Abstract
SARS-CoV-2 is spreading globally at a rapid pace. To contain its spread and prevent further fatalities, the development of a vaccine against SARS-CoV-2 is an urgent prerequisite. Thus, in this article, by utilizing the in-silico approach, a vaccine candidate for SARS-CoV-2 has been proposed. Moreover, the effectiveness and safety measures of our proposed epitopic vaccine candidate have been evaluated by in-silico tools and servers (AllerTOP and AllergenFP servers). We observed that the vaccine candidate has no allergenicity and successfully combined with Toll-like receptor (TLR) protein to elicit an inflammatory immune response. Stable, functional mobility of the vaccine-TLR protein binding interface was confirmed by the Normal Mode Analysis. The in-silico cloning model demonstrated the efficacy of the construct vaccine along with the identified epitopes against SARS-CoV-2. Taken together, our proposed in-silico vaccine candidate has potent efficacy against COVID-19 infection, and successive research work might validate its effectiveness in in vitro and in vivo models.
Keywords: SARS-CoV-2, COVID-19, Epitopic vaccine, Allergenicity, Inflammatory response
Abbreviations: SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; NMA, Normal Mode Analysis; TLR, Toll-like receptor; ACC, Auto Cross-Covariance; CAI, Codon Adaptation Index
Highlights
-
•
In-silico cloning of peptide based vaccine candidate SARS-CoV-2.
-
•
Validation of vaccine construct through computational biology.
-
•
Proposed In-silico vaccine construct help to develop vaccine against COVID-19.
1. Introduction
The epidemiology of coronavirus infection COVID-19 was first reported in the Wuhan City of China, and later, it has spread rapidly throughout China along with other countries, following a pandemic nature. On January 30, 2020, the World Health Organization (WHO) declared a public health emergency of international concern considering the quick outbreak of the disease [1]. To date, a total of 10,719,946 confirmed cases and 517,337 deaths have been reported by WHO. Infected patients develop symptoms like fever, cough, fatigue, myalgia, dyspnea, decreased leukocyte counts, etc. [[2], [3], [4]]. The established mode of transmission for this disease is through physical contact by any of the following means: cough, sneezes, and respiratory droplets [[5], [6], [7]].
The virus, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), earlier known as Novel Coronavirus (2019-nCoV), is the pathogenic identity for COVID-19 [[8], [9], [10]]. It is an enveloped virus consisting of positive-strand RNA as the genomic component [11]. The virus has a characteristic of crown-like appearance due to the spike glycoprotein on the viral envelope [12]. Spike glycoprotein could be the most suitable target for drug designing, vaccine development, and immunotherapy because of its outer surface localization. All the selected epitopes were either predicted or validated in the human immunological framework model. Such an approach could provide a practical method for designing and developing vaccine candidates against the SARS-CoV-2. Initially, the selection of the desired antigens or considering specific proteins as immunogens is a challenging job. Therefore, the epitope-based vaccine design, in-silico cloning and validation, can allow the evaluation of a particular vaccine for its novelty and effectiveness in a given time frame.
Recently, we have proposed an epitope-based in-silico peptide vaccine against the SARS-CoV-2 [13]. Herein, we have tried to perform in-silico characterizations and validations of these identified epitopes. An understanding of the allergenic nature is a must for the safety and effectiveness of any vaccine. Thus, we analyzed the allergenicity of the constructed vaccine candidate and verified whether these epitopes are feasible for developing a robust vaccine against SARS-CoV-2. Moreover, molecular docking was performed in a web-based docking server, and molecular dynamic simulation was executed to compare and validate the interactions between receptor-ligand complex. Additionally, codon adaptation and in-silico cloning were designed for future amplification of targeted vaccine into the expression vector as per requirement. Collectively, this in-silico validation has tried to prove the effectiveness of the developed vaccine candidate with identified epitopes to control the SARS-CoV-2 infection.
2. Materials and methods
2.1. Retrieval of spike protein multi epitopes and vaccine designing
The multi-epitopes of the SARS-CoV-2 spike protein were selected and retrieved from our previously published work [13]. These epitopes were used to construct a vaccine, and a similar methodology has been employed here for analyzing the effectiveness of the proposed vaccine candidate.
