Summary
Voltage-gated calcium channels (CaV) regulate numerous vital functions in nerve and muscle cells. To fulfill their diverse functions, the multiple members of the CaV channel family are activated over a wide range of voltages. Voltage sensing in potassium and sodium channels involves the sequential transition of positively charged amino acids across a ring of residues comprising the charge transfer center. In CaV channels, the precise molecular mechanism underlying voltage sensing remains elusive. Here we combined Rosetta structural modeling with site-directed mutagenesis to identify the molecular mechanism responsible for the specific gating properties of two CaV1.1 splice variants. Our data reveal previously unnoticed interactions of S4 arginines with an aspartate (D1196) outside the charge transfer center of the fourth voltage-sensing domain that are regulated by alternative splicing of the S3-S4 linker. These interactions facilitate the final transition into the activated state and critically determine the voltage sensitivity and current amplitude of these CaV channels.
Introduction
Voltage-gated calcium (CaV) channels contribute to the excitability of nerve and muscle cells and are the exclusive molecular agents to convert membrane depolarization into calcium-mediated cellular functions like muscle contraction, secretion of neurotransmitters and hormones, and gene regulation (Catterall, 2011). In mammals, CaV channels constitute a family of ten genes each with multiple splice variants. These give rise to functionally diverse channels, which activate at greatly different membrane potentials, precisely tuned to the specific requirements of the cell function they regulate (Lipscombe et al., 2013). Whereas voltage sensing in voltage-gated potassium (KV) and sodium (NaV) channels is well studied, the molecular details of the voltage-sensing mechanism in CaV channels are still elusive.
CaV α1 subunits are polypeptides of more than 1,800 amino acids folded into four homologous repeats (I–IV) with six transmembrane segments each (S1–S6). Segments S1–S4 of each repeat form the voltage-sensing domains (VSD), while segments S5 and S6 with the connecting P loop form the channel pore and selectivity filter (Figure 1A). The S4 transmembrane segments of CaV channels contain four to five positively charged amino acids (arginines or lysines) spaced at three-amino acid intervals that serve as the actual voltage sensors of the voltage-dependent gating mechanism (Bezanilla, 2000; Bezanilla et al., 1991; Catterall, 2010; Kontis and Goldin, 1997; Noceti et al., 1996; Papazian et al., 1991; Seoh et al., 1996). In response to membrane depolarization, the S4 segments translocate toward the extracellular side of the membrane, inducing a conformational change of the S5 and S6 segments that leads to the opening of the channel pore (Ahern and Horn, 2004; Catterall and Yarov-Yarovoy, 2010; Yarov-Yarovoy et al., 2006a). To facilitate the movement of the S4 segments through the dielectric interior of the plasma membrane, the S1–S3 helices of the VSD contain several negatively charged and polar residues that stabilize the positive charges of S4 (Bezanilla, 2000; Catterall, 2010; Lacroix et al., 2014; Pless et al., 2014). In KV and NaV channels, three highly conserved residues have been identified as a “gating charge transfer center” (CTC) including a phenylalanine in S2 and negatively charged residues in S2 and S3 (Payandeh et al., 2011, 2012; Tao et al., 2010). These residues form transient hydrogen bond (H-bond) pairs with the positive gating charges of S4 as they sequentially pass through the CTC (Cheng et al., 2013; Lin et al., 2011; Tao et al., 2010; Yarov-Yarovoy et al., 2012). In the VSD of CaV channels, corresponding residues are also highly conserved.
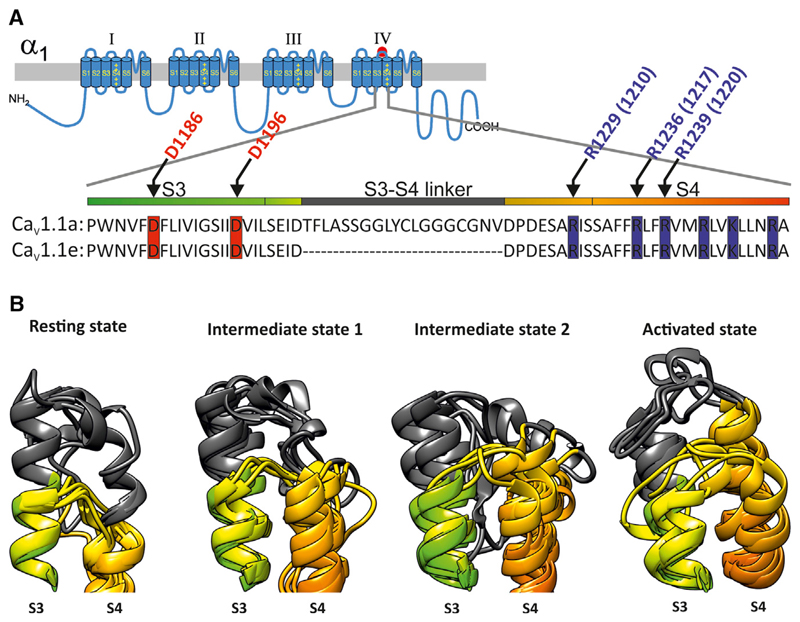
Figure 1. Structural Modeling of the IVS3-S4 Loop in Different Conformational States Depicts a Higher Variability of CaV1.1a Loop Compared with CaV1.1e.
(A) Transmembrane topology of the α1 subunit of the voltage-gated calcium channels consisting of four repeats with six transmembrane domains each. The red mark indicates the position of exon 29 in the extracellular loop connecting IVS3 and IVS4. The sequence alignment of IVS3, the IVS3-S4 extracellular loop, and IVS4 of the two CaV1.1 splice variants shows the position of the 19 amino acids encoded by exon 29 in the IVS3-S4 linker, the positions of the positive charges in IVS4 (blue) and potential negative counter charges in IVS3 (red). Note that the colors of the bar above the sequences match the ribbon models below (green, S3; gray, exon 29; orange, S4).
(B) Predicted conformations of both CaV1.1a and CaV1.1e IVS3-S4 loops in the resting state, intermediate states 1 and 2, and in the activated state. Gray highlights the extra 19 amino acids encoded by exon 29 present in the CaV1.1a loop, while the short IVS3-S4 linker of CaV1.1e is depicted in yellow. Note the substantial conformational changes of the CaV1.1a IVS3-S4 loop during gating and between different clusters, but less for the CaV1.1e IVS3-S4 loop. The images were generated using UCSF Chimera (Pettersen et al., 2004).
Previously we described a splice variant of CaV1.1 (CaV1.1e, α1S-ΔE29) lacking exon 29, which encodes 19 amino acids in the extracellular linker connecting transmembrane segments S3 and S4 in the fourth VSD (Figure 1A) (Flucher and Tuluc, 2011; Tuluc and Flucher, 2011; Tuluc et al., 2009). The shorter IVS3-S4 linker of CaV1.1e confers the channel a ~30-mV left shift in voltage sensitivity and a ~6-fold increased current amplitude compared with the long IVS3-S4 linker of CaV1.1a. This indicates that, in the full-length CaV1.1a channel, the VSD of the fourth repeat is limiting for voltage sensitivity and current amplitude. Since similar alternative splicing of the IVS3-S4 linker is found in most CACNA1 genes, regulation of gating properties by the fourth VSD may be a general principle for the specific gating properties of CaV channels (Liao et al., 2005; Lipscombe et al., 2013).
Here we addressed the molecular mechanism by which differences in the IVS3-S4 linker control the voltage sensitivity and amplitude of the two functionally distinct CaV1.1 channel variants. Using the Rosetta-Membrane method (Yarov-Yarovoy et al., 2006a, 2006b, 2012), we predicted the structure of the S3-S4 loop and the gating transitions of the VSD in both CaV1.1 channel splice variants. The structural models indicate that, in CaV1.1e but not in CaV1.1a, H-bonds are forming between D1196 (in IVS3) and R1 and R2 (in IVS4) during the final steps of the gating charge movement. Using site-directed mutagenesis, we confirmed that the D1196 negatively charged residue in IVS3 is critical for electrostatic compensation of S4 charges and therefore determines the channel voltage sensitivity. Inclusion of exon 29 in the IVS3-S4 linker or mutation of D1196 (in IVS3) weakens the H-bonds between S4 charges and D1196 thus inducing a rightward shift in the voltage sensitivity of CaV1.1a.
