Abstract
Cleavage factor “CFIm25”, as a key repressor at proximal poly (A) site, negatively correlates to cell proliferation and tumorigenicity in various cancers. Hence, understanding CFIm25 mechanism of action in breast cancer would be a great benefit. To this aim four steps were designed. First, potential miRNAs that target 3′-UTR of CFIm25 mRNA, retrieved from Targetscan web server. Second, screened miRNAs were profiled in 100 breast cancer and 100 normal adjacent samples. Third, miRNAs that their expression was inversely correlated to the CFIm25, overexpressed in MDA-MB-231 cell line, and their effect on proliferation and migration monitored via MTT and wound healing assays, respectively. Fourth, interaction of miRNAs of interest with 3′-UTR of CFIm25 confirmed via luciferase assay and western blot. Our results indicate that CFIm25 considerably down-regulates in human breast cancer tissue. qRT-PCR assay, luciferase test, and western blotting confirm that CFIm25 itself could be directly regulated by oncomiRs such as miR-23, -24, -27, -135, -182 and -374. Besides, according to MTT and wound healing assays of cell lines, CFIm25 knockdown intensifies cell growth, proliferation and migration. Our results also confirm indirect impact of CFIm25 on regulation of mRNA’s 3′–UTR length, which then control corresponding miRNAs’ action. miRNAs directly control CFIm25 expression level, which then tunes expression of the oncogenes and tumor proliferation. Therefore, regulation of CFIm25 expression level via miRNAs is expected to improve treatment responses in breast cancer.
Subject terms: Cancer, Cell biology, Computational biology and bioinformatics, Genetics
Introduction
Living organisms apply complicated mechanisms of gene regulation to generate different cell types, which result in various behaviors from a single genome1,2. Dynamic and highly polymorphic nature of mRNA polyadenylation, as a functional aspect of gene regulation, is one of the most recent discoveries in this field3,4. Cleavage and polyadenylation of nascent mRNA accomplishes via application of two large multimeric complexes, which named as cleavage–polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF), respectively. The CPSF recognizes polyadenylation site (PAS) element located ~ 19 nucleotides away from the polyA site in the 3′-UTR of mRNAs, while CstF directly interacts with CPSF and binds to GU-rich element at downstream of cleavage site5–7. Most human genes harbor multiple polyadenylation signals, which results in variable 3′-UTR length without changing the coding region, or produce different protein isoforms through the usage of poly(A) sites residing in internal introns/exons3,8–10.
The UTR-alternative polyadenylation1 site play critical roles in various biological networks through changes that occur in 3′-UTR length. For example, UTR-APA could influence the stability, translation efficiency and subcellular localization of mRNA. The UTR-APA also regulates protein transport by altering interaction of RNA binding proteins with the 3′-UTR. Therefore, UTR-APA site quantitatively affects corresponding protein expression through a complicated mechanism10,11. UTR-APA site controls length of the 3′-UTR, which then controls miRNAs’ binding sites availability at the 3′-UTR. The choice of poly (A) site can be influenced by a key factor in APA, which is human cleavage factor Im (CFIm)12.
The CFIm is an essential component in the pre-mRNA 3′-UTR processing complex. It contains three polypeptides of 25, 59 and 68 kDa that are designated as CFIm25 (CPSF5/NUDT21), CFIm59 (CPSF7, cleavage and polyadenylation specificity factor 7), and CFIm68 (or CPSF6). Diminution of CFIm25 level induces a global recruitment of weak polyA, placed completely proximal to the stop codon, while enhances cell proliferation. Previous studies confirmed down-regulation of CFIm25 in tumor tissue compared to the normal tissue. Besides, diminution of CFIm25 level comes up with shorter length of 3′-UTR in various oncogenes and induces proliferation of tumor cells13–15. However, mechanism that regulates CFIm25 expression, which then correlates to the occurrence and development of cancer, remains unknown.
miRNAs are tiny non-coding RNAs that are involved in post-transcriptional gene regulation. They target cognate mRNAs contain complementary sequences in their 3′-UTR, which either prevents mRNA translation or induce its degradation16–18. Numerous miRNAs have been shown to be aberrantly expressed, and act as oncogenes in several types of cancer such as breast cancer19–23. Besides, miRNAs mimic tumor suppressor roles in many cases. That’s why loss of miRNA expression correlates with inception or the aggressiveness of tumors24,25.
In this study, we checked if miRNAs are involved with regulation of CFIm25 expression. To examine regulatory impact of miRNAs on endogenously APA master regulator CFIm25, correlation of oncogenic miRNAs with CFIm25 investigated in clinical samples of breast cancer patients. Then, we explored impact of transfected miRNAs in breast cancer cell line, in a set of parallel separate experiments. Identification of numerous oncomiRs that suppress CFIm25, which then induce proximal polyadenylation and mediate in production of shortened oncogenes in breast cancer, confirms involvement of miRNAs in regulation of CFIm25.
Results
Correlation of CFIm25 expression level with oncogenic miRNAs in breast cancer clinical samples and cell lines
According to bioinformatics analysis, miR-23, miR-24, miR-27, miR-96, miR-135, miR-182, miR-221, miR-222 and miR-374 found to target CFIm25 beside their oncogenic potential. In order to determine whether CFIm25 expression is controlled by miRNAs of interest in breast cancer, we measured the expression levels of potentially involved miRNAs in breast cancer patients (n = 100) and normal adjacent tissues (n = 100) using miR-quantitative qRT–PCR analysis. Results showed considerable elevated level of miRNAs in samples acquired from patients (at least in 6 out of 9 monitored miRNAs), while CFIm25 expression level was diminished significantly (Fig. 1a, b). In the next step, patients categorized as low or high CFIm25‑expressing tumors, using the median expression for CFIm25 in the normal sample as a cut-off.
Figure 1.
