Abstract
Gene flow patterns and the genetic structure of domesticated crops like cotton are not well understood. Furthermore, marker-assisted breeding of cotton has lagged far behind that of other major crops because the loci associated with cotton traits such as fiber yield and quality have scarcely been identified. In this study, we used 19 microsatellites to first determine the population genetic structure and patterns of gene flow of superior germplasm resources in upland cotton. We then used association analysis to identify which markers were associated with 15 agronomic traits (including ten yield and five fiber quality traits). The results showed that the upland cotton accessions have low levels of genetic diversity (polymorphism information content = 0.427), although extensive gene flow occurred among different ecological and geographic regions. Bayesian clustering analysis indicated that the cotton resources used in this study did not belong to obvious geographic populations, which may be the consequence of a single source of domestication followed by frequent genetic introgression mediated by human transference. A total of 82 maker–trait associations were examined in association analysis and the related ratios for phenotypic variations ranged from 3.04% to 47.14%. Interestingly, nine SSR markers were detected in more than one environmental condition. In addition, 14 SSR markers were co-associated with two or more different traits. It was noteworthy that NAU4860 and NAU5077 markers detected at least in two environments were simultaneously associated with three fiber quality traits (uniformity index, specific breaking strength and micronaire value). In conclusion, these findings provide new insights into the population structure and genetic exchange pattern of cultivated cotton accessions. The quantitative trait loci of domesticated cotton identified will also be very useful for improvement of yield and fiber quality of cotton in molecular breeding programs.
Keywords: Domestication cotton, Fiber quality traits, Genetic exchange, Microsatellite markers, Yield
1. Introduction
Cotton is the most important renewable fiber and edible oil crop worldwide. The genus Gossypium L. contains more than 50 recognized species belonging to eight genome groups, of which four species, Gossypium herbaceum L. (A1), Gossypium arboretum L. (A2), Gossypium hirsutum L. (AD1), Gossypium barbadense L. (AD2), have been domesticated and cultivated in different regions worldwide (Wang et al., 2012). Cultivation of G. hirsutum, or upland cotton, contributes to over 95% of cotton production worldwide (Iqbal et al., 1997). Cotton traits such as high-yield and agroecological adaptability lead to significant economic benefits. Accordingly, most cotton breeding efforts have focused on improving these traits (e.g., fiber quality, lint yield, boll weight, seed weight) in upland cotton (Brubaker and Wendel, 1994, Ulloa and Meredith, 2000, Zhu et al., 2008).
Of the four cultivated Gossypium species, G. hirsutum shows the highest levels of genetic variation and gene flow (Wendel et al., 1992, Abdurakhmonov et al., 2009). However, some studies have indicated that the germplasm of G. hirsutum currently cultivated lacks genetic diversity and shows little sign of genetic introgression (Esbroeck et al., 1999, Campbell et al., 2010). This is likely because in addition to the initial bottleneck encountered during the domestication process, cotton breeding has frequently involved crossing and re-selection within small sets of breeding materials (May et al., 1995, Bradbury et al., 2007). The narrow genetic base of upland cotton has become a serious concern as limited genetic variation corresponds to limited allelic availability for continued genetic gain (Brown, 1983, Xu et al., 2001, Tyagi et al., 2014).
Cotton yield and fiber quality are typical quantitative traits, the phenotypes of which are the result of quantitative trait loci (QTLs), their interaction, the environment, and the interaction between QTLs and the environment. Understanding how QTLs and environmental conditions interact to influence phenotypes has generated numerous genetic models (Song et al., 2005, Dong et al., 2010). Traditionally, plant QTLs have been identified by linkage mapping, i.e., constructing a high-density molecular linkage map through limited parent hybridization, and localizing specific linkage segments associated with target traits (Fang et al., 2013). This approach was first used over twenty years ago to mine QTLs associated with agronomic and fiber traits of upland cotton (Shappley et al., 1998). In recent years, numerous QTLs in cotton varieties have been excavated through segregation analyses (Shen et al., 2005, Wang et al., 2006, Li et al., 2008). However, because of the limited number of markers used in upland cotton populations, the accuracy of QTLs is relatively low, which impedes further studies using map-based cloning and marker-assisted selection.
