Abstract
The ongoing global pandemic (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a huge public health issue. Hence, we devised a multiplex reverse transcription loop-mediated isothermal amplification (mRT-LAMP) coupled with a nanoparticle-based lateral flow biosensor (LFB) assay (mRT-LAMP-LFB) for diagnosing COVID-19. Using two LAMP primer sets, the ORF1ab (opening reading frame 1a/b) and N (nucleoprotein) genes of SARS-CoV-2 were simultaneously amplified in a single-tube reaction, and detected with the diagnosis results easily interpreted by LFB. In presence of FITC (fluorescein)-/digoxin- and biotin-labeled primers, mRT-LAMP produced numerous FITC-/digoxin- and biotin-attached duplex amplicons, which were determined by LFB through immunoreactions (FITC/digoxin on the duplex and anti-FITC/digoxin on the test line of LFB) and biotin/treptavidin interaction (biotin on the duplex and strptavidin on the polymerase nanoparticle). The accumulation of nanoparticles leaded a characteristic crimson band, enabling multiplex analysis of ORF1ab and N gene without instrumentation. The limit of detection (LoD) of COVID-19 mRT-LAMP-LFB was 12 copies (for each detection target) per reaction, and no cross-reactivity was generated from non-SARS-CoV-2 templates. The analytical sensitivity of SARS-CoV-2 was 100% (33/33 oropharynx swab samples collected from COVID-19 patients), and the assay's specificity was also 100% (96/96 oropharynx swab samples collected from non-COVID-19 patients). The total diagnostic test can be completed within 1 h from sample collection to result interpretation. In sum, the COVID-19 mRT-LAMP-LFB assay is a promising tool for diagnosing SARS-CoV-2 infections in frontline public health field and clinical laboratories, especially from resource-poor regions.
Keywords: SARS-CoV-2, COVID-19, Multiplex reverse transcription loop-mediated isothermal amplification, Lateral flow biosensor, Rapid diagnosis
Highlights
-
•
A COVID-19 mRT-LAMP-LFB assay was successfully devised for detecting SARS-CoV-2 infection..
-
•
mRT-LAMP-LFB assy only requires simple heating equipment to maintain a constant temperature of 63 °C for 40 min.
-
•
The total diagnostic test can be completed within 1 h from sample collection to result interpretation.
1. Introduction
In late December 2019, an unexpected outbreak of COVID-19 caused by a novel coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, also known as 2019-nCov) emerged in Wuhan, Hubei province, China (Xu et al., 2020). By May 11, 2020, the SARS-CoV-2 virus caused a huge epidemic in China and spread to over 140 countries/territories with 7,145,539 globally confirmed COVID-19 cases and 408,025 deaths (World Health Organization, COVID-19 Situation Report-142) (Lu et al., 2020). The current COVID-19 pandemic is a serious global health concern because of its possibly fatal disease progression and rapid escalation of new cases (Wang et al., 2020). Therefore, in order to control COVID-19, it is necessary to urgently develop new detection assays, which can provide an early, rapid and reliable diagnosis of COVID-19.
The diagnosis of COVID-19 based on clinical symptoms, especially in the early stages of disease, is extremely difficult as there are no initial characteristic symptoms of COVID-19 (Huang et al., 2020). Although genome sequencing is highly accurate for COVID-19 diagnosis, its use in rapid large scale diagnosis is not practical because of its long run time and requirement for expensive equipment (Wu et al., 2020). In the current COVID-19 outbreak, real-time reverse transcription polymerase chain reaction (rRT-PCR) was used to detect SARS-CoV-2 in public health and clinical laboratories because of its high specificity and sensitivity (Corman et al., 2020). However, the total run time for rRT-PCR is several hours from clinical sample collection to result reporting. rRT-PCR also requires complex equipment, skilled personnel and a stable power supply. Together, this limits their use in some COVID-19 outbreak regions where there is insufficient infrastructure to perform rRT-PCR such as field laboratories and other resource-limited settings (Cui and Zhou, 2020). Hence, there is an urgent requirement to develop a simple, easy to use and rapid detection technique for the diagnosis of COVID-19.
Loop-mediated isothermal amplification (LAMP) is the most popular isothermal amplification technique and is able to address the limitations of current diagnostic methods in resource-limited settings (Azizi et al., 2019; Obande and Singh, 2020; Wang et al., 2017). The details of the LAMP mechanism have been previously described (Notomi et al., 2000; Obande and Singh, 2020). In LAMP, nucleic acid amplification is conducted under isothermal conditions (e.g. in a heat block) with high efficiency, specificity and speed, eliminating the use of complex precision thermal cyclers for temperature cycling (Obande and Singh, 2020). LAMP is highly specific because it requires six independent primers to recognize the target sequence (Mukama et al., 2020). Thus far, LAMP combined with reverse transcription (RT-LAMP) assays have been developed for the detection of multiple respiratory RNA viruses (e.g. influenza viruses, middle east respiratory syndrome and severe acute respiratory syndrome coronavirus) (Wong et al., 2018). Regarding these traits of the LAMP technique, the development of a LAMP-based assay for SARS-CoV-2 detection facilitates the rapid diagnosis and public health surveillance of COVID-19.