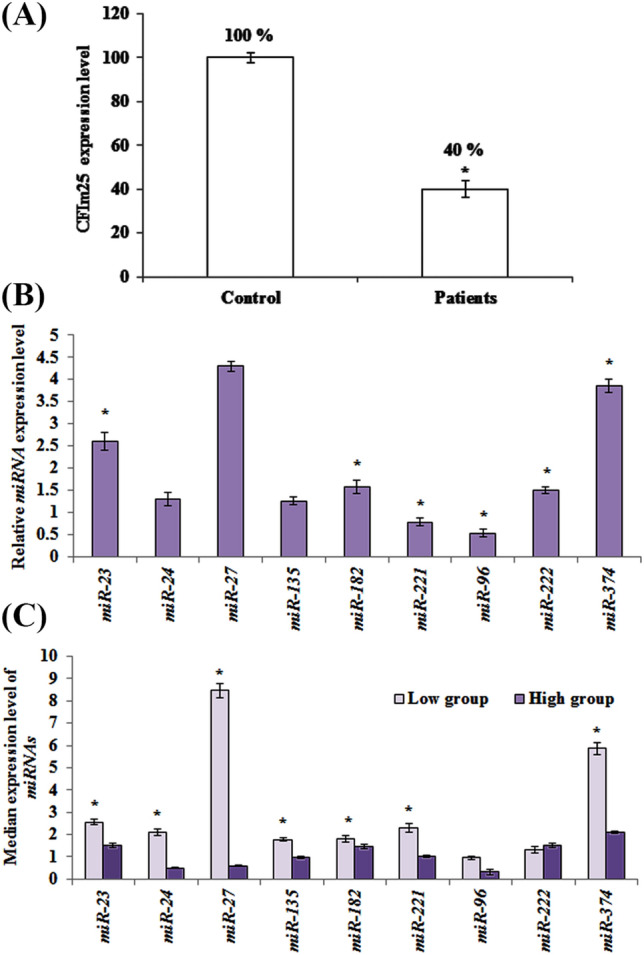
Comparative expression of CFIm25 vs involved miRNAs. (a) CFIm25 expression level in samples acquired from breast cancer patients and control group. CFIm25 is significantly lower in patients compared to controls. (b) Relative gene expression of miRNAs in patients. (c) Distribution of miRNA expression in patients according to low and high expression levels of CFIm25. Patients categorized as low or high CFIm25‑expressing tumors, using median expression of CFIm25 in normal samples as a cut-off. Data represent mean of at least three independent experiments. The star above the bars, represents significant difference respect to the corresponding control reference. The results are relative gene expression after normalization with β-actin for CFIm25 and SNORD47 for miRNA genes, using 2−ΔΔCt method.
Since CFIm25 level differs in samples acquired from patients, their correspondingly correlated miRNAs might be variable too. By this regard, distribution of the median expression of nine miRNAs of interest evaluated respect to both low and high levels of CFIm25 expression (Fig. 1c).
The median gene expression level of miR-23, miR-24, miR-27, miR-135, miR-182, miR-221 and miR-374 was significantly higher in the patients expressing lower levels of CFIm25, compared to those expressing higher levels (p < 0.05).
Intervention of CFIm25 in length determination of proto-oncogenes’ 3′-UTR
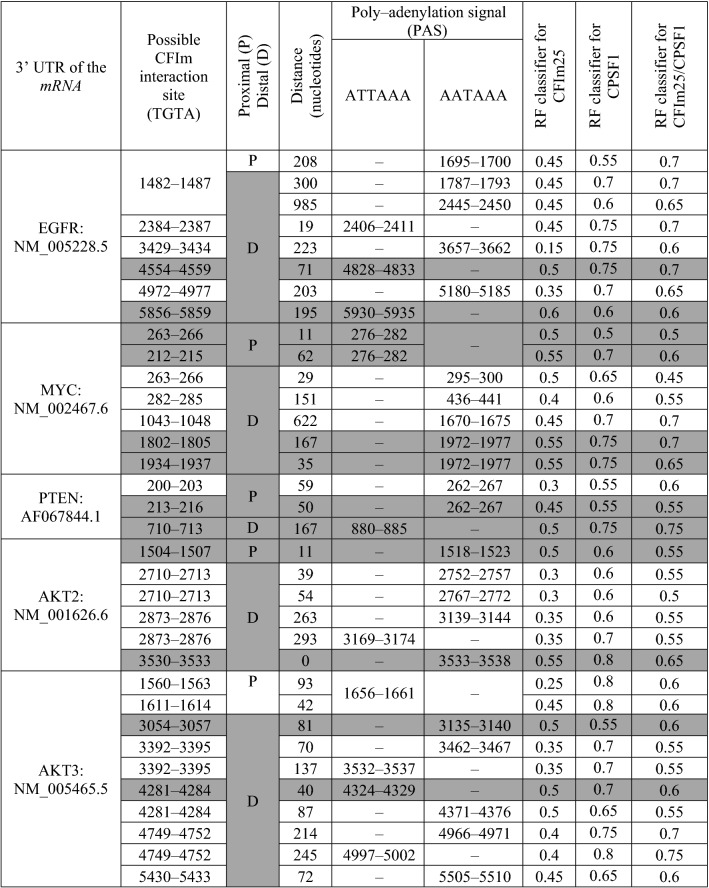
Three out of five proto-oncogenes interact efficiently with CFIm25 and CPSF1 either at proximal or distal of 3′-UTR while the other two proto-oncogenes (EGFR: NM_005228.5, AKT3: NM_005465.5) interact with both proteins merely at distal of the 3′-UTR (Table 1). Therefore, exclusive interaction of CFIm25/CPSF1 complex with distal region of EGFR and AKT3 3′-UTR coding mRNA, would guarantee suppression of breast cancer progression.
Table 1.
Proto-oncogenes that CFIm25 and CPSF1 target their 3′UTR mRNA.
The cases that are highlighted in dark grey represent proto-oncogenes with positive interaction probability. The RF (Random Forest) classifier is a probability index, which falls between 0 (the lowest probability) to 1 (the highest probability).
