Abstract
Aims
The molecular pathogenesis of COVID-19 is similar to other coronavirus (CoV) infections viz. severe acute respiratory syndrome (SARS) in human. Due to scarcity of the suitable treatment strategy, the present study was undertaken to explore host protein(s) targeted by potent repurposed drug(s) in COVID-19.
Materials and methods
The differentially expressed genes (DEGs) were identified from microarray data repository of SARS-CoV patient blood. The repurposed drugs for COVID-19 were selected from available literature. Using DEGs and drugs, the protein-protein interaction (PPI) and chemo-protein interaction (CPI) networks were constructed and combined to develop an interactome model of PPI-CPI network. The top-ranked sub-network with its hub-bottleneck nodes were evaluated with their functional annotations.
Key findings
A total of 120 DEGs and 65 drugs were identified. The PPI-CPI network (118 nodes and 293 edges) exhibited a top-ranked sub-network (35 nodes and 174 connectivities) with 12 hub-bottleneck nodes having two drugs chloroquine and melatonin in association with 10 proteins corresponding to six upregulated and four downregulated genes. Two drugs interacted directly with the hub-bottleneck node i.e. matrix metallopeptidase 9 (MMP9), a host protein corresponding to its upregulated gene. MMP9 showed functional annotations associated with neutrophil mediated immunoinflammation. Moreover, literature survey revealed that angiotensin converting enzyme 2, a membrane receptor of SARS-CoV-2 virus, might have functional cooperativity with MMP9 and a possible interaction with both drugs.
Significance
The present study reveals that between chloroquine and melatonin, melatonin appears to be more promising repurposed drug against MMP9 for better immunocompromisation in COVID-19.
Keywords: COVID-19, SARS-CoV-2, Coronavirus, PPI-CPI network, MMP9, Chloroquine, Melatonin
1. Introduction
The pandemic ‘coronavirus disease 2019’ (COVID-19) is a severe respiratory illness caused by human coronavirus (HCoV), also known as SARS (severe acute respiratory syndrome)-CoV-2 or novel coronavirus which contains a single-stranded RNA genome [[1], [2], [3]]. The virus can primarily produce influenza-like symptoms viz. fever and myalgia along with pneumonia-like symptoms viz. dry cough and shortness of breath; however in extreme condition acute respiratory distress syndrome (ARDS), multiorgan failure and death may result [2,[4], [5], [6], [7]]. The transmission of SARS-CoV-2 from human to human occurs mostly by physical contacts, nasal droplets, uncooked foods and excreta of conciliator animals [1,2].
The SARS-CoV-2 phylogenetically belongs to other HCoVs under the genera of beta-coronaviruses viz. epidemic SARS-CoV and MERS (Middle East respiratory syndrome)-CoV. The whole genome of SARS-CoV-2 has 79% nucleotide sequence homology to that of SARS-CoV. The surface envelope spike glycoprotein-S is the major antigen of both SARS-CoV and SARS-CoV-2 having 75% amino acid similarity. The glycoprotein-S binds to its receptor, a plasma membrane bound protein called angiotensin converting enzyme subtype 2 (ACE2) of human, allowing endocytosis of the complexes and entry of viruses into the host cells [1,2,8]. The host defense mechanism during HCoV infection gets compromised, as the immune defense responses against the interferons occur late which ensures enough time for replication and survival of the viruses [9]. Clinically, interferon supplementation in COVID-19 patients has been reported to reduce the virus load and inflammatory insult [10].
Extensive review analyses using non-randomized clinical studies [6,[11], [12], [13], [14], [15]] and in vitro/culture studies [[16], [17], [18]] reveal that several approved drugs including anti-viral (remdesevir) [13,14,16,17], anti-retroviral (ritonavir, darunavir, lopinavir) [13,15], anti-malarial (chloroquine, hydroxychloroquine) [13,14,16,17,19], anti-protozoal (nitazoxanide, ivermectin) [14,18] and immunosuppressive (tocilizumab) [13] agents may be the choice of treatment for COVID-19. Monoclonal antibody against the spike protein of SARS-CoV-2 has been raised in view of developing protection against COVID-19 [20]. Moreover, the network analysis based on the human coronavirus genome and host protein interactome has recently proposed that some repurposed drugs including anti-inflammatory agent (melatonin), non-steroidal selective estrogen receptor modulator (toremifene), angiotensin receptor blockers (e.g. irbesartan), immunosuppressants (e.g. sirolimus), anti-neoplastic drugs (e.g. mercaptopurine) and a natural plant product (emodin) having antiviral activities may be the likely candidates for treatment of COVID-19 [21]. However, all these pharmacologic agents suffer from certain limitations in their uses.
Due to the paucity of information about the molecular pathogenesis of COVID-19 and its best suitable treatment, the present study has been executed with potential systems biology approach using integrative protein-protein and chemo-protein network analyses to find out (a) the most effective host protein target(s) in human and (b) the putative drug(s) against the target(s), to establish a functional link between drug(s) vs. targeted host protein(s) for better understanding of treatment strategy in COVID-19.
2. Materials and methods
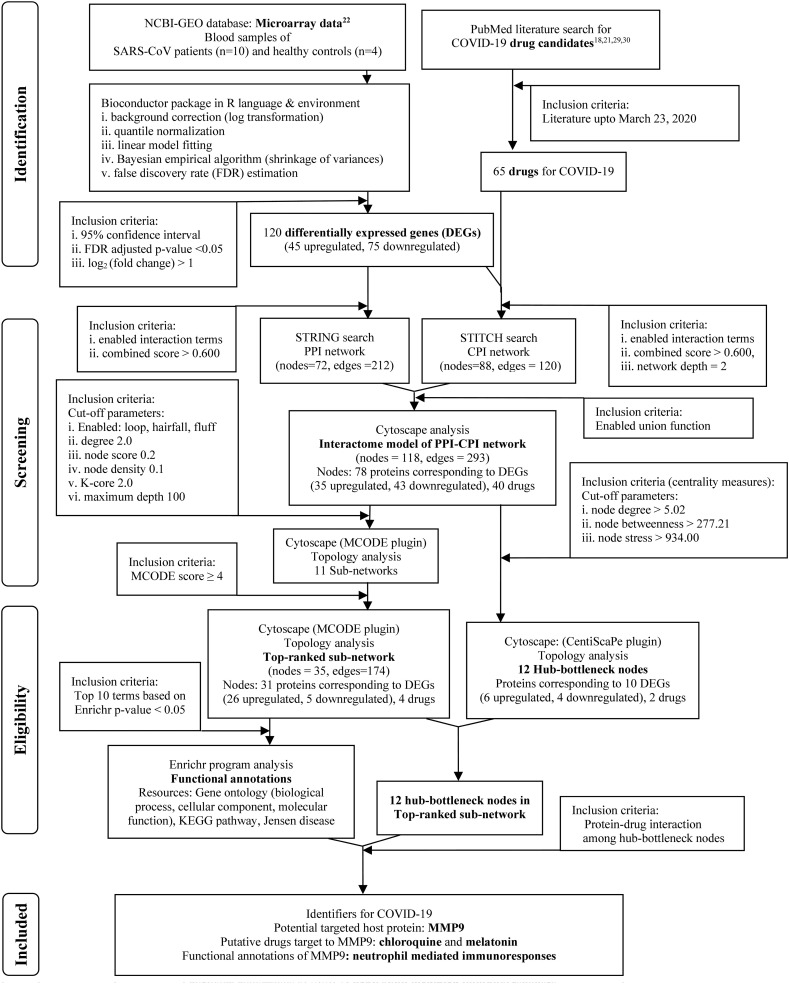
The systematic and stringent methodology with inclusion and exclusion criteria applied in the present study is given in the flow diagram (Fig. 1 ).
Fig. 1.
Flowchart of the systematic and stringent methodology applied and the results found in the mechanistic systems biology analysis to identify potential drugs against their targeted biomolecule(s) in COVID-19.
