Abstract
Neuroblastoma represents 8–10% of all childhood cancer cases and is responsible for 15% of all cancer-related deaths in infants. Even though patients with low- and intermediate-risk disease have a good prognosis, the 5-year survival rate of the vast majority of patients with high-risk neuroblastoma is 50%. Despite extensive research efforts to find a cure for neuroblastoma, current treatment options are still limited. The aim of our study was to identify novel therapeutic compounds using high-throughput drug screening of a small molecule kinase inhibitor library containing 960 compounds. This screening resulted in the identification of two compounds, ST013381 and ST022328, that showed pronounced cytotoxic effects in six human neuroblastoma cell lines in vitro while having reduced effects in the BJ-5ta control cell line. These effects were observed in both MYCN-amplified and -non-amplified cells, indicating that these compounds can affect a wide range of neuroblastomas. Our experiments also revealed that several signaling pathways underlie the selective elimination of neuroblastoma cells by the ST013381 and ST022328 compounds. In summary, we have identified two novel compounds with a strong cytotoxic effect in vitro as promising agents for the treatment of neuroblastoma.
Electronic Supplementary Material
The online version of this article (10.1007/s40487-020-00113-5) contains supplementary material, which is available to authorized users.
Keywords: Drug discovery, High throughput screening, Kinase inhibitors, Neuroblastoma
Key Summary Points
| Why carry out this study? |
| Neuroblastoma, although representing 8–10% of all childhood cancer cases, accounts for 15% of all cancer-related deaths in infants. |
| No treatment options other than chemotherapy exist for high-risk neuroblastoma, even though the 5-year survival rate is 50%. |
| We searched for novel therapeutic compounds using high-throughput drug screening of a small-molecule kinase inhibitor library containing 960 compounds. |
| What was learned from the study? |
| We identified a group of compounds with cytotoxic effects in six neuroblastoma cell lines. |
| Data from validation studies on two of the identified compounds suggest that the cytotoxic effects are attained through modulation of the PKA, PKC, ATM/ATR, AMPK, Akt and MAPK/CDK signalling pathways. |
Introduction
Neuroblastoma is the most common extracranial solid malignancy in infants and children, with origins in sympathetic structures, such as the adrenal glands or paraspinal sympathetic ganglia [1]. It is usually diagnosed in children between 1 and 2 years of age, with up to 95% of all cases being diagnosed before 10 years of age [2].
Although responsible for 8–10% of all childhood malignancies, neuroblastoma accounts for 15% of all pediatric cancer-related deaths [3]. Patients with low-risk and intermediate-risk neuroblastoma usually have a very good outcome, with 5-year overall survival rates of 90–100%. First-line treatment of these patients is surgical removal of the tumor, with chemotherapy being recommended only if residual tumor is present or excision of the tumor is difficult [4]. Further, a subset of these low-risk and intermediate-risk patients does not require any treatment or surgery at all, with spontaneous regression of the tumor even with metastases [5]. In contrast, survival rates are lower than 50% in patients with high-risk neuroblastoma [4] and, more specifically, in a small subset of high-risk patients characterized by high levels of either tyrosine hydroxylase (TH) or PHOX2B mRNA in the peripheral blood, the 5-year overall survival is 0% [6]. Despite the improvement in the 5-year overall survival for high-risk patients over the years, from 29% in 1990 to 50% in 2010, new and improved therapies are greatly needed [4].
Although neuroblastoma has a remarkably low mutational burden, genetic alterations in neuroblastoma have been extensively studied. Such studies have lead to the identification of a number of somatic mutations, including those in anaplastic lymphoma kinase (ALK), alpha thalassemia/mental retardation syndrome X-linked (ATRX), v-myc avian myelocytomatosis viral oncogene neuroblastoma-derived homolog (MYCN), and neuroblastoma RAS viral oncogene homolog (NRAS) [7–10]. Germline mutations in ALK are the main driver of familial neuroblastoma, and somatic mutations in the kinetic domain of this gene are found in 10% of the sporadic cases of high-risk neuroblastoma [9]. ALK is able to signal through the RAS-MAPK signal transduction pathway, a pathway that is also frequently active during relapses [11]. Loss-of-function mutations in ATRX account for 9.6% of the cases of high-risk neuroblastoma, increasing to 44% in adolescents or young adults with neuroblastoma with metastatic disease. Interestingly, ATRX mutations are mutually exclusive with MYCN amplification [9, 12].
Identification and characterization of the genetic landscape of neuroblastoma have lead to the development and clinical applications of various therapies targeting altered proteins. One example is the discovery of ALK-tyrosine kinase domain (TKI)-targeting therapies. However, although new ALK-TKI-targeting therapies for non-small cell lung cancer (NSCLC) have become available in recent years, studies on NCLC have lead to the conclusion that new combinatorial therapies are needed to target secondary mutations that may arise following the treatment of neuroblastoma with these drugs [13].