One alternative approach to studying complex quantitative traits and mapping target genes is association analysis (Thornsberry et al., 2001, Flint-Garcia et al., 2005, Shen et al., 2007, Li et al., 2016a, Li et al., 2016b). Previous studies have used association analysis in upland cotton to map QTLs to agronomic traits, including disease resistance (Mei et al., 2014, Zhao et al., 2014) and seed quality (Liu et al., 2015). Linkage disequilibrium-based association mapping and model-based Bayesian analysis of upland cotton has been used to investigate the population genetics and significant quality traits. For example, association analysis of G. hirsutum accessions has revealed the existence of potential genetic variation for primary fiber quality traits and shown that the development of fiber quality traits is influenced by environmental factors (Abdurakhmonov et al., 2009). Additional studies in cotton have used association analysis to detect associations between elite alleles and fiber quality traits (Cai et al., 2014, Islam et al., 2016, Nie et al., 2016, Ademe et al., 2017). Investigating both population structure and kinship has revealed that in upland cotton interspecific gene combinations appear to improve fiber length traits (Handi et al., 2017, Wang et al., 2017a, Wang et al., 2017b). Despite this progress, association mapping studies have been limited to single traits, e.g., lint yield or fiber quality and/or focused on either genetic structure or association analysis of the cotton quality traits (Islam et al., 2016, Ademe et al., 2017), while ignoring the effects of human transference on genetic exchange of superior cotton accessions.
In this study, we have evaluated genetic structure and gene flow pattern of 285 domestication cotton superior germplasm accessions and performed association analysis of nine yield components and six fiber quality traits using 19 polymorphic microsatellite markers. Our results will provide useful information for understanding the genetic base of upland cotton superior varieties and will also promote future high yield and excellent fiber quality breeding.
2. Materials and methods
2.1. Collection of upland cotton superior accessions
A total of 285 G. hirsutum accessions were selected for this study (Table S1). All selections were derived from five sources: Yangtze River region, China (136 varieties), Yellow River region, China (123 varieties), Northern China (8 varieties), Northwestern China (9 varieties), and the United States (9 varieties) (Table S1).
2.2. Field experiments and trait phenotyping
All 285 accessions of upland cotton were planted in each of four locations that have distinct environmental conditions: Jingzhou, Hubei Province, China in 2015 and 2016 (designated environments E1 and E2, respectively); Jiujiang, Jiangxi Province in China in 2015 and 2016 (designated E3 and E4, respectively); Xinjiang in China in 2015 and 2016 (designated E5 and E6, respectively); and Anyang, Henan Province in China in 2015, 2016 and 2017 (designated E7, E8, and E9, respectively). Each accession was grown in a plot having 40–45 plants in two rows, with 0.10 m between plants in each row and 0.45 m between rows. Field planting followed a randomized complete block design with three replications in each environment. Field management followed conventional standard field practices.
The ten plants in the middle of each row were tagged for scoring and harvesting cotton seed. The yield traits evaluated included growing period, plant height (cm), number of fruit branches per plant, height of first fruit (cm), number of first fruit node, number of bolls per plant, seed butter (g), lint percentage (%), boll weight (g), and unit area yield (g·m−2). The fiber quality traits evaluated were as follows: mean length of upper half fiber (mm), uniformity index (%), specific breaking strength (cN·tex−1), micronaire value, and elongation (%).
2.3. Marker screening and genotyping
Total genomic DNA was extracted from collected samples following the methods of Doyle and Doyle (1987) and stored at −20 °C. Amplification efficiency and polymorphism were randomly tested with 30 randomly selected microsatellite primer pairs (Nie et al., 2016). We found that 19 primer pairs generated polymorphic markers across all 285 G. hirsutum accessions (Table S2). Polymerase chain reaction (PCR) was performed in a volume of 10 μL containing 1 μL of DNA template, 0.3 μL of each primer (1000 × ), 5 μL of 2 × PCR Master Mix and 3.4 μL of ddH2O under the following conditions: 3 min at 94 °C, followed by 32 cycles of 30 s at 94 °C, 40 s at 53 °C, and 30 s at 72 °C, and then a final extension of 5 min at 72 °C. PCR products were firstly separated by 10% polyacrylamide gels and visualized by silver staining. Fragment sizes of each locus were estimated using Quantity One (Bio-Rad Laboratories, Berkeley city, CA, USA) and a 50 bp DNA ladders size standard (pBR322 DNA/MspI marker; Tiangen, Beijing, China). We then verified the polyacrylamide results by regenotyping all samples using an ABI3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA) and fluorescently labeled primers. We performed the PCR amplification using a Veriti 96-Well Thermal Cycler. The upper primers were labeled with 6-FAM, HEX, TAMRA, or ROX (Sangon, Shanghai, China) (Table S2). The PCR reaction included 1 μL DNA template, 0.3 μL each primer (1000 × ), 10 μL2 × PCR Master Mix and 8.4 μL ddH2O. ABI3730 DNA analyzer (Applied Biosystems) was used to score genotypes and GeneMarkers v. 2.0 was used for binning (Holland and Parson, 2011).