2.2. Allergenicity analysis of vaccine component
The novel vaccine should be non-allergenic for its compelling performance. Here, two servers AllerTOP and AllergenFP, were used for allergenicity assessment of our vaccine candidate [14,15]. Both the servers rely on the principle of Auto Cross-Covariance (ACC) and allergenic evaluation methods. These two methods utilize the physicochemical properties of amino acid residues.
2.3. Molecular docking, molecular dynamics (MD) simulation, and normal mode analysis mobility analysis
Molecular docking analysis was performed to study the stable protein-protein interaction and related sub-cellular functions. For molecular docking, the ClusPro 2.0: protein-protein docking server was used [16]. In the ClusPro server, the rigid body docking phase uses the PIPER docking program, which relies on the Fast Fourier Transform (FFT) correlation approach. PIPER represents the interaction energy between two proteins using an expression of form E;
| E = w1Erep+w2Eattr+w3Eelec+w4EDARS |
Where,
E rep and E attr denote the attractive and repulsive contributions to the van der Waals interaction energy, and E elec is electrostatic energy.
E DARS is a pairwise structure-based potential; it primarily represents desolvation contributions, i.e., free energy change by removal of the water molecules from the interface.
The coefficients w 1 , w 2 , w 3, and w 4 define the weights of the corresponding residues.
For the analysis and display of molecular assemblies of the TLR4/5 proteins and vaccine component complex, the visual molecular dynamics (VMD 1.8.3.) program was utilized [17]. The root mean square fluctuation (RMSF) is calculated for Cα atoms, and root mean square deviations (RMSD) of total protein is selected against the backbone Cα atom of the protein complex. Applying the present force field, we completed a 10 ps (pico-second) unrestrained MD simulation of the acid-unfolded state of TLR4/5 proteins and vaccine candidate complex. Primarily the magnitude within the state-of-the-art simulation can define the structural and dynamical features of the studied protein [18].
Normal mode analysis mobility allowed us to investigate the large-scale mobility and the stability of macromolecules. The iMODS server performed the internal coordinates analysis based on the protein-protein structural complex [19]. The server calculates a specific combined motion of large macro-molecule along with the NMA of dihedral coordinates of Cα atoms. Additionally, iMODS estimates B-factor (a disorder of an atom in a protein), structural deformability, and computes the eigenvalue.
2.4. In-silico cloning and physicochemical property assessments of the peptide-based vaccine contrast
Due to the dissimilarity between the codons of human and E. coli, codon adaptation tools were used. It is necessary to adapt the codon usage within the prokaryotic organism to boost the expression rate in the respective host system. For the cloning of the vaccine component, E. coli strain K12 was selected as a host. The Java Codon Adaptation Tool (JCat) was employed for codon optimization of our vaccine component [20]. Here, we selected the pET28a (+) expression vector for cloning, and its nucleotides sequences were collected from the ‘Addgene’ vector database [21]. The WebDSVver 2.0 (http://www.molbiotools.com/WebDSV/) was used for pursuing the in-silico cloning of peptide-based vaccine component against SARS-CoV-2. Solubility and physicochemical property assessments of the primary sequence of vaccine candidates are essential for determining the state, stability, and accessibility of a vaccine. The solubility of the construct vaccine was predicted against the average solubility of E. coli protein in the Protein-Sol webserver [22]. The ProtParam server was accessed for further analysis of various physicochemical properties of the designed vaccine candidate [23].
3. Result
3.1. Retrieval of spike protein multi-epitopes and vaccine designing
We collected 13 MHC-I (SQCVNLTTR, GVYYHKNNK, GKQGNFKNL, GIYQTSNFR, VSPTKLNDL, KIADYNYKL, KVGGNYNYL, EGFNCYFPL, GPKKSTNLV, SPRRARSVA, LGAENSVAY, FKNHTSPDV, and DEDDSEPVL) and 3 MHC-II (IHVSGTNGT, VYYHKNNKS, and FKNHTSPDV) 9mer epitopes. These epitopes were used to construct a peptide-based vaccine against SARS-CoV-2 (Table 1 ).
Table 1.