LAMP assays have been developed for diagnosis of COVID-19, and preliminarily evaluated using clinical or simulated respiratory samples (Lamb et al., 2020; Yu et al., 2020). However these reports used a genetic sequence where only the open reading frame 1a/b (ORF1ab) was amplification and detected. This may lead to an unreliable diagnostic result because the ORF1ab gene alone cannot ensure the sufficient sensitivity for SARS-CoV-2 test (Suo et al., 2020). These studies also utilized traditional monitoring techniques, such as agarose gel electrophoresis, SYBR dyes and pH indicators, to detect COVID-19 LAMP products. These techniques can detect any nucleic acids and are not specific for COVID-19 LAMP products. Additionally, electrophoresis is time-consuming and tedious, while for colored dyes and indicators, the judgment of color change in tubes by eye is potentially subjective (Lee et al., 2020). Therefore, there is an ongoing need to develop new LAMP-based assays that are capable of simultaneously detecting multiple SARS-CoV-2 targets to provide a rapid and more objective result, as well as to facilitate simpler diagnostic tests.
Here, a mRT-LAMP coupled with a nanoparticle-based lateral flow biosensor (LFB) assay (mRT-LAMP-LFB) was developed for the diagnosis of COVID-19. Two target sequences, including ORF1ab and the nucleoprotein gene (N), were simultaneously amplified in an isothermal reaction and detected in one test step. We described below the basic COVID-19 RT-LAMP principle and the optimized reaction parameters (e.g. amplification temperature) as well as demonstrate the feasibility of this new method.
2. Materials and methods
2.1. Construction of nanoparticles-based biosensor (LFB)
As shown in Fig. 1 C, the LFB contains four components (a sample pad, a conjugate pad, a nitrocellulose membrane and an absorbent pad) (Jie-Yi Biotechnology). Rabbit anti-fluorescein antibody (anti-FITC, 0.2 mg/mL, Abcam. Co. Ltd.), sheep anti-digoxigenin antibody (Anti-Dig, 0.25 mg/mL, Abcam. Co. Ltd.) and biotinylated bovine serum albumin (biotin-BSA, 4 mg/mL, Abcam. Co. Ltd.) were immobilized within the detection regions (nitrocellulose membrane, NC) of test line 1 (TL1), test line 2 (TL2) and control line (CL), respectively, with each line separated by 5 mm. Dye streptavidin coated polymer nanoparticles (SA-DNPs, 129 nm, 10 mg mL-1, 100 mM borate, pH 8.5 with 0.1% BSA, 0.05% Tween 20 and 10 mM EDTA; Bangs, Laboratories, Inc. Indiana, USA) were immobilized at the conjugated region. Thus, the LFB devised here can detect three targets (a chromatography control and two target amplicons). The assembled LFB were cut into 4-mm dipsticks, and was packaged in a plastic cassette (Jie-Yi Biotech, Shanghai, Beijing) according to the manual. The LFB was stored dry at room temperature until use.
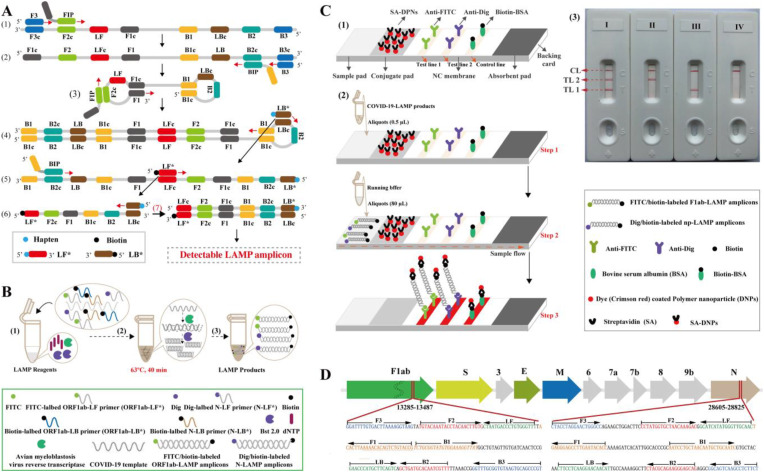
Fig. 1.