In the study reported here, we used a high-throughput screening technique to test a library of 960 small-molecule kinase modulators in a neuroblastoma cell line, followed by validation of selected compounds in a larger neuroblastoma cell line cohort. To better understand the biological effects behind the selected compounds, we analyzed phosphorylation of the kinome using the KinomeView® kit (Cell Signaling Technology, Beverly, MA, USA). Our data suggest that two of the compounds identified, ST013881 and ST022328, have cytotoxic effects in neuroblastoma through modulation of the PKA (protein kinase A), PKC (protein kinase C), ATM/ATR (ataxia-telangiectasia mutated/ATM- and Rad3-related), AMPK (5′ AMP-activated protein kinase), Akt (protein kinase B) and MAPK/CDK (mitogen-activated protein kinase/cyclin-dependent kinase) signaling pathways.
Methods
Cell Culture
Human neuroblastoma cell lines GI-ME-N and IGR-N-91 were kindly provided by Dr. Annick Mühlethaler-Mottet (Lausanne University Hospital, Lausanne, Switzerland); human neuroblastoma cell lines IMR-5, SH-SY5Y, and SK-N-AS were obtained from University Children’s Hospital Zurich; and human neuroblastoma cell line SK-N-BE(2) and human immortalized fibroblast cell line BJ-5ta were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Neuroblastoma cell lines were cultured in DMEM/F-12 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Dutscher, Strasbourg, France), 1× l-glutamine (Thermo Fisher Scientific), and 1× penicillin/streptomycin. hTERT (human telomerase reverse transcriptase) immortalized BJ-5ta fibroblasts were cultured in a mixture of 4 parts of DMEM-high glucose and 1 part of medium 199, supplemented with 0.01 mg/mL hygromycin B and 10% fetal bovine serum. All cells were maintained in a humidified incubator at 37 °C, 5% CO2.
High-Throughput Drug Screening
A cell-based screening was performed at NEXUS Personalized Health Technologies (Zurich, Switzerland). The IGR-N-91 neuroblastoma cell line was cultured on 384-well black plates (Greiner Bio-One GmbH, Kremsmünster, Austria) at a seeding density of 300 cells per well, with each well containing 30 µL DMEM complete media, using a BioTek EL406 peripump (BioTek Instruments Inc., Winooski, VT, USA). Cells were seeded on a total of eight plates, in two batches of four plates each, in order to avoid uneven plating of the cells along the plates. After seeding, the plates were incubated at 22 °C for 1 h and subsequently transferred to an incubator at 37 °C, 95% humidity, and 5% CO2. After 24 h, six plates were selected for further use based on the homogeneity and cell count of the plates. Compounds of the ActiTarg-K960 library (Timtec Inc., Newark, DE, USA) were diluted 0.5 µL in 125 µL of DMEM complete media, and 10-µL aliquots were transferred to the destination plates, to reach a final concentration of 1 and 10 µM, respectively. Control edges were prepared using doxorubicin and etoposide at 10 µM (Selleck Chemicals Llc, Houston, TX, USA) and DMSO (Sigma, St. Louis, MO, USA). After 72 h of incubation with these compounds, cell viability was measured using the CellTiter-Glo 2.0 assay (Promega Corp., Fitchburg, WI, USA) according to the manufacturer’s instructions. Briefly, CellTiter-Glo solution was brought to room temperature, and 40 µL was added to each well of all plates. The plates were then shaken for 5 min on a BioTek EL406 device and further incubated at 22 °C for 25 min, after which luminescence was measured using a Tecan M1000 Pro monochromator-based microplate reader (Tecan Group Ltd., Männedorf, Switzerland), with an integration time of 1000 ms. Data analysis was performed using a semi-automated workflow implemented in R [37].
Compound Validation
Six neuroblastoma cell lines and one immortalized fibroblast cell line (see "Cell Culture") were used to validate the results observed in the high-throughput drug screening. The cell lines were plated on 384-well black plates (Greiner Bio-One GmbH) at a seeding density of 3000 cells per well, with each well containing 30 μL of the respective medium, and on the following day treated with 16 compounds at final concentrations ranging from 0.01 and 10 µM. After 72 h of culture, cell viability was evaluated using the CellTiterGlo 2.0 assay, and the luminescence resulting from the reaction was measured using a Tecan Infinite 200 Pro with an integration time of 1000 ms, as described in "High-Throughput Drug Screening". Data visualization was performed using the software program GraphPad Prism (GraphPad Software, San Diego, CA, USA).
