Abstract
Cone-beam computed tomography (CBCT) scans are commonly used in diagnosing and planning surgical or orthodontic treatment to correct craniomaxillofacial (CMF) deformities. Based on CBCT images, it is clinically essential to generate an accurate 3D model of CMF structures (e.g., midface, and mandible) and digitize anatomical landmarks. This process often involves two tasks, i.e., bone segmentation and anatomical landmark digitization. Because landmarks usually lie on the boundaries of segmented bone regions, the tasks of bone segmentation and landmark digitization could be highly associated. Also, the spatial context information (e.g., displacements from voxels to landmarks) in CBCT images is intuitively important for accurately indicating the spatial association between voxels and landmarks. However, most of the existing studies simply treat bone segmentation and landmark digitization as two standalone tasks without considering their inherent relationship, and rarely take advantage of the spatial context information contained in CBCT images. To address these issues, we propose a Joint bone Segmentation and landmark Digitization (JSD) framework via context-guided fully convolutional networks (FCNs). Specifically, we first utilize displacement maps to model the spatial context information in CBCT images, where each element in the displacement map denotes the displacement from a voxel to a particular landmark. An FCN is learned to construct the mapping from the input image to its corresponding displacement maps. Using the learned displacement maps as guidance, we further develop a multi-task FCN model to perform bone segmentation and landmark digitization jointly. We validate the proposed JSD method on 107 subjects, and the experimental results demonstrate that our method is superior to the state-of-the-art approaches in both tasks of bone segmentation and landmark digitization.
Keywords: Cone-beam computed tomography, landmark digitization, bone segmentation, fully convolutional networks
1. Introduction
Craniomaxillofacial (CMF) deformities include acquired and congenital deformities of the head and the face. It is reported that approximately 16.8 million Americans require surgical or orthodontic treatment to correct CMF deformities based on computed tomography (CT) scans (De Vos et al., 2009). Due to nature of complex CMF anatomy, these treatments require an accurate and detailed pretreatment plan. Cone-beam CT (CBCT) scan is a commonly used imaging modality for this purpose because they have been widely used in doctors’ offices. Compared with the spiral multi-slide CT (MSCT) scan, CBCT scan also has the advantages of lower radiation exposure and cost (Loubele et al., 2009). To develop accurate treatment plans for patients, one essential step is to segment the CMF structures (e.g., midface and mandible) and digitize anatomical landmarks on CBCT images. However, due to severe image artifacts (e.g., imaging noise, inhomogeneity, and truncation), it is extremely challenging to accurately segment bony structures and digitize anatomical landmarks for CBCT images (Schulze et al., 2011; Loubele et al., 2006).
In current clinical practices, a gold standard is to manually perform bone segmentation and landmark digitization for CBCT images, which is very time-consuming and labor-intensive. In recent years, there have been reports on computer-aided methods for bone segmentation and landmark digitization with reasonable results in generating patient-specific jaw reference models for CMF surgery (Shahidi et al., 2014; Cheng et al., 2011; Wang et al., 2014; Zhang et al., 2016b). These methods can generally be divided into two categories: 1) multi-atlas based methods (Coupé et al., 2011; Rousseau et al., 2011; Wang et al., 2013; Shahidi et al., 2014), and 2) learning based methods (Cheng et al., 2011; Chen et al., 2014; Wang et al., 2014; Chen et al., 2015; Zhang et al., 2016b). In the first category, bone segmentation and landmark digitization are achieved by transferring the labeled regions and anatomical landmarks from multi-atlas images to the target image via image registration. However, it is often difficult to accurately perform nonlinear registration, thus eventually affecting the accuracy of bone segmentation and landmark digitization. In the second category, human-engineered features are first extracted from CBCT images, and then fed to a classifier or regressor for bone segmentation or landmark digitization. Since feature extraction and model training are performed separately in these learning based methods, the used features and the learned model may not necessarily be coordinated with each other, leading to sub-optimal performance.
Both tasks of bone segmentation and landmark digitization can be considered highly associated, because the anatomical landmarks generally lie on the boundaries of segmented bone regions. Based on this assumption, a number of learning based approaches have been proposed by using the detected anatomical landmarks to aid the organ segmentation (Seghers et al., 2007; Wang et al., 2014), or employing the results of bone segmentation as guidance for landmark digitization (Zhang et al., 2016b). However, these methods still treat bone segmentation and landmark digitization as two independent tasks without considering their underlying association. Recently, multi-task learning has demonstrated promising performance in different areas (Zhang et al., 2014; Yim et al., 2015; Liu et al., 2015b; Li et al., 2016; Dai et al., 2016). Ranjan et al. (Ranjan et al., 2017) proposed a multi-task learning method using convolutional neural networks (CNN) for face detection, landmark localization, pose estimation, and gender recognition. This work demonstrated that exploiting the synergy among multiple tasks could boost the individual performance of each task. Motivated by the recent success of deep learning, we propose a joint bone segmentation and landmark digitization (JSD) framework via a context-guided fully convolutional network (FCN). To our knowledge, this is the first report on the integration of bone segmentation and landmark digitization into a unified deep learning framework. The preliminary work of this approach was reported on MICCAI 2017 (Zhang et al., 2017c). In this journal version, we offered new contributions in the following aspects: 1) investigating the learned segmentation maps of midface and mandible, as well as the heatmaps for landmarks, 2) illustrating the segmentation results and the landmark digitization results visually, 3) comparing our method with two additional state-of-the-art methods, 4) studying the computational costs, and 5) analyzing the influence of the size of sliding window.
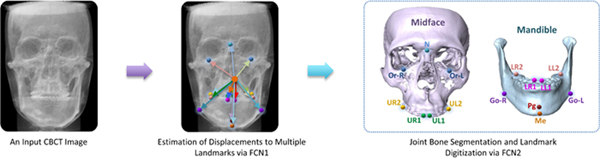
Figure 1 illustrates the schematic diagram of our proposed JSD framework. For a CBCT image, we first estimate the displacements from the voxels to the landmarks via the first-stage FCN (i.e., FCN1), to capture the spatial context information contained in the input image. Then, we simultaneously perform both bone segmentation and landmark digitization via the second-stage FCN (i.e., FCN2). In FCN2, the input contains both the displacement maps (estimated by FCN1) and the original image, while the output includes the segmentation maps and the heatmaps of landmarks. In this study, each element in the displacement map records the displacement from the certain voxel location to a respective landmark in a specific axis space, and hence the size of each displacement map is the same size as the input image.
Figure 1:
The schematic diagram of the proposed Joint craniomaxillofacial bone Segmentation and landmark Digitization (JSD) framework. There are two major components, including (1) estimation of displacements from voxels to landmarks via the first-stage fully convolutional network (FCN1), and (2) joint bone segmentation and landmark digitization via the second-stage FCN (FCN2). The locations of 15 anatomical landmarks are also illustrated in this figure. FCN: fully convolutional neural network.