2.1. Raw data acquisition and processing to identify differentially expressed genes (DEGs)
The microarray dataset (GSE1739) of gene expression profiles in the blood of 10 SARS-CoV patients along with that of four healthy individuals were collected from the NCBI Gene Expression Omnibus (GEO) database. Diagnostic profiles and experimental assay with the including and excluding selection criteria for human subjects had been described in the original publication [22]. The basic packages of the Bioconductor project [23] were used for retrieval, background correction (logarithmic transformation) and quantile normalization of the data in R language and environment [24]. A 95% confidence interval was applied to select the data for further analysis. Further, the R package limma [25] was implemented to analyze the expressions of genes in patients and healthy individuals. The lmFit function was used to fit the linear model using the least square method followed by another function eBayes (a hierarchical Bayesian model) for computation of moderated t-statistics on the linear model to find differential gene expression. The moderated t-statistics uses an empirical Bayesian shrinkage estimator to reduce the variance dependency for a specific gene on its mean expression values [26]. The differential expression of genes was computed using ‘false discovery rate’ (FDR) based on Benjamini and Hochberg method [27] with the criteria of adjusted p-value < 0.05 and |log2(FC)| > 1 (FC representing ‘fold change’) for each comparison.
2.2. In silico modelling of physical ‘protein-protein interaction’ (PPI) and ‘chemo-protein interaction’ (CPI) networks
The DEGs found in the present study, had been incorporated on the STRING 11.0 database [28] with the settings for active interaction from all enable sources (‘Textmining’, ‘Experiments’, ‘Databases’, ‘Co-expression’, ‘Neighborhood’, ‘Gene Fusion’, ‘Co-occurrence’). The reliable interaction strength was detected by applying parameter customized confidence score of 0.600 as threshold to filter out spurious interactions. The study included only the moderate and high probable interactions between the protein partners of differentially expressed genes to construct an integrative physical ‘protein-protein interaction’ (PPI) network model for the characterization of molecular interactions involved in SARS-CoV infection.
Then the systematic review had been performed to acquire recent relevant literatures associated with COVID-19 therapy in PubMed database (www.ncbi.nlm.nih.gov/pubmed/) using the following keywords: ‘SARS’,‘SARS-CoV’, ‘COVID-19’, ‘treatment’, ‘therapy’, ‘therapeutics’ and/or ‘Drug’ respectively. The search results were filtered with ‘Publication date 1 year’ for the latest updates on COVID-19 therapies to select drug compounds from the latest literatures [18,21,29,30]. The drugs (targeted to COVID-19/SARS-CoV-2 treatment) selected and DEGs (identified from the peripheral blood samples of SARS-CoV patients) found in the present study were submitted to STITCH 5.0 database [31] with the setting of active interactions from all enable sources (‘Textmining’, ‘Experiments’, ‘Databases’, ‘Co-expression’, ‘Neighborhood’, ‘Gene Fusion’, ‘Co-occurrence’). The known and predicted binding interactions between genes/proteins and small chemical compounds/drugs were detected by applying parameters including customized confidence score of 0.600, ‘interactor/query protein only’ and ‘network depth equal to 2’ to construct a ‘chemo-protein interaction’ (CPI) network that included moderate and strong target (protein)-drugs interactions for COVID-19.
Furthermore the CPI network model (developed in STITCH) and PPI network model (developed in STRING) were imported in Cytoscape 3.7.2 software [32] and merged by applying the ‘union function’ of the Cytoscape core plugin ‘Merge Networks’ based on attribute values of nodes and edges to get interactome model of merged physical ‘protein-protein and chemo-protein’ (PPI-CPI) network to identify both the potential drug targets (proteins) and potential drug candidates for COVID-19.
2.3. Topology analysis of interactome model of PPI-CPI network to find sub-networks and hub-bottleneck nodes
Molecular Complex Detection (MCODE) [33] (a Cytoscape plugin available at http://www.cytoscape.org/plugins2.php) module using cut-off parameters (scoring and cluster finding) and the MCODE score ≥ 4 was utilized to screen the significant/major sub-network of PPI-CPI interactome for COVID-19. The scoring cut-off parameters were ‘loop included’ (inclusion of loops i.e. self-edges in neighborhood density calculation) and ‘degree cut-off 2’ (number of connections necessary for a node to be scored). The cluster finding cut-off parameters were ‘node score cut-off 0.2’ (cluster size for selection of new members), hairfall included (deletion of all single connected nodes from clusters), fluff included (allowance of cluster expansion with one neighbor shells), node density cut-off 0.1 (controls the neighbor inclusion criteria during ‘fluffing’), K-core cut-off 2 (filters out clusters by eliminating small inter-connected sub-cluster) and maximum depth of network cut-off 100 (limiting the distance from the seed node within which the cluster member can be searched).
The chemo-protein components of overall PPI-CPI interactome were analyzed in CentiScaPe 2.2 [34] plugin of Cytoscape using key network centrality parameters like node degree (the number of neighboring nodes to which the node of interest is directly connected), shortest path betweenness (the number of information streams passing through a given node) and stress (the extent to which a node can hold network communications) to identify the most potential nodes in the PPI-CPI network. Most relevant nodes were selected using the numerical threshold values according to the mean centrality values for each of the topological centrality properties: ‘mean node degree’: 5.02; ‘mean betweenness’: 277.21; ‘mean stress’: 934.0. Nodes (protein/chemical) with high degree/connectivity centrality, high betweenness centrality, and high stress centrality (above mean threshold values) were considered as the hub-bottleneck nodes, which were finally taken into account to serve as the functionally essential nodes (genes/proteins/drugs) of this network model.
2.4. Functional enrichment analysis of the interactome model of the top-ranked sub-network
The list of genes of interest with their identifiers (ID) found in the top sub-network of PPI-CPI interactome was submitted to Enrichr web-tool platform (http://amp.pharm.mssm.edu/Enrichr) [35,36] for the enrichment (i.e. over representation of common annotated biological features) analysis of functional annotations implemented across a number of resources including ‘gene ontology’ (‘biological process’, ‘cellular component’, ‘molecular function’), ‘KEGG biological pathways’, ‘Jensen Disease’. The Enrichr analysis was performed using statistical parameters viz. p-value (Fisher exact test), q-value (adjusted p-value for false discovery rate), old p-value, adjusted old p-value, odd ratio, z-score and combined score (log(p-value) × z-value). The Enriched results (functional annotations) were ranked based on the levels of significance with p-values < 0.05 and corresponding combined scores followed by selection of terms with certain cut-off (top 10 terms).
3. Results
The results found in the present mechanistic systems biology analysis are systematically documented in the flow diagram with the findings of potential drugs against the targeted biomolecule(s) in COVID-19 (Fig. 1).
3.1. Identification of DEGs using microarray data of SARS-CoV patient blood
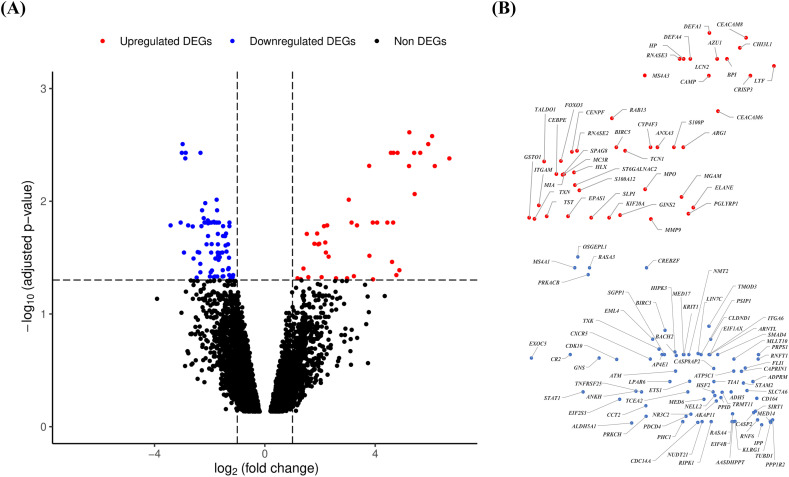
From in silico analysis of SARS microarray dataset we identified 120 differentially expressed genes among which 45 genes were upregulated and 75 genes were downregulated. Details of the differentially expressed genes including gene identifiers (ID), “false discovery rate” (FDR) adjusted p-values (<0.05) and log2(fold change) values (>1) are summarized in Fig. 2 .