Outline of COVID-19 RT-LAMP-LFB design. A. Outline of LAMP assay with LF* and LB*.Top row, outline of LAMP with LF* and LB*; Bottom row, schematic depiction of the new forward/backward loop primers (LF*/LB*). LF* was labeled with a hapten at the 5′ end, and LB* was labeled with biotin at the 5′ end. B. Mechanistic description of the COVID-19 RT-LAMP-LFB assay.(1), Preparing the amplification mixtures. (2), RT-LAMP reaction. (3), The detectable COVID-19 RT-LAMP products after reaction. ORF1ab-RT-LAMP products were simultaneously labeled with FITC and biotin, and the N-RT-LAMP products were labeled with Dig and biotin. C, The principle of LFB for visualization of COVID-19 RT-LAMP products. (1), The details of LFB. (2), The principle of LFB for COVID-19 RT-LAMP products. (3), Interpretation of the COVID-19 RT-LAMP results. I, a positive result for ORF1ab and N (Test line 1, Test line 2 and Control line appear on the LFB); II, a positive result for N (Test line 2 and Control line appear on the detection region); III, a positive result for ORF1ab (Test line 1 and Control line appear on the detection region); IV, negative (only the control line appears on the LFB). *Note, LFB I (Positive result for ORF1ab and N genes), LFB II (Positive for N gene) and LFB III (Positive result for ORF1ab gene) can be judged as positive for SARS-CoV-2. D, Primer design of COVID-19 mRT-MCDA-LFB assay. Up row, SARS-CoV-2 genome organization (GenBank: MN908947, Wuhan-Hu-1). The length of all genes are not displayed to scale. Bottom row, nucleotide sequence and location of ORF1ab and N gene used to design COVID-19 RT-LAMP primers. Part of the nucleotide sequences of ORF1ab (Left) and N (Right) are listed. The sites of primer sequence are underlined. Right arrows and Left arrows show the sense and complementary sequences that are used. * Note: ORF1ab (Open reading frame 1a/b); S (Spike protein); E (Envelope protein); M (Membrane protein); N (Nucleoprotein); Accessory proteins (3, 6, 7a, 7b, and 9b).
For reporting the COVID-19 mRT-LAMP results, a 0.5 aliquot of reaction mixtures was applied to the sample region of LFB. Then, a 80 μL aliquot of BF (running buffer, 10 mM PBS, PH 7.4 with 1% Tween 20) also was deposited to the same region of LFB, the LFB is able to absorb the whole BF. As a result, the presence of the COVID-19 mRT-LAMP products was indicated by the appearance of red lines on the reaction region (NC membrane) (Fig. 1C and Fig. S1). Usually, only 1 min was required for reporting the amplification products using LFB. To ensure the reliability of diagnosis test, we recommended a time of 2 min for COVID-19 mRT-LAMP-LAMP assay during the result indicating stage.
2.2. Primer design
Two RT-LAMP primer sets (ORF1ab-RT-LAMP and N-RT-LAMP) were designed according to the LAMP mechanism using a specialized software (PrimerExplore V5) according to the manual (http://primerexplorer.jp/e/v5_manual/index.html), which targeted the ORF1ab and N gene of SARS-CoV-2 (GenBank MN908947, Wuhan-Hu-1) (Fig. 1D). A BLAST analysis of the GenBank nucleotide database was performed for the ORF1ab- and N-LAMP primer sets to validate sequence specificity. More details of primer design, locations, sequences and modifications are shown in Fig. 1D and Table S1. All of the oligomers were synthesized and purified by RuiBo Biotech. Co. Ltd. (Beijing, China) at HPLC purification grade.
2.3. Reverse transcription LAMP reaction (RT-LAMP)
The conventional singleplex RT-LAMP (ORF1ab- and N-RT-LAMP) was carried out in a one-step 25 μl reaction mixture containing 12.5 μl 2 × isothermal reaction buffer [40 mM Tris-HCl (pH 8.8), 40 mM KCl, 16 mM MgSO4, 20 mM (NH4)2SO4, 2 M betaine and 0.2% Tween-20], 8 U of Bst 2.0 DNA polymerase (New England Biolabs), 5 U of avian myeloblastosis virus reverse transcriptase (Invitrogen), 1.4 mM dATP, 1.4 mM dCTP, 1.4 mM dGTP, 1.4 mM dTTP, 0.4 μM each of F3 and B3, 0.4 μM each of LF, LF*, LB and LB*, 1.6 μM each of FIP and BIP and template (1 μl for the standard ORF1ab or N plasmid). Notably, based on the genomic sequence (Wuhan-Hu-1) of SARS-CoV-2 (Genbank Number: MN908947), part sequences of ORF1ab (13075-14124) and N gene (28273-29519) of SARS-CoV-2 were used for constructing the standard ORF1ab-plasmid and N-plasmid, respectively.
The multiplex RT-LAMP was also performed in a one-step 25 μl reaction mixture containing 12.5 μl 2 × isothermal reaction buffer [40 mM Tris-HCl (pH 8.8), 40 mM KCl, 16 mM MgSO4, 20 mM (NH4)2SO4, 2 M betaine and 0.2% Tween-20], 8 U of Bst 2.0 DNA polymerase (New England Biolabs), 5 U of the avian myeloblastosis virus reverse transcriptase (Invitrogen), 1.4 mM dATP, 1.4 mM dCTP, 1.4 mM dGTP, 1.4 mM dTTP, 0.25 μM each of ORF1ab-F3 and ORF1ab-B3, 0.25 μM each of ORF1ab-LF, ORF1ab-LF*, ORF1ab-LB and ORF1ab-LB*, 1.0 μM each of ORF1ab-FIP and ORF1ab-BIP, 0.15 μM each of N–F3 and N–B3, 0.15 μM each of N-LF, N-LF*, N-LB and N-LB*, 0.6 μM each of N-FIP and N-BIP and template (1 μl for each standard plasmid, 5 μl for samples).