2.4. Data analysis
Popgene v. 1.32 (Yeh and Boyle, 1996) was used to calculate linkage disequilibrium (LD) of all SSR markers (Table S3). Genetic diversity parameters of G. hirsutum from each source region were evaluated per locus using the following descriptive summary statistics: number of alleles (NA), observed (Ho) and expected (He) heterozygosity, and inbreeding coefficient (FIS) using GenAlEx v. 6.5 (Peakall and Smouse, 2012). CERVUS v. 3.0 (Kalinowski et al., 2007) was used to calculate polymorphismin formation content (PIC). In addition, Arlequin v. 3.5 (Excoffier and Lischer, 2010) was used to test the Hardy–Weinberg equilibrium (HWE) for all SSR markers (Table S4). We also performed a hierarchical analysis of genetic differentiation using an analysis of molecular variance (AMOVA) with 1000 permutations as implemented in Arlequin v. 3.5 (Excoffier and Lischer, 2010). The significance of fixation indices was tested using 10,000 permutations.
We used Migrate-n v. 3.6.11 (Beerli and Felsenstein, 1999) to investigate long-term effective population sizes (θ) and migration rates (Nm) of G. hirsutum accessions from five geographic regions. We also calculated these measures for two subpopulations (P1 and P2) that had higher likelihood at K = 2 according to our evaluation of population structure. Subpopulation P1 contained 80 lines, including 43 cultivars from Yellow River region, 36 cultivars from Yangtze River region, and one line from Northern China region; subpopulation P2 contained 205 accessions including 80 lines from Yellow River regions, 100 cultivars from Yangtze River region, nine lines introduced from US, andeight lines both from Northern China and Northwestern China regions. Migrate-n uses coalescence theory to model population sizes and migration rates, and mutation models to explain change of alleles at sites over time. Bayesian analysis was run using the infinite allele option, which is recommended when the mutation model is unknown. Uniform prior for θ was set to min: 0.0, max: 100.0, delta: 10.0. Uniform prior for migration was set to 0.0, max: 1000.0, delta: 100.0. STRUCTURE analysis was run using 100,000 burn-in MCMC iterations, with a length of 1,000,000 iterations, and eight replicates per run for K = 1–8 clusters with admixture model (Pritchard et al., 2000). The website program STRUCTURE HARVESTER was used to calculate the optimal value of K (Earl and Vonholdt, 2012) using the delta K criterion (Evanno et al., 2005, Jakobsson and Rosenberg, 2007). The corresponding Q-matrix at K = 2 was obtained for further marker–trait association analysis.
Many cotton traits are complicated quantitative traits; therefore, the most stable markers are those that can be detected at the same time in multiple populations and multiple environments (Li et al., 2016a, Li et al., 2016b). General linear model (GLM) and mixed linear model (MLM) are two prevalent statistical models in association mapping (Cardon and Palmer, 2003). To generate more accurate correlations with less-inflated type I errors (Yu et al., 2006), the MLM (+K + Q) method was employed in the present study (Cardon and Palmer, 2003). Considering the cultivation history of upland cotton and the relatively simple population structure in this panel, GLM (+Q) was also employed, and the results derived from the GLM and MLM were compared. TASSEL v. 2.1 was used to determine relative kinship among the individuals of experimental materials (Yu et al., 2006), and the relative coefficient matrix (K-matrix), which was obtained for subsequent association analysis. General Linear Model (GLM) and Mixed Linear Model (MLM) were used to construct markers-yield and fiber quality traits association tests within TASSEL v. 2.1 (Yu et al., 2006); we set the number of permutations in “define F tests” to 1000.