The technical contributions of this work are summarized as follows. First, a displacement map is used to explicitly model the spatial context information in CBCT images. Second, using the estimated displacement maps as the guidance information, we introduce a joint deep learning framework for both bone segmentation and landmark digitization, through which the inherent association between these two tasks can be seamlessly incorporated into the learning process.
The rest of the paper is organized as follows. We first introduce relevant studies in Section 2. In Section 3, we describe the materials used in this study and present the proposed method in detail. In Section 4, we introduce the competing methods, experimental settings, experimental results, and influence of parameters. We further compare our method with previous studies and discuss the limitations and possible future work in Section 5. We finally conclude this paper in Section 6.
2. Related Work
2.1. Bone Segmentation and Landmark Digitization
Since manual bone segmentation and landmark digitization for CBCT images is tedious and time-consuming, many computer-aided automatic approaches have been proposed in the previous studies (Shahidi et al., 2014; Cheng et al., 2011; Wang et al., 2014; Zhang et al., 2016b; Liu et al., 2017), which is clinically significant. For instance, in multi-atlas (MA) based methods (Shahidi et al., 2014; Coupé et al., 2011; Rousseau et al., 2011; Wang et al., 2013; Liu et al., 2016), the segmented bony regions (or landmark locations) are transferred from multi-atlas images to the target image via nonlinear image registration. This method is usually computationally expensive (i.e., taking hours) due to the required nonlinear registration between multi-atlas images and the target image. In addition, because of morphological variations among different subjects, it is also challenging to accurately perform bone segmentation and landmark digitization by simply using nonlinear registration results.
In contrast, learning based methods generally construct classifiers and regressors for bone segmentation and landmark detection, respectively, based on CBCT images. The commonly used classifiers/regressors for bone (or organ) segmentation and landmark digitization include random forest classifier (Cheng et al., 2011; Zhang et al., 2017a; Cuingnet et al., 2012; Lindner et al., 2013; Mitra et al., 2014; Zhu et al., 2017), regression forest (Criminisi et al., 2010; Zhang et al., 2016b,a), sparse representation (Wang et al., 2014), and AdaBoost (Zhan et al., 2011). Although learning based approaches usually acquire better results than multi-atlas based methods, their performances are highly dependent on the feature representations for CBCT images. Since steps of human-engineered feature extraction and classifier/regressor training are independent to each other in these learning based methods, the final performances of bone segmentation and landmark digitization could be sub-optimal.
2.2. Deep Learning Methods
There also have been reports on deep learning based methods in which both the feature learning and the classifier/regressor training are incorporated into a unified framework (Ronneberger et al., 2015; Payer et al., 2016; Lian et al., 2018b,a). For instance, Ronneberger et al. (Ronneberger et al., 2015) developed a U-Net framework to perform image segmentation, achieving remarkable performance in biomedical image segmentation. Payer et al. (Payer et al., 2016) proposed a fully convolutional network (FCN) for landmark heatmap regression and yielded good result in landmark localization using even the limited training data. Zhang et al. (Zhang et al., 2017b) developed a two-stage task-oriented deep learning method to jointly detect large-scale (e.g., 1000) landmarks in real time. Baumgartner et al. (Baumgartner et al., 2017) proposed a SonoNet for real-time localization of fetal standard scan planes in freehand ultrasound. Farag et al. (Farag et al., 2017) proposed a bottom-up strategy for pancreas segmentation by classifying image patches at different resolutions and cascading superpixels. Alansary et al. (Alansary et al., 2019) evaluated deep reinforcement learning for landmark localization, where several deep Q-network architectures were employed for detecting landmarks in fetal head ultrasound and adult brain and cardiac magnetic resonance imaging (MRI). The limitation of these methods is that they simply focus on one single task, i.e., image segmentation or landmark localization, without considering the inherent association between the two tasks. In particular, the two tasks of bone segmentation and landmark digitization for CBCT images are highly associated, since the majority of the anatomical landmarks lie on the boundaries of segmented bones.
Currently, several studies have focused on taking advantage of the inherent association of the tasks of bone segmentation and landmark digitization, and have achieved reasonable results (Wang et al., 2014; Zhang et al., 2016b). Wang et al. (Wang et al., 2014) proposed a landmark-guided sparse representation (LSR) method for bone segmentation, using the results of landmark digitization as guidance for segmenting CBCT images. Zhang et al. (Zhang et al., 2016b) developed an automated landmark digitization framework, called segmentation-guided partially joint regression forest (SPRF), with the aid of results of bone segmentation for CBCT images. Recently, Torosdagli et al. (Torosdagli et al., 2018) proposed a dental CBCT analysis framework using deep geodesic learning, achieving state-of-the-art performance in both mandible segmentation and landmark digitization. Specifically, the mandible segmentation was segmented with a segmentation network, and then a geodesic learning network was proposed with the distance transform based on the segmentation. Finally, typical landmarks were localized using a classification model, and all the others were further estimated using a recurrent neural network(RNN). Unfortunately, these methods still treat the tasks of bone segmentation and landmark digitization separately. Multi-task learning has achieved impressive performance to assist each correlated task. For example, Xu et al. (Xu et al., 2018) proposed a multi-task model for landmark detection and view classification in abdominal ultrasound images. Liu et al. (Liu et al., 2018) proposed a joint classification and regression CNN model, and achieved promising results in computer-aided brain disease diagnosis. Cao et al. (Cao et al., 2018) performed joint hippocampus segmentation and clinical score regression to boost the performance of both tasks. Motivated by all these studies, we propose a joint bone segmentation and landmark digitization framework via fully convolutional networks. Experimental results on 107 subjects demonstrate the effectiveness of the proposed method.
3. Materials and Methods
In this section, we first introduce the materials used in this study, and then present the proposed method in detail.
3.1. Data Description
This study was approved by Institute Review Board prior to the data collection, and the clinical target is to help clinicians plan surgical or orthodontic treatment to correct craniomaxillofacial (CMF) deformities. There are a total of 77 CBCT images (with the spatial resolution of 0.40 × 0.40 × 0.40 mm3 or 0.30 × 0.30 × 0.30 mm3) from patients with non-syndromic dentofacial deformities. According to different types of deformities, those patients with dentofacial deformities were categorized into three classes. 1) Skeletal Class I, where the mandible is retrognathic caused by mandibular retrusion, maxillary protrusion or the combination. 2) Skeletal Class II, where the mandible is prognathic caused by mandibular protrusion, or maxillary retrusion, or the combination. 3) Skeletal Class III, where the profile is orthognathic by either double-jaw protrusion, retrusion or vertical deformity. Among these 77 patients, 20 patients were Skeletal Class I, 21 were Skeletal Class II, and 36 were Skeletal Class III. Considering that the number of CBCT images is limited, to augment the training samples, we further employ an additional dataset with 30 MSCT images (0.488 × 0.488 × 1.25 mm3) from normal control subjects which were collected in an unrelated study. In this work, we use these MSCT images as additional training data for network optimization in the experiments.