KinomeView
Cells were plated, treated for 2, 4, 6, or 12 h with 10 µM ST013881 and ST022328, two compounds identified from the high-throughput screening as having cytotoxic activity in neuroblastoma cells, and protein extracted using RIPA buffer (Thermo Scientific, Thermo Fisher Scientific) with 1× PhosSTOP phosphatase inhibitors, and 1× Omplete ULTRA protease inhibitor cocktail (Roche AG, Basel, Switzerland). Cell lysates were centrifuged at 14,000 rpm for 30 min at 4 °C and the supernatant collected. Protein was quantified using the Pierce BCA assay (Thermo Scientific). Protein (30-μg samples) obtained from the cell lysate was denatured by adding Laemmli buffer (Bio-Rad, Hercules, CA, USA) and boiling the samples at 95 °C for 5 min. The samples were then loaded into Mini-PROTEAN TGXTM 4–20% precast gel (Bio-Rad), electrophoresed, and transferred onto Trans-Blot Turbo PVDF membranes (Bio-Rad) using a Trans-Blot Turbo Transfer System (Bio-Rad). The PVDF membranes were subsequently blocked using 5% milk for 1 h at room temperature, followed by overnight incubation with primary antibodies from the KinomeView kit (1:1000, Cell Signaling Technology) and tubulin (1:1000; Sigma) diluted in TBS-T (TBS with 0.1% Tween-20). The membranes were then washed with TBS-T and incubated for 1 h at room temperature with secondary horseradish peroxidase-coupled antibody (1:2000; BioLegend, San Diego, CA, USA). After washing with TBS-T, the membranes were developed using Clarity Western ECL Substrate (Bio-Rad) on a Fusion Fx imaging system (Vilber Lourmat Sté, Collégien, France).
Compliance with Ethics Guidelines
No ethical approval was required for this research. The human cell lines used are commercially available and, therefore, their use did not require specific approval.
Results
High-Throughput Screening of Small-Molecule Kinase Modulators
To identify new compounds with cytotoxic activity in neuroblastoma cells, we, in collaboration with NEXUS Personalized Health Technologies (ETH Zürich), screened the ActiTarg-K960 small-molecule library on the MYCN-amplified IGR-N-91 cell line, as MYCN amplification represents about 25% of neuroblastoma cases and correlates with high-risk cases (Fig. 1a). Doxorubicin and etoposide, two chemotherapeutics commonly used to treat neuroblastoma, were used as positive controls and added to both edges of the 384-well black plates. To reduce false positives that could arise from the treatment with only one concentration, treatments were performed at final concentrations of 1 and 10 µM, respectively (Fig. 1b), and cell viability was determined after 72 h using the CellTiter-Glo 2.0 assay (Fig. 1c; Electronic Supplementary Material [ESM] Table 1). The quality of the assay was verified using the Z′-factor, and only assays with a Z′-factor > 0.5 were accepted [14]. To improve the quality of the results, plate-to-plate systemic variations, edge effects, and within-plate strip patterns were removed, the first by standard normalization procedures and the latter two by subtracting a smooth polynomial using the LOESS function (ESM Fig. 1, ESM Table 1) [15–17]. From the 1920 hits observed when testing the 960 compounds at both concentrations, 141 were significantly active (false discovery rate [FDR] < 0.01) and 63 presented significantly strong activity (FDR < 0.01; normalized effect size > 0.5), corresponding to 7.3 and 3.2% of total hits, respectively. Significantly and strongly active hits were clustered according with their cytotoxic activity at both concentrations (Fig. 1d). From 55 compounds that were considered to be significant and strongly active (5.7% of total compounds), seven (0.7%) showed strong cytotoxic activity at both concentrations (green cluster), nine (0.9%) showed strong cytotoxic activity at 10 µM (blue cluster), and 39 (4.0%) had mild cytotoxic activity at 10 µM (red cluster) (Fig. 1d). Sixteen compounds found in the green and blue clusters were chosen for further validation due to their strong cytotoxic activity.
Fig. 1.