Results
Structure Prediction of the Fourth VSD of CaV1.1a and CaV1.1e Channels
We generated structural models of the fourth VSD of CaV1.1a and CaV1.1e channels using the Rosetta-Membrane method to reveal the structural basis for the critical role of the IVS3-S4 linker in determining voltage sensitivity (Yarov-Yarovoy et al., 2006a, 2006b, 2012). In order to identify the critical gating transition affected by the IVS3-S4 linker lengths (with and without the exon 29 sequence), the position of the S4 segment in relation to the CTC was modeled in four different conformational states (resting, intermediate 1, intermediate 2, and activated) as described in the Experimental Procedures. After several steps of selection and refinement, the top 10% of the best scoring models of the VSD in each conformational state are clustered and chosen for further visual evaluation and analysis. Although we observed a certain degree of variability between the different top clusters, a common feature of all top cluster models of the long CaV1.1a IVS3-S4 linker is that the exon 29 sequence further extends the alpha helix of IVS3 toward the extracellular side of the membrane (Figure 1B). In the resting and intermediate states 1 and 2, all top cluster models show the CaV1.1a IVS3-S4 linker orientated toward the outer vestibule of the VSD, while in the activated state, the IVS3-S4 linker bends away from the vestibule. Theoretically, these structural differences between the S3-S4 loops of the two CaV1.1 splice variants could alone alter the position or restrict the movement of the VSD transmembrane segments and thus explain the differences in voltage sensitivity.
Relaxed Homology Structure Models Reveal Differences in Packing and H-Bond Formation of the CaV1.1a and CaV1.1e VSDs
In order to test if the structurally divergent IVS3-S4 linkers differentially affect the VSD structure, and thus the voltage sensitivity of the two CaV1.1 splice variants, we relaxed the backbone and side-chain atoms in our homology models of the fourth VSD with the short and long IVS3-S4 linker. After full-atom relax, the VSD models of both splice variants in each conformational state reach similar low energy levels, indicating that the IVS3-S4 linker does not restrict the S4 movement. Previously it has been demonstrated that gating of Shaker potassium channels requires the formation of salt bridges of the basic residues in S4 with several acidic residues in the transmembrane helices S1, S2, and S3. These residues are E1 (E283) and E2 (E293) in the transmembrane helix S2 and D3 (D316) in the transmembrane helix S3 (Ma et al., 2006; Pless et al., 2011; Seoh et al., 1996). In CaV channels, the key residues of the CTC (E2, F, and D3) are conserved with those previously described for KV (Lacroix et al., 2014; Tao et al., 2010) and NaV channels (DeCaen et al., 2008, 2009, 2011; Yarov-Yarovoy et al., 2012), but in the fourth VSD of CaV1.1 channels, the counter charge corresponding to Shaker E1 is missing. The sequential movement of the positively charged S4 residues past the negative counter charges (E2, D3) and over the hydrophobic cap (phenylalanine) of the CTC is considered essential for the voltage-sensing process (Lacroix et al., 2014; Tao et al., 2010; Yarov-Yarovoy et al., 2012). The superimposed structural models (Figure 2A) show that the different size IVS3-S4 linkers do not dramatically change the general position of the IVS4 helix relative to the other helices of the VSD. However, especially in the intermediate state 2 and in the activated state, the outer parts of the IVS3 and IVS4 helices are displaced in CaV1.1a relative to CaV1.1e, indicative of the impact of the IVS3-S4 linker on the structure of the VSD. Two morphs were generated to facilitate the visualization of the structural changes in the fourth VSD of CaV1.1 variants during gating (see Movies S1 and S2 related to Figure 2).
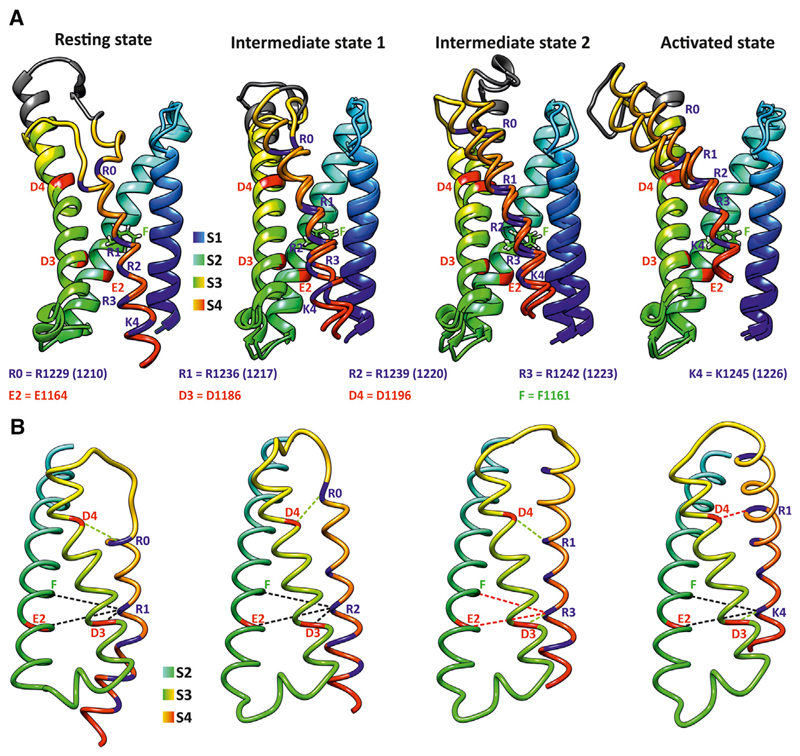
Figure 2. Homology Rosetta Modeling of the CaV1.1 Splice Variants Fourth Repeat VSD Gating States.
(A) Superimposed structures of CaV1.1a and CaV1.1e VSD at the resting state, intermediate states 1 and 2, and the activated state. S1–S3 are shown as ribbons while the S4 transmembrane domain is shown as licorice to facilitate observation of similarities and differences in the structures of CaV1.1a and CaV1.1e. The phenylalanine “cap” is shown to indicate the position of the CTC. In the fourth VSD of CaV1.1, the CTC comprises a phenylalanine (F1161 = F) and a glutamate (E1164 = E2) in IVS2, and an aspartate (D1186 = D3) in IVS3. S4 positively charged arginines are highlighted in blue, while the negatively charged amino acids that could participate to the charge transfer are highlighted in red. The extra 19 amino acids encoded by exon 29 in CaV1.1a are shown in gray.
(B) Distance measurements between the Cα atoms of IVS4-charged amino acids and the Cα atoms of F and E2 in IVS2, D3, and D4 (D1196) in IVS3. Only S2, S3, and S4 of CaV1.1e are shown for clarity. Black dotted lines indicate that the distances are the same in CaV1.1a and CaV1.1e in a given state, green dotted lines indicate that the distance is larger in CaV1.1e compared with CaV1.1a, while red dotted lines indicate that the distance is larger in CaV1.1a compared with CaV1.1e by at least 0.2 Å. Distances in the CTC differ in the intermediate state 2 and the activated state, whereas S3-S4 distances near the IVS3-S4 linker (as indicated by D4 to the closest arginine in S4 substantially differ in all states. Distances are measured using UCSF Chimera (Pettersen et al., 2004) and the values are presented in Table 1.
To quantitatively describe differences in the structure of the CTC of CaV1.1a and CaV1.1e, we measured the distances between the Cα atoms of the positive charges in IVS4 relative to F, E2, and D3 in all four conformational states (Figure 2B and Table 1). In the resting and intermediate state 1, the intramolecular distances within the CTC were similar for both CaV1.1 splice variants. In intermediate state 2, the distances from F and E2 to R3 were shorter, while D3 to R3 was longer in CaV1.1e compared with CaV1.1a. Because the effect of the IVS3-S4 linker was more pronounced near the extracellular side of the VSD, we also measured the distance between the R1, R2, and R3 charges and a negatively charged residue (D4 = D1196) further outside in IVS3. Here the models show an increase of the distance in the resting state and in both intermediate states and a reduced distance in the activated state of CaV1.1e relative to CaV1.1a. The increased variability between clusters of the same variant and between variants are expected to alter the transient H-bond formation between IVS4-positive charges and potential counter charges in IVS2 and IVS3 during the sequential steps of the voltage-sensing process.