3. Results
3.1. Phenotypic variation of G. hirsutum accessions grown in different environments
G. hirsutum yield and fiber quality traits displayed broad variation when grown under different environmental conditions (Table 1). For example, the average phenotypic values of ten upland cotton yield traits for accessions grown innine environments are as follows: Growth period (GP) was 131.93 d (range 126.31–142.19 d), plant height (PH) was 93.04 cm (range 58.80–117.97 cm), the number of fruit branches per plant (NFB) was 11.75 (range 8.00–17.39), the height of first fruit (HF) was 18.74 cm (range 14.59–24.33 cm), the number of first fruit node (ND) was 6.74 (range 5.20–8.11), the number of bolls per plant (NB) was 19.22 (range 6.59–33.89), seed butter (SB) was 25.35 g (range 9.87–37.22 g), lint percentage (LP) was 37.61% (range 35.99–41.60%), boll weight (BW) was 5.22 g (range 4.38–6.00 g) and unit area yield (UAY) was 300.36 g m−2 (range 149.05–541.71 g m−2). The average phenotypic values of five upland cotton quality traits for accessions grown in nine environments are as follows: the mean length of upper half fiber (LF) was 28.97 mm (range 27.73–29.54 mm), uniformity index (UI) was 84.57% (range 82.20–85.62%), specific breaking strength (BS) was 28.31 cN·tex−1 (range 26.24–30.39 cN·tex−1), miconaire value (MV) was 4.98 (range 4.71–5.26) and fiber elongation (FE) was 6.71% (range 6.67–6.76%).
Table 1.
Statistical analysis for yield and fiber quality traits of 285 upland cotton accessions.
| Trait | GP |
PH |
NFB |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Min | Max | CV (%) | Mean | SD | Min | Max | CV (%) | Mean | SD | Min | Max | CV (%) | |
| E1 | 126.31 | 3.67 | 110 | 132 | 2.90% | 104.32 | 13.07 | 74.60 | 142.80 | 12.53% | 13.44 | 2.76 | 8.40 | 36.20 | 20.52% |
| E2 | 127.48 | 2.81 | 114 | 132 | 2.20% | 116.21 | 12.64 | 78.00 | 194.20 | 10.88% | 13.25 | 1.55 | 8.60 | 17.20 | 11.73% |
| E3 | 142.19 | 7.76 | 123 | 152 | 5.45% | 102.90 | 11.73 | 69.38 | 138.88 | 11.40% | 16.17 | 1.50 | 12.50 | 25.38 | 9.29% |
| E4 | 141.44 | 6.93 | 123 | 152 | 4.90% | 117.97 | 15.22 | 78.63 | 163.38 | 12.90% | 17.39 | 1.64 | 13.13 | 22.13 | 9.42% |
| E5 | 126.75 | 5.16 | 108 | 138 | 4.07% | 62.33 | 11.13 | 34.60 | 148.40 | 17.86% | 8.17 | 1.12 | 5.30 | 11.10 | 13.67% |
| E6 | 127.39 | 5.93 | 110 | 139 | 4.65% | 58.80 | 9.36 | 36.30 | 85.10 | 15.92% | 8.00 | 1.14 | 5.10 | 11.80 | 14.27% |
| E7 | 89.28 | 13.99 | 54.17 | 123.33 | 15.67% | 9.28 | 2.30 | 3.50 | 14.83 | 24.78% | |||||
| E8 | 91.55 | 12.16 | 60.83 | 129.17 | 13.29% | 9.59 | 1.90 | 5.17 | 13.83 | 19.79% | |||||
| E9 | 93.99 | 13.19 | 54.17 | 129.17 | 14.04% | 10.46 | 2.30 | 3.67 | 28.67 | 21.98% | |||||
| Mean | 131.93 | 4.03% | 93.04 | 13.83% | 11.75 | 16.16% | |||||||||
| HF |
ND |
