11.002 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouzounov DG et al. In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nat Methods 14, 388–390, doi: 10.1038/nmeth.4183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T et al. Three-photon imaging of mouse brain structure and function through the intact skull. Nat Methods 15, 789–792, doi: 10.1038/s41592-018-0115-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrar MJ, Wise FW, Fetcho JR & Schaffer CB In vivo imaging of myelin in the vertebrate central nervous system using third harmonic generation microscopy. Biophys J 100, 1362–1371, doi: 10.1016/j.bpj.2011.01.031 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park HC et al. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol 227, 279–293, doi: 10.1006/dbio.2000.9898 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Satou C, Kimura Y & Higashijima S Generation of multiple classes of V0 neurons in zebrafish spinal cord: progenitor heterogeneity and temporal control of neuronal diversity. J Neurosci 32, 1771–1783, doi: 10.1523/JNEUROSCI.5500-11.2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bae Y-K et al. Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Developmental Biology 330, 406–426, doi: 10.1016/j.ydbio.2009.04.013 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Corbo CP, Othman NA, Gutkin MC, Alonso Adel C & Fulop ZL Use of different morphological techniques to analyze the cellular composition of the adult zebrafish optic tectum. Microsc Res Tech 75, 325–333, doi: 10.1002/jemt.21061 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diotel N et al. Comprehensive expression map of transcription regulators in the adult zebrafish telencephalon reveals distinct neurogenic niches. J Comp Neurol 523, 1202–1221, doi: 10.1002/cne.23733 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britz R, Conway KW & Ruber L Spectacular morphological novelty in a miniature cyprinid fish, Danionella dracula n. sp. Proc Biol Sci 276, 2179–2186, doi: 10.1098/rspb.2009.0141 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video of nonlinear damage induced by excessive laser intensity
Blood flow imaged with THG at 8.5 Hz
Functional imaging with GCaMP6s 300 μm deep into the optic tectum for approximately 6 minutes in a region with densely packed neurons from the same optical section and data shown in Fig. 2B.
Video from a montage of Z stacks through the depth of telencephalon on one entire side of the brain from superficial to deep
3D video of one half of the telencephalon made from the Z stacks shown in Supp. video 3.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.