The monitoring techniques, including real-time turbidity (LA-320C), visual detection reagents (VDR, Haitai-Zhengyuan biotech, Co. Ltd. Beijing, China) and LFB, were employed for confirming the RT-LAMP reactions and for optimizing the reaction parameters (e.g. reaction temperature and isothermal amplification time). Particularly, the plasmid concentration at the lowest limit of detection was employed for optimizing the duration time required for the COVID-19 RT-LAMP-LFB assay.
2.4. Sensitivity of the RT-LAMP-LFB assay
Two standard plasmids (ORF1ab-plasmid and N-plasmid) were commercially constructed by Tianyi-Huiyuan Biotech. Co. Ltd. (Beijing, China), which contain the ORF1ab and N sequences, respectively. Ten-fold serial dilutions (1.2 × 104 to 1.2 × 10−2 copies) of ORF1ab-plasmid and N-plasmid were used to evaluate the assay's sensitivity.
2.5. Specificity of the COVID-19 RT-LAMP-LFB assay
The specificity of the COVID-19 RT-LAMP-LFB assay was examined by detecting the various templates, including synthetic nucleic acid sequences, viruses, bacteria and fungi (Table S2).
2.6. Feasibility of COVID-19 RT-LAMP-LFB using clinical samples
A total of 33 respiratory samples were collected from COVID-19 patients (Sanya People's Hospital, Hainan), and COVID-19 patients were defined according to standard diagnostic and treatment criteria of COVID-19 (Trial Version 6). The RNA templates were rapidly extracted using Daan Nucleic Acid Isolation Kit (Daanene Co. LtD.) according to the manufacture's instructions. These templates were firstly used for clinical and laboratory diagnosis, which was conducted using two RT-qPCR kits (Daangene Co. LtD. and BGI Co. LtD.) as recommended by the China CDC and Hannan CDC. A volume of 5 μl of RNA was used as the input template for COVID-19 RT-LAMP-LFB test. Collection and analysis of these RNA templates were approved by Sanya People's Hospital (Ethical approval: SYPH-2019[41]-2020-03-06).
3. Results
3.1. COVID-19 RT-LAMP-LFB design
In the LAMP system (Fig. 1A), FIP (forward inner primer) initiates the isothermal amplification, and the new strand derived from FIP primer is displaced by the F3 (forward primer) synthesis (Step 1). Then, 2 primers, including BIP (backward inner prime) and B3 (backward primer), anneals to the newly produced strand (Step 2). The displacement polymerase Bst 2.0, then extends the sequence in tandem generating a dumbbell shaped product (Step 3). This stem-loop product can then serve as the template for the second stage of the LAMP reaction (exponential amplification). The LB* primer (backward loop primer), which is labeled with biotin at the 5′ end, can anneal to a distinct product derived from the exponential LAMP reaction stage (Step 4). The LB* product also serves as the template for the next amplification step by LF* (forward loop primer), which is modified at the 5′ end with hapten (Step 5). As a result, a double-labeled detectable product (LF*/LB* product) is formed with one end of the LF*/LB* product labeled with hapten, and the other end with biotin (Step 6, 7). A hapten is assigned to one primer set which allows for multiplex LAMP detection.
A representative schematic of COVID-19 mRT-LAMP-LFB assay is displayed in Fig. 1B. In the COVID-19 mRT-LAMP system, one hapten, fluorescein (FITC) was attached to the ORF1ab primer set, and another, digoxigenin (Dig) was attached to the N primer set. Hence, ORF1ab-LF* and ORF1ab-LB* primers were labeled at the 5′ end with FITC and biotin, and N-LF* and N-LB* with Dig and biotin, respectively (Fig. 1B, step 1). With the assistance of AMV (avian myeloblastosis virus) reverse transcriptase, the RNA (SARS-CoV-2 template) was converted to cDNA, and acted as the initial template for subsequent LAMP amplification (Fig. 1B, Step 2). After 40 min at 63 °C, ORF1ab-LAMP products were simultaneously labeled with FITC and biotin, and N-LAMP products with Dig and biotin (Fig. 1B, Step 3).
3.2. The principle of LFB visualization of COVID-19 mRT-LAMP results
As shown in Fig. 1C, the result of COVID-19 mRT-LAMP assay was read out using LFB. The details of LFB is shown in Fig. 1C (Up row). For visualization of the COVID-19 RT-LAMP results using LFB, aliquots (0.5 μl) of the RT-LAMP reaction mixtures were deposited into the sample well (Fig. 1C, Bottom row, Step 1), along with 80 μl of running buffer (Fig. 1C, Bottom row, Step 2). At the detection stage, running buffer moves along the LFB through capillary action, and rehydrates the SA-DNPs which are immobilized in the conjugate pad. One end of the ORF1ab-RT-LAMP products are labeled with FITC which can be captured by the anti-FITC antibody located in the TL1 region (Test line 1), while the end of the N-RT-LAMP products are labeled with Dig which can be captured by the anti-Dig antibody located in the TL2 region (Test line 2). The other ends of the ORF1ab- and N-RT-LAMP products are labeled with biotin and binds streptavidin-conjugated colored nanoparticles for visualization (Fig. 1C, Bottom row, Step 3). Excess streptavidin-conjugated colored nanoparticles are captured by biotinylated bovine serum albumin immobilized in the CL region (Control line), which demonstrates the complete flow of products through the LFB (Fig. 1C, Bottom row, Step 3). The interpretation of the COVID-19 RT-LAMP results using LFB is shown in Fig. 1C (Right row) and Fig. S1.