Some of the potentially involved miRNAs in breast cancer, down-regulate CFIm25 in MDA-MB-231 cell line
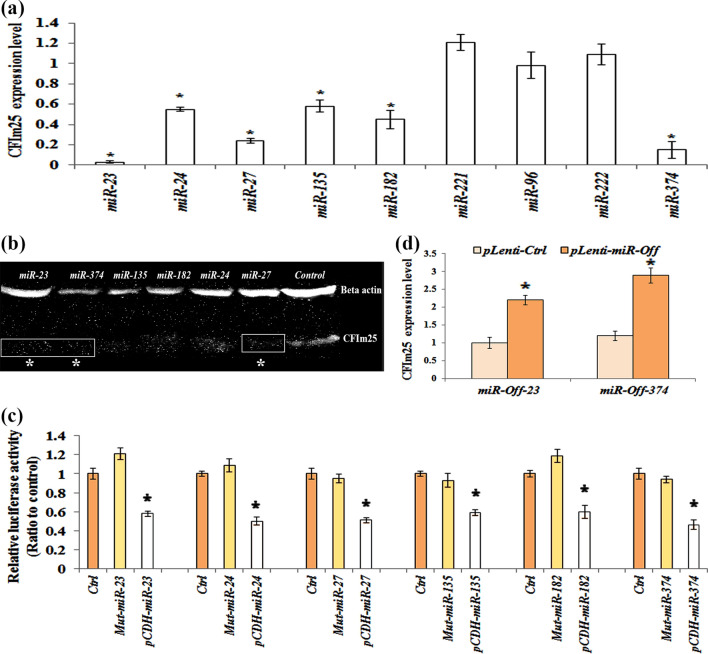
To confirm the impact of miRNAs on CFIm25 regulation in clinical samples, MDA-MB-231 stable cell lines were generated using miRNA containing lentivirus transduction. We determined that enforced expressions of miR-23, miR-24, miR-27, miR-135, miR-182 and miR-374 significantly down-regulated CFIm25 (p < 0.05), while made no change in cells that were transducted with Ctrl-vector, miR-96, miR-221 and miR-222 (Fig. 2a). Up-regulation of miR-23 and miR-374 down-regulated CFIm25 the most, compared to other miRNAs including miR-24, miR-27, miR-135 and miR-182 (Fig. 2a). According to image analysis of western blot gels via Image J software, CFIm25 protein level notably decreased while miR-23, 27, and 374 were up-regulated (Fig. 2b).
Figure 2.
Some of predicted miRNAs directly regulate CFIm25 expression. (a) CFIm25 expression level in cell line with enforced expressions of miRNAs of interest, monitored by Real-time PCR and (b) Western blot technique. The western blot bonds cropped from a same gel and membrane. The original image of the membrane and gel could be found in supplementary Figs. 1 and 2. (c) Luciferase assay of the HEK293 cells that are transiently transfected with CFIm25-pSICHECK2 construct and WT-miRNAs/mut-miRNAs. (d) Expression level of CFIm25 in cells transducted with either of miR-Off-23 or miR-Off-374 constructs. Protein normalization performed against β-actin expression. Columns are mean of three different experiments and error bars are standard deviation (p < 0.05). The star represents significant difference respect to the corresponding control reference.
In the next step, we investigated whether 3′-UTR of CFIm25 has functional targets for predicted miRNAs. To this aim, full-length sequence of CFIm25 3′-UTR cloned to psiCHECK-2 vector, which is placed at downstream of Renilla luciferase reporter gene (CFIm25-psiCHECK2). In a parallel experiment, as negative controls, the seed sequence of miR-23, miR-24, miR-27, miR-135, miR-182 and miR-374 mutated intentionally at certain points. The stem-loop structures that formed following introduction of mutations, cloned to pCDH-TurboGFP vector (Mut-miRNAs) in shRNA format. The HEK293 cells transiently transfected with the CFIm25-pSICHECK2 construct and WT-miRNAs, which led to a significant decrease of reporter activity compared to the control (Fig. 2c). Activity of the reporter construct was unaffected by a simultaneous transfection with Mut-miRNAs carried a mutated seed sequence. Taken together, these data strongly suggest that miR-23, miR-24, miR-27, miR-135, miR-182 and miR-374 directly bind to 3′-UTR of CFIm25’s mRNA.
According to our findings that suggested miR-23 and miR-347 had something to do with CFIm25 down-regulation, we used miR-Off-23 and miR-Off-374 to silence miR-23 and miR-374. Now, by using miR-Off-23 and miR-Off-374, we can confirm if silencing of miR-23 and miR-374 directly controls overexpression of CFIm25 in breast cancer’s cell line or not. To this aim, miR-Off-23 and miR-Off-374 transducted to MDA-MB-231 cell line and efficiency of transduction monitored using qRT–PCR analysis. Transduction of miR-Off-23 and miR-Off-374 resulted in significant overexpression of CFIm25 as demonstrated in Fig. 2d (by 3.3 and 2.89 fold increase in case of miR-Off-23 and miR-Off -374, respectively).
Selected miRNAs can regulate cell proliferation in breast cancer cell line partly by modulating CFIm25 mRNA
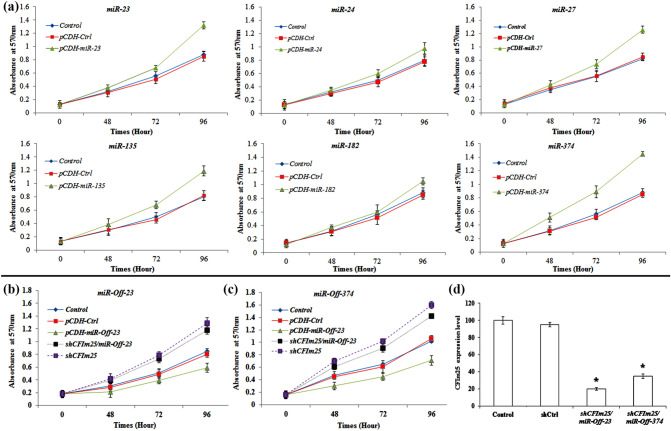
To determine importance of CFIm25 in regulation of MDA-MB-231 proliferation, we intentionally performed MTT and wound healing assays in miRNAs transduced MDA-MB-231 cells. Result of the MTT assay confirmed increment of “cell proliferation index” in transduced cells as compared with that of the control groups (p < 0.05; Fig. 3a).