Fig. 2.
Volcano plot analysis to identify the differentially expressed genes (DEGs). The expressions of genes are evaluated by analysis of microarray data of the peripheral blood samples of SARS-CoV patients (n = 10) versus healthy controls (n = 4) collected from the data source (GSE1739) using Bayesian algorithm in limma Bioconductor package of bioinformatics tools in R language and environment. A: The volcano plot of the expressions of genes using the logarithmic values of fold changes (log2 fold change) in x-axis and ‘false discovery rate’ (FDR) adjusted p-values (−log10 adjusted p-value) in y-axis. Dotted lines parallel to x-axis and y-axis indicate the threshold values using FDR adjusted p-value < 0.05 and | log2 fold change | > 1 respectively to identify the upregulated and downregulated DEGs. The red, blue and black color dots indicate the upregulated DEGs, downregulated DEGs and non DEGs respectively. B: The gene IDs of upregulated (upper panel) and downregulated (lower panel) DEGs found in volcano plot. The position of dots of DEGs (45 upregulated and 75 downregulated) are same as those appeared in volcano plot. The scales of upregulated and downregulated DEGs are adjusted manually for proper presentation of the gene IDs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Construction of interactome model of PPI-CPI network for COVID-19
The interactome model of PPI network was constructed using proteins corresponding to the respective DEGs of SARS-CoV patients in STRING webtool. A total of 72 protein nodes and 212 connections had been found in PPI network (data not shown). Literature survey provided a total of 65 potential drugs proposed for treatment of COVID-19 (Table 1 ) and these drugs were included in further analysis for selection of most potent drug(s) against putative protein target(s). The interactome model of CPI network was constructed using COVID-19 drug candidates and DEGs of SARS-CoV patients in STITCH webtool. Total 88 nodes (proteins and drugs) and 120 connections had been found in the CPI network (data not shown).
Table 1.
Summary of the potential drug candidates selected from recent literature on COVID-19. Drugs are categorized on the basis of their mode of actions.
| Category | Drug candidate(s) |
|---|---|
| Analgesic | Diperodon, Phenazopyridine, Tetrandrine |
| Anti-bacterial | Dihydrocelastryl diacetate, Monensin sodium, Oligomycin, Salinomycin sodium, Valinomycin |
| Anti-depressant | Desipramine |
| Anti-fungal | Antimycin A, Exalamide, Phenylmercuric acetate |
| Anti-helminthic | Pyrviniumpamoate, Ivermectin |
| Anti-histamine | Chloropyramine |
| Anti-hypertensive | Alprenolol, Berbamine, Carvedilol, Doxazosinmesylate, Irbesartan, Propranolol |
| Anti-infective | Cetylpyridinium chloride, Camphor |
| Anti-inflammatory | Colchicine, Emodin, Mesalazine |
| Anti-malarial | Chloroquine, Conessine, Hydroxychloroquine, Quinacrine |
| Anti-neoplastic | Dactinomycin, Hydroxychalcone, Lycorine, Mercaptopurine, Mycophenolate mofetil, Mycophenolic acid, Pristimerin, Toremifene |
| Anti-Parkinsonian | Harmine |
| Anti-protozoal | Nitazoxanide |
| Anti-psychotic | Promazine |
| Anti-viral | Acyclovir, Favipiravir, Ganciclovir, Lopinavir, Oseltamivir, Penciclovir, Remdesivir, Ribavirin, Ritonavir, Tilorone |
| Ca2+ channel blocker | Loperamide |
| Diuretic | Eplerenone |
| Estrogen steroid | Equilin |
| Hormone | Melatonin |
| IL-6 inhibitor | Tocilizumab |
| Immunosuppresants | Sirolimus |
| Muscle relaxant | Papaverine, Zoxazolamine |
| PAF inhibitor | Ticlopidine |
| Protein synthesis blocker | Cycloheximide, Emetine |
| Selective serotonin reuptake inhibitor | Paroxetine |
| Steroid hormone | Oxymetholone |
| Serine protease inhibitor | Nafamostat |
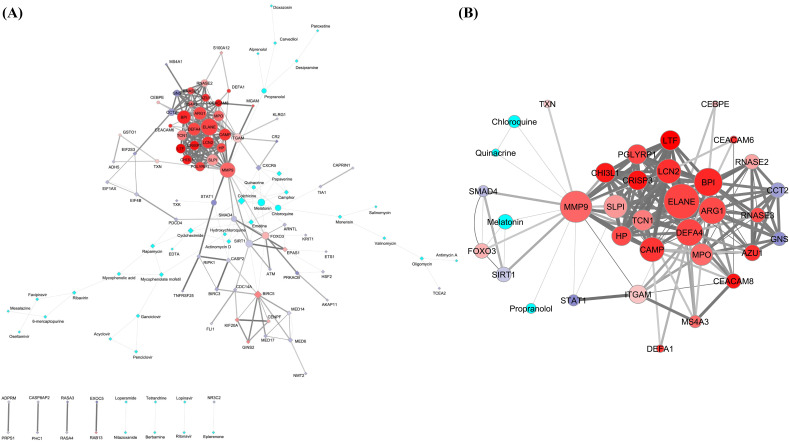
The characterization of protein-drug interactions in COVID-19 had been executed through development of interactome model of PPI-CPI network in Cytoscape by merging the already identified PPI and CPI network and that was found to be composed of 118 nodes (proteins and drugs) interconnected by 293 interactions (Fig. 3A). The identification of nodes of gene products/proteins and drugs has been designated by the corresponding gene IDs and name of the drugs respectively in the present study. Accordingly, the respective gene IDs have been used as the node (protein) identifiers in the further descriptions.
Fig. 3.
The interactome models of PPI-CPI network (A) and its top ranked sub-network (B) obtained in the Cytoscape software. The networks are developed using DEGs of the SARS patients and the promising COVID-19 repurposed drug candidates. A: The PPI-CPI network consists of 118 nodes of gene products/proteins and drugs corresponding to their respective gene IDs (35 upregulated and 43 downregulated DEGs) and name of drugs (40 in number). The 293 edges correspond to the functional connectivities between nodes. The upper panels of the images represent networks having continuous connections. The lower panels of the same designate discrete networks having only single connections. B: The top ranked sub-network was identified in the MCODE module of the Cytoscape considering MCODE score ≥ 4. The sub-network consists of 35 nodes of gene products/proteins and drugs of corresponding gene IDs (26 upregulated and 5 downregulated DEGs) and name of drugs (4 in number) respectively. The 174 edges correspond to the functional connectivities between nodes. A and B: The shape of the nodes in interactome models (A and B) depicted as circles represent the nodes associated with the top ranked sub-network. The rest of the nodes in PPI-CPI network (A) are manually altered to diamond shapes to identify the top ranked sub-network in the main network for better interpretation. The intrinsic attributes of nodes and edges are kept intact in interactome models (A and B). The border of the nodes is illustrated with 2 pts. grey color. The sizes of the nodes indicate their connectivities (higher the value, higher will be the size) adjusted by the ‘continuous mapping of node size’ in ranges between 25 and 60 pts. for the lowest and highest node size respectively. The color of the nodes is represented as cyan for the drugs. The color gradients of red:white:blue are denoted as the expression pattern (log2 fold change) of genes specific for gene products/proteins (nodes), whereby the gradients of the colors have been adjusted to the expression values over the ranges of 6.68:0:−6.68 from the ‘continuous mapping of node color’ in the node network style of the Cytoscape. The widths of the edges are based on the combined scores (0.600 to 1) obtained as interaction data in the STRING for PPI network and STITCH for CPI network which are adjusted by 0.5 to 5 pts. of ‘continuous mapping of edge width’ in the edge network style of the Cytoscape. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Identification of top-ranked sub-network and hub-bottleneck nodes within the interactome model of PPI-CPI network for COVID-19
From the interactome model of PPI-CPI network, the MCODE module in Cytoscape identified only one top-ranked sub-network (cluster having MCODE score 9.33) that comprised 35 nodes (proteins and drugs) with 174 interconnections (Fig. 3B) among which 31 nodes appeared to be protein molecules and four viz. chloroquine, melatonin, propranolol and quinacrine were found to be drug candidates. The sub-network (Fig. 3B) contributed the 59.38% major connections and 29.66% nodes in the overall interactome model of PPI-CPI network (Fig. 3A).