High-throughput drug screening in the IGR-N-91 neuroblastoma cell line. a Schematic of the screening procedure with the Acti-TargK960 small-molecule library. b Overview of the plates in the final screen, after 72 h of treatment with the compounds at 1 and 10 µM. c Volcano plot of all wells (control, doxorubicin, etoposide and test compounds). Horizontal line corresponds to a false discovery rate (FDR ) = 0.01, and compounds under this line are considered to be ‘not significant’ in terms of cytotoxic activity. The two vertical lines correspond to a change of 0.5 in cytotoxic activity, and compounds outside this range are considered to have ‘strong’ cytotoxic activity. d Heatmap of the activity of 55 compounds that were considered to have significant and strong cytotoxic activity. Doxorubicin, a chemotherapeutic used to treat neuroblastoma, and dimethyl sulfoxide (DMSO) were used as controls
Compound Validation
Amplification of MYCN and mutations in ALK are two of the most frequent genetic aberrations found in neuroblastoma, present in up to 25 and 14% of sporadic cases, respectively [18]. To achieve compound validation that would be representative of these aberrations, we chose a cohort of six independent neuroblastoma cell lines (IGR-N-91, SK-N-AS, SK-N-BE(2), SH-SY5Y, IMR-5, GI-ME-N) with varying levels of ALK and MYCN expression. Although only SH-SY5Y presents an activating ALK F1174L mutation, SK-N-BE(2) and IMR-5 show high expression of the wildtype ALK gene. Cell lines SK-N-AS, IGR-N-91, and GI-ME-N carry the wildtype ALK gene and present no or low expression of ALK protein. In terms of MYCN amplification, SK-N-BE(2), IGR-N-91, and IMR-5 are characterized by MYCN gene amplification, while SK-N-AS, SH-SY5Y, and GI-ME-N present only a single copy of MYCN. To avoid general cytotoxicity at the cellular level, we used one hTERT immortalized fibroblast cell line (BJ-5ta) as a normal cell line control for compound validation. Following treatment of these cell lines with the test compounds at concentrations of 0.01, 0.1, 1, and 10 µM, respectively, we observed that all nine compounds belonging to the blue cluster, i.e., ST003148, ST012339, ST013855, ST025226, ST033390, ST036501, ST053680, ST056969, and ST058459, presented a lower toxicity than doxorubicin in the BJ-5ta control cell line (Fig. 2a). However, overall these compounds presented a half-maximal inhibitory concentration (IC50) in the six neuroblastoma cell lines tested that was similar to IC50 observed in the control cell line BJ-5ta, indicating possible toxicity for normal tissues at effective treatment concentrations (Fig. 2a; ESM Fig. 2). Following the treatment of BJ-5ta cells with the seven compounds from the green cluster, we observed that the cytotoxicity of these compounds was similar to that of doxorubicin (Fig. 2b). Also, overall, these seven compounds showed similar or lower IC50 in the neuroblastoma cell lines when compared with doxorubicin, with ST013881 presenting the best response among all the compounds, followed in order of decreasing IC50 by ST022328, ST024334, ST012435, ST029265, ST024333, and ST033385 (Fig. 2b; ESM Fig. 2). Interestingly, we found that these seven compounds could be divided into four groups based on their structural similarity, as follows: (1) ST012435 and ST013881, which share a common phenanthroline core, differing only on the position of the methyl groups; (2) ST024333 and ST024334, which differ by the addition of two chlorine atoms symmetrically to the structure of ST024334; (3) ST029265 and ST033385, which share the same core chemical structure and differ by having either two chlorine or two bromine groups in the same position; and (4) ST022328 (Fig. 2c). To study the effects on the alterations on cellular phosphorylation, we selected ST013881 and ST022328 for further studies, since overall these two compounds presented the lowest IC50 of all the compounds analyzed.
Fig. 2.
Compound validation of blue and green clusters in six neuroblastoma cell lines (IGR-N-91, SK-N-AS, SK-N-BE(2), SH-SY5Y, IMR-5, GI-ME-N) and one control immortalized fibroblast cell line (BJ-5ta). a Validation of compounds from the blue cluster, b validation of compounds from the green cluster. Data is shown as the mean of three independent experiments ± standard deviation. The dashed line represents the half-maximal inhibitory concentration (IC50). c Chemical structure of compounds from the green cluster
Dissecting Cellular Phosphorylation with KinomeView
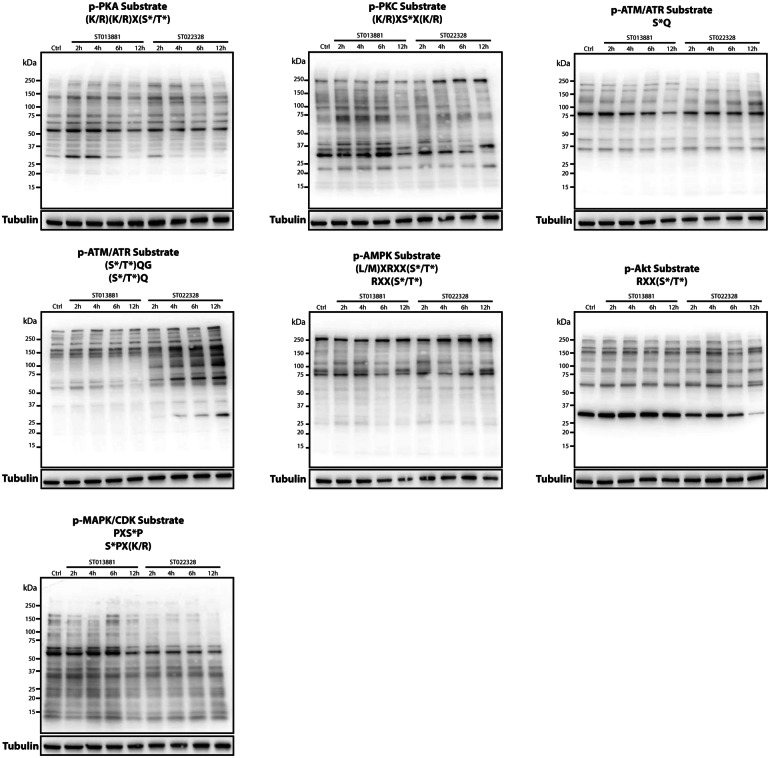
Small-molecule kinase inhibitors can act on one or several different kinases. A small molecule with multiple biological targets can sometimes be advantageous to achieve a certain therapeutic aim. Therefore, with the identification of these novel small-molecule kinase inhibitors, it is important to better understand the biology behind these compounds and how ST013881 and ST022328 affect signaling through specific kinase families. Different kinase families are able to recognize and phosphorylate specific amino acid motifs in their downstream effectors. The KinomeView kit can be used to identify not only these motifs but also a specific serine, threonine, and tyrosine phosphorylation pattern in a wide range of kinase families, allowing dissection of the alterations in the phosphoproteome following experimental treatment. The different motifs identified by the antibodies in the kit are specified in Fig. 3 and ESM Fig. 2.