List showing the epitopes with encountering MHC-I and MHC‐II alleles.
| Sl. no | Epitope encountering MHC-I allele | Sl. No. | Epitope encountering MHC-II allele |
|---|---|---|---|
| 1 | SQCVNLTTR | 1 | IHVSGTNGT |
| 2 | GVYYHKNNK | 2 | VYYHKNNKS |
| 3 | GKQGNFKNL | 3 | FKNHTSPDV |
| 4 | GIYQTSNFR | ||
| 5 | VSPTKLNDL | ||
| 6 | KIADYNYKL | ||
| 7 | KVGGNYNYL | ||
| 8 | EGFNCYFPL | ||
| 9 | GPKKSTNLV | ||
| 10 | SPRRARSVA | ||
| 11 | LGAENSVAY | ||
| 12 | FKNHTSPDV | ||
| 13 | DEDDSEPVL |
3.2. Allergenicity analysis of vaccine component
Both AllerTOP and AllergenFP servers confirmed the non-allergenic nature of the proposed vaccine component. Therefore, the vaccine might not show any harmful allergic reaction after administration.
3.3. Molecular docking, molecular dynamics simulation, and normal mode analysis mobility analysis
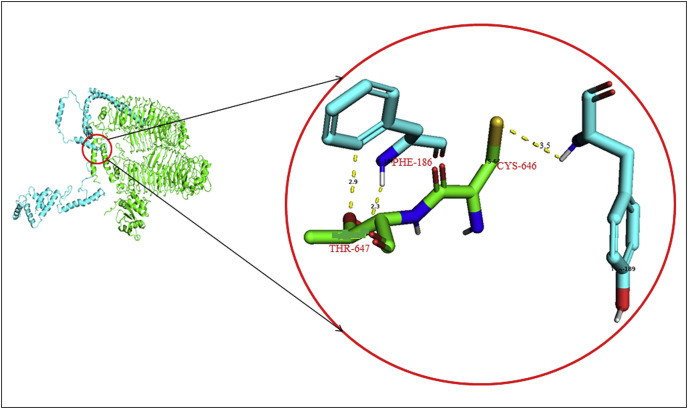
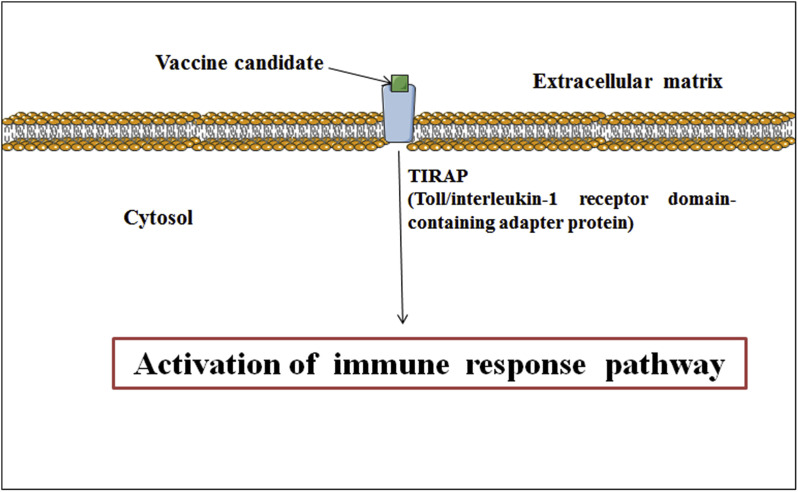
The outputs of molecular docking in ClusPro 2.0 showed high negative energy (−1362.3 kcal/mol) for the docking between TLR4/5 proteins and the vaccine component. The cysteine 646 of TLR4/TLR5 formed a non-covalent bond (3.5 Å) with tyrosine 189 of the vaccine construct, whereas, threonine 647 of TLR4/TLR5 formed two non-covalent bonds (2.9 Å and 2.3 Å) with phenylalanine 186 (Fig. 1 ). The probable immune cascade mechanism of the vaccine candidate in the TIRAP receptor protein has been depicted in Fig. 2 [24].
Fig. 1.
Molecular docking shows non-covalent interactions between TLR protein and construct vaccine candidate.
Fig. 2.
Schematic diagram of TLR induced immune response pathway.