Figure 3.
Viability test of MDA-MB-231 cells that were transducted with different cloned miRNAs. (a) MTT assay of MDA-MB-231 cells that are transducted with miRNAs of interest. MTT assay of transducted MDA-MB-231 cells by (b) miR-Off-23 and (c) miR-Off-374 that transfected by shCFIm25 vector. Each time point was expressed as total absorbance at 570 nm after background subtraction (Y axis). Points are mean of three experiments and error bars represent standard deviation. (d) CFIm25 expression level in shCFIm25 transfected cells. The star above the bar, represents the significant difference respect to the corresponding control reference.
To clarify direct impact of miRNAs on cell proliferation through regulation of 3′-UTR of CFIm25 mRNA, MDA-MB-231 cells once transduced with miR-Off-23/miR-Off-374 and assessed by MTT assay. In the second approach, transduced cells transfected by shCFIm25 vector and evaluated by MTT assay. When miR-Off-23 and miR-Off-374 transduced to cells and transiently co-transfected with shCFIm25 vector (Fig. 3b, c), CFIm25 expression level diminished and cell proliferation significantly increased (Fig. 3d). Collectively, these results suggest that miR-Off-23/-374 vectors up-regulated CFIm25 protein level, which eventually suppressed proliferation of cancer cells.
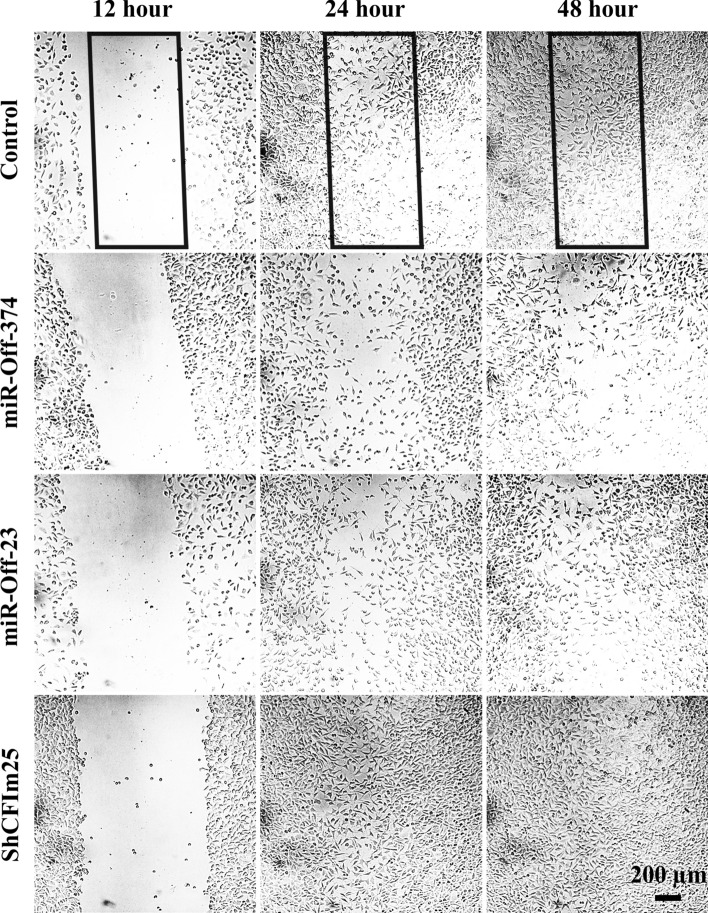
Wound healing assay exemplifies CFIm25 balance with miR-23, -374
The wound healing assay designed to evaluate role of miR-Off-23/miR-Off-374 in MDA-MB-231 in cell migration. As what is shown in Fig. 4, down-regulation of miR-23 and miR-374 (by miR-off-23, -374 transfection) significantly decreased migration ability of transduced cells. However, shCFIm25 vector improved migration.
Figure 4.
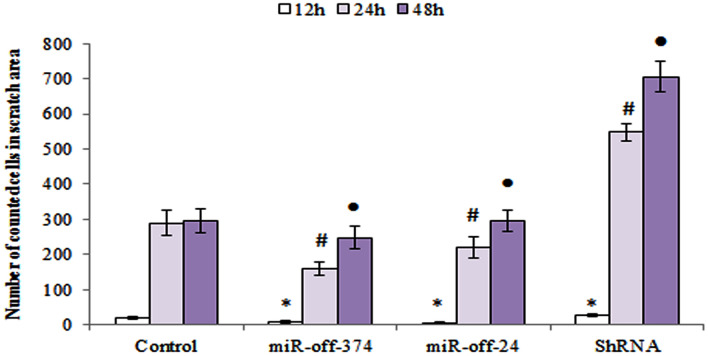
Wound healing assay of transducted MDA-MB-231 cells. Migration of transducted MDA-MB-231 cells toward the scratched area (rectangle) is monitored by 12-h intervals.
Results of migration assay showed that number of migrated cells in the miR-Off-23/miR-Off-374 transduced group, was less than that in the shCFIm25 group. This last finding confirmed that the migration ability of MDA-MB-231 cells was inhibited by miR-Off-23/miR-Off-374, which had been found previously to be under influence of CFIm25 expression level (Fig. 5).
Figure 5.
Time course of MDA-MB-231 migration from scratch borders toward the empty area. Diagram of migrated cells to the area of interest that are counted using ImageJ software. Columns are mean of three experiments and error bars represent standard deviation. The (asterisk, hash and filled circle) above the bars, represents the significant difference respect to the control reference at 12, 24 and 48 h, respectively.