Table 1. Distance Measurements between Cα of S4 Charged Amino Acids and Different Amino Acids from S2 and S3 during Gating.
| F-R1 | E2-R1 | D3-R1 | D4-R0 | ||
| Resting state | CaV1.1a | 10.63 ± 0.00 | 12.44 ± 0.00 | 6.23 ± 0.00 | 7.50 ± 0.30a |
| CaV1.1e | 10.63 ± 0.00 | 12.44 ± 0.00 | 6.23 ± 0.00 | 8.01 ± 0.37a | |
| F-R2 | E2-R2 | D3-R2 | D4-R0 | ||
| Intermediate State 1 | CaV1.1a | 10.55 ± 0.30 | 12.13 ± 0.45 | 6.39 ± 0.40 | 5.52 ± 0.33a |
| CaV1.1e | 10.47 ± 0.24 | 12.08 ± 0.29 | 6.23 ± 0.00 | 6.23 ± 0.47a | |
| F-R3 | E2-R3 | D3-R3 | D4-R1 | ||
| Intermediate State 2 | CaV1.1a | 10.86 ± 0.36b | 12.73 ± 0.67b | 6.61 ± 0.21a | 6.81 ± 0.68a |
| CaV1.1e | 10.64 ± 0.08b | 12.17 ± 0.15b | 6.83 ± 0.20a | 7.24 ± 0.76a | |
| F-K4 | E2-K4 | D3-K4 | D4-R1 | ||
| Activated State | CaV1.1a | 10.29 ± 0.17 | 12.05 ± 0.24 | 5.83 ± 0.18a | 5.89 ± 0.16b |
| CaV1.1e | 10.21 ± 0.34 | 11.86 ± 0.60 | 6.04 ± 0.24a | 5.67 ± 0.22b |
All distances are expressed in Ångström (Å) ± SD. The exact amino acid in the fourth VSD of CaV1.1 are: F=F1161, E2= E1164, D3=D1186, D4=D1196, R0=R1229 (CaV1.1a) or R1210 (CaV1.1e), R1=R1236 (CaV1.1a) or R1217 (CaV1.1e), R2=R1239 (CaV1.1a) or R1220 (CaV1.1e), R3=R1242 (CaV1.1a) or R1223 (CaV1.1e), K4=K1245 (CaV1.1a) or K1226 (CaV1.1e).
The difference in distance (of at least 0.2 Å) if the distance is larger in CaV1.1e compared with CaV1.1a.
The difference in distance (of at least 0.2 Å) if the distance is larger in CaV1.1a compared with CaV1.1e.
In the resting state of both CaV1.1 variants (Figure 3A), the outer arginines of IVS4 (R1 and R2) reside in the CTC forming H-bonds with D3. A smaller number of H-bonds and distances >2 Å indicate weaker interactions in CaV1.1a. The outer aspartate D4 resides near arginine (R0) at the extracellular end of the IVS4 helix, and three of the five top model clusters indicate an H-bond between R0 and D4 in CaV1.1e. In the intermediate state 1 (Figure 3B), the number and length of the H-bonds of R0, R1, and R2 and partners in IVS2 and IVS3 are very similar in both splice variants. However, in the intermediate state 2 (Figure 3C), the changed distances between the transmembrane helices of CaV1.1a or CaV1.1e profoundly affect H-bond formation especially between D4 and R1. Whereas in CaV1.1a only two of the top five clusters form H-bonds between R1 and D4 in IVS3, the CaV1.1e models indicate stronger interactions between these two residues (Table S1 related to Figures 3 and 4). In three of the top five analyzed clusters, R1 forms two H-bonds with D4 and, in one cluster, one H-bond. In the activated state of both CaV1.1e and CaV1.1a (Figure 3D), D4 forms H-bonds with the next arginine of the IVS4 helix, R2. However, overall the number of D4-R2 H-bonds is higher and their distance shorter in the models of CaV1.1e compared with CaV1.1a, consistent with stronger interactions in CaV1.1e. In addition, in two of the five top CaV1.1e clusters, also the interaction between R1 and D4 is maintained in the activated state. This is not found in any of the CaV1.1a models, in which R0 and R1 form several H-bonds with different acceptors in IVS3. Collectively our modeling indicates that, in CaV1.1e stronger, R1-D4 interactions in the intermediate state 2 may facilitate the transition toward activation, thus increasing the voltage sensitivity. The multiple interactions of both R1 and R2 with D4 may stabilize the voltage sensor of CaV1.1e in the activated state, possibly causing the increased CaV1.1e channel current amplitude. Therefore, we hypothesize that the aspartate D1196 (D4) is a critical residue for voltage sensing of CaV1.1 channels, potentially involved in facilitating the transition from intermediate state 2 to activation, and in stabilizing the activated state of the CaV1.1e splice variant.
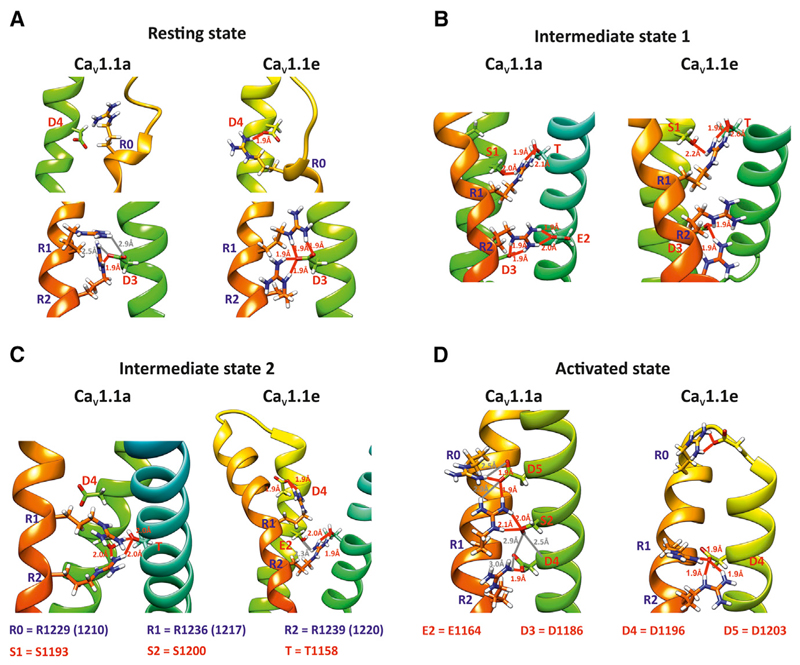
Figure 3. Structural Differences in the Fourth VSD of CaV1.1e and CaV1.1a Affect the Number and Strengths of H-bonds Sequentially Formed During Gating.
(A) In the resting state, R1 and R2 of CaV1.1e form several H-bonds with D3 with distances ≤2 Å, while CaV1.1a R1 and R2 interaction with D3 results in less and weaker H-bonds. An additional arginine on the extracellular side of IVS4 (R0) forms one H-bond with an IVS3 aspartate (D4) in CaV1.1e but not in CaV1.1a.
(B) H-bonds in intermediate state 1 are similar in both splice variants.
(C) In intermediate state 2, R1 of CaV1.1e interacts with D4, and R2 with E2 and T, while in CaV1.1a, both R1 and R2 interact exclusively with E2 and T.
(D) The activated state of CaV1.1e is characterized by multiple interactions of R1 and R2 with D4, while R0, R1, and R2 of CaV1.1a form interactions with different H-bond acceptors from IVS3 (D4, D4, S2). The H-bonds and their distances are illustrated using UCSF Chimera (Pettersen et al., 2004). Relevant H-bonds <2.0 Å are colored in red while distances >2.0 Å are shown in gray.