3.3. Confirmation and detection of ORF1ab-, N-, and COVID-19 RT-LAMP products
The reaction tubes with positive results of ORF1ab-, N- and COVID-19 RT-LAMP assays were visualized as light green using VDR (Visual detection reagent), while the reaction tubes of negative results and blank controls remained colorless (Fig. S2, top row). Using LFB, TL1 and CL were observed in the detection region for positive ORF1ab-RT-LAMP results, and TL2 and CL for positive N-RT-LAMP results. TL1, TL2 and CL simultaneously appeared on the detection region of LFB for positive COVID-19 mRT-LAMP results. Only CL appeared on the analysis area of LFB for negative and blank controls of ORF1ab-, N- and COVID-19 RT-LAMP results (Fig. S2, bottom row). These results indicated that the COVID-19 mRT-LAMP-LFB assay using ORF1ab- and N-LAMP primer sets designed in this study can be used for the rapid and reliable detection of SARS-CoV-2.
3.4. Optimization of COVID-19 mRT-LAMP-LFB assay
The optimal temperature for COVID-19 RT-LAMP was tested with 63 °C shown to be the best for COVID-19 RT-LAMP amplification (Figs. S3 and S4). Then, the optimal isothermal amplification time for the COVID-19 mRT-LAMP-LFB assay was also determined. It was found that the plasmid template at LoD (limit of detection) level (12 copies), an incubation time of 30 min at 63 °C produced positive results with three red lines (TL1, TL2 and CL) (Fig. S5). For the RNA template detection, a reverse transcription process (approximate 10 min) was essential, thus a COVID-19 mRT-LAMP reaction time of 40 min was recommended for the detection of clinical samples. Therefore, the entire diagnostic process for COVID-19 mRT-LAMP-LFB can be completed within 1 h with sample collection (3 min), rapid RNA extraction (15 min), mRT-LAMP reaction (40 min) and result interpretation (<2 min) (Fig. 2 ).
Fig. 2.
The workflow of COVID-19 mRT-LAMP-LFB assay. Four steps, including sample collection (3 min), rapid RNA extraction (15 min), mRT-LAMP reaction (40 min) and result reporting (<2 min), were required for conduct the COVID-19 RT-LAMP-LFB diagnosis test, and the whole process could be completed within 60 min.
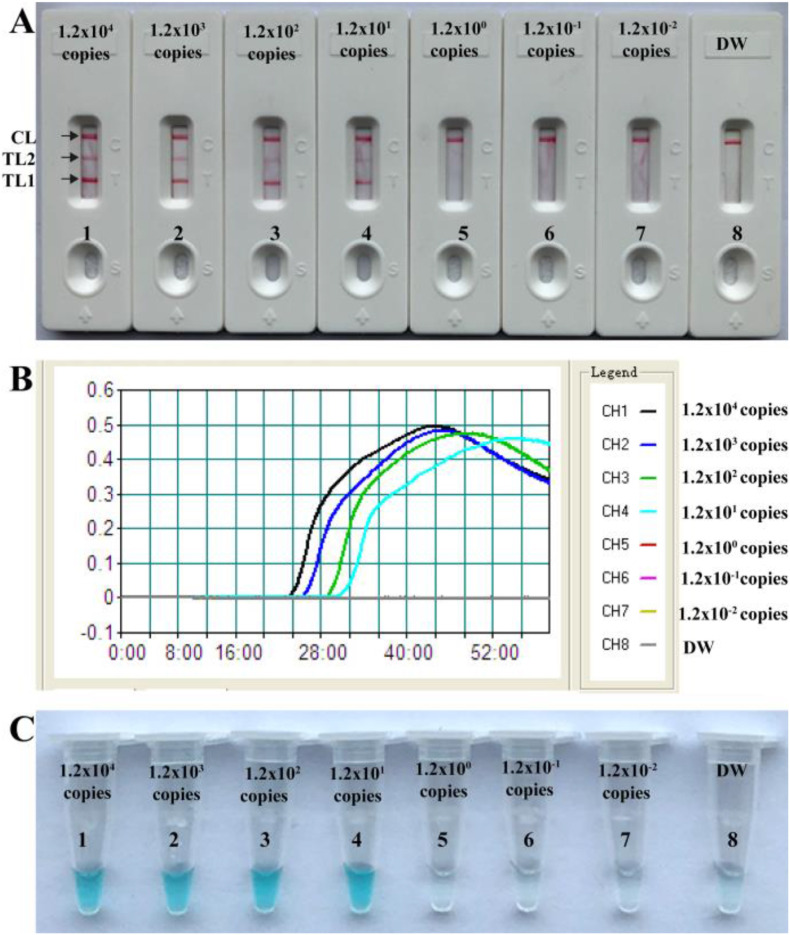
3.5. Sensitivity of COVID-19 COVID-19 mRT-LAMP-LFB assay
Dilutions of templates (ORF1ab-plasmid and N-plasmid constructs) from 1.2 × 104 to 1.2 × 10−2 copies were used to determine the sensitivity of COVID-19 mRT-LAMP-LFB. As shown in Fig. 3 , the limit of detection (LoD) for our assay was 12 copies per reaction for both the ORF1ab-plasmid and N-plasmid (Fig. 3). The two target genes were able to be detected and identified in a single-tube reaction (Fig. 3A). The COVID-19 mRT-LAMP results using LFB were consistent with turbidity and VDR detection (Fig. 3B and C), however, it should be noted that traditional visual monitoring techniques (VDR and turbidity) cannot differentiate target genes for multiplex analysis. The sensitivity of COVID-19 mRT-LAMP-LFB assay was also consistent with singleplex ORF1ab- and N-RT-LAMP assays (Fig. 3, Fig. S6 and Fig. S7).