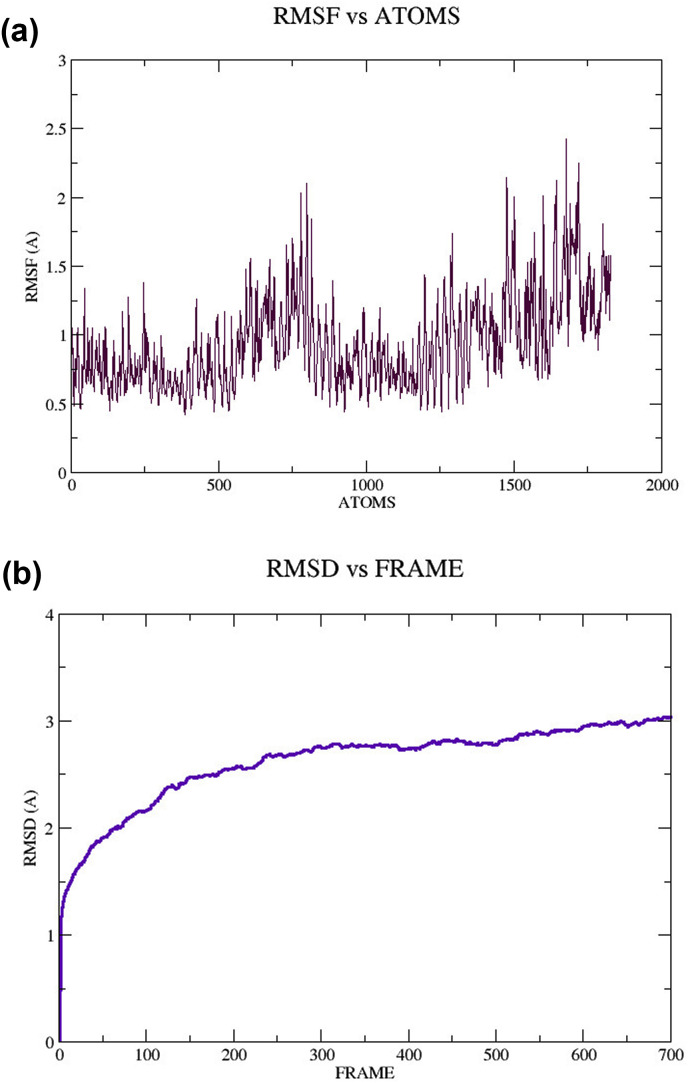
The molecular dynamic simulation of TLR4/5 proteins and the vaccine candidate was performed by the Visual Molecular Dynamics (VMD 1.8.3.) program, and it produced 700 frames (100 frame = 5000 TS = 10 ps) and RMSD plot (Fig. 3 a and b). RMSF was calculated for Cα atoms, and in RMSD, total protein was selected against the backbone of Cα. The plot displayed that the protein complex was in a steady-state.
Fig. 3.
Molecular Dynamics (MD) simulation of TLR protein and vaccine candidate complex (time against distance) a) Root Mean Square Fluctuation (RMSF) plot, b) Root mean square deviations (RMSD) plot.
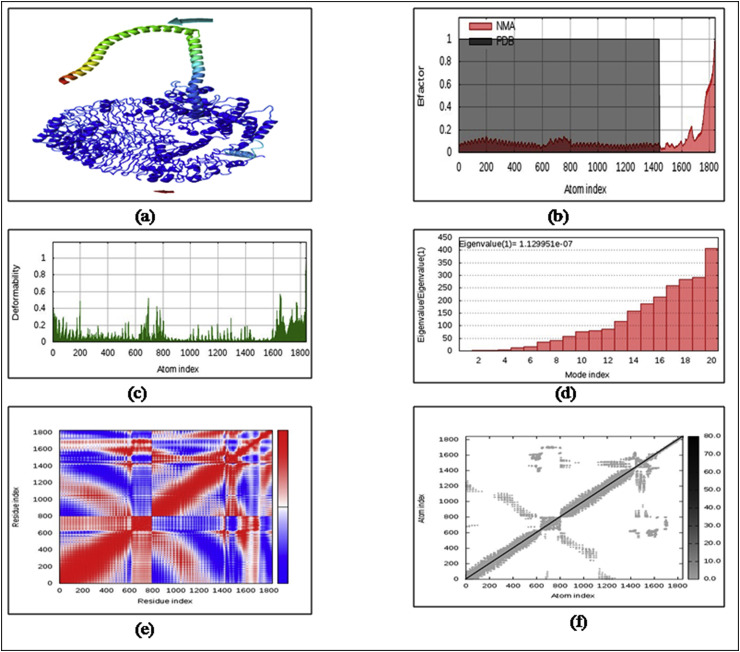
As per the normal mode analysis mobility, upon binding, the vaccine component and the receptor proteins (TLR4/TLR5) were significantly directed to each other (Fig. 4 a) [25]. The deformability plot showed a little bit of fluctuation (Fig. 4c), whereas; the normal mode analysis B-factor was highly minimized from its PDB B-factor (Fig. 4b). The eigenvalue showed an inverse relationship with the variance of the protein-protein docking complex, and the estimated eigenvalue was 1.129951e−07 (Fig. 4d). In the present work, the covariance matrix is illustrated through the graphical representation via white, red and blue color variations indicating the correlated, uncorrelated, and anti-correlated pairs of amino acid residues, respectively (Fig. 4e). Springs of atomic contact are plotted as grey dots in the elastic network model, where the stiffness of interaction was proportional to the gradient of the grey color (Fig. 4f).