NB |
SB |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Min | Max | CV (%) | Mean | SD | Min | Max | CV (%) | Mean | SD | Min | Max | CV (%) | Mean | SD | Min | Max | CV (%) |
| 15.12 | 2.12 | 7.20 | 21.20 | 14.05% | 7.94 | 1.32 | 5.20 | 12.00 | 16.62% | 18.56 | 3.26 | 9.60 | 29.80 | 17.56% | 13.15 | 1.30 | 10.10 | 17.80 | 9.87% |
| 15.36 | 2.54 | 6.60 | 24.60 | 16.51% | 8.11 | 1.36 | 4.60 | 12.40 | 16.74% | 20.39 | 2.45 | 10.40 | 28.20 | 12.00% | 13.18 | 1.30 | 9.10 | 16.70 | 9.84% |
| 14.87 | 2.61 | 9.13 | 22.88 | 17.56% | 7.16 | 1.19 | 4.25 | 11.00 | 16.63% | 33.63 | 7.43 | 15.33 | 57.29 | 22.10% | 35.09 | 6.99 | 0.00 | 44.40 | 19.93% |
| 14.59 | 2.34 | 9.13 | 23.25 | 16.03% | 6.79 | 0.94 | 4.50 | 9.75 | 13.89% | 33.89 | 7.18 | 15.88 | 55.00 | 21.18% | 35.99 | 4.47 | 0.00 | 44.55 | 12.42% |
| 19.44 | 3.57 | 10.80 | 28.90 | 18.34% | 5.24 | 0.55 | 3.50 | 6.50 | 10.48% | 6.59 | 1.79 | 2.10 | 12.90 | 27.13% | 9.87 | 1.12 | 7.05 | 14.02 | 11.36% |
| 18.20 | 3.58 | 7.90 | 30.30 | 19.69% | 5.20 | 0.55 | 3.60 | 6.90 | 10.59% | 6.80 | 1.66 | 2.60 | 13.80 | 24.38% | 10.10 | 1.07 | 7.40 | 15.00 | 10.59% |
| 24.33 | 3.84 | 13.67 | 35.83 | 15.79% | 15.79 | 3.86 | 3.67 | 26.67 | 24.43% | 37.22 | 3.89 | 19.98 | 55.16 | 10.45% | |||||
| 23.60 | 5.15 | 9.50 | 44.50 | 21.81% | 17.72 | 4.01 | 6.50 | 29.17 | 22.62% | 36.72 | 4.47 | 15.33 | 77.48 | 12.16% | |||||
| 23.09 | 4.58 | 6.17 | 39.17 | 19.81% | 19.58 | 4.79 | 7.50 | 37.17 | 24.47% | 36.87 | 3.95 | 17.93 | 62.91 | 10.73% | |||||
| 18.74 | 17.73% | 6.74 | 14.16% | 19.22 | 21.76% | 25.35 | 11.93% | ||||||||||||
| LP |
BW |
UAY |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Min | Max | CV (%) | Mean | SD | Min | Max | CV (%) | Mean | SD | Min | Max | CV (%) |
| 36.14 | 4.39 | 9.35 | 44.61 | 12.15% | 4.50 | 0.70 | 1.28 | 6.02 | 15.56% | 327.20 | 82.81 | 85.12 | 609.52 | 25.31% |
| 36.41 | 4.20 | 16.42 | 39.59 | 11.54% | 4.51 | 0.70 | 1.61 | 5.39 | 15.42% | 348.51 | 86.96 | 101.24 | 629.69 | 24.95% |
| 36.11 | 3.69 | 8.10 | 17.40 | 12.15% | 4.38 | 0.62 | 2.40 | 6.43 | 14.16% | 185.98 | 58.79 | 34.58 | 351.63 | 31.61% |
| 35.99 | 3.93 | 9.90 | 18.30 | 11.54% | 4.51 | 0.59 | 3.08 | 6.39 | 12.99% | 209.64 | 66.05 | 46.72 | 510.32 | 31.51% |
| 41.40 | 3.70 | 0.20 | 0.50 | 10.22% | 5.54 | 0.67 | 2.63 | 7.71 | 12.03% | 488.30 | 164.55 | 51.96 | 1033.86 | 33.70% |
| 41.60 | 3.24 | 0.30 | 0.48 | 10.92% | 5.64 | 0.61 | 3.34 | 7.48 | 10.85% | 541.71 | 152.78 | 49.73 | 915.95 | 28.20% |
| 37.22 | 3.89 | 8.65 | 15.43 | 8.94% | 5.91 | 0.64 | 4.23 | 8.20 | 10.78% | 149.05 | 66.34 | 7.63 | 402.71 | 44.51% |
| 36.72 | 4.47 | 8.36 | 15.68 | 7.79% | 6.00 | 0.65 | 3.08 | 7.89 | 10.90% | 227.82 | 98.33 | 34.76 | 574.59 | 43.16% |
| 36.87 | 3.95 | 8.98 | 14.28 | 10.71% | 5.96 | 0.64 | 4.11 | 7.42 | 10.80% | 225.01 | 100.68 | 17.30 | 560.39 | 44.75% |