Discussion
It is conformed that most of the human genes have multiple polyadenylation signals, which dynamically regulate their variations in 3′-UTR length26. The CFIm25, as a newly discovered repressor of proximal poly (A) site usage, has a significant role in UTR-APA site regulation. The CFIm25, as a subunit of cleavage factor complex, recruits CPSF6 and/or CPSF7 along with CPSF5, and activates 3′-mRNA cleavage site and polyadenylation processing machinery3,11. Down-regulation of CFIm25, significantly diminishes “distal poly-A signal” length, and enhances tumorigenic properties and size of the tumor cells. However, CFIm25 overexpression reduces aforementioned properties and inhibits tumor growth. An explanation for this is that by choosing a proximal PAS, the 3′-UTR length of the mRNAs shortens, which then eliminates miRNAs binding sites and revokes regulation through miRNAs11,27–29. Here, we approved CFIm25 is indirectly involved in regulation of many genes through changing 3′-UTR length, which then eliminates binding sites of their corresponding miRNAs. Nonetheless, more studies are still needed to clarify comprehensive regulatory mechanism of CFIm25 gene by miRNAs.
Although breast cancer is the most common cancer diagnosed in women, survival rates of breast cancer have improved recently, which is mostly due to factors such as a new personalized approach to cancer treatment and a better understanding of the molecular mechanism of the disease30. In this study, we used human breast cancer specimens to analyze their CFIm25 expression profile. We confirmed that CFIm25 is significantly down-regulated in breast cancer samples, compared to the controls. This phenomenon is consistent with the previous study15 that introduced CFIm25 as APA regulator that was also down-regulated in glioblastoma and came up with cancer proliferation and tumorigenicity16. Besides, it was reported that expression level of CFIm25 in hepatocellular carcinoma (HCC) was negatively correlated to the metastatic potential of HCC cell line through increasing E-cadherin level31, while glutaminase mRNA isoforms, which contain distinct 3′-UTR (KGA and GAC), showed a complex interplay between RNA processing and microRNA repression in controlling glutamine metabolism in cancer cells14.
In the next step of the current study, we selected nine oncomiRs using bioinformatics analysis. They all confirmed to have recognition sites in the 3′-UTR of the CFIm25 (miR-23: 2 sites, miR-24: 2 sites, miR-27: 3 sites, miR-96: 1 site, miR-135: 2 sites, miR-182: 1 site, miR-221: 2 sites, miR-222: 2 sites and miR-374: 4 sites). So far, several studies investigated role of oncomiRs in different cancers including breast cancer20,24. However, no experimental work has performed to clarify effects of these miRNAs on CFIm25 expression level and, as a result, in regulation of UTR-APA site. When clinical samples divided into two groups, whose CFIm25 expression was more or lower than the median expression of control samples, a negative correlation between level of miR-23, miR-24, miR-27, miR-135, miR-182 and miR-374 and CFIm25’s expression was found. This negative correlation in turn suggests that CFIm25 is being regulated by aforementioned miRNAs. Besides, in vitro data from transduced MDA-MB-231 cells confirmed that high levels of miR-23, miR-24, miR-27, miR-135, miR-182 and miR-374 was negatively correlated with either CFIm25 mRNA or its protein content.
According to previous reports, low level of CFIm25 leads to cancer proliferation and migration14,28,31, which is in agreement with our MTT and wound healing assay results. Importantly, functional regulatory role of miR-23 and miR-374 on CFIm25 mRNA, approved via MTT assay. This assay confirmed increased growth of MDA-MB-231 cells, which were co-transduced with miR-Off-23/miR-Off-374 and CFIm25 knock-down vectors. CFIm25 is known to be crucial in controlling invasion and metastasis of HCC32. It is also confirmed that activation of APA sites can affect the migratory capacity of cancer cells33 as well as fibroblasts34. According to the results of wound healing assay, regulation of CFIm25 expression by miR-23 and 374 directly controls breast cancer cells’ migration and invasion. Since invasion and metastasis are the underlying causes of poor long-term survival of breast cancer patients35, regulation of CFIm25 via miRs could be considered as a promising strategy for inhibition of metastasis in future clinical studies.
Parental genes of mRNAs with shorter 3′-UTRs, are more prone to up-regulation during tumorigenesis due to increased escaping from miRNA repression. It is determined that almost 67% genes with shorter 3′-UTRs in tumors, have lost at least one predicted miRNA-binding site36. In some cases, overexpression of oncogenic proteins comes up without regulatory intervenience of the proto-oncogenes. One example of this phenomenon is cancer cell lines (such as MDA-MB-231) that express substantial amounts of mRNA isoforms with shortened 3′-UTRs30,31. CFIm25 is a regulatory protein that controls key factors such as EGFR and MYC that are involved in NF-κB and MAPK/ERK pathways31,37,38. Our finding suggests that regulation of proto-oncogenes by CFIm25, could be mediated via APA and shortening of 3′UTR of their corresponding mRNAs, which eventually result in progression of breast cancer. The MYC, PTEN and AKT2 proto-oncogenes are more responsive to CFIm25 than EGFR and AKT3, since they have several potential interaction sites either at proximal or distal of their mRNA’s 3′-UTRs (Table 1). Tuning impact of CFIm25 on MYC, PTEN and AKT2 will then control downstream phenomena such as glutamine metabolism in cancer cells14, which could be applied as an strategy for targeted therapy39. Besides, 3′-UTR of EGFR’s mRNA contain multiple microRNA target sites, which are associated with cell cycle arrest and cell death. As long as CFIm25 interacts with distal region of EGFR’s 3′-UTR mRNA (Table 1), suppression of breast cancer tumorigenicity by miRNAs is highly plausible40. In a similar way, interaction of CFIm25 with distal region of AKT3 3′-UTR mRNA, facilitates down-regulation of AKT3, which then regulates migration and metastasis in breast cancer cells41. These two examples can clarify how CFIm25 interaction with distal region of EGFR and AKT3 3′-UTR mRNAs, can facilitates suppression of breast cancer tumorigenicity and metastasis, respectively.