Fig. 4.
Outputs of NMA study a) NMA mobility of protein domains b) B-factor plot of PDB and NMA c) deformability plot of atomic fluctuation d) eigenvalue plot e) covariance matrix plot f) elastic network plot.
3.4. In-silico cloning and physicochemical property assessments of peptide-based vaccine construct
In-silico cloning is a rapid method to assess the possibility of developing a potent multi-epitopic vaccine in a given time frame. Here, the JCat server calculated the codon adaptation index (CAI) and GC content of the optimized nucleotides sequence of constructed SARS-CoV-2 peptide-based vaccine. The value was noted as 1.0 and 46.61654, respectively. These are expected values for possible efficient expression of the construct vaccine into the host. Finally, the WebDSV tool was applied for inserting the adapted codon sequences of the peptide-based vaccine candidate within the pET28a(+) vector for expression (Fig. 5 ). The physiochemical properties and details of solubility of the peptide-based-epitope vaccine candidate are listed in Table 2 . Our vaccine candidate showed an instability index of 16.64 (<40, Stable) and a solubility score of 0.698. The estimated half-life was determined as 1 h (mammalian reticulocytes, in vitro) and >10 h (Escherichia coli, in vivo).
Fig. 5.
Schematic representation of our in-silico cloning of vaccine candidate within pET28a(+) expression vector.
Table 2.
Different physicochemical properties of our vaccine candidate.
| Sl. No. | Physicochemical property | Analytical values |
|---|---|---|
| 1 | Solubility | 0.698 (Soluble) |
| 2 | Number of amino acids | 399 |
| 3 | Molecular weight | 40198.87 Da |
| 4 | Theoretical Isoelectric point (pI) | 8.94 |
| 5 | Total number of atoms | 5667 |
| 6 | Formula | C1740H2838N510O577S2 |
| 7 | Estimated half-life | 1 h (mammalian reticulocytes, in vitro) >20 h (yeast, in vivo) >10 h (Escherichia coli, in vivo) |
| 8 | Instability index | 16.64 (Stable) |
| 9 | Aliphatic index | 61.03 |
| 10 | Grand average of hydropathicity (GRAVY) | −0.552 |
4. Discussion
The SARS-CoV-2 has become a global concern and is rapidly emerging as a threat to human civilization. COVID-19, as a pandemic, has already caused many deaths around the globe and still is affecting the human population at a rapid rate. Hence, the new therapeutic approach by targeting TLRs (TLR4/TLR5) modulation may serve as a better choice for vaccine or adjuvant development against the infection of SARS-CoV-2 [26].
Kaur et al. developed a multi-epitope peptide-based chimeric vaccine against the Taenia solium by in-silico approaches [26]. The structural vaccinological analysis and computational validation of the vaccine candidate were also performed against the Mayaro virus [27]. In considerate to the human pathogenic viruses (West Nile virus and Ebola virus) specific peptide-based vaccine has been developed by employing immuno-pharmacoinformatic techniques [28,29].