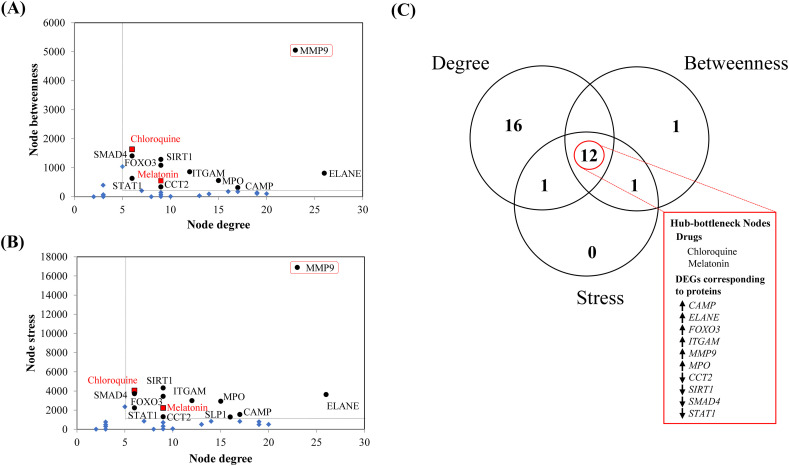
The top-ranked sub-network showed hub-bottleneck nodes consisting of 12 proteins and drugs with high topological centrality index values (high node connectivity, high betweenness, and high stresses, each with greater mean cut-off threshold values) that include 10 proteins corresponding to genes viz. CAMP, CCT2, ELANE, FOXO3, ITGAM, MMP9, MPO, SIRT1, SMAD4, STAT1 and two drug candidates, namely chloroquine and melatonin (Fig. 4A–C). The hub-bottleneck nodes were identified using the cut-off thresholds of node degree (value 5.02), node betweenness (value 277.21) and node stress (value 934.0) applied to the top-ranked sub-network (Fig. 4A–B).
Fig. 4.
Pictorial summary of the topological properties and the centrality analyses for the top ranked sub-network to identify the hub-bottleneck nodes (A, B, C) using the CentiScaPe module of Cytoscape software. The identification of nodes of gene products/proteins and drugs are designated by corresponding gene IDs and name of the drugs. Graphical plots represent the dot plots of values of (A) node degree (x-axis) vs. node betweenness (y-axis), (B) node degree (x-axis) vs. node stress (y-axis) and (C) Venn diagram of high node degree/connectivity, high node betweenness and high node stress. Here, the term ‘high’ indicates higher than the mean cut-off thresholds for node degree/connectivity, betweenness and stress, which have been obtained from the CentiScaPe module of the Cytoscape software. Mean centrality values are presented as dotted lines in the graphs (A, B). The black round dots are hub-bottleneck protein nodes, the blue diamond shapes are non-hub-bottleneck nodes (proteins and drugs) and red boxes with black borders are the hub-bottleneck drug nodes in the (A, B) graphs. MMP9 in the red outlined boxes in the graph (A, B) represent the only target of chloroquine and melatonin among other hub-bottleneck nodes of the top-ranked sub-network. (C) Venn diagram indicates the common nodes that have the topological centrality indices viz. node degree/connectivity, betweenness and stress with the values higher than the mean cut-off respective threshold values obtained from the CentiScaPe module of the Cytoscape software. The upward and downward arrows indicate the expressions of upregulated and downregulated genes (gene IDs right to the arrows) corresponding to respective gene products/proteins in the hub-bottleneck nodes of COVID-19. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Among the 10 hub proteins (Fig. 4C) six proteins corresponding to respective upregulated genes (CAMP, ELANE, FOXO3, ITGAM, MMP9, MPO) and four proteins corresponding to respective downregulated genes (CCT2, SIRT1, SMAD4, STAT1) (as evident in their expression profile in Fig. 2), only matrix metallopeptidase 9 (MMP9) protein had been found to interact with both hub drug nodes, namely chloroquine and melatonin, in the top ranked sub-network (Fig. 3B). MMP9 had been found as the second largest hub node consisting of connectivities with 23 nodes (19 protein and four drug partners). Notably, the first largest hub node ELANE had connectivities with 26 nodes without having any interacting drug partner (Fig. 3B). Furthermore, the other hub node FOXO3 appeared to have connectivity with melatonin as the only interacting drug partner (Fig. 3B). However, there is no evidence in support of the involvement of gene product of FOXO3 with the respiratory viral infection. Therefore our present results clearly indicated that MMP9 could be the most potent host protein target of chloroquine and melatonin as interacting drugs in COVID-19.
3.4. Functional enrichment analysis of the interactome model of the top-ranked sub-network for COVID-19
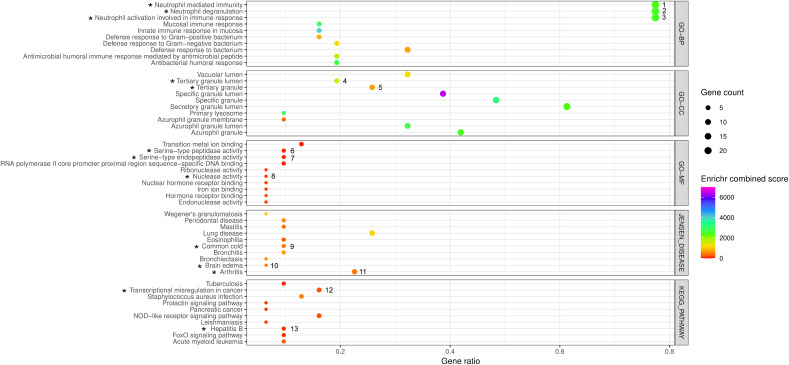
Gene enrichment analysis study (Fig. 5 ) indicated that the gene products of the top-ranked sub-network were associated with biological processes including neutrophil related processes (neutrophil activation, neutrophil degranulation, neutrophil mediated immunity and immune responses), innate immune responses in mucosa, humoral immune responses mediated by antimicrobial peptides, antibacterial humoral responses, defense responses to bacteria with high statistical significances (Enrichr p-value < 0.05 and combined score). The association of the gene products of this sub-network were found to be statistically significant (Enrichr p-value < 0.05 and combined score) for the cellular components involving subcellular and granular secretary functions. Statistically significant (Enrichr p-value < 0.05 and combined score) molecular functions like nuclear hormone receptor bindings, iron and transition metal ion bindings, serine-type endopeptidase activities, endonuclease and ribonuclease activities and RNA polymerase-II core promoter proximal region sequence-specific DNA bindings were also associated with the gene products of this sub-network. In addition, KEGG pathway enrichment study included certain signaling pathways (FoxO, NOD-like receptor and prolactin), several types of microbial infections (exerted by Staphylococcus aureus, Mycobacterium tuberculosis, Leishmania donovani and Hepatitis-B virus) and cancer (pancreatic and acute myeloid leukemia) in association with the gene products of the sub-network. Jensen disease enrichment data indicated that the respiratory system disorders (lung diseases, bronchitis and common colds), immunoinflammation related complications (Wegener's granulomatosis, Mastitis, Arthritis), eosinophilia, brain edema, periodontal disease were highly enriched in association with the gene products of the top-ranked sub-network.
Fig. 5.