To obtain the ground-truth results of bone segmentation, two experienced CMF surgeons manually segmented all CBCT and MSCT images into midface and mandible, using the Mimics software (Materialise, Leuven, Belgium). In addition, as shown in Fig. 1(right), the most clinically relevant 15 anatomical landmarks (Zhang et al., 2016b; Wang et al., 2014) were also manually digitized by the same CMF surgeons, including N, Or-R, Or-L, UR2, UL2, UR1, UL1, LR2, LL2, LR1, LL1, Go-R, Go-L, Pg, and Me.
3.2. Displacement Estimation via FCN1
Similar to (Pfister et al., 2015), we adopt the displacement maps to model the context information of an input image. Different from (Pfister et al., 2015), given a 3D image Xn with V voxels, we represent a displacement map by a 3D volume of the same size as Xn, where each element denotes the displacement from a voxel to a certain landmark in a specific axis space. Since the Euclidean distance can only provide the distance (magnitude) information that cannot be used to estimate the actual positions of landmarks, we use 3L displacement maps to capture both orientation and distance information. That is, for the l-th landmark in Xn, there are 3 displacement maps (i.e., ) corresponding to x, y, and z axes, respectively. Given L landmarks, we have 3L displacement maps for each image.
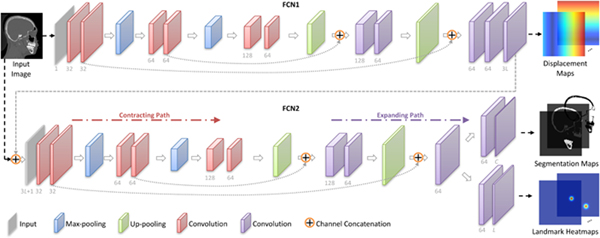
To construct the mapping function between an input image and its 3L displacement maps, we develop a first-stage fully convolutional network (i.e., FCN1), with its architecture shown in Fig. 2 (left). Using a set of training images and their corresponding target displacement maps, FCN1 (with a U-Net architecture (Ronneberger et al., 2015)) is used to capture both the global and the local structural information of input images. Specifically, there are a contracting path and an expanding path in FCN1. 1) The contracting path follows the typical architecture of CNN. Every step in the contracting path consists of two 3 × 3 × 3 convolutions, followed by a rectified linear unit (ReLU) and a 2 × 2 × 2 max pooling operation with the stride 2 for down-sampling. 2) Each step in the expanding path consists of a 3 × 3 × 3 up-convolution, followed by a concatenation with the corresponding feature map from the contracting path, and two 3 × 3 × 3 convolutions (each followed by a ReLU function). Due to the use of the contracting path and the expanding path, FCN1 can grasp a large image area using small kernel sizes while still keeping high localization accuracy. In the experiments, we normalize the output of the last layer in FCN1 into [−1,1].
Figure 2:
Overview of the proposed context-guided multi-task fully convolutional network (FCN), including two sub-networks (i.e., FCN1, and FCN2). FCN1 estimates the displacement maps, while FCN2 performs joint bone segmentation and landmark digitization. Each sub-network contains a contracting patch and an expanding path.
Let Xn,v represent the v-th voxel of the image Xn. In the a-th (a ∈ {x,y,z}) axis space, we denote the l-th displacement map of Xn as and its v-th element as The target of FCN1 is to learn a nonlinear mapping function to transform the original input image onto its corresponding 3L displacement maps, by minimizing the following loss function:
| (1) |
where f(Xn,v;W1) is the estimated displacement by using the network coefficients W1, and N is the number of training images in a batch.
3.3. Joint Bone Segmentation and Landmark Digitization via FCN2
Based on the estimated displacement maps and the original CT image, we further propose the second-stage FCN (i.e., FCN2) with a U-Net architecture to jointly perform bone segmentation and landmark digitization. As shown in Fig. 2 (right), FCN2 adopts a stacked representation of displacement maps and the original image as the input, through which the spatial context information of images provided by displacement maps is explicitly incorporated into the learning process. Also, such representation could guide the network to focus on informative regions in the image, and may thus help alleviate the negative influence of image artifacts. Besides, 1) for bone segmentation, the output is transformed to the probability scores by using the softmax function; 2) for landmark digitization, the output is normalized to [0,1].
Denote as the ground-truth segmentation map of the n-th subject belonging to the c-th category, with its v-th element as Here, a CT image is segmented into C = 3 categories (i.e., midface, mandible, and background). We denote as the ground-truth landmark heatmap of the l-th landmark in Xn, with its v-th element as The objective function of FCN2 is as follows:
| (2) |
where the first term is the cross-entropy error for bone segmentation and the second term is the mean squared error for landmark digitization. Here, 1 {·} is an indicator function, with 1 {·} = 1 if {·} is true; and 0, otherwise. indicates the probability of the v-th voxel in the image Xn being correctly classified as the category using the network coefficients W2. The second term in Eq. (2) is used to compute the loss between the estimated landmark location g(Xn,v;W2) and the ground-truth location in the l-th landmark heatmap.
3.4. Implementation Details
As shown in Fig. 2, the proposed two cascaded sub-networks (i.e., FCN1, and FCN2) are included in a unified framework. The input is a CT image, while the output includes segmentation probability maps (for midface, mandible, and background) and landmark heatmaps. Also, the displacement maps are intermediate outputs of the whole network, providing context information of input image to guide the joint learning of bone segmentation and landmark digitization. For each landmark, we generate a heatmap by using a Gaussian filtering with the standard derivation of 2 mm, and then stretch the values to the range of [0,1]. For optimizing the network coefficients, we adopt the stochastic gradient descent (SGD) algorithm (Boyd and Vandenberghe, 2004) combined with the back-propagation algorithm. The computer we used in the experiments contains a single GPU (i.e., NVIDIA GTX TITAN 12GB), and the implementation of FCN is based on Tensorflow (Abadi et al., 2016).
In the training stage, we first train FCN1 using CT images and their corresponding target displacement maps as the input and output, respectively. With FCN1 fixed, we then train FCN2 for joint bone segmentation and landmark digitization, by using the stacked representation of the estimated displacement maps from FNC1 and the original image as the joint input, while segmentation maps and landmark heatmaps as the output. Finally, using the learned coefficients of FCN1 and FCN2 as initialization, we further train both FCN1 and FCN2 jointly. Besides, the training process is done in a sliding window fashion (with the fixed window size of 96 × 96 × 96). To speed up the training process, we down-sample the original CT image using a 2 × 2 × 2 filter, since the displacement map only provides the rough context information. In this way, via FCN1, we can obtain the estimated displacement maps for the down-sampled input image. We then up-sample the displacement maps to their original sizes for training FCN2.