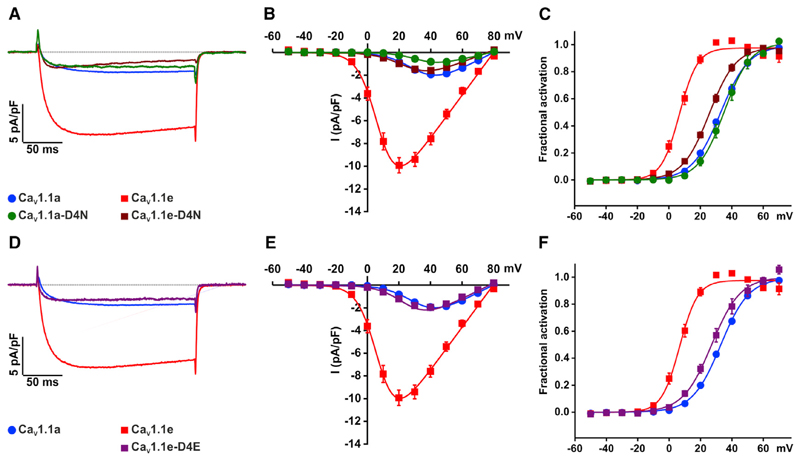
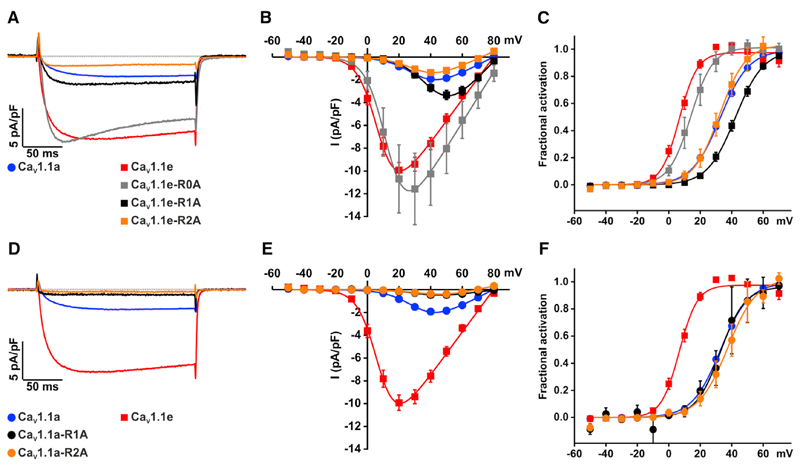
Figure 4. Aspartate at Position 1196 Is Critical in Determining the Voltage Sensitivity of CaV1.1 but Not CaV1.1a.
(A–C) Representative calcium currents recorded from myotubes expressing CaV1.1a (blue), CaV1.1e (red), CaV1.1a-D4N (green), and CaV1.1e-D4N (wine) during a 200-ms step depolarization to the maximum current amplitude (A). The CaV1.1e splice variant has a ∼6-fold higher current amplitude and ∼26 mV left shift in voltage dependence compared with CaV1.1a. Mutating the negatively charged aspartate (D4) at position 1196 to the neutral asparagine (CaV1.1e-D4N, wine) reverts CaV1.1e calcium current properties to those of CaV1.1a, as illustrated by (B) the I/V curve and (C) voltage dependence of current activation. Mutating the same amino acid in CaV1.1a (CaV1.1a-D4N, green) has no effect on voltage sensitivity but slightly reduces the current amplitude.
(D–F) Mutating the CaV1.1e aspartate (D4) to a glutamate results in the same reduction in amplitude (CaV1.1e-D4E, purple) and shift in the voltage dependence as observed for the aspartate to asparagine mutation. All data are presented as means ± SEM. Currents were analyzed as previously described (Tuluc et al., 2007) and the calcium current parameters and statistics are given in Table 2.
Site-Directed Mutagenesis Confirms the Importance of D1196 for Determining the Voltage Sensitivity of CaV1.1e
To test the hypothesis that D4 is a critical determinant of the distinct voltage sensitivities and current amplitude of CaV1.1 variants, we neutralized the charge of the aspartate D1196 by mutating it to asparagine in both splice variants. We expressed wild-type and mutant GFP-tagged CaV1.1a or CaV1.1e constructs in dysgenic (CaV1.1-null) myotubes and analyzed their current properties using whole-cell patch-clamp recording. Wild-type CaV1.1e showed a 6- to 8-fold increased calcium current amplitude and a 26-mV left shift in the voltage dependence of current activation compared with full-length CaV1.1a, as previously described (Tuluc et al., 2009) (Figures 4A–4C; Table 2). Consistent with the structural modeling predictions, neutralizing the negatively charged D4 in CaV1.1e (CaV1.1e-D4N) reversed the effects of exon 29 exclusion almost completely. Its voltage dependence of activation was right shifted by 19 mV, and the peak amplitude was reduced to about the same level as that of CaV1.1a. In contrast, the identical mutation in the full-length CaV1.1a (CaV1.1a-D4N) had no effect on voltage dependence, and only a small effect on the current amplitude. Thus, D1196 (D4) is essential for the characteristic gating properties of the CaV1.1e channel by acting as counter charge for IVS4 gating charges, but it does not contribute substantially to the gating of CaV1.1a.
Table 2. Calcium Currents Biophysical Properties.
| Ipeak (pA/pF) | Vhalf (mV) | Vhalf Significance | Vrev (mV) | kact (mV) | n | |
|---|---|---|---|---|---|---|
| CaV1.1e | –10.4 ± 0.7 | 7.0 ± 1.1 | – | 79.2 ± 1.1 | 4.7 ± 0.2 | 27 |
| CaV1.1e-D4N | –1.6 ± 0.1 | 26.4 ± 1.0 | <0.0001 | 75.8 ± 1.2 | 8.8 ± 0.4 | 16 |
| CaV1.1e-D4E | –1.9 ±0.2 | 27.8 ± 1.7 | 0.0002 | 77.6 ± 2.9 | 9.3 ± 0.9 | 6 |
| CaV1.1e-R0A | –12.1 ± 2.3 | 14.8 ± 2.6 | 0.0288 | 83.8 ± 1.2 | 5.7 ± 0.6 | 9 |
| CaV1.1e-R1A | –3.4 ± 0.4 | 43.9 ± 0.8 | <0.0001 | 82.8 ± 1.4 | 9.4 ± 0.7 | 10 |
| CaV1.1e-R2A | –1.4 ± 0.1 | 32.3 ± 1.4 | <0.0001 | 72.1 ± 1.3 | 8.1 ± 0.7 | 8 |
| CaV1.1a | –1.9 ± 0.2 | 33.2 ± 0.8 | – | 79.8 ± 1.1 | 8.9 ± 0.3 | 21 |
| CaV1.1a-D4N | –0.9 ± 0.2 | 35.2 ± 1.9 | 0.4261 | 76.6 ± 1.9 | 7.8 ± 0.6 | 7 |
| CaV1.1a-R1A | –0.5 ± 0.1 | 34.8 ± 5.6 | 0.8614 | 77.5 ± 3.7 | 7.7 ± 1.7 | 3 |
| CaV1.1a-R2A | –0.4 ± 0.1 | 37.0 ± 3.9 | 0.4813 | 70.7 ± 2.4 | 7.8 ± 0.5 | 4 |
All data are presented as means ± SEM. The significance levels were calculated using the Mann-Whitney U test between each construct and the original channel splice variant used as backbone.
To examine whether the negative charge or the predicted H-bonds of the D1196 are critical for the leftwards shift in the voltage sensitivity of CaV1.1e, we generated a second mutant in which the aspartate was substituted by the negatively charged but longer side-chain glutamate (CaV1.1e-D4E). Interestingly, this mutation (CaV1.1e-D4E) also reduced the voltage sensitivity and current amplitude to values near those of CaV1.1a (Figures 4D–4F). Evidently the presence of the negatively charged carboxyl group at D4 is not sufficient to left shift the voltage sensitivity of CaV1.1e, but it also has to be positioned at the exact distance from the protein backbone to properly interact with the basic side chain of the arginines in IVS4.