The principal focus of our current study was the in-silico evaluation of potent vaccine candidates from identified common epitopes. Primarily, we have chosen 16 common B-cell and T-cell epitopes from our previous published work [13]. Afterward, an analysis of the allergenic property was carried out on the designed vaccine candidate. Safe efficacy is a prerequisite for a vaccine, for successful human administration [30]. From the analysis in the AllerTOP and AllergenFP servers, it was found that the constructed vaccine candidate is non-allergenic and safe for future in vivo and in vitro work. Further, molecular docking of vaccine candidates with TLR proteins was performed in the ClusPro 2.0 server to compare and validate the accuracy, stable protein-protein binding, and molecular interactions [31,32]. It was observed that the molecular docking was significant with a high negative energy value (lowest energy value −1362.3 kcal/mol) of the top ordered protein-protein docking complex [33,34]. The molecular dynamics simulation based refinement of the TLR4/TLR5 and vaccine candidate protein complex established the fact that the conformational model was in a steady state. The residues in the protein complex accelerated conformation transitions among the local energy state in 700 frames (5000 TS = 10 ps), time against distance trajectory. The mobility of the protein complex formed by molecular docking was justified through the normal mode analysis mobility study [35]. Outputs of the normal mode analysis study using iMODS revealed that both TLR4/TLR5 and vaccine candidates have stable interaction movement towards each other upon molecular binding.
Additionally, the complex was not easily deformable as there were significantly lower peaks in the deformability plot. Furthermore, codon adaptation within the prokaryotic system was analyzed to obtain a better adaptation and high expression profile in a eukaryotic expression system. When the construct vaccine candidate was cloned into the pET28 (+) expression vector with the help of WebDSVver 2.0, higher expression of the cloned codons was projected.
The pI value predicted in the ProtParam tool indicates the essential (cation rich) character of the vaccine candidate. The prediction also helps to compute the stability of the vaccine into the host body. An aliphatic index of 61.03 predicted that the vaccine is thermostable while the instability index lower than 40 (16.60) indicated higher stability of the vaccine structure after expression.
5. Conclusion
Our in-silico work reveals that the proposed vaccine candidate is non-allergenic, and has efficient as well as stable molecular interaction with the TLR proteins of humans. Results also indicate a high potential and effectiveness of the selected epitopes against SARS-CoV-2. The codon adaptation and in-silico cloning established the impending amplification ability of the vaccine construct within the expression vector as per the conditional requirement. Furthermore, our in-silico analysis might be encouraging to the researchers who are trying to develop efficient therapy against COVID-19. However, constructed vaccine candidates will require successive laboratory validation in in vitro and in vivo models.
Ethics approval and consent to participate
Not Applicable.
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Consent for publication
Not applicable.
Author contributions
MB and ARS designed the model of the computational framework, in-silico analysis, and wrote the manuscript. PP, PG, and GS carried out the implementation and validations., BCP and RPS helped with the analysis editing the manuscript. SSL and CC conceived the study and were in charge of overall direction and planning.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This research was supported by Hallym University Research Fund and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1A2B4012944 & NRF-2020R1C1C1008694).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2020.100394.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Team E.E. Note from the editors: World Health Organization declares novel coronavirus (2019-nCoV) sixth public health emergency of international concern. Euro Surveill. 2020;25(5) doi: 10.2807/1560-7917.ES.2020.25.5.200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Jama. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. 11th ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlos W.G. Novel Wuhan (2019-nCoV) coronavirus. Am J Respir Crit Care Med. 2020;201(4):7–8. doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]
- 5.Riou J., Althaus C.L. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill. 2020;25(4) doi: 10.2807/1560-7917.ES.2020.25.4.2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F.-W. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty C. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur Rev Med Pharmacol Sci. 2020;24(7):4016–4026. doi: 10.26355/eurrev_202004_20871. [DOI] [PubMed] [Google Scholar]
- 8.Liu T. 2020. Transmission dynamics of 2019 novel coronavirus (2019-nCoV) [Google Scholar]