The bubble plot to demonstrate the summary of the functional enrichment analysis of DEGs under the top ranked sub-network cluster of the PPI-CPI network in COVID-19 identified by the Enrichr program. The results include the top ten enriched terms corresponding to each of the Enrichr functional annotations viz. GO terms [Biological Process (GO-BP), Cellular Component (GO-CC), Molecular Function(GO-MF)], KEGG pathways and Jensen Diseases obtained from Enrichr analyses using 31 genes corresponding to the gene products/proteins nodes of top ranked sub-network of the PPI-CPI network in COVID-19. The abscissa represents the ‘Enriched gene ratio’. The sizes of bubbles indicate the ‘Enriched gene count’ adjusted with 0.5 to 4 pts. based upon the gene counts of respective functional annotations. The colors of the bubbles indicate the ‘Enrichr combined scores’ (log(p-value) × z-value) which are adjusted in a color (VIBGYOR) gradient, ranging from 0 to 7000 on the basis of values of the scores. The p-value < 0.05 of Fisher's exact test is considered for significant result. The black colored asterisks indicate the enriched functional annotations of the gene products/proteins including MMP9 that correspond to their DEGs denoted by numbers (1 to 13). 1–3: ARG1, AZU1, BPI, CAMP, CCT2, CEACAM6, CEACAM8, CHI3L1, CRISP3, DEFA1, DEFA4, ELANE, GNS, ITGAM, LCN2, LTF, MMP9, MPO, MS4A3, PGLYRP1, RNASE2, RNASE3, SLPI, TCN1; 4: CAMP, CRISP3, LTF, MMP9, PGLYRP1, TCN1; 5: CAMP, CEACAM8, CRISP3, ITGAM, LTF, MMP9, PGLYRP1, TCN1; 6–7: AZU1, ELANE, MMP9; 8: RNASE2, RNASE3; 9: MMP9, MPO, RNASE3; 10: MMP9, MPO; 11: ITGAM, STAT1, HP, CHI3L1, MPO, ELANE, MMP9; 12: CEBPE, ELANE, ITGAM, MMP9, MPO; 13: MMP9, SMAD4, STAT1.
4. Discussion
The present study developed a chemo-protein interactome network (Fig. 3A) on basis of differentially expressed genes in SARS-CoV infection (Fig. 2) and repurposed drugs for COVID-19 (Table 1). A sub-network (Fig. 3B) was identified that appeared to be involved in neutrophil activation and degranulation pathways (Fig. 5). The functionally important hub node MMP9, an immunogenic protease linked to the degradation of cellular matrix, was revealed as the target (Fig. 5) of two repurposed promising drug candidates (Figs. 3B and 4C) i.e. chloroquine (antimalarial drug) and melatonin (pineal hormone) in COVID-19. Here, the systematic analytical approach using data integration reveals the global systems-level relationship (drug vs. target vs. functional annotation) in COVID-19.
ARDS is the main cause of morbidities in COVID-19 and infections of other coronaviruses [[5], [6], [7],37,38]. Cytokine storm is a key mechanism of ARDS materialization leading to multiorgan failure and death [5,6,39,40]. Till date, the molecular pathogenesis of COVID-19 is unclear. Therefore, the similar mechanism of SARS-CoV/MERS-CoV infection can confer a lot to the molecular level of understanding in pathogenesis of COVID-19 [6]. The present study identified a sub-network (Fig. 3B) that includes the statistically significant pathways of both neutrophil activation and degranulation (Fig. 5). The sub-network comprised 29.66% nodes (proteins) and provided 59.38% connectivities of the overall network, which indicated the colossal importance of this sub-network in the whole network model (Fig. 3A–B). The global and local topological analyses provided ten potential protein molecules consisting of six proteins corresponding to respective upregulated genes (CAMP, ELANE, FOXO3, ITGAM, MMP9 and MPO) and four proteins corresponding to respective downregulated genes (CCT2, SIRT1, SMAD4 and STAT1). These ten nodes appeared to be the hub nodes of the PPI-CPI interactome network and were chosen for further study (Fig. 4C). All these molecules had been found to be highly enriched in blood after antimicrobial-induced neutrophil-mediated humoral and innate immunity responses in COVID-19 (Fig. 5).
In the present study, MMP9 was found as the functionally important hub node (protein) and target of the drugs in the central hub node of the PPI-CPI interactome network (Figs. 3B and 4C). Matrix metallopeptidases (MMPs), the zinc-containing and calcium-dependent proteolytic endopeptidases, are released from the intracellular stores and become active extracellularly [41]. MMPs cause deterioration of a number of extracellular matrix proteins that help in the extracellular matrix remodelling in various physiological and pathological processes including inflammation [[41], [42], [43], [44]]. MMPs are widely distributed in tissues and their expressions are controlled by cytokines, growth factors and hormones. MMP9 (83kD), a glycoprotein with type-IV collagenase activity, is found in neutrophils, lymphocytes and dendritic cells that are involved in angiogenesis and inflammatory cytokine generation [41,44]. Notably, the Enrichr functional analysis in the current study revealed MMP9 as to be associated with transition ion binding (molecular function under gene ontology) and diverse pathophysiological conditions viz. neutrophil activation and related immunity (biological process under gene ontology), transcriptional misregulation in cancer and hepatitis B infection (KEGG pathways), arthritis, brain edema and common cold infection (Jensen disease) in human (Fig. 5). The computational study using functional pathways enrichment analysis has also reported the bidirectional regulation of MMP9 gene expression in diverse viral infections and predicted MMP9 as the repurposing drug target [45].
MMP9 is constitutively expressed at low level under physiological condition. The MMP9 action depends on the balance between its pro-peptide activation by a serine protease (plasmin) and inhibition through formation of complexes with ‘tissue inhibitor of matrix metalloprotease 1’ (TIMP1). Therefore, the dysregulations of MMP9 systems (gene expressions, post-translational modifications, levels of activators vs. inhibitors) lead to the development of pathological conditions [46,47]. The low level of TIMP1 and persistent MMP9 activity in neutrophils which bring about airway remodelling and congestion in association with the acute inflammatory responses are implicated in development of both ARDS and acute asthma [[48], [49], [50], [51]]. Recently the gene expressions of TIMP1 in the bronchoalveolar lavage fluid of COVID-19 patients have been found to undergo upregulation in one victim and downregulation in another one as well as upregulation in the blood mononuclear cells of the other three victims [52]. In the present communication, the Enrichr functional analysis (cellular components database under gene ontology) exhibited the association of MMP9 with functional aspects of lumens releasing tertiary granules (Fig. 5). MMP9 is reported to relate with tertiary lumens for release of cytokines during neutrophil activation [53]. In monocytes and macrophages, MMP9 is over-activated by plasmin and reacts with ‘toll like receptor 9’ (TLR9) signaling to induce formation of tumour necrosis factor (TNF) that leads to the development of pro-inflammatory cytokine storm [54,55]. The involvement of MMP9 with serine protease activities was also noticed in Enrichr functional analysis with molecular function under gene ontology (Fig. 5). MMP9 deficiency is found to be protective against severe H1N1 influenza virus A infection in mice model [56]. Therefore, our findings and other reports support the view that the approach to MMP9 inhibition and/or alterations of its activators (plasmin) and inhibitors (TIMP1) may prevent the deadly cytokine storm and the life-risk of COVID-19 patients.
Here, both chloroquine and melatonin were identified as the repurposed drug candidates that appeared to be the interacting partners of MMP9 (Figs. 3B and 4). Chloroquine treatment decreases serum MMP9 level in systemic lupus erythematosus [57] and suppresses MMP9 activity and its mRNA expression in breast cancer [58]. Chloroquine can also lower the TNF-mediated neutrophil apoptosis, neutrophil degranulation and cytokine burst [[59], [60], [61], [62], [63]]. Chloroquine/hydroxychloroquine efficiently inhibits the human retrovirus activities in vitro [64]. Recently, a number of studies [17,65,66] and a clinical trial [67] support the use of chloroquine/hydroxychloroquine for the treatment of COVID-19. Conversely, certain clinical studies strongly disagree with its uses [[68], [69], [70]], as it develops cardiac failure in COVID-19 patients [68], indicating the clinical data about chloroquine/hydroxychloroquine are not conclusive [71]. Notably, hydroxychloroquine did not appear in the sub-network in the present study (Fig. 3B) and remained unattained for further consideration.