Conclusion
In summary, CFIm25 is down-regulated in human breast cancer tissues compared to the adjacent non-cancerous breast tissues. Besides, CFIm25 mRNA is also regulated by several miRNAs including miR-23, miR-24, miR-27, miR-135, miR-182 and miR-374. Since CFIm25 is a key factor in cancer proliferation, identification of its modulators, such as miRNAs, facilitates development of molecular-targeted therapeutics for various cancers including breast cancer. Our in-vitro results confirm functionality of certain miRNAs toward regulation of 3′-UTR via CFIm25 in breast cancer cells, which then introduce it as an emerging field of study in pre-clinical studies of breast cancer.
Methods
Cell lines and patient sample collection
Hundred fresh frozen samples of human breast cancer tumors and their normal adjacent tissue obtained from the Iran national tumor bank of Cancer Institute (Imam Khomeini hospital, Tehran University of medical sciences, Tehran, Iran). Sample collection performed according to the international tumor bank SOP protocol and written informed consent obtained from patients. This study was approved by the cancer institute of Imam Khomeini Hospital, Tehran, Iran. SOP protocol and consent designed in accordance with ethical standards of Tehran University of medical sciences and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Human embryonic kidney cell line HEK293T (RRID:CVCL_0063) and human breast cancer cell line MDA-MB-231 (RRID:CVCL_0062) were obtained from Pasteur Institute of Iran (IPI) and Iranian biological resource center, respectively. Aforementioned institutes check the authenticity of human cell lines via DNA profiling annually. The obtained cell lines cultured in Dulbecco’s modified Eagle’s Medium (DMEM; Gibco), supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin–streptomycin (Gibco) at 37 °C with 5% CO2 incubation.
Bioinformatic analysis
The miRNAs that were susceptible to target CFIm25 mRNA 3′-UTR acquired using prediction web servers such as TargetScan, miRWalk (version 3.0) and DianaTool. The standard cutoff for screening predicted miRNAs was 8 mer, at the most, while the percentage of context ++ score (CS) did not exceed 95%. Predicted miRNAs with the high score were investigated in the miRCancer database. Finally, miRNAs with the oncogenic roles were selected for the rest of the research, including miR-23, miR-24, miR-27, miR-96, miR-135, miR-182, miR-221, miR-222 and miR-374.
In order to find out if CFIm25 efficiently targets 3′-UTR of proto-oncogenes’ mRNAs, three steps were followed. First, 3′-UTR mRNA of twenty proto-oncogenes that known to be involved in breast cancer42, retrieved from UTR database43. Second, twenty retrieved proto-oncogenes including; NM_053056.2, NM_001759.4, NM_000077.4, JX391994.1, S67388.1, NM_005163.2, NM_001626.6, NM_005465.5, BC040540.1, AF067844.1, L78833.1, U43746.1, KX710182.1, XM_011512894.2, XM_024450643.1, NM_002467.6, NM_002745.4, NM_005343.4, NM_005228.5 and NM_005417.4, were tested as a matter of proximal or distal placement of CPSF interaction site at polyadenylation signal (A[A/T]TAAA) and their neighbor CFIm25 interaction site (TGTA). The PAS that is closest to the 5′ end of the 3′-UTR is considered as “proximal” and the other PAS(s) in the downstream of 3′-UTR considered as distal44. Third, the retrieved 3′-UTRs beside CFIm25 (O43809) and CPSF1 (Q10570) submitted to the RNA–Protein Interaction Prediction (RPISeq) server to estimate their interaction probabilities45. Interaction probabilities ranged from zero to one. Optimal RNA/protein interactions were the ones with interaction probabilities of more than 0.5, which represented accuracy of the Random Forest (RF) classifiers in cross-validation evaluation experiments on benchmark datasets.
RNA extraction and qRT-PCR
Total RNA extraction of cell line, human breast cancer tissues and normal specimens was performed using Trizol (Invitrogen) in accordance with manufacturer’s protocol and previous study22. Then, 500 ng of total RNA was reverse transcribed to synthesize cDNA with random hexamer primers and Fermentas kit (for CFIm25 gene) or BON RT adaptor primer and BONmiR kit (for miRNA genes). A total of 1 μL of cDNA was amplified using SYBR Green Real-time PCR Master Mix (Takara) and performed on ABI 157 PRISM 7,500 real-time PCR System (Applied Biosystems). The relative expression levels of the CFIm25 and miRNA genes calculated using the 2−∆∆Ct method, with normalization against β-actin and SNORD47 internal controls, respectively. Three biological replicates were performed for each set of experiments. The qRT-PCR primer sequences are listed in supplementary Table 1 and 2.
In clinical samples, the mean of CFIm25 and β-actin Cts between all of the normal samples was determined and compared with the obtained Cts of each patient. This procedure adopted from our previous studies with minor modifications20,46.
Based on the results, tumors were classified as low and high/equal CFIm25‑expressing tumors using the median of the normal samples as a cut-off. Next, the relative gene expression of predicted miRNAs in two groups of patients was calculated by comparing obtained Cts with the mean Cts of normal samples.
Plasmids, viral vectors construction and luciferase assay
For the construction of miRNA-expressing vectors, genomic fragment consists of the stem-loop structure of each miRNA and theirs flanking genomic sequences, cloned into the mammalian expression vector pLEX.jred and pCDH.TurboGFP at downstream of the cytomegalovirus (CMV) promoter. The PCR primers used for miRNAs cloning were listed in supplementary Table 2. Empty vector without any cloned sequence (pLEX-Ctrl/ pCDH-Ctrl) was used as control in accordance with Furukawa et al. method47.
HEK293T cells were transiently co-transfected by miRNA-expressing lentivectors (or empty backbones), pPAX2 plasmid (packaging plasmid), and pMDG plasmid (containing vsv-G) using calcium phosphate. Lentivirus supernatants were harvested for two or three times, every 12 h, concentrated by ultracentrifuge at 47,000×g for 2 h at 4 °C. Lentivirus titer examined by flow cytometry analysis of reporter positive 293 T cells (Attune Acoustic Focusing Flow Cytometer, ABI, FlowJo software)48.