Fig. 3.
Results of analysis using the KinomeView kit upon treatment of IGR-N-91 with the compounds ST013881 and ST022328 identifies alterations in the PKA, PKC, ATM/ATR, AMPK, Akt, and MAPK/CDK signaling pathways. Immunoblotting was performed using 30 µg of protein lysates from the IGR-N-91 cell line treated with the ST013881 and ST022328 compounds for 2, 4, 6, and 12 h. Membranes were blocked and primary antibodies were diluted 1:1000 in TBS-T. Upon horseradish peroxidase (HRP)-coupled secondary antibody incubation, membranes were developed using the Clarity Western ECL Substrate (Bio-Rad). PKA Protein kinase A, PKC protein kinase C, ATM/ATR ataxia-telangiectasia mutated/ATM- and Rad3-related, AMPK 5′ AMP-activated protein kinase, Akt protein kinase B, MAPK/CDK mitogen-activated protein kinase/cyclin-dependent kinase
In this context, we treated the IGR-N-91 cell line with the ST013881 and ST022328 compounds. To ensure a sufficient inhibition of the target kinase(s), we used the 10 µM concentration for both compounds. We evaluated the effects of the treatment using the KinomeView kit after 2, 4, 6, and 12 h of incubation time. Alterations in phosphorylation were observed as early as 2 h post-treatment. We noted that the substrates of PKA and PKC had changed following ST013881 treatment and that the substrates of ATM/ATR (S*Q and [S*/T*]QG motifs), AMPK, Akt, and MAPK/CDK had been modified following ST022328 treatment (Fig. 3). In the case of phosphorylation in ATM/ATR substrates (S*Q motif) by ST022328, alterations were found as early as after 1 h of treatment (data not shown). Interestingly, phosphorylation alterations in MAPK/CDK substrates by ST013881 and in PKC substrates by ST022328 were found after 6 and 12 h, respectively (Fig. 3). No other phosphorylation alterations were observed in the other motifs analyzed using the KinomeView kit (ESM Fig. 3). These observations indicate that alterations in the phosphorylation of MAPK/CDK substrates by ST013881 are likely a secondary outcome of the inhibition observed after 2 h in the PKA and PKC substrates. Similarly, alterations observed in the phosphorylation of PKC substrates 12 h after treatment with ST022328 are probably the result of the early inhibition of one or the inhibition of a combination of the phosphorylation of ATM/ATR, AMPK, Akt, and MAPK/CDK substrates.
Discussion
There has been little progression in developing new treatments for neuroblastma during the last few decades, and this is most clearly reflected by the slow improvement in the 5-year overall survival of high-risk patients (from 29% in 1990 to 50% in 2010) [4]. Patients with low- and intermediate-risk neuroblastoma have 5-year survival rates of 90–100% [4, 19], and often the first-line treatment of surgical intervention is sufficient in these patients, with adjuvant chemotherapy restricted to patients with unreachable tumors, tumors with unfavorable histology, and tumors associated with genetic aberrations [4, 20]. In high-risk patients, the options for tumor management include surgery, multimodal chemotherapy with conventional dosage (induction therapy) followed by high-dosage chemotherapy combined with autologous stem cell rescue (consolidation therapy), external beam radiation therapy, or immunotherapy [21]. However, these options do not deliver ideal outcomes to high-risk neuroblastoma patients: up to 10% of the patients who reach the criteria to initiate consolidation therapy show signs of disease progression, and even though consolidation therapy has demonstrated good event-free survival rates, the overall survival of these patients remains unchanged [22, 23]. Radiotherapy is used as an adjuvant therapy following stem cell transplant, but even with new technologies that restrict the exposure of the tumor to radiation therapy, treating young patients with such therapies can lead to growth problems or to the development of other malignancies later in life [24, 25]. Immunotherapy is another therapeutic approach that has resulted in successful outcomes in other cancers. In recent years, anti-PD-1/PD-L1 or anti-CTLA-4 antibodies have revolutionized cancer therapy for many tumors, from melanoma to lung cancer [26]—but this fruitful outcome has not yet reached neuroblastoma patients. Only one anti-disialoganglioside antibody (dinutuximab) has come on the market for neuroblastoma patients, and although this monoclonal antibody has improved the outcome in high-risk patients, it is only prescribed to patients who have successfully completed both induction and consolidation therapies [27].