In the testing stage, ideally, we can feed a new testing image of any size into the trained model, since FCN only contains the convolutional computation. But, in practice, due to the limited memory, we partition each testing image into multiple 128 × 128 × 128 sub-images with a certain overlap to perform a valid prediction. After predicting the segmentation maps and landmark heatmaps via FCN2, the center 64 × 64 × 64 patches (it can be up to 88 × 88 × 88) are used to reconstruct the whole image. Note that even the testing performance would not be affected by the size of sub-images, the computational time will be increased if smaller (e.g., < 128 × 128 × 128) sub-images were used for prediction. For instance, given a testing image (400 × 400 × 400) with a spatial resolution of 0.40 × 0.40 × 0.40 mm3, if we select the size of sub-images as 64 × 64 × 64 and the valid size as 24 × 24 × 24, our proposed FCN2 model costs ~1.5min for prediction in the testing stage. In contrast, if we select the size of sub-images as 168 × 168 × 168 and the valid size as 128 × 128 × 128, our proposed FCN2 model costs ~ 20 s for prediction in the testing stage. Considering the image size and computational power in this work, we set the size of sub-images as 128 × 128 × 128 and the valid size of 64 × 64 × 64 in the experiments.
4. Experiments
4.1. Methods for Comparison
We first compare our JSD method with two baseline methods that can be directly used for both bone segmentation and landmark digitization, which include 1) multi-atlas (MA) based method (Shahidi et al., 2014; Coupé et al., 2011), and 2) random forest (RF) based method (Cheng et al., 2011). We further compare our method with two state-of-the-art methods for bone segmentation and landmark digitization, respectively, such as 1) landmark-guided sparse representation (LSR) (Wang et al., 2014) for bone segmentation, and 2) segmentation-guided partially-joint regression forest (SPRF) (Zhang et al., 2016b) for landmark digitization. Besides, we also compare our JSD method with its own three variants. We now briefly describe these methods as follows.
Multi-Atlas (MA) based method (Shahidi et al., 2014). In the experiments, we implement multi-atlas based models for bone segmentation and landmark digitization using nonlinear alignment. For landmark digitization, we map anatomical landmarks from corresponding positions in the nonlinearly aligned atlases, by using the majority voting strategy. Similar to the landmark digitization, we also transfer the labeled regions (bones) from multi-atlas images to the target image using the majority voting strategy (Schapire et al., 1998; Artaechevarria et al., 2009; Liu et al., 2015a).
Random Forest (RF) based method (Cheng et al., 2011). In this method, we first extract Harr-like features from CBCT images. Here, we use the random forest classifier for bone segmentation (Schroff et al., 2008) and the random forest regressor for landmark digitization (Criminisi et al., 2013). Note that the RF based method treats bone segmentation and landmark digitization as independent tasks.
Landmark-guided Sparse Representation (LSR) (Wang et al., 2014) for bone segmentation. There are three main elements in LSR, including region-specific registration with the guidance of landmarks, estimating a patient-specific atlas, and convex segmentation based on maximum a posteriori (MAP). Specifically, a region-specific landmark-guided registration strategy is first proposed to warp all atlases to a testing subject. Here, the same 15 anatomical landmarks as we used in this study are used to initialize the multiple atlases. Then, a sparse representation based label propagation strategy is employed to estimate a patient-specific atlas from all aligned atlases. Finally, the patient-specific atlas is integrated into a MAP probability based convex segmentation framework for accurate bone segmentation. In brief, the LSR method adopts the landmark digitization results to aid the task of bone segmentation for CBCT images.
Segmentation-guided Partially-joint Regression Forest (SPRF) (Zhang et al., 2016b) for landmark digitization. In SPRF, a regression voting strategy is first adopted to localize each landmark by aggregating evidence from context locations. The bone segmentation results (via multi-atlas based method) for CBCT image is then utilized to remove uninformative voxels caused by morphological variations across subjects. Third, a partially joint model is used to separately localize landmarks. In addition, this method adopts a fast vector quantization method to extract high-level multiscale statistical features to describe the appearance of voxels. That is, the results of bone segmentation are used as the guidance to aid the process of landmark digitization in SPRF (Zhang et al., 2016b).
In addition, there are two new strategies utilized in our proposed JSD method, i.e., using displacement maps as guidance, and joint learning of two tasks. To evaluate their specific contributions, we further compare JSD with its three variants, called JSD1, JSD2, and JSD3, respectively. Specifically, 1) JSD1 only adopts FCN2 in Fig. 2 to separately perform bone segmentation and landmark digitization, without using the joint learning strategy and displacement maps as guidance. That is, JSD1 is actually a simple U-Net (Ronneberger et al., 2015) framework. 2) JSD2 only adopts FCN2 for the jointly learning of two tasks, without using displacement maps as guidance. 3) JSD3 performs bone segmentation and landmark digitization independently via FCN2, but using the displacement maps estimated by FCN1 as guidance for both tasks.
It is worth noting that, among all comparison methods, four approaches (i.e., MA, RF, JSD1, and JSD3) can perform the tasks of bone segmentation and landmark digitization separately, two methods (i.e., JSD2, and JSD) can jointly perform these two tasks, while LSR and SPRF can only perform bone segmentation and landmark digitization, respectively.
4.2. Experimental Settings
Before training the model, all images are spatially normalized to have the same resolution (i.e., 0.40 × 0.40 × 0.40 mm3), and are also intensity-normalized to have similar intensity distributions via a histogram matching technique (Rother et al., 2006). For 77 CBCT images from patients with CMF deformities, we adopt a 5-fold cross-validation strategy (Zhang, 1993). The 30 MSCT images from normal controls are used as additional training samples for model learning in each of 5 folds. We report the mean and the standard deviation of results yielded by different methods.
To evaluate the results of bone segmentation (separating bony structures between the mandible and the midface), we use three metrics, including 1) Dice similarity coefficient (DSC), 2) sensitivity (SEN), and 3) positive predictive value (PPV). Specifically, DSC measures the overlap ratio between automatic and manual segmentation results, defined as Here, Vs and Vm denote the bone-labeled voxel sets automatically segmented by an automated method and manually segmented by a clinical expert, respectively, while denotes the cardinality of a set. The term SEN measures the percentage of manual segmentation that overlaps with automatic segmentation, defined as And PPV is defined as to measure the rate of automatic segmentation that overlaps with manual segmentation. To quantitatively evaluate the results of landmark digitization, we adopt the detection error (via Euclidean distance) as the evaluation criterion, to measure the displacement from estimated landmark locations to predetermined locations.
4.3. Experimental Results
4.3.1. Segmentation Maps and Landmark Heatmaps
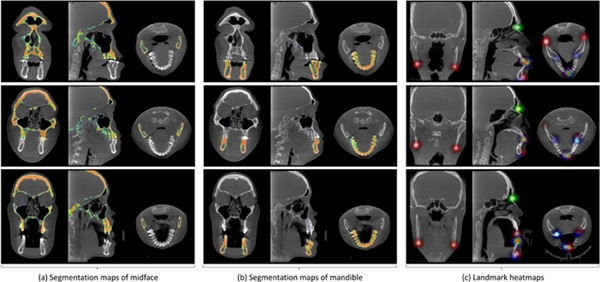
We first visually illustrate the segmentation maps and landmark heatmaps achieved by our JSD method on two patients with CMF deformities in Fig. 3. Each row in Fig. 3 reports the results for a specific subject. For the convenience of visualization, we show the 2D probability maps for the segmented midface and mandible in three views in Fig. 3 (a) and Fig. 3 (b), respectively. In addition, we overlap the heatmaps of 15 anatomical landmarks onto a single 3D image, and illustrate the results in three views in Fig. 3 (c). Their corresponding 3D renderings are also provided in the online Supplementary Materials. From Fig. 3 (a)–(b), we can see that for the task of bone segmentation, our method can accurately separate midface and mandible. Also, as can be seen from Fig. 3 (c), our JSD method can estimate clear and smooth heatmaps for landmarks on three typical subjects.