To further examine this possibility, we modeled CaV1.1e-D4E in the four different conformational states and compared it with CaV1.1e. Indeed, structural modeling indicated that mutating the aspartate to glutamate at position 1196 in CaV1.1e may well reduce the H-bonds transiently formed with R1, especially in the intermediate state 2 (Table S1 and Figure S2 related to Figures 3 and 4). Whereas in CaV1.1e four of five clusters R1 interacted with D4 (three clusters two H-bonds and one cluster one H-bond) in CaV1.1-D4E, the number of clusters showing interactions between R1 and E1196 was reduced to one with two H-bonds plus two with a single H-bond. Such a drop in the number of H-bonds and in the fraction of model clusters where these bonds occur is consistent with the experimentally observed shift of the voltage sensitivity toward more depolarizing potentials. Together these mutagenesis experiments and related structural models suggest that D4 facilitates voltage-sensitive transitions by interacting with positive IVS4 charges. This raises the question as to which interactions of D4 with which of the arginines represents the limiting gating transition for current activation?
The Interactions of R1 and R2 with D4 Determine the Voltage Sensitivity of CaV1.1e
Our structural modeling (Figure 3) indicated that R0, R1, and R2 differentially interact with D4 in CaV1.1e compared with CaV1.1a. To determine which of these putative interactions are important for the ~26-mV left shift in the voltage sensitivity of CaV1.1e, we mutated R0, R1, or R2 to alanine and analyzed their current properties. According to structural modeling, the outermost arginine R0 forms an H-bond with D4 of CaV1.1e but not of CaV1.1a in the resting state. Mutation of R0 in CaV1.1e (CaV1.1e-R0A) caused a slight increase in current density and only a 7-mV right shift of the voltage dependence of activation (Figures 5A–5C). These modest effects are very different from the properties of CaV1.1a or from the changes effected by mutations of D4. Therefore the R0-D4 interaction is not the major determinant for the specific voltage sensitivity and current amplitude of CaV1.1e.
Figure 5. R1 and R2 Are Functionally Relevant Interaction Partners for D4 in CaV1.1e but Not in CaV1.1a.
(A) Representative calcium currents recorded during a 200-ms step depolarization to the maximum current amplitude from myotubes expressing CaV1.1a (blue), CaV1.1e (red), or CaV1.1e with arginine-to-alanine mutations of R0 (CaV1.1e-R0A, gray), R1 (CaV1.1e-R1A, black), and R2 (CaV1.1e-R2A, orange).
(B and C) Neutralizing R2 of CaV1.1e (CaV1.1e-R2A) converts the amplitude and voltage-dependence of CaV1.1e to that of CaV1.1a. R1 neutralization in CaV1.1e (CaV1.1e-R1A) elicits an even further right-shifted voltage dependence compared with CaV1.1a. In contrast, R0 neutralization of CaV1.1e (CaV1.1e-R0A) shifts the voltage dependence of CaV1.1e by only ∼7 mV to the right and slightly increases the current amplitude.
(D–F) Neutralizing R1 and R2 charges in CaV1.1a (CaV1.1a-R1A, black and CaV1.1a-R2A, orange) does not affect the voltage dependence, but results in reduced calcium current amplitude. All data are presented as means ± SEM. The recording conditions and analysis were identical to those described in the legend of Figure 4. The calcium current parameters and statistics are given in Table 2.
Our structural models further predicted that, in the intermediate state 2, R1 forms more H-bonds with D4 in CaV1.1e, but not in CaV1.1a, and that this interaction is maintained in the activated state. Thus, the R1-D4 interaction might facilitate the final transition toward activation in CaV1.1e and then contribute to the stabilization of the activated state. Consistent with its importance for voltage sensing, mutating R1 in CaV1.1e (CaV1.1e-R1A) caused a substantial right shift (35 mV) of the voltage dependence of activation to even higher voltages than CaV1.1a (Figures 5A–5C). Also, the CaV1.1e-R1A channel shows a decreased current amplitude compared with CaV1.1e. However, CaV1.1e-R1A currents are still higher than in CaV1.1a and about half of the decline can be explained by the decreased driving force at the higher activation voltages. Overall, neutralizing R1 identifies the R1-D3 interaction as a major determinant of the voltage sensitivity of CaV1.1e and reveals a differential role of this interaction in determining the voltage dependence and current amplitude of CaV1.1.
Finally, our structural modeling indicated that, in the activated state, R2 forms a greater number of H-bonds with D4 in CaV1.1e compared with CaV1.1a. These H-bonds may facilitate the transition to the open state and stabilize the voltage sensor in the activated state, thus shifting the voltage sensitivity toward lower potentials and increasing the amplitude. Indeed, mutation of R2 in CaV1.1e (CaV1.1e-R2A, Figures 5A–5C) reduced the current density and right shifted the voltage dependence by 30 mV adopting current properties identical to those of CaV1.1a, confirming the importance of the R2-D4 interaction for determining the gating properties of CaV1.1e.
If R1A and R2A mutations in CaV1.1e reverse the facilitating effects of exon 29 deletion, then the corresponding mutations in CaV1.1a should not have an additional effect on its current-voltage dependence. Indeed, R1A and R2A mutations in CaV1.1a (Figures 5D–5F) did not affect the voltage dependence of activation. This also excludes the possibility that the effects observed with the corresponding mutations in CaV1.1e simply resulted from the reduced number of total charges in the voltage sensor. Only if the voltage sensitivity of the CaV1.1a R1A mutation had also been right shifted compared with the wild-type would this have been an indication that R1 interactions with other counter charges besides D4 were responsible for the voltage sensitivity. But this was not the case.
As it stands, the strikingly similar effects of mutating D4, R1, or R2 identify these oppositely charged residues in IVS3 and IVS4 as interaction partners that determine the ~26-mV leftward shift in the voltage sensitivity and the high current amplitude of CaV1.1e splice variant. Thus, structure modeling and mutagenesis experiments indicate that inclusion of exon 29 in the IVS3-S4 linker of CaV1.1a perturbs interactions between D4 in IVS3 and the two outer IVS4 arginines R1 and R2. This possibly increases the energy necessary for the transition between intermediate state 2 and the activated state and reduces the stability of the activated state.
Discussion
Here we used structural modeling, site-directed mutagenesis and electrophysiology in a native cell system to identify amino acid interactions responsible for charge transfer in the CaV channel VSD, which critically determine the gating properties of CaV channels. Our study elucidates the role of the extracellular IVS3-S4 linker on the gating mechanism responsible for the dramatic difference in the voltage sensitivity and current amplitude of two naturally occurring CaV1.1 channel splice variants. The gating behavior of the long CaV1.1a splice variant has been a conundrum in the ion channel field for over three decades, because the voltage dependence of its gating currents and of excitation-contraction coupling occurs at about 30 mV lower voltage than the voltage sensitivity of its ionic conductance. Several previous studies demonstrated that the specific slow speed of CaV1.1 current activation is determined by the S3 segment plus S3-S4 linker in the first repeat (Nakai et al., 1994; Tanabe et al., 1991); nevertheless, the mechanism responsible for the right-shifted voltage sensitivity of CaV1.1a ionic current has not been identified. The characterization of the embryonic CaV1.1e splice variant (Tuluc et al., 2009), which exhibits the same voltage sensitivity as gating currents, excitation-contraction coupling, and ionic conductance, indicated that the limiting voltage-sensitive step required for CaV1.1 channel gating occurs in the VSD of the fourth repeat. Therefore, the naturally occurring CaV1.1 splice variants represent an excellent model to study the voltage sensor transitions responsible for the differential voltage sensitivities of CaV channels.
How does the inclusion or exclusion of 19 amino acids encoded by exon 29 in the IVS3-S4 linker cause the striking differences in voltage sensitivity of the two CaV1.1 splice variants? Conceptually, a very short IVS3-S4 linker in CaV1.1e could lock this voltage sensor in the activated position, and consequently the voltage sensitivity of the channel would be determined by the other three voltage sensors. However, immobilizing one of the four voltage sensors would be expected to result in a measurable reduction of the gating charge currents (QON) in CaV1.1e compared with CaV1.1a. This has not been observed in our previous study where the voltage dependence and maximum amplitude of the gating charge currents (QON) were found to be identical for both splice variants, while the expression level (determined by RyR calcium release) was equal (Tuluc et al., 2009). Furthermore, here we show that neutralization of R1, R2, or D4 in the fourth VSD of CaV1.1e similarly right shifted the voltage dependence of current activation, which is inconsistent with a model assuming a locked fourth voltage sensor.