| 37.61 | 10.54% | 5.22 | 12.61% | 300.36 | 34.19% | |||||||||
| LF |
UI |
BS |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Min | Max | CV (%) | Mean | SD | Min | Max | CV (%) | Mean | SD | Min | Max | CV (%) |
| 29.54 | 1.44 | 23.50 | 33.70 | 4.87% | 85.52 | 1.41 | 77.20 | 88.40 | 1.65% | 30.39 | 0.51 | 24.10 | 38.20 | 1.69% |
| 29.51 | 1.58 | 23.50 | 33.80 | 5.36% | 85.58 | 1.21 | 78.90 | 88.40 | 1.42% | 29.85 | 2.17 | 23.60 | 35.50 | 7.28% |
| 29.08 | 1.52 | 22.90 | 33.00 | 5.22% | 84.71 | 1.45 | 78.00 | 88.00 | 1.71% | 28.19 | 1.66 | 23.50 | 33.70 | 5.87% |
| 29.36 | 1.49 | 22.40 | 33.50 | 5.09% | 84.91 | 1.53 | 78.70 | 87.80 | 1.80% | 28.30 | 1.66 | 22.80 | 32.60 | 5.85% |
| 27.73 | 1.13 | 24.40 | 31.20 | 4.09% | 82.20 | 1.72 | 75.30 | 86.20 | 2.09% | 26.24 | 1.38 | 22.60 | 33.70 | 5.27% |
| 27.84 | 1.08 | 24.70 | 31.00 | 3.87% | 82.22 | 1.38 | 78.60 | 86.10 | 1.68% | 26.70 | 1.26 | 23.40 | 33.20 | 4.70% |
| 29.18 | 1.24 | 24.10 | 33.00 | 4.26% | 85.05 | 1.42 | 80.60 | 88.90 | 1.67% | 28.31 | 1.70 | 22.90 | 34.80 | 6.02% |
| 29.30 | 1.47 | 22.80 | 33.30 | 5.03% | 85.62 | 1.44 | 79.50 | 88.90 | 1.69% | 28.57 | 1.89 | 21.80 | 36.10 | 6.60% |
| 29.23 | 1.50 | 22.00 | 33.60 | 5.14% | 85.36 | 1.26 | 80.50 | 88.30 | 1.48% | 28.25 | 1.94 | 22.50 | 35.60 | 6.85% |
| 28.97 | 4.77% | 84.57 | 1.69% | 28.31 | 5.57% | |||||||||
| MV |
FE |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Min | Max | CV (%) | Mean | SD | Min | Max | CV (%) |
| 5.21 | 2.01 | 3.80 | 6.30 | 38.63% | 6.76 | 0.09 | 6.40 | 6.90 | 1.36% |
| 5.17 | 0.59 | 3.60 | 6.10 | 11.43% | 6.74 | 0.10 | 6.40 | 7.00 | 1.42% |
| 5.26 | 0.45 | 4.00 | 6.30 | 8.48% | 6.75 | 0.10 | 6.50 | 7.00 | 1.52% |
| 5.13 | 0.47 | 3.20 | 6.10 | 9.16% | 6.72 | 0.09 | 6.40 | 6.90 | 1.29% |
| 4.71 | 0.50 | 2.30 | 5.80 | 10.66% | 6.67 | 0.09 | 6.40 | 6.90 | 1.40% |
| 4.74 | 0.45 | 2.60 | 5.80 | 9.44% | 6.69 | 0.08 | 6.40 | 6.90 | 1.25% |
| 4.88 | 0.43 | 3.10 | 5.80 | 8.87% | 6.68 | 0.10 | 6.40 | 7.00 | 1.46% |
| 4.86 | 0.41 | 3.60 | 6.00 | 8.36% | 6.69 | 0.11 | 6.30 | 7.00 | 1.66% |
| 4.84 | 0.44 | 3.70 | 6.10 | 9.11% | 6.69 | 0.12 | 6.20 | 7.30 | 1.79% |
Note: GP, growth period (d); PH, plant height (cm); NFB, the number of fruit branches per plant; HF, the height of first fruit (cm); ND, the number of first fruit node; NB, the number of bolls per plant; SB, seed butter (g); LP, lint percentage (%); BW, boll weight (g); UAY, unit area yield (g·m−2); LF, the mean length of upper half fiber (mm); UI, uniformity index (%); BS, specific breaking strength (cN·tex−1); MV, micronaire value (%); FE, fiber elongation (%). E1-E9 indicate Jingzhou in 2015, 2016; Jiujiang in 2015, 2016; Xinjiang in 2015, 2016, and Anyang in 2015, 2016 and 2017, respectively.