Kinases have long been recognized to be important drug targets due to their essential role in the regulation of virtually all cellular functions. Since 2001, more than 30 small-molecule kinase inhibitors have been approved for the treatment of several diseases, with a special focus on cancer therapies [28]. However, although these therapies have revolutionized the treatment of several cancers, such as BRAF/MEK inhibitors in melanoma and ALK inhibitors in NSCLC, new targeted inhibitors for neuroblastoma remain unavailable to clinics. Efforts are ongoing to use crizotinib, an ALK-TKI-targeting small-molecule kinase inhibitor currently used in clinics in the treatment of NSCLC, for the treatment of ALK-mutated neuroblastoma patients (ClinicalTrials.gov Identifier: NCT03126916). However, as observed in melanoma patients treated with BRAF and MEK inhibitors, the treatment of ALK-TKI-mutated NSCLC with crizotinib eventually leads to the acquisition of resistance through the development of secondary mutations in, for example, ALK-TKI or alternative pathways. In addition, despite second-generation ALK-TKI-targeting inhibitors, such as alectinib, ceritinib and lorlatinib, having been developed to circumvent these secondary mutations in ALK-TKI, it is expected that they or similar mutations will appear upon the treatment of neuroblastoma with these same inhibitors [13, 29]. To overcome these problems and fill the current lack of therapeutic options, new alternatives need to be developed that can be used as single drugs or in combination with other therapies, namely immunotherapy.
Several studies have been performed in the search for possible target kinases to be used in the treatment of neuroblastoma, but very few of these actually evolved into clinical trials. The results of a phase I trial on the inhibition of aurora kinase A in combination with irinotecan and temozolomide in relapsed or refractory neuroblastoma patients suggest that this combination has significant anti-tumor activity [30]. Also, the inhibition of both aurora kinase A and B appears to provide a relatively stronger antitumor activity in a panel of neuroblastoma cell lines with acquired drug resistance, opening doors for an improved clinical trial [31]. Trials are currently being carried out on conpalisib, a phosphatidylinositol-3-kinase inhibitor (ClinicalTrials.gov Identifier: NCT03458728), and RAS-MAPK inhibitors (ClinicalTrials.gov Identifier: NCT02124772, NCT02780128, and NCT03698994) in refractory or relapsed neuroblastoma. Interestingly, about 80% of relapsed neuroblastomas show activating mutations of the RAS-MAPK pathway, suggesting the important role of this pathway as a therapeutic target in these tumors [11].
In an effort to identify new small-molecule kinase inhibitors targeting neuroblastoma, we performed a high-throughput screening using a commercially available library of 960 compounds and subsequently identified and validated 16 compounds with cytotoxic properties in a larger cohort of neuroblastoma cell lines. Further evaluation of the effects of two of these compounds, ST013881 and ST022328, revealed that they are able to modulate the phosphorylation of the Akt, AMPK, ATM/ATR, MAPK/CDK, PKA, and PKC pathways. ST013881 (2,9-dimethylpyridino[3,2-h]quinoline), also commonly known as neocuproine, has been previously reported to have cytotoxic properties in medulloblastoma, melanoma, and breast cancer cell lines [32–34]. Interestingly, neocuproine and similar compounds have been described as having copper (Cu)(II)-chelating properties, and the cytotoxic activity of neocuproine has been attributed to the capacity to form such complexes. In fact, the cytotoxicity of complexes of Cu(II)-neocuproine is higher than that of cisplatin in MCF-7 breast cancer cell line [34]. Moreover, Cu(II) has been shown to enhance the phosphorylation of Erk by Mek1, which can be reverted by treatment with neocuproine, affecting the signaling through the MAPK pathway [35]. The cytotoxic activity we observed following the treatment of neuroblastoma cell lines with neocuproine could be explained by Cu(II)-neocuproine complexes as well as by deregulation of the MAPK/CDK pathway. However, the mechanism of the cytotoxic activity remains to be fully elucidated. ST022328 (6-methyl-3-[(2E)-2-(pyridin-2-ylmethylidene)hydrazinyl]-4H-1,2,4-triazin-5-one) has been identified as an inhibitor of ATPase family AAA domain-containing protein 5 (ATAD5) (AID: 504467), bromodomain-adjacent to zinc finger domain protein (BAZ2B9) (AID: 504333), lysine-specific demethylase 4A (KDM4A) (AID: 504339), and 4D proteins (KDM4D) (AID: 2147). Interestingly, the results of one assay, although inconclusive, suggest that ST022328 might have a role as an activator of ATM protein (AID: 485349). Reports on yeast studies reveal that ATAD5 (also known as ELG1) is important for the regulation of DNA damage response pathways, namely the ATM/ATR pathway [36]. It is possible that the inhibition of ATAD5, as well as of other proteins that regulate DNA damage response, result in the cytotoxicity observed in neuroblastoma cell lines when treated with ST022328. Although this study does not identify specific targets for the compounds analyzed nor addresses in vivo validation and toxicity, it presents new treatment possibilities that should be further analyzed in future studies while shining new light on important pathways that regulate the development and survival of neuroblastomas.