Figure 3:
Results achieved by our JSD method on three typical CMF patients of (a) segmentation maps of midface, (b) segmentation maps of mandible, and (c) landmarks heatmaps. Each row denotes a particular subject, with three views shown.
4.3.2. Results of Bone Segmentation and Landmark Digitization
In Table 1, we report the experimental results achieved by the proposed JSD method and 7 comparison methods for the tasks of bone segmentation and landmark digitization. From Table 1, we can make the following observations. First, compared with two baseline methods (i.e., MA, and RF), our JSD method consistently achieves the best performance in both bone segmentation and landmark digitization. For instance, compared with MA and RF, JSD achieves 12.05% and 6.33% improvements (in terms of DSC), respectively, in the segmentation of midface. Second, the proposed JSD method consistently outperforms two state-of-art methods (i.e., LSR, and SPRF) for bone segmentation and landmark digitization. For instance, the average error of landmark digitization by our JSD method is 1.10 mm which is lower than the error of SPRF (1.52 mm). Moreover, compared with the other 6 methods that treat two tasks independently, the methods that jointly learn two tasks (i.e., JSD2, and JSD) usually yield more accurate segmentation results and much lower digitization errors. This implies that the proposed jointly learning strategy improves the learning performances of two tasks, by modeling the inherent association between the two tasks of bone segmentation and landmark digitization. Finally, for the joint learning methods, JSD consistently outperforms JSD2 in both tasks of bone segmentation and landmark digitization; note that JSD2 does not employ displacement maps as guidance information. This suggests that the guidance provided by displacement maps can further promote the performance of our joint learning model.
Table 1:
Results achieved by 8 different methods in bone segmentation (i.e., midface, and mandible) and landmark digitization (i.e., average error for 15 landmarks).
| Bone Segmentation | Landmark | ||||||
|---|---|---|---|---|---|---|---|
| Methods | Midface | Mandible | Digitization | ||||
| DSC (%) | SEN (%) | PPV (%) | DSC (%) | SEN (%) | PPV (%) | Average Error (mm) | |
| MA | 81.14 ± 2.54 | 80.17 ± 3.27 | 82.48 ± 2.85 | 83.82 ± 2.21 | 84.31 ± 2.21 | 83.29 ± 2.30 | 3.05 ± 1.54 |
| RF | 86.86 ± 1.63 | 87.36 ± 2.98 | 85.92 ± 2.28 | 88.21 ± 1.52 | 88.54 ± 2.77 | 88.01 ± 1.95 | 2.67 ± 1.58 |
| LSR | 92.27 ± 1.31 | 91.96 ± 2.86 | 92.64 ± 1.83 | 89.19 ± 1.75 | 89.55 ± 2.34 | 89.03 ± 1.62 | - |
| SPRF | - | - | - | - | - | - | 1.52 ± 1.25 |
| JSD1 | 91.83 ± 1.06 | 90.05 ± 2.35 | 93.72 ± 1.24 | 91.66 ± 1.07 | 91.35 ± 2.13 | 91.99 ± 1.01 | 1.78 ± 1.31 |
| JSD2 | 92.20 ± 1.02 | 92.73 ± 2.50 | 91.78 ± 2.14 | 92.17 ± 0.99 | 93.30 ± 2.29 | 91.13 ± 1.41 | 1.33 ± 0.92 |
| JSD3 | 91.89 ± 1.15 | 91.50 ± 2.63 | 92.02 ± 1.99 | 92.03 ± 1.08 | 93.14 ± 2.17 | 91.15 ± 1.52 | 1.49 ± 1.28 |
| JSD | 93.19 ± 0.89 | 92.82 ± 1.91 | 93.61 ± 1.40 | 93.27 ± 0.97 | 93.63 ± 1.37 | 92.93 ± 1.09 | 1.10 ± 0.71 |
4.3.3. Digitization Error for Each Landmark
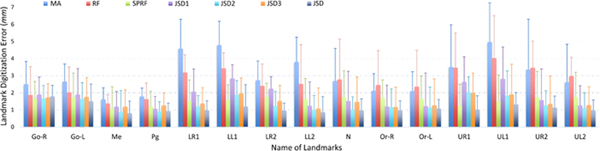
In Fig. 4, we further show digitization errors for each of 15 anatomical landmarks achieved by 7 different methods. From Fig. 4, we can see that, compared with MA, RF, and SPRF, our proposed methods (JSD, JSD1, JSD2, and JSD3) generally achieve the lower errors in detecting these 15 landmarks, especially for the landmarks located at the lower teeth and upper teeth (e.g., LR1, LL1, LR2, LL2, UR1, UL1, UR2, and UL2, see Fig. 1). It is worth noting that, because of large inter-subject variations in the local appearance of the tooth, it is very challenging to accurately localize tooth landmarks. These results demonstrate that our proposed two strategies (i.e., using displacement maps as guidance information, and joint learning) can help accurately locate anatomical landmarks in CBCT images. Also, it is clinically acceptable if the digitization error of CMF landmarks for CBCT images is below 1.50 mm. Table 1 and Fig. 4 clearly demonstrate that the average digitization error achieved by our JSD method is below 1.50 mm, indicating that JSD has a great value in real clinical applications.
Figure 4:
The digitization errors (mm) for each of 15 landmarks, achieved by 7 different methods.
4.3.4. Visualization Results
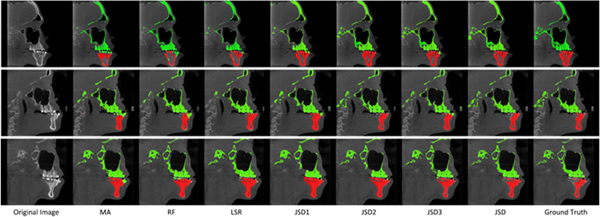
To visually compare the segmentation results of different methods, we also show the bone segmentation results for both midface and mandible on three subjects in Fig. 5. In Fig. 5, the first and the last columns denote the original CT images and the ground-truth segmentation results, respectively. From this figure, we can see that MA, RF, and LSR method can not clearly segment the mandible from the midface, while the results yielded by our proposed JSD method and its three variants (i.e., JSD1, JSD2, and JSD3) are very close to the ground truth. For instance, for the first subject (corresponding to the first row of Fig. 5), JSD can accurately separate the mandible, especially for the lower teeth position, while MA and RF cannot complete the task well. It implies that context information of images captured by the displacement maps in our JSD method helps improve the learning performance.
Figure 5:
Results of bone segmentation achieved by 7 different methods, where each row denotes a specific subject.