For yield traits, the coefficientof variance ranged from 4.03% (growth period) to 34.19% (unit area yield). Phenotypic variation in fiber quality traits was lower, with the coefficient of variance ranging from 1.46% (fiber elongation) to 12.68% (miconaire value).
3.2. Genetic diversity
We used 19 SSR markers to examine the genetic diversity of G. hirsutum accessions from different geographic regions and their relationships. The expected heterozygosity (He) and observed heterozygosity (Ho) ranged from 0.417 and 0.715 in G. hirsutum accessions from Northwestern China to 0.493 and 0.782 in the accessions from the Yangtze River region, with an average of 0.469 and 0.759, respectively. Moreover, the effective number of alleles per locus (Ne) varied from 1.903 in the accessions from Northwestern China to 2.121 in the accessions from the Yangtze River region, with a mean of 2.032 (Table 2). The average polymorphism information content (PIC) was 0.427 (ranging from 0.071 to 0.712) (Table S5). The average fixation indices values (F) were lower than zero for all accessions. These negative values indicate an excess of heterozygotes. The relatively small values of PIC suggest that the genetic diversity of the upland cotton cultivars examined in this study is relatively low.
Table 2.
Genetic diversity of the tested cotton accessions revealed by simple sequence repeat (SSR) markers.
| Group | N | Na | Ne | I | Ho | He | F | PPL (%) |
|---|---|---|---|---|---|---|---|---|
| YtRr | 136 | 4.421 | 2.121 | 0.812 | 0.782 | 0.493 | −0.524 | 100.00% |
| YRr | 122 | 4.526 | 2.077 | 0.804 | 0.757 | 0.481 | −0.487 | 100.00% |
| US | 10 | 2.579 | 2.074 | 0.761 | 0.798 | 0.488 | −0.527 | 100.00% |
| NC | 8 | 2.316 | 1.987 | 0.713 | 0.743 | 0.467 | −0.526 | 100.00% |
| NWC | 9 | 2.105 | 1.903 | 0.626 | 0.715 | 0.417 | −0.661 | 84.21% |
| Total | 285 | 3.189 | 2.032 | 0.743 | 0.759 | 0.469 | −0.541 | 96.84% |
Note: N, the number of individuals for the group; Na, the mean number of alleles per locus; Ne,theeffective number of alleles per locus; I, Shannon's Information Index; Ho, the observed heterozygosity; He, the expected heterozygosity; F, fixation index; PPL, proportion of polymorphic loci. YtRr, Yangtze River region; YRr, Yellow River region; US, the United States; NC, Northern China; NWC, Northwestern inland.