Conclusions
High-risk neuroblastoma presents a worst outcome than low- and intermediate-risk neuroblastoma, mostly due to the inefficient treatment options. Here we present new compounds for which clinical implications should be further addressed. We also present an overview of kinome alterations induced by these compounds, opening doors for the study of these pathways as possible therapeutic targets.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank the team from NEXUS Personalized Health Technologies from ETH Zürich, in particular to Tijmen Booij, for the help and support on the development and execution of the high-throughput drug screening. We also thank our laboratory colleagues for the technical support.
Funding
Olga Shakhova is funded by Stiftung Kinderkrebsforschung Grant, FORCE Foundation, Oncosuisse Grant (KFS 3607-02-2015-R), Krebsliga Schweiz, UBS Promedica Foundation, and Novartis Foundation for Biomedical Research. No Rapid Service Fee was received by the journal for the publication of this article.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
André Serra-Roma and Olga Shakhova have nothing to disclose.
Compliance with Ethics Guidelines
Due to the nature of this research, no ethical approval was required. The human cell lines used were commercially available so did not require additional approval.
Data Availability
All data generated or analyzed during this study are included in this published article.
Footnotes
Enhanced Digital Features
To view enhanced digital features for this article go to 10.6084/m9.figshare.11982231.
References
- 1.Heck JE, Ritz B, Hung RJ, Hashibe M, Boffetta P. The epidemiology of neuroblastoma: a review. Paediatr Perinat Epidemiol. 2009;23(2):125–143. doi: 10.1111/j.1365-3016.2008.00983.x. [DOI] [PubMed] [Google Scholar]
- 2.Matthay KK, Maris JM, Schleiermacher G, et al. Neuroblastoma. Nat Rev Dis Prim. 2016;2:16078. 10.1038/nrdp.2016.78. [DOI] [PubMed]
- 3.Park JR, Eggert A, Caron H. Neuroblastoma: biology, prognosis, and treatment. Hematol Oncol Clin North Am. 2010;24(1):65–86. doi: 10.1016/j.hoc.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Pinto NR, Applebaum MA, Volchenboum SL, et al. Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol. 2015;33(27):3008–3017. doi: 10.1200/JCO.2014.59.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park JR, Bagatell R, London WB, et al. Children’s Oncology Group’s 2013 blueprint for research: neuroblastoma. Pediatr Blood Cancer. 2013;60(6):985–993. doi: 10.1002/pbc.24433. [DOI] [PubMed] [Google Scholar]
- 6.Viprey VF, Gregory WM, Corrias MV, et al. Neuroblastoma mRNAs predict outcome in children with stage 4 neuroblastoma: a European HR-NBL1/SIOPEN study. J Clin Oncol. 2014;32(10):1074–1083. doi: 10.1200/JCO.2013.53.3604. [DOI] [PubMed] [Google Scholar]
- 7.Mossé YP, Laudenslager M, Longo L, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455(7215):930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George RE, Sanda T, Hanna M, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455(7215):975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45(3):279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohl NE, Kanda N, Schreck RR, et al. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983;35(2 part 1):359–367. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- 11.Eleveld TF, Oldridge DA, Bernard V, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet. 2015;47(8):864–871. doi: 10.1038/ng.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung NKV, Zhang J, Lu C, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. J Am Med Assoc. 2012;307(10):1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trigg RM, Turner SD. ALK in neuroblastoma: biological and therapeutic implications. Cancers (Basel). 2018;10(4):113. 10.3390/cancers10040113 [DOI] [PMC free article] [PubMed]
- 14.Prummer M. Hypothesis testing in high-throughput screening for drug discovery. J Biomol Screen. 2012;17(4):519–529. doi: 10.1177/1087057111431278. [DOI] [PubMed] [Google Scholar]
- 15.Boutros M, Brás LP, Huber W. Analysis of cell-based RNAi screens. Genome Biol. 2006;7(7):R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tukey BH, John W. Exploratory data analysis. Addison-Wesley Publishing Company Reading, Mass.—Menlo Park, Cal., London, Amsterdam, Don Mills, Ontario, Sydney 1977, XVI, 688 S. Biom J. 1981;23(4):413–414. [Google Scholar]