To down-regulate miR-23 and miR-374 expression, pLenti-III-miR-Off constructs containing GFP marker purchased from ABM Company and packaged into lentiviruses particles. For luciferase assay, full length of CFIm25 3′-UTR amplified by PCR using the forward 5′-GCAAGCTTAGCTGTTCTTCTGCC-3′ and reverse 5′-GCACTAGTGTCATCAAAATATTTATTAA-3′, respectively. The CFIm25 3′-UTR was then cloned to downstream of the luciferase gene in the pSICHECK2 vector (Promega). Mutant stem-loop forms of each miRNA, harboring two mutations in seed sequence, cloned in the lentiviral vector and named Mut-miR, which was considered as negative control. The HEK293 cells were transiently transfected with wild type or mutant miRNA-expressing vector and CFIm25-pSICHECK2 vector using lipofectamine 2000 (Invitrogen)49.
Luciferase assay performed after 48 h of post-transfection, using the dual-luciferase reporter assay system (Promega). Renilla luciferase signal normalized against firefly luciferase activity to monitor transfection efficiency. Data are the means of experiments performed in triplicate.
Western blot
The MDA-MB-231 cells were lysed using cell lysis buffer (50 Mm Tris, pH = 8.0, 150 mM NaCl,1% NP-40, 0.5% sodium deoxycholat, 1 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM EDTA). The polypeptide component were resolved by electrophoresis at 200 V using 12% SDS–polyacrylamide gel electrophoresis (PAGE) transferred into poly vinyl den fluoride membrane and immersed in 5% non-fat milk powder over one hour at room temperature. Upon completion of the transfer, membranes were incubated with CFIm25 and β-actin primary antibodies (Abcam). After washing, membranes probed with horseradish peroxidase-conjugated secondary antibodies (1:1,000, Abcam) and developed for detection by chemiluminescence on Kodak X-film. In order to quantify bands in western blot gel, acquired images of CFIm25 and beta actin bands with quality of 75 out of 100 compared as a matter of expression level, using Image J software. Higher number of averaged white pixels considered as higher amount of expression.
Short hairpin RNA (shRNA) design and experiments
For knockdown endogenous CFIm25, target sequence cloned as shRNA format in the pCDH-turboGFP. The shCFIm25 target sequence is 5′-GCCTCATTCTTATTTCAAGAT-3′. An empty shRNA vector (shCtrl) used as a negative control. The MDA-MB-231cells seeded into a six-well plate and transfected by pCDH-shCFIm25 or pCDH-shCtrl and lipofectamine 3,000 (Invitrogen) according to the manufacturer instruction.
Cell viability assay
For investigation of cell proliferation, experiments performed into three groups. In the first group, MDA-MB-231 cells transduced with miR-Off-23/miR-Off-374 expressing vectors. In the second group, MDA-MB-231 cells that were co-transduced with miR-Off-23/miR-Off-374 expressing vectors were transfected by pCDH-shCFIm25. In the third group, MDA-MB-231 cells transfected merely by pCDH-shCFIm25. At 48, 72 and 96 h after transduction/transfection, 10 μL of MTT reagent added into wells and incubated for next three hours. Next, 100 μL DMSO was added and the resulting absorbance measured at 550 nm using a multi-well spectrophotometer (Bio-Tek).
Wound healing assay
Wound healing assay performed to evaluate MDA-MB-231 cell migration. The MDA-MB-231 cells seeded in 24-well tissue culture plate until their growth rate reached a confluence of ~ 80%. The monolayer scratched gently with a 20-μL pipette tip across the center of the well. After scratching, wells washed twice with medium to remove the detached cells. Cell migration to the scratch area monitored for additional 48 h and photographed every 12 h. Number of cells moved from the border of scratch toward the empty area considered as the “migration index”. Related calculations performed by ImageJ software.
Supplementary information
Abbreviations
- CPSF
Cleavage and polyadenylation specificity factor
- CstF
Cleavage stimulation factor
- PAS
Polyadenylation site
- 3′-UTR
3′-Untranslated region
- UTR-APA
Untranslated region-alternative polyadenylation
- miRNA/miR
MicroRNA
- CFIm25
Cleavage factor Im 25
- RF
Random forest classifiers
- shRNA
Short hairpin RNA
- HCC
Hepatocellular carcinoma
Author contributions
F.K. and S.M.A.H. designed the study, M.T., G.S., performed the experiments. I.R. analyzed the data, provided intellectual support and expertise toward interpretation of results. À.E. collaborated in cell line preparation and culture set up. All authors contributed to the edition and the critical review of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-68406-3.