Melatonin is responsible for normal sleep and maintenance of the ‘biological clock’ in human [72]. It has anti-inflammatory properties [73,74], exerts its potential role in anti-viral mechanism [[74], [75], [76], [77], [78]] and has also been used in Ebola virus infection [78]. The pathophysiological observation in COVID-19 patients supports the chronobiological uses of melatonin in its treatment [79,80] and a therapeutic algorithm in this regard has also been proposed recently [81]. In addition, melatonin is reported as the promising adjuvant for COVID-19 treatment [21,82] and its deficiencies may enhance susceptibility of diabetic and hypertensive elderly patients in SARS-CoV-2 infection [80]. The first clinical trial of melatonin in a small cohort indicates its involvement in abating ARDS in COVID-19 patients [83]. Interestingly, melatonin can bind to the active site of MMP9 and inhibit the latter to arrest immunoinflammation [84]. Thus, the present network based meta-analysis justified the involvement of melatonin interaction with MMP9 in immunocompromised COVID-19.
The cell culture study (kidney cell line Vero E6) and network analysis reveal that chloroquine [85] and melatonin [21] respectively may have potential anti-viral role in targeting human cell membrane-bound ACE2 receptor, a zinc dependent carboxypeptidase, and other associated protein partners in COVID-19. Interestingly, separate studies report that MMP9 level increases in ACE2-knockout mice model [86] and ACE inhibitors (lisinopril and imidapril) also target MMP9 along with ACE2 [87]. Therefore, both metallopeptidases i.e. ACE2 and MMP9 may have cooperativities for immunoinflammation in COVID-19 and can be potential targets of both chloroquine and melatonin therapies.
The glycoprotein-S of surface spikes of corona viruses including SARS-CoV-2 recognize the membrane-bound ACE2 receptors in airways and lungs for entry into the body. ACE2 along with transmembrane serine protease (TMPRSS2) triggers conformational changes in the glycoprotein-S and releases S-fragments that enable the fusion of viral envelopes with the host cell membrane followed by internalization of viruses through the formation of endosomes. The viral RNA is processed within the host cells and replica of viruses are released out of the cells. The viral infected cells present antigenic peptides through major histocompatibility complexes and elicit humoral and cellular immunities along with cytokine storm [6,[88], [89], [90], [91], [92], [93]]. The ectodomain of membrane-bound ACE2 is shed into the extracellular fluid as its soluble form [88,[94], [95], [96]]. In vitro studies also indicate that the soluble ACE2 can bind to SARS-CoV/SARS-CoV-2 and is supposed to limit the availability of viruses to interact with the membrane-bound ACE2 receptors thus minimizing the chances of viral load [88,92,94]. Notably, ACE2 converts angiotensin-II to angiotensin(1-7), a peptide which causes modification of inflammatory processes [97]. Angiotensin(1-7) also has a vasodilatory effect [98] and enhances the heart rate acting on the caudal ventrolateral medulla in the brain [99], which may contribute to the development of hypotension and tachycardia as found in SARS/COVID-19 patients [100]. Moreover, the interaction of SARS/SARS-CoV-2 with the ACE2 receptors of the support cells in olfactory mucosa also results in inflammation-induced anosmia [101]. Further experimental studies are needed to elaborate the regulatory role of ACE2 receptor and MMP9 in therapeutic management of COVID-19.
5. Conclusion
The present study using network-based systems biology approach clearly demonstrates that (a) the upregulation of MMP9 gene and status of MMP9 protein with its activity/activity regulators, as potentiators of cytokine storms, may be the key point of etiopathophysiology of COVID-19 and (b) chloroquine and melatonin may be the drugs that can target MMP9 to reduce the immunoinflammatory cascades associated with ARDS symptoms in COVID-19 patients. Consistent with the present findings, it would also be worthy to mention that the therapeutic actions of both chloroquine and melatonin may have converged onto the activities of cell membrane ACE2 receptors of SARS-CoV-2. Recently the use of chloroquine does not appear attractive as it produces cytotoxicity at doses used in the clinical trials. In this respect melatonin could be a better promising safe drug that needs more clinical trials in large cohorts to ensure its efficacy in treatment of COVID-19 patients.
Declaration of competing interest
The authors declare that there is no conflict of interest.
Acknowledgments
Acknowledgments
The present study was funded by the Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India, under BINC (Bioinformatics National Certification) scheme [No.: BT/BI/10/078/2014]. We are thankful to Professor Pritha Mukhopadhyay, coordinator of CPEPA-UGC centre [“Centre for Electrophysiological and Neuro-imaging studies including Mathematical Modelling” (CPEPA) through “University Grants Commission” (UGC)], under the University of Calcutta, India.
Funding
The Senior Research Fellowship grant was funded by the Department of Biotechnology (DBT), BINC (Bioinformatics National Certification), Ministry of Science and Technology, Government of India, to Suvojit Hazra (Grant No.: BT/BI/10/078/2014).
CRediT authorship contribution statement
Suvojit Hazra: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Resources, Data Curation, Writing - Original Draft. Alok Ghosh Choudhuri: Conceptualization, Validation, Resources, Writing - Review & Editing. Basant K. Tiwary: Validation, Resources, Writing - Review & Editing, Project administration. Nilkanta Chakrabarti: Conceptualization, Validation, Resources, Writing - Review & Editing, Supervision, Project administration.
Data statement
All data associated with this study are present in the paper.