Alternatively, our structural models of the long IVS3-S4 linker indicate a substantial conformational change between the intermediate state 2 and the activated state. Since voltage-sensing transitions are expected to be very fast, the movement of the extra 19 amino acids encoded by exon 29 away from the outer vestibule could affect the response speed of this voltage sensor and therefore the current activation kinetics. Indeed, in our previous study, we report that CaV1.1e currents activate faster than CaV1.1a currents (Tuluc et al., 2009). Although we cannot exclude that the structural rearrangement of the IVS3-S4 linker might also increase the energy required to gate this VSD of CaV1.1a, our experimental finding that mutations of D4 (D4N and D4E) in CaV1.1e right shifts the voltage dependence in the absence of exon 29 contradicts this possibility.
Another possibility is that inclusion of exon 29 in the IVS3-S4 linker causes displacement of IVS3 and IVS4 relative to each other and consequently alters the intramolecular interactions required for the transfer of the IVS4 gating charges upon depolarization. Indeed our structural models of the fourth VSD of CaV1.1a and CaV1.1e reveal differences in the relative positioning of IVS2, IVS3, and IVS4. These differences dramatically affect H-bond formation of several IVS4 arginines, particularly in intermediate state 2 and the activated state. Our experimental results showing that mutations of R1 and R2 in IVS4 or of the pivotal aspartate in IVS3 (D4) of CaV1.1e revert voltage sensitivity and current amplitude to values similar to those found in CaV1.1a strongly support this model. In NaV and Kv channels, negatively charged residues in the transmembrane helices S2 (E2) and S3 (D3) and a non-polar residue in S2 (F) form the CTC, which supports the voltage-dependent transition of the positively charged residues in S4 (Payandeh et al., 2011, 2012; Tao et al., 2010). Here we show that the corresponding amino acids in the fourth VSD of CaV1.1 channels appear to serve the same function but are not much affected by the inclusion of exon 29. Instead we identified a previously unnoticed negatively charged amino acid in IVS3 (D1196), which is essential for proper gating of the CaV1.1e channel. Our structural models predict that during the gating process D1196 sequentially interacts with R1 and R2, and that these transitions are dramatically altered by inclusion of exon 29 in CaV1.1a. Our experimental analysis demonstrates that mutation of D1196 to asparagine or glutamate in CaV1.1e reverts both the voltage sensitivity and the current density to that of CaV1.1a. Thus, D1196 is the key determinant of the dramatically different gating properties of the two CaV1.1 splice variants. Moreover, mutational analysis of the IVS4 arginines confirms the importance of D1196 interactions with R1 and R2 in determining the leftward shift by 26 mV of CaV1.1e voltage sensitivity. Thus, in CaV1.1 channels, an additional charged residue in IVS3 contributes to S4 charge compensation, and deviations of its relative position to IVS4 accomplish dramatic changes of the channel’s voltage sensitivity and current amplitude.
Our experimental data support a model according to which the interactions of R1 and R2 with D4 contribute to the final steps of the charge transfer to different degrees in CaV1.1a and CaV1.1e. In the intermediate state 2 of CaV1.1a, both R1 and R2 form H-bonds with a single residue in IVS2 (T). Although these interactions might stabilize this state, our experimental data showing that neutralization of R1 or R2 does not improve CaV1.1a gating do not support this notion. In contrast, in the intermediate state 2 of CaV1.1e, the R1-D4 interaction (Figure 3C) most probably facilitates the final transition to the activated state as the mutation of R1 in CaV1.1e shifts its voltage sensitivity to the right to potentials even higher than CaV1.1a. The notion that CaV1.1a R1 and D4 do not interact is supported by our data showing that, in CaV1.1a, neutralization of R1 or D4 does not alter the voltage dependence. In the activated state, R1 and R2 of CaV1.1a form H-bonds with three separate IVS3 residues, whereas, in CaV1.1e, both R1 and R2 form multiple H-bonds with D4 (Figure 3D). This convergence on a single residue most probably stabilizes the activated state resulting in the observed higher current density of CaV1.1e. Our experimental data showing that, in the absence of the R1-D4 interactions in CaV1.1e-R1A, the current amplitude is substantially higher than that of CaV1.1a (Figure 5B) confirms that the stronger R2-D4 interaction in CaV1.1e significantly contributes to the stability of the open state.
Our current and previous observation that the deletion of exon 29 dramatically alters the voltage dependence and amplitude of CaV1.1a calcium currents demonstrates that voltage-sensitive transitions in the fourth repeat VSD are necessary for CaV1.1 channel opening, but are not sufficient. As has been previously shown, our data also support the idea that gating of the calcium channel requires different contributions from all VSDs (Beyl et al., 2012; Dirksen et al., 1997; Garcia et al., 1997; Hamid et al., 2006; Hohaus et al., 2005; Kudrnac et al., 2009; Li et al., 2004; Nakai et al., 1994; Pantazis et al., 2014; Tanabe et al., 1991; Wall-Lacelle et al., 2011). Recently published voltage-clamp fluorometry experiments demonstrate that, in the CaV1.2 channel, VSD II and III control the channel opening with no apparent contribution of the fourth VSD (Pantazis et al., 2014). Nevertheless, this does not exclude the possibility that alternative splicing in the fourth VSD of CaV1.2 would change the contribution of this VSD to channel gating. Our data support a gating model according to which the transitions of the first three VSDs and the early transitions of the fourth VSD are similar in both CaV1.1 variants, but, in the CaV1.1a variant, the final transitions of the fourth VSD require more energy and thus determine the voltage sensitivity of channel activation. The voltage dependence of channel gating in the CaV1.1e variant is probably determined by the other VSDs similarly to the CaV1.2 channel previously described (Pantazis et al., 2014). The quantification of the slope factor (kact see Table 2) of the Bolzmann fit to the voltage dependence of the conductance (Bezanilla, 2000; Noceti et al., 1996) also supports our model that the critical difference between CaV1.1 variants is the final gating of the fourth VSD, because the slope factor of CaV1.1a mirrors the movement of ~3.2 charges, whereas the slope factor for CaV1.1e channel mirrors the movement of ~5.4 charges. Previous evidence supports a cooperative gating model for CaV channels according to which the VSDs have different contributions to channel opening and affect each other (Beyl et al., 2012; Demers-Giroux et al., 2013; Pantazis et al., 2014). Whether the increased number of charges responsible for gating in CaV1.1e reflect the biophysical changes in the fourth VSD alone or altered gating of the other VSDs remains to be further studied.
Together our structural modeling and mutagenesis data identified a molecular mechanism in the fourth VSD of CaV1.1 channels that explains the specific current amplitude and voltage sensitivity of this L-type CaV channel. The molecular mechanism hinges on the newly discovered interaction between D4 and two IVS4 arginines, and it is exquisitely sensitive to structural rearrangements between the IVS3 and IVS4 helices imposed by alternative splicing of the IVS3-S4 linker. Importantly, the D4 aspartate is highly conserved in the fourth VSD of all high voltage-activated CaV channels (Figure S1 related to Figure 1). Because all but one CACNA1 gene encoding CaV channels also contain a physiologically relevant alternative splicing site in the extracellular IVS3-S4 linker of this domain (Lipscombe et al., 2013), the molecular mechanism described here may be relevant for fine-tuning the voltage sensitivity throughout the CaV channel family.