- 9.Nguyen T.M., Zhang Y., Pandolfi P.P. 3rd. Vol. 30. SPRINGER- NATURE; 2020. Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses; pp. 189–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai C.-C. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorusso A. 1st. Vol. 56. Veterinaria Italiana; 2020. Novel coronavirus (SARS-CoV-2) epidemic: a veterinary perspective; pp. 5–10. [DOI] [PubMed] [Google Scholar]
- 12.Chung M. 3rd. Vol. 30. Radiological Society of North America; 2020. CT imaging features of 2019 novel coronavirus (2019-nCoV) pp. 189–190. Radiology. [Google Scholar]
- 13.Bhattacharya M. Development of epitope‐based peptide vaccine against novel coronavirus 2019 (SARS‐COV‐2): immunoinformatics approach. J Med Virol. 2020;92(6):618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitrov I. AllerTOP v. 2—a server for in-silico prediction of allergens. J Mol Model. 2014;20(6):2278. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 15.Dimitrov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30(6):846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- 16.Kozakov D. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12(2):255. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphrey W. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 18.Lindorff-Larsen W. Structure and dynamics of an unfolded protein examined by molecular dynamics simulation. J Am Chem Soc. 2012;134(8):3787–3791. doi: 10.1021/ja209931w. [DOI] [PubMed] [Google Scholar]
- 19.López-Blanco J.R. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42(W1):W271–W276. doi: 10.1093/nar/gku339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grote A. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33(suppl_2):W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamens J. The Addgene repository: an international nonprofit plasmid and data resource. Nucleic Acids Res. 2015;43(D1):D1152–D1157. doi: 10.1093/nar/gku893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hebditch M. Protein-Sol: a web tool for predicting protein solubility from sequence. Bioinformatics (Oxford, England) 2017;33(19):3098–3100. doi: 10.1093/bioinformatics/btx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garg V.K. MFPPI - multi FASTA ProtParam interface. Bioinformation. 2016;12(2):74–77. doi: 10.6026/97320630012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakraborty C. Consider TLR5 for new therapeutic development against COVID‐19. J Med Virol. 2020 doi: 10.1002/jmv.25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer J.A., Pavlović J., Bauerová-Hlinková V. Normal mode analysis as a routine part of a structural investigation. Molecules. 2019;24(18):3293. doi: 10.3390/molecules24183293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaur R. Development of multi-epitope chimeric vaccine against Taenia solium by exploring its proteome: an in-silico approach. Expet Rev Vaccine. 2020;19(1):105–114. doi: 10.1080/14760584.2019.1711057. [DOI] [PubMed] [Google Scholar]
- 27.Khan S. Immunoinformatics and structural vaccinology driven prediction of multi-epitope vaccine against Mayaro virus and validation through in-silico expression. Infect Genet Evol. 2019;73:390–400. doi: 10.1016/j.meegid.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Hossain M.U. An immunopharmacoinformatics approach in development of vaccine and drug candidates for West Nile virus. Front Chem. 2018;6:246. doi: 10.3389/fchem.2018.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwofie S.K. Pharmacoinformatics-based identification of potential bioactive compounds against Ebola virus protein VP24. Comput Biol Med. 2019;113:103414. doi: 10.1016/j.compbiomed.2019.103414. [DOI] [PubMed] [Google Scholar]
- 30.Linhart B., Valenta R. Vaccines for allergy. Curr Opin Immunol. 2012;24(3):354–360. doi: 10.1016/j.coi.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kharisma V.D., Ansori A.N. Construction of epitope-based peptide vaccine against SARS-CoV-2: immunoinformatics study. J Pure Appl Microbiol. 2020;14(suppl 1):999–1005. [Google Scholar]
- 32.Gupta M., Sharma R., Kumar A. Docking techniques in pharmacology: how much promising? Comput Biol Chem. 2018;76:210–217. doi: 10.1016/j.compbiolchem.2018.06.005. 1. [DOI] [PubMed] [Google Scholar]
- 33.Pandey R.K. Immunoinformatics approaches to design a novel multi-epitope subunit vaccine against HIV infection. Vaccine. 2018;36(17):2262–2272. doi: 10.1016/j.vaccine.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 34.Kant K., Lal U.R., Kumar A., Ghosh M. A merged molecular docking, ADME-T and dynamics approaches towards the genus of Arisaema as herpes simplex virus type 1 and type 2 inhibitors. Comput Biol Chem. 2019;78:217–226. doi: 10.1016/j.compbiolchem.2018.12.005. 1. [DOI] [PubMed] [Google Scholar]
- 35.Hayward S., Kitao A., Berendsen H. J. Model-free methods of analyzing domain motions in proteins from simulation: a comparison of normal mode analysis and molecular dynamics simulation of lysozyme. Proteins: Struct Funct Bioinf. 1997;27(3):425–437. doi: 10.1002/(sici)1097-0134(199703)27:3<425::aid-prot10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.