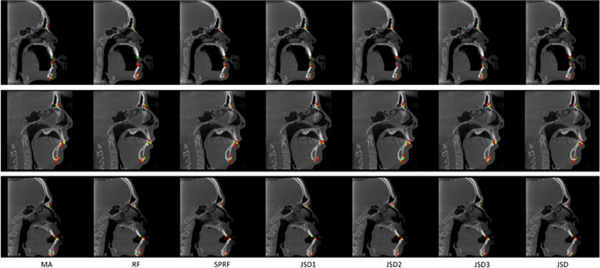
In addition, to visually illustrate the landmark digitization results achieved by different methods, Fig. 6 shows a comparison of three randomly selected subjects, with each row denoting a particular subject. In Fig. 6, the red points indicate the detected landmarks by 7 different methods, and the green points represent the ground-truth landmarks. Figure 6 shows that the landmark locations estimated by our JSD method are usually very close to the ground truth, while MA, RF, and SPRF can not achieve excellent results for these subjects.
Figure 6:
Results of landmark digitization achieved by 7 different methods, where each row denotes a specific subject. The red points denote the detected landmarks by different methods, while the green points represent the ground-truth landmarks.
4.4. Comparison of Computational Cost
Different methods were implemented using different programming languages (e.g., MATLAB, Python, and C++) and computing environments (e.g., CPU and GPU). Here, we only roughly compare the computational costs of our method and those competing methods in bone segmentation and landmark digitization in the testing stage (e.g., for a 400×400×400 image with a spatial resolution of 0.4 mm3), with the results reported in Table 2. From Table 2, we can see that our JSD method requires approximate 1min to complete the two tasks jointly, which is faster than MA, RF, and LSR. MA is slow because of time-consuming registrations between multiple atlases and the target image. LSR for landmark digitization is very slow because many iterations were used to improve the segmentation results. Although SPRF is efficient in landmark digitization (i.e., 20 s), this step relies on previous segmentation process which is usually very time-consuming.
Table 2:
Computational costs of different methods in bone segmentation and landmark digitization.
| Method | Environment | Bone Segmentation | Landmark Digitization |
|---|---|---|---|
| MA | CPU | ~ 3 hours | |
| RF | CPU | ~ 4 min | ~ 15 s |
| LSR | CPU | ~ 5 hours | – |
| SPRF | CPU | – | Time for segmentation+20 s |
| JSD1 | GPU | ~ 35 s | ~ 35 s |
| JSD2 | GPU | ~ 40 s | |
| JSD3 | GPU | ~ 55 s | ~ 55 s |
| JSD | GPU | ~ 60 s | |
Besides, our JSD method is slower than RF in landmark digitization. The reason could be that RF based method only samples thousands of patches for estimating landmark positions, which is more efficient than JSD. Currently, we cannot predict the whole image via JSD because of limited GPU memory. To perform a valid prediction, we have to partition each testing image into multiple 128 × 128 × 128 sub-images with a certain overlap. Based on the segmentation maps and landmark heatmaps for these sub-images, the center 64 × 64 × 64 patches are used to reconstruct the whole image. In this way, we can predict the maps accurately with limited memory. However, since these sub-images do not share convolutional computations, the proposed JSD method is not very fast in prediction. This problem can be avoided by using a prediction strategy based on the whole image other than sub-images, using a GPU with larger memory. Besides, Table 2 suggests that the computational time of our JSD is comparable to its three variants (i.e., JSD1, JSD2, and JSD3). The possible reason is that we employ down-sampled images to generate the displacement map (helping to reduce the use of GPU memory) and the GPU is successively used for FCN1 and FCN2 in JSD.
4.5. Influence of the Size of Sliding Window
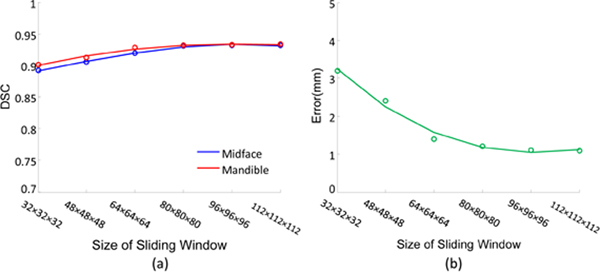
In the experiments mentioned above, we adopt the fixed size (i.e., 96 × 96 × 96) for the sliding window in the proposed JSD method. To investigate the influence of the size of sliding window, we perform an additional group of experiments by varying the size of sliding windows in the set {32 × 32 × 32,48 × 48 × 48,64 × 64 × 64,80 × 80 × 80,96 × 96 × 96,112 × 112 × 112}. However, due to the limited GPU memory, we could not use an even larger size of the sliding window in a 3D manner. The experimental results are shown in Fig. 7.
Figure 7:
Results of (a) bone segmentation for midface and mandible, and (b) landmark digitization, achieved by our JSD method using different sizes of sliding window.
Figure 7 (a)–(b) shows that JSD can yield relatively stable performance when the size of sliding window is larger than 80 × 80 × 80 in both tasks of bone segmentation and landmark digitization. Particularly, the performance of landmark digitization is poor when the size of the sliding window is small (e.g., < 64×64×64), as shown in Fig. 7 (b). The possible reason is that, if the window size is small, the sampled sub-images may only contain a small number of informative voxels (i.e., landmarks) but many uninformative voxels. In this case, a large number of sampled sub-images are less informative, and thus we cannot effectively train the proposed fully convolutional network based on these sampled sub-images via min-batch using SGD strategy. On the contrary, as shown in Fig. 7 (a), the segmentation results are not largely affected by the size of sliding windows. The underlying reason is that there are usually more informative voxels in the regions of midface/mandible in sampled sub-images, because of the large areas of midface and mandible.
5. Discussion
5.1. Comparison with Previous Studies
In this work, we propose a joint bone segmentation and landmark digitization (JSD) framework via a multi-task FCN model. Compared with previous multi-atlas based approaches (Shahidi et al., 2014; Coupé et al., 2011; Rousseau et al., 2011; Wang et al., 2013) for bone segmentation and landmark digitization, the proposed JSD method does not need the time-consuming nonlinear registration between a target image and multi-atlas images. Compared with previous learning based approaches (Cheng et al., 2011; Wang et al., 2014; Zhan et al., 2011) that require human-engineered features for CT images and pre-defined classifiers/regressors, our method learns an end-to-end model that can automatically perform feature extraction and segmentation/digitization. Since the feature representations for CT images and the subsequent classifiers/regressor are well coordinated, our method is expected to yield better results than the conventional learning based methods. The experimental results in Table 1 and Fig. 4 demonstrate the effectiveness of our proposed method. Compared with state-of-the-art methods for bone segmentation (Wang et al., 2014) and landmark digitization (Zhang et al., 2016b, 2017b), JSD jointly performs two tasks, by actively considering the underlying association between two tasks during the learning process. Besides, we develop the first-stage FCN (FCN1) to explicitly capture the context information of input images, by learning the displacement from each voxel to a specific landmark.