Experimental Procedures
Structural Modeling
Homology, de novo, and full-atom modeling of the CaV1.1e and CaV1.1a VSDs was performed using the Rosetta-Membrane method (Yarov-Yarovoy et al., 2006a, 2006b, 2012) and NaVAb VSD structure (Payandeh et al., 2011) as a template. For modeling of resting and intermediate states, we used multiple alignments between S4 segments of CaV1.1e or CaV1.1a and NaVAb guided by experimental data on specific pairwise interactions and positions of S4 arginines in different states of KV and NaV VSDs (Broomand and Elinder, 2008; Campos et al., 2007; DeCaen et al., 2008, 2009, 2011; Henrion et al., 2012; Lacroix et al., 2014; Pathak et al., 2007; Starace and Bezanilla, 2004; Tombola et al., 2005; Vargas et al., 2012; Yarov-Yarovoy et al., 2012). The S4 alignment where R1 in CaV1.1 variants is aligned to R1 in NaVAb represents the activated state. The intermediate state 2, intermediate state 1, and resting state were modeled by aligning R1 in CaV1.1 variants to NaVAb R2, R3, and R4 respectively. In the resting state, the CTC is occupied by R1 (CaV1.1a-R1236; CaV1.1e-R1217); in the intermediate state 1 by R2 (CaV1.1a-R1239; CaV1.1e-R1220), and in the intermediate state 2 by R3 (R1242-CaV1.1a; CaV1.1e-R1223). In the activated state, K4 (CaV1.1a-K1245; CaV1.1e-K1226) occupies the CTC. The S1-S2, S2-S3, and S3-S4 loops were built using Rosetta cyclic coordinate descent and kinematic closure loop modeling applications (Mandell et al., 2009; Wang et al., 2007), and guided by membrane environment specific energy function (Barth et al., 2007; Yarov-Yarovoy et al., 2012). A total of 10,000 models were generated for each state and ranked by total Rosetta score (Barth et al., 2007; Rohl et al., 2004; Yarov-Yarovoy et al., 2012). The top 10% of best scoring models are clustered (Bonneau et al., 2002) using the root mean square deviation threshold, which generates at least 150–200 models in the largest cluster. Models representing centers of the top 20 clusters were used as inputs for full-atom relax. Models representing five largest clusters for each of the VSD states were chosen for further visual evaluation and analysis. All structural figures in the article were generated using UCSF Chimera (Pettersen et al., 2004).
Mutagenesis and Electrophysiology
The point mutations were introduced using splicing by overhang extension PCR into the GFP-α1S or GFP-α1S-ΔE29 plasmids previously described (Tuluc et al., 2009). All mutants were verified by sequencing performed by Eurofins Genomis. The plasmids were transfected in myotubes of the dysgenic (mdg/mdg) cell line GLT using FuGene transfection reagent (Roche Diagnostics) (Tuluc et al., 2007). This restores the normal composition of the channel with auxiliary α2δ-1, β1a, and γ1 subunits enabling recording of the otherwise undetectable CaV1.1a currents (Flucher et al., 2005; Rios and Brum, 1987; Tanabe et al., 1988). Calcium currents were recorded with the ruptured whole-cell patch-clamp technique in voltage-clamp mode using an Axopatch 200B amplifier (Axon Instruments). Patch pipettes (borosilicate glass, Harvard Apparatus) had a resistance of 1.5–3 MΩ when filled with 145 mM Cs-aspartate, 2 mM MgCl2, 10 mM HEPES, 0.1 mM Cs-EGTA, 2 mM Mg-ATP (pH 7.4 with CsOH). The bath solution contained 10 mM CaCl2, 145 mM tetra-ethyl ammonium chloride, 10 mM HEPES (pH 7.4 with tetra-ethyl ammonium hydroxide). Data acquisition and command potentials were controlled by pClamp software (version 8.0; Axon Instruments); analysis was performed using Clampfit 8.0 (Axon Instruments) and SigmaPlot 8.0 (SPSS Science) software. The current-voltage dependence was fitted according to: I = Gmax·(V − Vrev)/1(1 + exp(−(V − V1/2)/k)). The conductance was calculated using G = (−I ﹡ 1000)/(V − Vrev) and its voltage dependence was fitted according to a Boltzmann distribution: G = Gmax/(1 + exp(−(V − V1/2)/kact)), where Gmax is the maximum conductance of the L-type calcium currents, Vrev is the extrapolated reversal potential of the calcium current, V1/2 is the potential for half maximal conductance, and kact is the slope. The statistical significance was calculated using the Mann-Whitney U test, and the values are present in Table 2.
Supplementary Material
Supplemental Information includes two figures, one table, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.str.2015.11.011.
Highlights.
S3-S4 linker lengths in CaV channels control their voltage sensitivity
Rosetta modeling predicts that the S3-S4 linker determines the charge transfer
Mutagenesis has identified a new counter charge in S3 of CaV critical for gating
Acknowledgments
We thank A. Benedetti, R. Egger, and Dr. M. Campiglio for excellent technical support. This study was funded by grants from the Austrian Science Fund (FWF) P23479 and F4406 to B.E.F., and from the University of Innsbruck P7400-027-011 and P7400-027-012 to P.T.
Footnotes
Author Contributions
Conceptualization, P.T. and B.E.F.; methodology, P.T. and V.Y-Y.; investigation, P.T., V.Y.-Y., B.B., and B.E.F.; formal analysis, P.T. and B.B.; writing the original draft, P.T., V.Y.-Y., B.B., and B.E.F.; Writing, review, and editing, P.T. and B.E.F.; visualization, P.T. and B.B.; funding acquisition, P.T. and B.E.F.
References
- Ahern CA, Horn R. Stirring up controversy with a voltage sensor paddle. Trends Neurosci. 2004;27:303–307. doi: 10.1016/j.tins.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Barth P, Schonbrun J, Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc Natl Acad Sci USA. 2007;104:15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyl S, Depil K, Hohaus A, Stary-Weinzinger A, Linder T, Timin E, Hering S. Neutralisation of a single voltage sensor affects gating determinants in all four pore-forming S6 segments of Ca(V)1.2: a cooperative gating model. Pflugers Arch. 2012;464:391–401. doi: 10.1007/s00424-012-1144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F. The voltage sensor in voltage-dependent ion channels. Physiol Rev. 2000;80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- Bezanilla F, Perozo E, Papazian DM, Stefani E. Molecular basis of gating charge immobilization in Shaker potassium channels. Science. 1991;254:679–683. doi: 10.1126/science.1948047. [DOI] [PubMed] [Google Scholar]
- Bonneau R, Strauss CE, Rohl CA, Chivian D, Bradley P, Malmstrom L, Robertson T, Baker D. De novo prediction of three-dimensional structures for major protein families. J Mol Biol. 2002;322:65–78. doi: 10.1016/s0022-2836(02)00698-8. [DOI] [PubMed] [Google Scholar]
- Broomand A, Elinder F. Large-scale movement within the voltage-sensor paddle of a potassium channel-support for a helical-screw motion. Neuron. 2008;59:770–777. doi: 10.1016/j.neuron.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Campos FV, Chanda B, Roux B, Bezanilla F. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc Natl Acad Sci USA. 2007;104:7904–7909. doi: 10.1073/pnas.0702638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron. 2010;67:915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA. Voltage-gated calcium channels. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003947. a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, Yarov-Yarovoy V. Helical motion of an S4 voltage sensor revealed by gating pore currents. Channels (Austin) 2010;4:75–77. doi: 10.4161/chan.4.2.10998. [DOI] [PubMed] [Google Scholar]
- Cheng YM, Hull CM, Niven CM, Qi J, Allard CR, Claydon TW. Functional interactions of voltage sensor charges with an S2 hydrophobic plug in hERG channels. J Gen Physiol. 2013;142:289–303. doi: 10.1085/jgp.201310992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc Natl Acad Sci USA. 2008;105:15142–15147. doi: 10.1073/pnas.0806486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc Natl Acad Sci USA. 2009;106:22498–22503. doi: 10.1073/pnas.0912307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen PG, Yarov-Yarovoy V, Scheuer T, Catterall WA. Gating charge interactions with the S1 segment during activation of a Na+ channel voltage sensor. Proc Natl Acad Sci USA. 2011;108:18825–18830. doi: 10.1073/pnas.1116449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers-Giroux PO, Bourdin B, Sauve R, Parent L. Cooperative activation of the T-type CaV3.2 channel: interaction between domains II and III. J Biol Chem. 2013;288:29281–29293. doi: 10.1074/jbc.M113.500975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen RT, Nakai J, Gonzalez A, Imoto K, Beam KG. The S5-S6 linker of repeat I is a critical determinant of L-type Ca2+ channel conductance. Biophys J. 1997;73:1402–1409. doi: 10.1016/S0006-3495(97)78172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flucher BE, Obermair GJ, Tuluc P, Schredelseker J, Kern G, Grabner M. The role of auxiliary dihydropyridine receptor subunits in muscle. J Muscle Res Cell Motil. 2005;26:1–6. doi: 10.1007/s10974-005-9000-2. [DOI] [PubMed] [Google Scholar]