References
- 1.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO WHO; March 11, 2020. We have therefore made the assessment that COVID-19 can be characterized as a pandemic. WHO Director-General's opening remarks at the media briefing on COVID-19 on 11 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020
- 4.Fu L., Wang B., Yuan T., Chen X., Ao Y., Fitzpatrick T., Li P., Zhou Y., Lin Y., Duan Q., Luo G. Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: a systematic review and meta-analysis. J. Inf. Secur. 2020;80:656–665. doi: 10.1016/j.jinf.2020.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.03.001. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Q., Chen V., Shannon C.P., Wei X.S., Xiang X., Wang X., Wang Z.H., Tebbutt S.J., Kollmann T.R., Fish E.N. Interferon-α2b treatment for COVID-19. Front. Immunol. 2020;11:1061. doi: 10.3389/fimmu.2020.01061. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holshue M.L., DeBolt C., Lindquist S., Lofy K., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M., Weldon W., Biggs H., Uyeki T., Pillai S. First case of 2019 novel coronavirus in the united state. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicastri E., Petrosillo N., Ippolito G., D’Offizi G., Marchioni L., Bartoli T.A., Lepore L., Mondi A., Murachelli S., Antinori A., National Institute for the Infectious Diseases “L. Spallanzani” IRCCS Recommendations for COVID-19 clinical management. Infect. Dis. Rep. 2020;12 doi: 10.4081/idr.2020.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Y., Cao Q., Hong Z., Tan Y., Chen S., Jin H., Tan K., Wang D., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-an update on the status. Mili. Med. Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim J., Jeon S., Shin H., Kim M., Seong Y., Lee W., Choe K., Kang Y., Lee B., Park S. Case of the index patient who caused tertiary transmission of coronavirus disease 2019 in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 pneumonia monitored by quantitative RT-PCR. J. Korean Med. Sci. 2020;35:e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L., Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J. Med. Virol. 2020;92:479–490. doi: 10.1002/jmv.25707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caly L., Druce J., Catton M., Jans D., Wagstaff K. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Favalli E.G., Ingegnoli F., De Lucia O., Cincinelli G., Cimaz R., Caporali R. COVID-19 infection and rheumatoid arthritis: faraway, so close! Autoimmun. Rev. 2020;19 doi: 10.1016/j.autrev.2020.102523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reghunathan R., Jayapal M., Hsu L.Y., Chng H.H., Tai D., Leung B.P., Melendez A.J. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2013. R: A Language and Environment for Statistical Computing; p. 201.https://www.R-project.org/ [Google Scholar]
- 25.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 28.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Mering C. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skariyachan S., Challapilli S., Packirisamy S., Kumargowda S., Sridhar V. Recent aspects on the pathogenesis mechanism, animal models and novel therapeutic interventions for Middle East respiratory syndrome coronavirus infections. Front. Microbiol. 2019;10:569. doi: 10.3389/fmicb.2019.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N., Deng Y., Wang H., Ye F., Cen S., Tan W. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J. Virol. 2019;93 doi: 10.1128/JVI.00023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhn M., von Mering C., Campillos M., Jensen L.J., Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2007;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bader G.D., Hogue C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scardoni G., Petterlini M., Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss S.R., Leibowitz J.L. Coronavirus pathogenesis. Adv. Virus Res. 2011;81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams A.E., Chambers R.C. The mercurial nature of neutrophils: still an enigma in ARDS? Am. J. Physiol. Lung Cell Mol. Physiol. 2014;306:L217–L230. doi: 10.1152/ajplung.00311.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapoor C., Vaidya S., Wadhwan V., Kaur G., Pathak A. Seesaw of matrix metalloproteinases (MMPs) J. Cancer Res. Ther. 2016;12:28. doi: 10.4103/0973-1482.157337. [DOI] [PubMed] [Google Scholar]
- 42.Klein T., Bischoff R. Physiology and pathophysiology of matrix metalloproteases. Amino Acids. 2011;41:271–290. doi: 10.1007/s00726-010-0689-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Djuric T., Zivkovic M. Overview of MMP biology and gene associations in human diseases. Role Matrix Met. Hum. Body Pathol. 2017;1:3–33. doi: 10.5772/intechopen.70265. [DOI] [Google Scholar]
- 44.Vafadari, Salamian A., Kaczmarek L. MMP-9 in translation: from molecule to brain physiology, pathology, and therapy. J. Neurochem. 2016;139:91–114. doi: 10.1111/jnc.13415. [DOI] [PubMed] [Google Scholar]
- 45.Smith S.B., Dampier W., Tozeren A., Brown J.R., Magid-Slav M. Identification of common biological pathways and drug targets across multiple respiratory viruses based on human host gene expression analysis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez D.E., Alonso D.F., Yoshiji H., Thorgeirsson U.P. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur. J. Cell Biol. 1997;74:111–122. [PubMed] [Google Scholar]
- 47.Löffek S., Schilling O., Franzke C.W. Series “matrix metalloproteinases in lung health and disease” edited by J. Müller-Quernheim and O. Eickelberg number 1 in this series: biological role of matrix metalloproteinases: a critical balance. Eur. Resp. J. 2011;38:191–208. doi: 10.1183/09031936.00146510. [DOI] [PubMed] [Google Scholar]
- 48.Chung F.T., Huang H.Y., Lo C.Y., Huang Y.C., Lin C.W., He C.C., He J.R., Sheng T.F., Wang C.H. Increased ratio of matrix metalloproteinase-9 (MMP-9)/tissue inhibitor metalloproteinase-1 from alveolar macrophages in chronic asthma with a fast decline in FEV1 at 5-year follow-up. J. Clin. Med. 2019;8:1451. doi: 10.3390/jcm8091451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potey P.M., Rossi A.G., Lucas C.D., Dorward D.A. Neutrophils in the initiation and resolution of acute pulmonary inflammation: understanding biological function and therapeutic potential. J. Pathol. 2019;247:672–685. doi: 10.1002/path.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ray, Kolls J.K. Neutrophilic inflammation in asthma and association with disease severity. Trends Immunol. 2017;38:942–954. doi: 10.1016/j.it.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hassan N., Mohamed-Hussein A., Mohamed E., Mohamed O., Mohamed H., Tammam M. Matrix metalloproteinase-9 (MMP-9) and tissue inhibitor of metalloproteinase-1 (TIMP-1) as non-invasive biomarkers of remodelling in asthma. Eur. Respir. J. 2015;46 doi: 10.1183/13993003.congress-2015.OA1467. [DOI] [Google Scholar]
- 52.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infec. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cassatella M.A., Östberg N.K., Tamassia N., Soehnlein O. Biological roles of neutrophil-derived granule proteins and cytokines. Trends Immunol. 2019;40:648–664. doi: 10.1016/j.it.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 54.Shimazu H., Munakata S., Tashiro Y., Salama Y., Dhahri D., Eiamboonsert S., Ota Y., Onoda H., Tsuda Y., Okada Y., Nakauchi H. Pharmacological targeting of plasmin prevents lethality in a murine model of macrophage activation syndrome. Blood. 2017;130:59–72. doi: 10.1182/blood-2016-09-738096. [DOI] [PubMed] [Google Scholar]
- 55.Gomez-Salinero J.M., Rafii S. Plasmin regulation of acute cytokine storm. Blood. 2017;130:5–6. doi: 10.1182/blood-2017-04-776385. [DOI] [PubMed] [Google Scholar]
- 56.Rojas-Quintero J., Wang X., Tipper J., Burkett P.R., Zuñiga J., Ashtekar A.R., Polverino F., Rout A., Yambayev I., Hernández C., Jimenez L. Matrix metalloproteinase-9 deficiency protects mice from severe influenza A viral infection. JCI Insight. 2018;3 doi: 10.1172/jci.insight.99022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lesiak, Narbutt J., Sysa-Jedrzejowska A., Lukamowicz J., McCauliffe D.P., Wózniacka A. Effect of chloroquine phosphate treatment on serum MMP-9 and TIMP-1 levels in patients with systemic lupus erythematosus. Lupus. 2010;19:683–688. doi: 10.1177/0961203309356455. [DOI] [PubMed] [Google Scholar]
- 58.Tuomela J., Sandholm J., Kauppila J.H., Lehenkari P., Harris K.W., Selander K.S. Chloroquine has tumor inhibitory and tumor promoting effects in triple negative breast cancer. Oncol. Lett. 2013;6:1665–1672. doi: 10.3892/ol.2013.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pliyev B.K., Menshikov M. Differential effects of the autophagy inhibitors 3-methyladenine and chloroquine on spontaneous and TNF-α-induced neutrophil apoptosis. Apoptosis. 2012;17:1050–1065. doi: 10.1007/s10495-012-0738-x. [DOI] [PubMed] [Google Scholar]