5.2. Limitations and Future Work
Although the proposed JSD method achieves promising results, there are still few limitations. First, there are only 107 images at hand, and we need more clinical data for the model learning. It is interesting to augment the training images by using synthetic data to improve the robustness of the proposed method. For instance, we may employ deformable transformation or Generative Adversarial Networks (Goodfellow et al., 2014) to generate a large number of synthetic data. Second, we treat the tasks of bone segmentation and landmark digitization equally, without considering their specific contributions. A possible solution may be to learn the optimal weights for different tasks automatically from the data. Besides, we do not consider the spatial relationships among landmarks. For instance, landmarks in the midface and landmarks in the mandible can be regarded as two subgroups, according to their spatial locations. Such prior information can be employed to further improve the performance of our method.
6. Conclusion
We have proposed a joint CMF bone segmentation and landmark digitization (JSD) framework via a context-guided multi-task FCN. Specifically, to capture the spatial context information of images, we propose to use displacement maps for modeling the displacement information from voxels to anatomical landmarks in input images. We further develop a context-guided FCN model, by using the first sub-network to learn a nonlinear mapping from an image onto its displacement maps, and employing the second sub-network to perform joint tasks of bone segmentation and landmark digitization. Experimental results on 107 subjects with CBCT/MSCT images suggest that JSD is superior to several state-of-the-art methods in both tasks of bone segmentation and landmark digitization.
Supplementary Material
Highlights.
A joint learning framework for both bone segmentation and landmark digitization
A displacement map is used to explicitly model the spatial context information
Results achieved by our method are clinically acceptable
Only 1 min to complete both tasks of bone segmentation and landmark digitization
7. Acknowledgements:
This work was supported in part by NIH grants R01 DE022676, R01 DE021863, and R01 DE027251
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, et al. , 2016. Tensorflow: A system for large-scale machine learning, in: Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation. [Google Scholar]
- Alansary A, Oktay O, Li Y, Le Folgoc L, Hou B, Vaillant G, Kamnitsas K, Vlontzos A, Glocker B, Kainz B, et al. , 2019. Evaluating reinforcement learning agents for anatomical landmark detection. Medical Image Analysis 53, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artaechevarria X, Munoz-Barrutia A, Ortiz-de Solorzano C, 2009. Combination strategies in multi-atlas image segmentation: Application tó brain MR data. IEEE Transactions on Medical Imaging 28, 1266–1277. [DOI] [PubMed] [Google Scholar]
- Baumgartner CF, Kamnitsas K, Matthew J, Fletcher TP, Smith S, Koch LM, Kainz B, Rueckert D, 2017. Sononet: Real-time detection and localisation of fetal standard scan planes in freehand ultrasound. IEEE Transactions on Medical Imaging 36, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd S, Vandenberghe L, 2004. Convex Optimization. Cambridge University Press. [Google Scholar]
- Cao L, Li L, Zheng J, Fan X, Yin F, Shen H, Zhang J, 2018. Multi-task neural networks for joint hippocampus segmentation and clinical score regression. Multimedia Tools and Applications 77, 29669–29686. [Google Scholar]
- Chen C, Belavy D, Yu W, Chu C, Armbrecht G, Bansmann M, Felsenberg D, Zheng G, 2015. Localization and segmentation of 3d intervertebral discs in mr images by data driven estimation. IEEE Transactions on Medical Imaging 34, 1719–1729. [DOI] [PubMed] [Google Scholar]
- Chen C, Xie W, Franke J, Grutzner P, Nolte LP, Zheng G, 2014. Automatic x-ray landmark detection and shape segmentation via data-driven joint estimation of image displacements. Medical Image Analysis 18, 487–499. [DOI] [PubMed] [Google Scholar]
- Cheng E, Chen J, Yang J, Deng H, Wu Y, Megalooikonomou V, Gable B, Ling H, 2011. Automatic dent-landmark detection in 3-D CBCT dental volumes, in: EMBC, IEEE; pp. 6204–6207. [DOI] [PubMed] [Google Scholar]
- Coupé P, Manj JV, Fonov V, Pruessner J, Robles M, Collins DL, 2011. Patch-based segmentation using expert priors: Application tó hippocampus and ventricle segmentation. NeuroImage 54, 940–954. [DOI] [PubMed] [Google Scholar]
- Criminisi A, Robertson D, Konukoglu E, Shotton J, Pathak S, White S, Siddiqui K, 2013. Regression forests for efficient anatomy detection and localization in computed tomography scans. Medical Image Analysis 17, 1293–1303. [DOI] [PubMed] [Google Scholar]
- Criminisi A, Shotton J, Robertson D, Konukoglu E, 2010. Regression forests for efficient anatomy detection and localization in CT studies, in: International MICCAI Workshop on Medical Computer Vision, Springer; pp. 106–117. [Google Scholar]
- Cuingnet R, Prevost R, Lesage D, Cohen LD, Mory B, Ardon R, 2012. Automatic detection and segmentation of kidneys in 3D CT images using random forests, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer; pp. 66–74. [DOI] [PubMed] [Google Scholar]
- Dai J, He K, Sun J, 2016. I nstance-aware semantic segmentation via multi-task network cascades, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3150–3158. [Google Scholar]
- De Vos W, Casselman J, Swennen G, 2009. Cone-beam computerized tomography (CBCT) imaging of the oral and maxillofacial region: A systematic review of the literature. International Journal of Oral and Maxillofacial Surgery 38, 609–625. [DOI] [PubMed] [Google Scholar]
- Farag A, Lu L, Roth HR, Liu J, Turkbey E, Summers RM, 2017. A bottom-up approach for pancreas segmentation using cascaded superpixels and (deep) image patch labeling. IEEE Transactions on Image Processing 26, 386–399. [DOI] [PubMed] [Google Scholar]
- Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y, 2014. Generative adversarial nets, in: Advances in Neural Information Processing Systems, pp. 2672–2680. [Google Scholar]
- Li X, Zhao L, Wei L, Yang MH, Wu F, Zhuang Y, Ling H, Wang J, 2016. Deepsaliency: Multi-task deep neural network model for salient object detection. IEEE Transactions on Image Processing 25, 3919–3930. [DOI] [PubMed] [Google Scholar]