Fig. 3.
Sensitivity of COVID-19 mRT-LAMP-LFB assay. A, LFB applied for reporting the results; B, Real-time turbidity applied for reporting the results; C, Visual detection reagent applied for reporting the results. LFB (A)/Signals (B)/Tubes (C) 1–8 represented the plasmid levels (each of ORF1ab-plasmid and N-plasmid) of 1.2 × 104, 1.2 × 103, 1.2 × 102, 1.2 × 101, 1.2 × 100, 1.2 × 10−1, 1.2 × 10−2 copies per reaction and blank control (DW). The plasmid levels of 1.2 × 104 to 1.2 × 101 copies per reaction produced positive reactions.
3.6. Specificity of COVID-19 mRT-LAMP-LFB assay
The COVID-19 mRT-LAMP-LFB showed 100% specificity with positive results observed in all positive controls (120 copies each of ORF1ab-plasmid and N-plasmid) (Table S2). Similarly, a negative result was observed in all samples which contained non-SARS-CoV-2 templates (synthetic nucleic acid sequences, virus, bacteria and fungi). In these samples, only a single red band in the CL region was present. This indicates no cross-reaction with non-SARS-CoV-2 templates (Table S2).
3.7. Application of the RT-LAMP-LFB assay in clinical samples
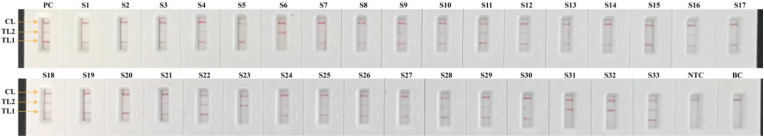
Of the total of 129 respiratory samples, which were initially analyzed using rRT-PCR in Sanya People's Hospital in 2020, 33 were COVID-19 positive samples and 96 were COVID-19 negative samples. The COVID-19 negative patient samples were diagnosed with pneumonia and confirmed to be caused by other pathogens (e.g., Klebsiella pneumoniae, Mycoplasma pneumoniae, Pseudomonas aerugiosa, infuenza virus A and B etc.) by the clinical laboratory in Sanya People's Hospital (Table S3 and Table S4). The samples were leftover RNA extracts from patients after rRT-PCR diagnosis. Among the 33 COVID-19 samples, the sensitivity of the COVID-19 RT-LAMP-LFB assay was 100% (33/33 samples) (Fig. 4 and Table S3). The specificity was also 100% (96/96 COVID-19 negative samples) for the COVID-19 RT-LAMP-LFB assay (Table S4). These results revealed that the proposed COVID-19 RT-LAMP-LFB assay had a high sensitivity and specificity for the diagnosis of SARS-CoV-2 infection.
Fig. 4.
COVID-19 mRT-LAMP-LFB results on 33 COVID-19 infected patient samples. Results were analyzed in accordance with the guidance displayed in Fig. S1. 33 samples (S1 to S33) were collected from 33 patients. PC (Positive control), 1.2 × 102 copies each of ORF1ab-plasmids and N-plasmids were added into COVID-19 mRT-LAMP reaction mixture. NTC (Negative control) and BC (Blank control), RNA template of H1N1 and distilled water were added into COVID-19 mRT-LAMP reaction mixtures, respectively.
4. Discussion
The ongoing SARS-CoV-2 pandemic which started in December 2019 in Wuhan, China, is a current worldwide public health concern (Heymann and Shindo, 2020). As of today (May 11, 2020), there has been 7,145,539 total confirmed cases, with 84,659 cases in China and the remaining distributed in 140 other countries/regions throughout every continent (WHO, COVID-19 Situation Report-142) (Liu et al., 2020). Hence, there is an immediate and urgent need for early, rapid and reliable diagnostic tests to detect SARS-CoV-2 infections. Such detection techniques are required not only in countries where SARS-CoV-2 infection is already spreading but also in countries/regions where COVID-19 has not yet emerged.