- 17.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24(2):167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 18.Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med. 2013;3(10):a014415. doi: 10.1101/cshperspect.a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuchtern JG, London WB, Barnewolt CE, et al. A prospective study of expectant observation as primary therapy for neuroblastoma in young infants: a children’s oncology group study. Ann Surg. 2012;256(4):573–580. doi: 10.1097/SLA.0b013e31826cbbbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strother DR, London WB, Lou SM, et al. Outcome after surgery alone or with restricted use of chemotherapy for patients with low-risk neuroblastoma: results of Children’s Oncology Group study P9641. J Clin Oncol. 2012;30(15):1842–1848. doi: 10.1200/JCO.2011.37.9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macfarland S, Bagatell R. Advances in neuroblastoma therapy. Curr Opin Pediatr. 2019;31(1):14–20. doi: 10.1097/MOP.0000000000000711. [DOI] [PubMed] [Google Scholar]
- 22.Ladenstein R, Pötschger U, Pearson ADJ, et al. Busulfan and melphalan versus carboplatin, etoposide, and melphalan as high-dose chemotherapy for high-risk neuroblastoma (HR-NBL1/SI OPEN): an international, randomised, multi-arm, open-label, phase 3 trial. Lancet Oncol. 2017;18(4):500–514. doi: 10.1016/S1470-2045(17)30070-0. [DOI] [PubMed] [Google Scholar]
- 23.Park JR, Kreissman SG, London WB, et al. A phase III randomized clinical trial (RCT) of tandem myeloablative autologous stem cell transplant (ASCT) using peripheral blood stem cell (PBSC) as consolidation therapy for high-risk neuroblastoma (HR-NB): a Children’s Oncology Group (COG) study. J Clin Oncol. 2016;34(18):LBA3. [Google Scholar]
- 24.Applebaum MA, Henderson TO, Lee SM, Pinto N, Volchenboum SL, Cohn SL. Second malignancies in patients with neuroblastoma: the effects of risk-based therapy. Pediatr Blood Cancer. 2015;62(1):128–133. doi: 10.1002/pbc.25249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu JI, Lim DH, Jung SH, Sung KW, Yoo SY, Nam H. The effects of radiation therapy on height and spine MRI characteristics in children with neuroblastoma. Radiother Oncol. 2015;114(3):384–388. doi: 10.1016/j.radonc.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Kruger S, Ilmer M, Kobold S, et al. Advances in cancer immunotherapy 2019—latest trends. J Exp Clin Cancer Res. 2019;38(1):1–11. doi: 10.1186/s13046-019-1266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu AL, Gilman AL, Ozkaynak MF, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med. 2010;363(14):1324–1334. doi: 10.1056/NEJMoa0911123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu P, Nielsen TE, Clausen MH. Small-molecule kinase inhibitors: an analysis of FDA-approved drugs. Drug Discov Today. 2016;21(1):5–10. doi: 10.1016/j.drudis.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Haratake N, Seto T, Takamori S, et al. Short progression-free survival of ALK inhibitors sensitive to secondary mutations in ALK-positive NSCLC patients. Thorac Cancer. 2019;10(9):1779–1787. doi: 10.1111/1759-7714.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DuBois SG, Marachelian A, Fox E, et al. Phase I study of the aurora A kinase inhibitor alisertib in combination with irinotecan and temozolomide for patients with relapsed or refractory neuroblastoma: a nant (new approaches to neuroblastoma therapy) trial. J Clin Oncol. 2016;34(12):1368–1375. doi: 10.1200/JCO.2015.65.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michaelis M, Selt F, Rothweiler F, et al. Aurora kinases as targets in drug-resistant neuroblastoma cells. PLoS ONE. 2014;9(9):1–9. doi: 10.1371/journal.pone.0108758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calderon-Aparicio A, Cornejo A, Orue A, Rieber M. Anticancer response to disulfiram may be enhanced by co-treatment with MEK inhibitor or oxaliplatin: modulation by tetrathiomolybdate, KRAS/BRAF mutations and c-MYC/p53 status. Ecancermedicalscience. 2019;13:1–13. doi: 10.3332/ecancer.2019.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagerweij T, Hiddingh L, Biesmans D, et al. A chemical screen for medulloblastoma identifies quercetin as a putative radiosensitizer. Oncotarget. 2016;7(24):35776–35788. doi: 10.18632/oncotarget.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganeshpandian M, Ramakrishnan S, Palaniandavar M, Suresh E, Riyasdeen A, Akbarsha MA. Mixed ligand copper(II) complexes of 2,9-dimethyl-1,10-phenanthroline: tridentate 3N primary ligands determine DNA binding and cleavage and cytotoxicity. J Inorg Biochem. 2014;140:202–212. doi: 10.1016/j.jinorgbio.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 35.Turski ML, Brady DC, Kim HJ, et al. A novel role for copper in ras/mitogen-activated protein kinase signaling. Mol Cell Biol. 2012;32(7):1284–1295. doi: 10.1128/MCB.05722-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sau S, Kupiec M. A role for the yeast PCNA unloader Elg1 in eliciting the DNA damage checkpoint. Curr Genet. 2019;10(3):1–15. doi: 10.1007/s00294-019-01020-7. [DOI] [PubMed] [Google Scholar]
- 37.Huber W, Carey VJ, Gentleman R, et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods. 2015;12(2):115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.