References
- 1.Sotiriou C, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 2.Ong C-T, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12:283. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017;18:18. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ustyantsev I, Golubchikova J, Borodulina O, Kramerov D. Canonical and noncanonical RNA polyadenylation. Mol. Biol. 2017;51:226–236. doi: 10.7868/S0026898417010189. [DOI] [PubMed] [Google Scholar]
- 6.Li W, et al. Distinct regulation of alternative polyadenylation and gene expression by nuclear poly (A) polymerases. Nucleic Acids Res. 2017;45:8930–8942. doi: 10.1093/nar/gkx560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 8.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013;14:496. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 9.Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip. Rev. RNA. 2011;2:22–31. doi: 10.1002/wrna.47. [DOI] [PubMed] [Google Scholar]
- 10.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayr C, Bartel DP. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misra A, Green MR. From polyadenylation to splicing: Dual role for mRNA 3'end formation factors. RNA Biol. 2016;13:259–264. doi: 10.1080/15476286.2015.1112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan S, et al. NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene. 2018;37:4887–4900. doi: 10.1038/s41388-018-0280-6. [DOI] [PubMed] [Google Scholar]
- 14.Masamha CP, et al. CFIm25 regulates glutaminase alternative terminal exon definition to modulate miR-23 function. RNA. 2016;22:830–838. doi: 10.1261/rna.055939.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masamha CP, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 17.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tasharrofi N, et al. Survival improvement in human retinal pigment epithelial cells via Fas receptor targeting by miR-374a. J. Cell. Biochem. 2017;118:4854–4861. doi: 10.1002/jcb.26160. [DOI] [PubMed] [Google Scholar]
- 19.Mobarra N, et al. Overexpression of microRNA-16 declines cellular growth, proliferation and induces apoptosis in human breast cancer cells. Vitro Cell. Dev. Biol. Anim. 2015;51:604–611. doi: 10.1007/s11626-015-9872-4. [DOI] [PubMed] [Google Scholar]
- 20.Kouhkan F, et al. MicroRNA-129-1 acts as tumour suppressor and induces cell cycle arrest of GBM cancer cells through targeting IGF2BP3 and MAPK1. J. Med. Genet. 2016;53:24–33. doi: 10.1136/jmedgenet-2015-103225. [DOI] [PubMed] [Google Scholar]
- 21.Soufi-Zomorrod M, et al. MicroRNAs modulating angiogenesis: miR-129-1 and miR-133 act as angio-miR in HUVECs. Tumor Biol. 2016;37:9527–9534. doi: 10.1007/s13277-016-4845-0. [DOI] [PubMed] [Google Scholar]
- 22.Pirooz HJ, et al. Functional SNP in microRNA-491-5p binding site of MMP9 3′-UTR affects cancer susceptibility. J. Cell. Biochem. 2018;119:5126–5134. doi: 10.1002/jcb.26471. [DOI] [PubMed] [Google Scholar]
- 23.Zomorrod MS, Kouhkan F, Soleimani M, Aliyan A, Tasharrofi N. Overexpression of miR-133 decrease primary endothelial cells proliferation and migration via FGFR1 targeting. Exp. Cell Res. 2018;369:11–16. doi: 10.1016/j.yexcr.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 24.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corsini LR, et al. The role of microRNAs in cancer: diagnostic and prognostic biomarkers and targets of therapies. Expert Opin. Therap. Targets. 2012;16:S103–S109. doi: 10.1517/14728222.2011.650632. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, et al. Alternative polyadenylation: methods, findings, and impacts. Genom. Proteom. Bioinform. 2017;15:287–300. doi: 10.1016/j.gpb.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lembo A, Di Cunto F, Provero P. Shortening of 3′ UTRs correlates with poor prognosis in breast and lung cancer. PLoS ONE. 2012;7:e31129. doi: 10.1371/journal.pone.0031129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3'untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Y, et al. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, et al. Cell cycle regulation by alternative polyadenylation of CCND1. Sci. Rep. 2018;8:6824. doi: 10.1038/s41598-018-25141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lou J-C, et al. Silencing NUDT21 attenuates the mesenchymal identity of glioblastoma cells via the NF-κB pathway. Front. Mol. Neurosci. 2017;10:420. doi: 10.3389/fnmol.2017.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, et al. CFIm25 inhibits hepatocellular carcinoma metastasis by suppressing the p38 and JNK/c-Jun signaling pathways. Oncotarget. 2018;9:11783. doi: 10.18632/oncotarget.24364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu ZJ, et al. MicroRNA-181a promotes proliferation and inhibits apoptosis by suppressing CFIm25 in osteosarcoma. Mol. Med. Rep. 2016;14:4271–4278. doi: 10.3892/mmr.2016.5741. [DOI] [PubMed] [Google Scholar]
- 34.Mitra M, et al. Alternative polyadenylation factors link cell cycle to migration. Genome Biol. 2018;19:176. doi: 10.1186/s13059-018-1551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heimann R, Lan F, McBride R, Hellman S. Separating favorable from unfavorable prognostic markers in breast cancer: the role of E-cadherin. Can. Res. 2000;60:298–304. [PubMed] [Google Scholar]
- 36.Xia Z, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat. Commun. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abadi MHJN, et al. CFIm25 and alternative polyadenylation: conflicting roles in cancer. Cancer Lett. 2019;459:112–121. doi: 10.1016/j.canlet.2019.114430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Zhang W. Knockdown of NUDT21 inhibits proliferation and promotes apoptosis of human K562 leukemia cells through ERK pathway. Cancer Manag. Res. 2018;10:4311. doi: 10.2147/CMAR.S173496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoerner, C. R., Chen, V. J. & Fan, A. C. The ‘Achilles Heel’of metabolism in renal cell carcinoma: glutaminase inhibition as a rational treatment strategy. Kidney Cancer, 1–17 (2019). [DOI] [PMC free article] [PubMed]
- 40.Webster RJ, et al. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J. Biol. Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 41.Grottke A, et al. Downregulation of AKT3 increases migration and metastasis in triple negative breast cancer cells by upregulating S100A4. PLoS ONE. 2016;11:e0146370. doi: 10.1371/journal.pone.0146370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harbor Perspect. Biol. 2010;2:a003236. doi: 10.1101/cshperspect.a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grillo G, et al. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2009;38:D75–D80. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y, et al. Molecular mechanisms for CFIm-mediated regulation of mRNA alternative polyadenylation. Mol. Cell. 2018;69:62–74. doi: 10.1016/j.molcel.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muppirala UK, Honavar VG, Dobbs D. Predicting RNA-protein interactions using only sequence information. BMC Bioinform. 2011;12:489. doi: 10.1186/1471-2105-12-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torabi S, et al. miR-455-5p downregulation promotes inflammation pathways in the relapse phase of relapsing-remitting multiple sclerosis disease. Immunogenetics. 2019;71:87–95. doi: 10.1007/s00251-018-1087-x. [DOI] [PubMed] [Google Scholar]
- 47.Furukawa N, et al. Optimization of a microRNA expression vector for function analysis of microRNA. J. Control. Release. 2011;150:94–101. doi: 10.1016/j.jconrel.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 48.Mohammadi Z, et al. A lentiviral vector expressing desired gene only in transduced cells: an approach for suicide gene therapy. Mol. Biotechnol. 2015;57:793–800. doi: 10.1007/s12033-015-9872-3. [DOI] [PubMed] [Google Scholar]
- 49.Parsons CJ, et al. Mutation of the 5′-untranslated region stem-loop structure inhibits α1 (I) collagen expression in vivo. J. Biol. Chem. 2011;286:8609–8619. doi: 10.1074/jbc.M110.189118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.