In this report, we developed a novel LAMP-based test that provides a simple, rapid and reliable diagnosis for COVID-19, named COVID-19 mRT-LAMP-LFB. Our assay integrates LAMP amplification, reverse transcription and multiplex analysis with a nanoparticles-based biosensor, to facilitate the diagnosis of COVID-19 in a one-step, single-tube reaction. Our test only requires a simple apparatus (e.g. a water bath or a heat block) to maintain a constant temperature (63 °C) for 40 min. Thus, various portable user-friendly apparatus adapted for RT-LAMP amplification exist, the dry block heater (Bioer, Hanzhou, People's Republic of China) being one example (Fig. 2). The portable (16 × 18 cm), $500 USD instrument supports 96 LAMP reactions per assay. Currently, the RT-LAMP reaction can be conducted using the commercial isothermal amplification kits (e.g., Eiken Loopamp Kit and NEB Loopamp Kit), and a RT-LAMP reaction only costs ~$3 USD. The cost of LFB designed in our protocol is estimated to be $2 USD per test. Herein, we deem that a COVID-19 RT-LAMP-LFB test would cost ~$5.5 USD per disposable, and only $500 USD or less for a dedicated instrument. In addition, compared with the published COVID-19 RT-LAMP assays, our protocol directly analyzed the mRT-LAMP results using LFB, which is a simple, objective, less error-prone and easy-to-use platform, and avoids the requirements of complex processes (e.g. electrophoresis), special reagents (e.g. pH indicators) and expensive instruments (e.g. real-time turbidity) (Lamb et al., 2020; Quesada-González and Merkoçi, 2015; Yu et al., 2020). Particularly, the total analysis procedure can be completed within approximately 1 h including sample collection (3 min), rapid RNA extraction (15 min), RT-LAMP reaction (40 min) and result interpretation (<2 min). Considering these advantages, COVID-19 RT-LAMP-LFB assay is a rapid, economical and technically simple method which offers practical solutions for field and clinical laboratories, especially in resource-limited settings.
Two RT-LAMP primer sets (ORF1ab-RT-LAMP and N-RT-LAMP primer sets) were specifically designed and recognize eight regions within the target genes (Fig. 1D). Our specificity tests revealed that no false-positive results were produced from non-SARS-CoV-2 templates, and positive results were obtained from positive control and SARS-CoV-2 templates (Table S2). Moreover, two targets (ORF1ab and N genes) could be simultaneously amplified and detected in a ‘one-step’ RT-LAMP reaction, which further increased the assay's reliability. Our data also demonstrated that 27 out of 33 COVID-19 patients were diagnosed as positive when only ORF1ab gene was employed as diagnostic marker, and 28 out of 33 patient samples were detection with SARS-CoV-2 infection when only N gene was used as diagnostic sequence (Table S3). Particularly, all 33 COVID-19 patients were diagnosed with SARS-CoV-2 infection by COVID-19 RT-LAMP-LFB assay, in which ORF1ab and N genes were simultaneously used as the diagnostic markers (Table S3). Thus, our approach could effectively avoid any undesirable false negative results from current COVID-19 RT-LAMP assays that can only amplify and detect a single gene target (e.g. ORF1ab) (Lamb et al., 2020; Yu et al., 2020).
The sensitivity of our RT-LAMP-LFB assay is sufficient for the diagnosis of COVID-19. Using our protocol, the limit of detection for COVID-19 RT-LAMP-LFB assay was 12 copies each of the ORF1ab-plasmid and N-plasmid constructs, which is consistent with the sensitivity test results generated from ORF1ab-RT-LAMP-LFB and N-RT-LAMP-LFB singleplex detection (Fig. 3, Fig. S6 and Fig. S7). The COVID-19 RT-LAMP-LFB duplex assay did not improve or decrease the sensitivity when compared with the simplex assays (ORF1ab-RT-LAMP and N-RT-LAMP assays). For the detection of RNA templates extracted from respiratory samples, our COVID-19 RT-LAMP-LFB assay was highly sensitive and specific and able to correctly diagnose 100% (33/33) of SARS-CoV-2 clinical samples examined by rRT-PCR and 100% (96/96) of samples from non-SARS-CoV-2 infected patients.
The COVID-19 RT-LAMP LFB assay designed in this report has advantages, some of which are shared with other LAMP-based diagnosis techniques. For example, the COVID-19 RT-LAMP-LFB assay was less sensitive to various inhibitors, or was less affected by the presence of various salts, or could tolerate the inhibitory effect of large amounts of biological substances (Kaneko et al., 2007). Contributing to these merits, the concordance of high sensitivity between COVID-19 RT-LAMP-LFB and COVID-19 RT-PCR for detection of SARS-CoV-2 was successfully validated in this report (Table S3). In addition, it is well known that RNA templates are sensitive to degradation by inadequate sample handing, postmortem processes, or storage, thus the quality of SARS-CoV-2 RNA (e.g. purity and integrity) is also a element for the success of COVID-19 RT-LAMP-LFB analysis. Other factors may affect COVID-19 RT-LAMP-LFB testing results including the specimen's source (lower or upper respiratory tract) and sampling timing (different periods of the disease development). Particularly, the limitations of this report include that the number of clinical samples was small, and that other types of clinical samples (e.g., sputum, blood, urine, feces and nasal samples) were not evaluated. Further investigation of the assay's feasibility using more or different types of specimens is warranted.