- 60.Aitken E.H., Alemu A., Rogerson S.J. Neutrophils and malaria. Front. Immunol. 2018;9:3005. doi: 10.3389/fimmu.2018.03005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wark P.A.B., Johnston S.L., Moric I., Simpson J.L., Hensley M.J., Gibson P.G. Neutrophil degranulation and cell lysis is associated with clinical severity in virus-induced asthma. Eur. Respir. J. 2002;19:68–75. doi: 10.1183/09031936.02.00226302. [DOI] [PubMed] [Google Scholar]
- 62.Camp J.V., Jonsson C.B. A role for neutrophils in viral respiratory disease. Front. Immunol. 2017;8:550. doi: 10.3389/fimmu.2017.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Labro M.T., Babin-Chevaye C. Effects of amodiaquine, chloroquine, and mefloquine on human polymorphonuclear neutrophil function in vitro. Antimicrob. Agents Chemother. 1988;32:1124–1130. doi: 10.1128/aac.32.8.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Romanelli F., Smith K.M., Hoven A.D. Chloroquine and hydroxychloroquine as inhibitors of human immunodeficiency virus (HIV-1) activity. Curr. Pharm. Des. 2004;10:2643–2648. doi: 10.2174/1381612043383791. [DOI] [PubMed] [Google Scholar]
- 65.Cortegiani, Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care. 2020;45:50. doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patil V.M., Singhal S., Masand N. A systematic review on use of aminoquinolines for the therapeutic management of COVID-19: efficacy, safety and clinical trials. Life Sci. 2020;254(2020) doi: 10.1016/j.lfs.2020.117775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;4:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 68.Mahévas M., Tran V.T., Roumier M., Chabrol A., Paule R., Guillaud C., Fois E., Lepeule R., Szwebel T.A., Lescure F.X., Schlemmer F. Clinical efficacy of hydroxychloroquine in patients with covid-19 pneumonia who require oxygen: observational comparative study using routine care data. BMJ. 2020;369 doi: 10.1136/bmj.m1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boulware D.R., Pullen M.F., Bangdiwala A.S., Pastick K.A., Lofgren S.M., Okafor E.C., Skipper C.P., Nascene A.A., Nicol M.R., Abassi M., Engen N.W. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vinetz J.M. Lack of efficacy of hydroxychloroquine in covid-19. BMJ. 2020;369 doi: 10.1136/bmj.m2018. [DOI] [PubMed] [Google Scholar]
- 71.Hernandez A.V., Roman Y.M., Pasupuleti V., Barboza J.J., White C.M. Hydroxychloroquine or chloroquine for treatment or prophylaxis of COVID-19: a living systematic review. Ann. Intern. Med. 2020;2020 doi: 10.7326/M20-2496. [DOI] [PubMed] [Google Scholar]
- 72.Pandi-Perumal S.R., Zisapel N., Srinivasan V., Cardinali D.P. Melatonin and sleep in aging population. Exp. Gerontol. 2005;40:911–925. doi: 10.1016/j.exger.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 73.Radogna F., Diederich M., Ghibelli L. Melatonin: a pleiotropic molecule regulating inflammation. Biochem. Pharmacol. 2010;80:1844–1852. doi: 10.1016/j.bcp.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 74.Huang S.H., Liao C.L., Chen S.J., Shi L.G., Lin L., Chen Y.W., Cheng C.P., Sytwu H.K., Shang S.T., Lin G.J. Melatonin possesses an anti-influenza potential through its immune modulatory effect. J. Funct. Foods. 2019;58:189–198. doi: 10.1016/j.jff.2019.04.062. [DOI] [Google Scholar]
- 75.Silvestri M., Rossi G.A. Melatonin: its possible role in the management of viral infections-a brief review. Ital. J. Pediatr. 2013;39:61. doi: 10.1186/1824-7288-39-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boga J.A., Coto-Montes A., Rosales-Corral S.A., Tan D.X., Reiter R.J. Beneficial actions of melatonin in the management of viral infections: a new use for this “molecular handyman”? Rev. Med. Virol. 2012;22:323–338. doi: 10.1002/rmv.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nunes O.D.S., Pereira R.D.S. Regression of herpes viral infection symptoms using melatonin and SB-73: comparison with Acyclovir. J. Pineal Res. 2008;44:373–378. doi: 10.1111/j.1600-079X.2007.00538.x. [DOI] [PubMed] [Google Scholar]
- 78.Tan D.X., Korkmaz A., Reiter R.J., Manchester L.C. Ebola virus disease: potential use of melatonin as a treatment. J. Pineal Res. 2014;57:381–384. doi: 10.1111/jpi.12186. [DOI] [PubMed] [Google Scholar]
- 79.Shneider A., Kudriavtsev A., Vakhrusheva A. Can melatonin reduce the severity of COVID-19 pandemic? Int. Rev. Immunol. 2020:1–10. doi: 10.1080/08830185.2020.1756284. [DOI] [PubMed] [Google Scholar]
- 80.Simko F., Reiter R.J. Is melatonin deficiency a unifying pathomechanism of high risk patients with COVID-19? Life Sci. 2020;256:117902. doi: 10.1016/j.lfs.2020.117902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reiter R.J., Abreu-Gonzalez P., Marik P.E., Dominguez-Rodriguez A. Therapeutic algorithm for use of melatonin in patients with COVID-19. Front. Med. 2020;7:226. doi: 10.3389/fmed.2020.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang R., Wang X., Ni L., Di X., Ma B., Niu S., Liu C., Reiter R.J. COVID-19: melatonin as a potential adjuvant treatment. Life Sci. 2020;250 doi: 10.1016/j.lfs.2020.117583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Castillo R.R., Quizon G.R.A., Juco M.J.M., Roman A.D.E., de Leon D.G., Punzalan F.E.R., Guingon R.B.L., Morales D.D., Tan D.X., Reiter R.J. Melatonin as adjuvant treatment for coronavirus disease 2019 pneumonia patients requiring hospitalization (MAC-19 PRO): a case series. Melatonin Res. 2020;3:297–310. doi: 10.32794/mr11250063. [DOI] [Google Scholar]
- 84.Rudra D.S., Pal U., Maiti N.C., Reiter R.J., Swarnakar S. Melatonin inhibits matrix metalloproteinase-9 activity by binding to its active site. J. Pineal Res. 2013;54:398–405. doi: 10.1111/jpi.12034. [DOI] [PubMed] [Google Scholar]
- 85.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clarke N.E., Turner A.J. Angiotensin-converting enzyme 2: the first decade. Int. J. Hypertens. 2012;2012 doi: 10.1155/2012/307315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jin Y., Han H.C., Lindsey M.L. ACE inhibitors to block MMP-9 activity: new functions for old inhibitors. J. Mol. Cell. Cardiol. 2007;43:664–666. doi: 10.1016/j.yjmcc.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., Henni B.P. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020 doi: 10.15252/embj.20105114. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Batlle, Wysocki J., Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin. Sci. 2020;134:543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 93.Patel V.B., Zhong J.C., Grant M.B., Oudit G.Y. Role of the ACE2/angiotensin 1-7 axis of the renin–angiotensin system in heart failure. Circ. Res. 2016;118:1313–1326. doi: 10.1161/CIRCRESAHA.116.307708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hofmann H., Geier M., Marzi A., Krumbiegel M., Peipp M., Fey G.H., Gramberg T., Pöhlmann S. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem. Biophys. Res. Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ferrario C.M., Ahmad S., Joyner J., Varagic J. Advances in the renin angiotensin system: focus on angiotensin-converting enzyme 2 and angiotensin-(1–7) Adv. Pharmacol. 2010;59:197–233. doi: 10.1016/S1054-3589(10)59007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iwata M., H. Greenberg B. Ectodomain shedding of ACE and ACE2 as regulators of their protein functions. Curr. Enzym. Inhib. 2011;7:42–55. doi: 10.2174/157340811795713756. [DOI] [Google Scholar]
- 97.Simoes e Silva A.C., Silveira K.D., Ferreira A.J., Teixeira M.M. ACE2, angiotensin-(1-7) and M as receptor axis in inflammation and fibrosis. Br. J. Pharmacol. 2013;169:477–492. doi: 10.1111/bph.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schindler C., Bramlage P., Kirch W., Ferrario C.M. Role of the vasodilator peptide angiotensin-(1-7) in cardiovascular drug therapy. Vasc. Health Risk Manag. 2007;3:125–137. [PMC free article] [PubMed] [Google Scholar]
- 99.Cangussu L.M., de Castro U.G., do Pilar Machado R., Silva M.E., Ferreira P.M., dos Santos R.A., Campagnole-Santos M.J., Alzamora A.C. Angiotensin-(1-7) antagonist, A-779, microinjection into the caudal ventrolateral medulla of renovascular hypertensive rats restores baroreflex bradycardia. Peptides. 2009;30:1921–1927. doi: 10.1016/j.peptides.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 100.Yu C.M., Wong R.S., Wu E.B., Kong S.L., Wong J., Yip G.W., Soo Y.O., Chiu M.L., Chan Y.S., Hui D., Lee N. Cardiovascular complications of severe acute respiratory syndrome. Postgrad. Med. J. 2006;82:140–144. doi: 10.1136/pgmj.2005.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brann D., Tsukahara T., Weinreb C., Logan D.W., Datta S.R. Non-neural expression of SARS-CoV-2 entry genes in the olfactory epithelium suggests mechanisms underlying anosmia in COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.03.25.009084. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]