- Lian C, Liu M, Zhang J, Shen D, 2018a. Hierarchical fully convolutional network for joint atrophy localization and Alzheimer’s disease diagnosis using structural MRI. IEEE Transactions on Pattern Analysis and Machine Intelligence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian C, Zhang J, Liu M, Zong X, Hung SC, Lin W, Shen D, 2018b. Multi-channel multi-scale fully convolutional network for 3D perivascular spaces segmentation in 7T MR images. Medical Image Analysis 46, 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner C, Thiagarajah S, Wilkinson J, Consortium T, Wallis G, Cootes T, 2013. Fully automatic segmentation of the proximal femur using random forest regression voting. IEEE Transactions on Medical Imaging 32, 1462–1472. [DOI] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D, 2015a. View-centralized multi-atlas classification for Alzheimer’s disease diagnosis. Human Brain Mapping 36, 1847–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D, 2016. Relationship induced multi-template learning for diagnosis of Alzheimer’s disease and mild cognitive impairment. IEEE Transactions on Medical Imaging 35, 1463–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang J, Adeli E, Shen D, 2018. Joint classification and regression via deep multi-task multi-channel learning for Alzheimer’s disease diagnosis. IEEE Transactions on Biomedical Engineering 66, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang J, Yap PT, Shen D, 2017. View-aligned hypergraph learning for Alzheimer’s disease diagnosis with incomplete multi-modality data. Medical Image Analysis 36, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Mei T, Zhang Y, Che C, Luo J, 2015b. Multi-task deep visual-semantic embedding for video thumbnail selection, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3707–3715. [Google Scholar]
- Loubele M, Bogaerts R, Van Dijck E, Pauwels R, Vanheusden S, Suetens P, Marchal G, Sanderink G, Jacobs R, 2009. Comparison between effective radiation dose of CBCT and MSCT scanners for dentomaxillofacial applications. European Journal of Radiology 71, 461–468. [DOI] [PubMed] [Google Scholar]
- Loubele M, Maes F, Schutyser F, Marchal G, Jacobs R, Suetens P, 2006. Assessment of bone segmentation quality of cone-beam CT versus multislice spiral CT: A pilot study. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontology 102, 225–234. [DOI] [PubMed] [Google Scholar]
- Mitra J, Bourgeat P, Fripp J, Ghose S, Rose S, Salvado O, Connelly A, Campbell B, Palmer S, Sharma G, et al. , 2014. Lesion segmentation from multimodal MRI using random forest following ischemic stroke. NeuroImage 98, 324–335. [DOI] [PubMed] [Google Scholar]
- Payer C, Stern D, Bischof H, Urschler M, 2016. Regressing heatmaps for multiple landmark localization using CNNs, in: Internationalˇ Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 230–238. [Google Scholar]
- Pfister T, Charles J, Zisserman A, 2015. Flowing convnets for human pose estimation in videos, in: ICCV, pp. 1913–1921. [Google Scholar]
- Ranjan R, Patel VM, Chellappa R, 2017. Hyperface: A deep multi-task learning framework for face detection, landmark localization, pose estimation, and gender recognition. IEEE Transactions on Pattern Analysis and Machine Intelligence, DOI: 10.1109/TPAMI.2017.2781233. [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, Brox T, 2015. U-Net: Convolutional networks for biomedical image segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 234–241. [Google Scholar]
- Rother C, Minka T, Blake A, Kolmogorov V, 2006. Cosegmentation of image pairs by histogram matching-incorporating a global constraint into MRFs, in: IEEE Computer Society Conference on Computer Vision and Pattern Recognition, IEEE; pp. 993–1000. [Google Scholar]
- Rousseau F, Habas PA, Studholme C, 2011. A supervised patch-based approach for human brain labeling. IEEE Transactions on Medical Imaging 30, 1852–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapire RE, Freund Y, Bartlett P, Lee WS, et al. , 1998. Boosting the margin: A new explanation for the effectiveness of voting methods. The Annals of Statistics 26, 1651–1686. [Google Scholar]
- Schroff F, Criminisi A, Zisserman A, 2008. Object class segmentation using random forests., in: BMVC, pp. 1–10. [Google Scholar]
- Schulze R, Heil U, Grob D, Bruellmann D, Dranischnikow E, Schwanecke U, Schoemer E, 2011. Artefacts in CBCT: A review. Dentomaxillofacial Radiology 40, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghers D, Slagmolen P, Lambelin Y, Hermans J, Loeckx D, Maes F, Suetens P, 2007. Landmark based liver segmentation using local shape and local intensity models, in: MICCAI Workshop on 3D Segmentation in the Clinic: A Grand Challenge, pp. 135–142. [Google Scholar]
- Shahidi S, Bahrampour E, Soltanimehr E, Zamani A, Oshagh M, Moattari M, Mehdizadeh A, 2014. The accuracy of a designed software for automated localization of craniofacial landmarks on CBCT images. BMC Medical Imaging 14, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torosdagli N, Liberton DK, Verma P, Sincan M, Lee JS, Bagci U, 2018. Deep geodesic learning for segmentation and anatomical landmarking. IEEE Transactions on Medical Imaging 38, 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA, 2013. Multi-atlas segmentation with joint label fusion. IEEE Transactions on Pattern Analysis and Machine Intelligence 35, 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chen KC, Gao Y, Shi F, Liao S, Li G, Shen SG, Yan J, Lee PK, Chow B, et al. , 2014. Automated bone segmentation from dental CBCT images using patch-based sparse representation and convex optimization. Medical Physics 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Huo Y, Park J, Landman B, Milkowski A, Grbic S, Zhou S, 2018. Less is more: Simultaneous view classification and landmark detection for abdominal ultrasound images, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer; pp. 711–719. [Google Scholar]
- Yim J, Jung H, Yoo B, Choi C, Park D, Kim J, 2015. Rotating your face using multi-task deep neural network, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 676–684. [Google Scholar]
- Zhan Y, Dewan M, Harder M, Krishnan A, Zhou XS, 2011. Robust automatic knee MR slice positioning through redundant and hierarchical anatomy detection. IEEE Transactions on Medical Imaging 30, 2087–2100. [DOI] [PubMed] [Google Scholar]
- Zhang J, Gao Y, Gao Y, Munsell BC, Shen D, 2016. a. Detecting anatomical landmarks for fast Alzheimer’s disease diagnosis. IEEE Transactions on Medical Imaging 35, 2524–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gao Y, Park SH, Zong X, Lin W, Shen D, 2017a. Structured learning for 3D perivascular spaces segmentation using vascular features. IEEE Transactions on Biomedical Engineering 64, 2803–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gao Y, Wang L, Tang Z, Xia JJ, Shen D, 2016b. Automatic craniomaxillofacial landmark digitization via segmentation-guided partially-joint regression forest model and multiscale statistical features. IEEE Transactions on Biomedical Engineering 63, 1820–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu M, Shen D, 2017b. Detecting anatomical landmarks from limited medical imaging data using two-stage task-oriented deep neural networks. IEEE Transactions on Image Processing 26, 4753–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu M, Wang L, Chen S, Yuan P, Li J, Shen SGF, Tang Z, Chen KC, Xia JJ, Shen D, 2017c. Joint craniomaxillofacial bone segmentation and landmark digitization by context-guided fully convolutional networks, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, 1993. Model selection via multifold cross validation. The Annals of Statistics, 299–313. [Google Scholar]
- Zhang Z, Luo P, Loy CC, Tang X, 2014. Facial landmark detection by deep multi-task learning, in: European Conference on Computer Vision, Springer; pp. 94–108. [Google Scholar]
- Zhu Y, Wang L, Liu M, Qian C, Yousuf A, Oto A, Shen D, 2017. MRI-based prostate cancer detection with high-level representation and hierarchical classification. Medical Physics 44, 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.