5. Conclusion
A COVID-19 mRT-LAMP-LFB assay was successfully devised for detecting SARS-CoV-2 infection, and preliminarily validated using standard plasmids and clinical samples. This protocol allowed multiplex detection of ORF1ab and N genes of SARS-CoV-2 in a diagnostic test within 1 h, and did not rely on complex apparatus. The use of LFB provided a rapid, convenient and easily interpretable readout of COVID-19 mRT-LAMP results. The data of analytical sensitivity and specificity demonstrated that COVID-19 mRT-LAMP-LFB method was a sensitive, reliable and feasible technology for the diagnosis of SARS-CoV-2 infection. Herein, the COVID-19 mRT-LAMP-LFB method is a valuable diagnostic tool for detection of SARS-CoV-2 infection in clinical, field, point-of-care and resource-poor settings, and will be important in controlling this pandemic.
Funding
This work was supported by grants from the Key Research and Development Program of Hainan Province (ZDYF2019149, ZDYF2017163), and Youth Science Foundation of Natural Science Fund of Hainan Province (818QN326).
Ethical approval
This study was approved by the Ethics Committee of the Sanya People’s Hospital (SYPH-2019(41)-2020-03-06).
Data sharing
No additional data available.
Transparency declaration
The lead author and guarantor affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned and registered have been explained.
CRediT authorship contribution statement
Xiong Zhu: Data curation, Formal analysis, Funding acquisition, Resources, Validation. Xiaoxia Wang: Data curation, Formal analysis, Validation, Writing - original draft. Limei Han: Data curation, Resources, Formal analysis. Ting Chen: Data curation, Resources, Formal analysis. Licheng Wang: Data curation, Resources, Formal analysis. Huan Li: Data curation, Resources, Formal analysis. Sha Li: Data curation, Resources, Formal analysis. Lvfen He: Data curation, Resources, Formal analysis. Xiaoying Fu: Data curation, Resources, Formal analysis. Shaojin Chen: Data curation, Resources, Formal analysis. Mei Xing: Data curation, Resources, Formal analysis. Hai Chen: Data curation, Resources, Formal analysis. Yi Wang: Conceptualization, Data curation, Formal analysis, Methodology, Software, Writing - original draft.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgement:
We would like to thank Prof. Ruiting Lan, Laurence Luu and Michael Payne (University of New South Wales, Sydney, Australia) for linguistic assistance during the preparation of this manuscript.
Footnotes
Supplementary data to this article can be found online at Uhttps://doi.org/10.1016/j.bios.2020.112437.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Azizi M., Zaferani M., Cheong S.H., Abbaspourrad A. ACS Sens. 2019;4(4):841–848. doi: 10.1021/acssensors.8b01206. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Euro Surveill. 2020;25(3) [Google Scholar]
- Cui F., Zhou H.S. Biosens. Bioelectron. 2020;165:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann D.L., Shindo N. 2020. The Lancet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E., Suzutani T. J. Biochem. Biophys. Methods. 2007;70(3):499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Lamb, L.E., Bartolone, S.N., Ward, E., Chancellor, M.B., 2020. Available at: SSRN 3539654.
- Lee J.-E., Mun H., Kim S.-R., Kim M.-G., Chang J.-Y., Shim W.-B. Biosens. Bioelectron. 2020;151:111968. doi: 10.1016/j.bios.2019.111968. [DOI] [PubMed] [Google Scholar]
- Liu Q., Liu Z., Li D., Gao Z., Zhu J., Yang J., Wang Q. 2020. medRxiv. [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukama O., de Dieu Habimana J., Meng X., Ting Y., Songwe F., Al Farga A., Mugisha S., Rwibasira P., Li Z., Zeng L. 2020. Analytical Biochemistry; p. 113762. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28(12) doi: 10.1093/nar/28.12.e63. e63-e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obande G.A., Singh K.K.B. Infect. Drug Resist. 2020;13:455. doi: 10.2147/IDR.S217571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada-González D., Merkoçi A. Biosens. Bioelectron. 2015;73:47–63. doi: 10.1016/j.bios.2015.05.050. [DOI] [PubMed] [Google Scholar]
- Suo T., Liu X., Guo M., Feng J., Hu W., Yang Y., Zhang Q., Wang X., Sajid M., Guo D., Huang Z., Deng L., Chen T., Liu F., Xu K., Liu Y., Zhang Q., Liu Y., Xiong Y., Guo G., Chen Y., Lan K. Emerg. Micro. Infect. 2020;1(30) doi: 10.1080/22221751.2020.1772678. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li H., Wang Y., Zhang L., Xu J., Ye C. Front. Microbiol. 2017;8:192. doi: 10.3389/fmicb.2017.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.P., Othman S., Lau Y.L., Radu S., Chee H.Y. J. Appl. Microbiol. 2018;124(3):626–643. doi: 10.1111/jam.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. 2020. Nature; pp. 1–5. [Google Scholar]
- Xu X.-W., Wu X.-X., Jiang X.-G., Xu K.-J., Ying L.-J., Ma C.-L., Li S.-B., Wang H.-Y., Zhang S., Gao H.-N. 2020. Bmj 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Wu S., Hao X., Li X., Liu X., Ye S., Han H., Dong X., Li X., Li J. medRxiv. 2020 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.