- Flucher BE, Tuluc P. A new L-type calcium channel isoform required for normal patterning of the developing neuromuscular junction. Channels (Austin) 2011;5:518–524. doi: 10.4161/chan.5.6.17951. [DOI] [PubMed] [Google Scholar]
- Garcia J, Nakai J, Imoto K, Beam KG. Role of S4 segments and the leucine heptad motif in the activation of an L-type calcium channel. Biophys J. 1997;72:2515–2523. doi: 10.1016/S0006-3495(97)78896-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid J, Peloquin JB, Monteil A, Zamponi GW. Determinants of the differential gating properties of Cav3.1 and Cav3.3 T-type channels: a role of domain IV? Neuroscience. 2006;143:717–728. doi: 10.1016/j.neuroscience.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Henrion U, Renhorn J, Borjesson SI, Nelson EM, Schwaiger CS, Bjelkmar P, Wallner B, Lindahl E, Elinder F. Tracking a complete voltage-sensor cycle with metal-ion bridges. Proc Natl Acad Sci USA. 2012;109:8552–8557. doi: 10.1073/pnas.1116938109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohaus A, Beyl S, Kudrnac M, Berjukow S, Timin EN, Marksteiner R, Maw MA, Hering S. Structural determinants of L-type channel activation in segment IIS6 revealed by a retinal disorder. J Biol Chem. 2005;280:38471–38477. doi: 10.1074/jbc.M507013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontis KJ, Goldin AL. Sodium channel inactivation is altered by substitution of voltage sensor positive charges. J Gen Physiol. 1997;110:403–413. doi: 10.1085/jgp.110.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudrnac M, Beyl S, Hohaus A, Stary A, Peterbauer T, Timin E, Hering S. Coupled and independent contributions of residues in IS6 and IIS6 to activation gating of CaV1.2. J Biol Chem. 2009;284:12276–12284. doi: 10.1074/jbc.M808402200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix JJ, Hyde HC, Campos FV, Bezanilla F. Moving gating charges through the gating pore in a Kv channel voltage sensor. Proc Natl Acad Sci USA. 2014;111:E1950–E1959. doi: 10.1073/pnas.1406161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Stevens L, Klugbauer N, Wray D. Roles of molecular regions in determining differences between voltage dependence of activation of CaV3.1 and CaV1.2 calcium channels. J Biol Chem. 2004;279:26858–26867. doi: 10.1074/jbc.M313981200. [DOI] [PubMed] [Google Scholar]
- Liao P, Yong TF, Liang MC, Yue DT, Soong TW. Splicing for alternative structures of Cav1.2 Ca2+ channels in cardiac and smooth muscles. Cardiovasc Res. 2005;68:197–203. doi: 10.1016/j.cardiores.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Lin MC, Hsieh JY, Mock AF, Papazian DM. R1 in the Shaker S4 occupies the gating charge transfer center in the resting state. J Gen Physiol. 2011;138:155–163. doi: 10.1085/jgp.201110642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D, Andrade A, Allen SE. Alternative splicing: functional diversity among voltage-gated calcium channels and behavioral consequences. Biochim Biophys Acta. 2013;1828:1522–1529. doi: 10.1016/j.bbamem.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Lou XJ, Horrigan FT. Role of charged residues in the S1-S4 voltage sensor of BK channels. J Gen Physiol. 2006;127:309–328. doi: 10.1085/jgp.200509421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J, Adams BA, Imoto K, Beam KG. Critical roles of the S3 segment and S3-S4 linker of repeat I in activation of L-type calcium channels. Proc Natl Acad Sci USA. 1994;91:1014–1018. doi: 10.1073/pnas.91.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noceti F, Baldelli P, Wei X, Qin N, Toro L, Birnbaumer L, Stefani E. Effective gating charges per channel in voltage-dependent K+ and Ca2+ channels. J Gen Physiol. 1996;108:143–155. doi: 10.1085/jgp.108.3.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazis A, Savalli N, Sigg D, Neely A, Olcese R. Functional heterogeneity of the four voltage sensors of a human L-type calcium channel. Proc Natl Acad Sci USA. 2014;111:18381–18386. doi: 10.1073/pnas.1411127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian DM, Timpe LC, Jan YN, Jan LY. Alteration of voltage-dependence of Shaker potassium channel by mutations in the S4 sequence. Nature. 1991;349:305–310. doi: 10.1038/349305a0. [DOI] [PubMed] [Google Scholar]
- Pathak MM, Yarov-Yarovoy V, Agarwal G, Roux B, Barth P, Kohout S, Tombola F, Isacoff EY. Closing in on the resting state of the shaker k(+) channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pless SA, Galpin JD, Niciforovic AP, Ahern CA. Contributions of counter-charge in a potassium channel voltage-sensor domain. Nat Chem Biol. 2011;7:617–623. doi: 10.1038/nchembio.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless SA, Elstone FD, Niciforovic AP, Galpin JD, Yang R, Kurata HT, Ahern CA. Asymmetric functional contributions of acidic and aromatic side chains in sodium channel voltage-sensor domains. J Gen Physiol. 2014;143:645–656. doi: 10.1085/jgp.201311036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios E, Brum G. Involvement of dihydropyridine receptors in excitation-contraction coupling in skeletal muscle. Nature. 1987;325:717–720. doi: 10.1038/325717a0. [DOI] [PubMed] [Google Scholar]
- Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- Tanabe T, Adams BA, Numa S, Beam KG. Repeat I of the dihydropyridine receptor is critical in determining calcium channel activation kinetics. Nature. 1991;352:800–803. doi: 10.1038/352800a0. [DOI] [PubMed] [Google Scholar]
- Tanabe T, Beam KG, Powell JA, Numa S. Restoration of excitation-contraction coupling and slow calcium current in dysgenic muscle by dihydropyridine receptor complementary DNA. Nature. 1988;336:134–139. doi: 10.1038/336134a0. [DOI] [PubMed] [Google Scholar]
- Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F, Pathak MM, Isacoff EY. Voltage-sensing arginines in a potassium channel permeate and occlude cation-selective pores. Neuron. 2005;45:379–388. doi: 10.1016/j.neuron.2004.12.047. [DOI] [PubMed] [Google Scholar]
- Tuluc P, Flucher BE. Divergent biophysical properties, gating mechanisms, and possible functions of the two skeletal muscle Ca(V)1.1 calcium channel splice variants. J Muscle Res Cell Motil. 2011;32:249–256. doi: 10.1007/s10974-011-9270-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuluc P, Kern G, Obermair GJ, Flucher BE. Computer modeling of siRNA knockdown effects indicates an essential role of the Ca2+ channel alpha2delta-1 subunit in cardiac excitation-contraction coupling. Proc Natl Acad Sci USA. 2007;104:11091–11096. doi: 10.1073/pnas.0700577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuluc P, Molenda N, Schlick B, Obermair GJ, Flucher BE, Jurkat-Rott K. A CaV1.1 Ca2+ channel splice variant with high conductance and voltage-sensitivity alters EC coupling in developing skeletal muscle. Biophys J. 2009;96:35–44. doi: 10.1016/j.bpj.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas E, Yarov-Yarovoy V, Khalili-Araghi F, Catterall WA, Klein ML, Tarek M, Lindahl E, Schulten K, Perozo E, Bezanilla F, et al. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J Gen Physiol. 2012;140:587–594. doi: 10.1085/jgp.201210873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall-Lacelle S, Hossain MI, Sauve R, Blunck R, Parent L. Double mutant cycle analysis identified a critical leucine residue in the IIS4S5 linker for the activation of the Ca(V)2.3 calcium channel. J Biol Chem. 2011;286:27197–27205. doi: 10.1074/jbc.M111.237412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Baker D, Catterall WA. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K(+) channels. Proc Natl Acad Sci USA. 2006a;103:7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006b;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Decaen PG, Westenbroek RE, Pan CY, Scheuer T, Baker D, Catterall WA. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci USA. 2012;109:E93–E102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.