3.3. Population structure and gene flow pattern
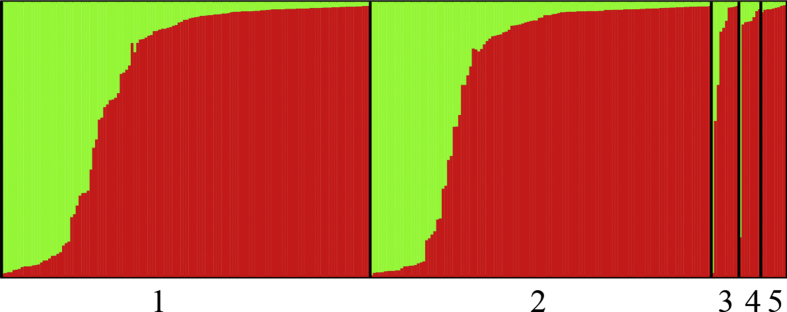
Bayesian clustering analysis revealed that G. hirsutum accessions were not clustered together according to geographic area or pedigree origin. The most likely value showed a much higher likelihood at K = 2, suggesting that the total panel could be divided into two subpopulations, designated P1 and P2 (Fig. S1; Fig. S2, Fig. 1). The P1 subpopulation contained 80 lines, including 43 cultivars (43/123, 35.0%) from the Yellow River region, 36 cultivars (36/136, 26.5%) from the Yangtze River region, and one line (1/9, 11.1%) from the Northern China region. The P2 subpopulation consisted of 205 accessions, including 80 lines (80/123, 65%) from the Yellow River region, 100 (100/136, 73.5%) cultivars from the Yangtze River region, nine lines (9/9100%) introduced from US, and eight lines both (8/8, 100%; 8/9, 88.9%) from the Northern and Northwestern China regions (Fig. S1). The corresponding Q matrix (at k = 2) was further used for marker–trait association mapping (Fig. S3). In addition, based on the results of the relatedness analysis, a K-matrix was also constructed for the association mapping.
Fig. 1.
Population structure of 285 upland cotton acccessions based on 19 polymorphic SSR markers.1–5 indicate Yangtze River region, Yellow river region, US, Northern China, Northwestern China, respectively.
Population structure analysis showed that the among-groups component of genetic variance was 0.05%, −98.06% among individuals within groups, indicating an excess of heterozygotes in accessions of the two subpopulations. The within-individuals component was 198.01% (Table 3). These results indicate that the variation among different cotton individuals contributed most to the overall variation. In addition, the Migrate-n analysis of the five geographic groups produced θ and M values greater than zero (Table 4). θ values did not vary among range sectors. Moreover, scaled immigration rates (M) revealed the existence of extensive historical gene flow between all five sectors. Gene movements occurred predominantly from northernto northwestern China, northwestern China to northern China (40.3744 vs 40.1705) and the Yellow River region (33.6081 vs. 21.9127), followed by the Yangtze River region to the US (34.9493 vs. 13.0342). These results illustrate that frequent human-induced gene flow may have occurred between the northern and northwestern China regions, the northwestern China region and the Yangtze River region, as well as into the Yellow River region. Additionally, the population Migrate-n analysis showed that there was a subtler deviation in the direction of migration from subpopulation P2 to subpopulation P1 (11.2123) than from subpopulation P1 to subpopulation P2 (7.8593) (Table 5).
Table 3.
Analysis of molecular variance (AMOVAs) for upland cotton accessions.
| Source of variation | nSSR |
||||
|---|---|---|---|---|---|
| d.f. | SS | VC | Variation (%) | Fixation Indices | |
| Among groups | 4 | 0.058 | 0.00012 Va | 0.05 | 0.00049 |
| Among individuals within groups | 280 | 1.294 | −0.23979 Vb | −98.06 | −0.98109 |
| Within individuals | 285 | 138 | 0.48421 Vc | 198.01 | −0.98012 |
| Total | 569 | 139.353 | 0.24454 | ||
Note: d.f, degree of freedom; SS, sum of squares; VC, variance of component.
Table 4.
Historical gene flow among five geographical groups estimated by Migrate-n.
| M (m/μ) | ||||||
|---|---|---|---|---|---|---|
| Group | θ | YtRr→ | YRr→ | US→ | NC→ | NWC→ |
| YtRr | 0.6701 (0.6491–0.6917) | 11.7276 (10.6690–12.7475) | 13.034 2 (12.0322–14.0861) | 5.8929 (5.2196–6.6921) | 5.2464 (4.6233–5.9303) | |
| YRr | 0.3927 (0.3787–0.4075) | 15.4807 (14.0052–17.0597) | 27.4881 (25.4093–29.6343) | 11.9983 (10.5221–13.4984) | 33.6081 (31.3326–35.9713) | |
| US | 0.3382 (0.3131–0.3671) | 34.9493 (32.2913–37.7283) | 22.2793 (20.1943–24.4996) | 21.0072 (18.9624–23.1736) | 9.0469 (7.7550–10.4807) | |
| NC | 0.2118 (0.1940–0.2308) | 18.8081 (16.6861–21.0671) | 10.1301 (8.5951–11.8135) | 27.9258 (25.2810–30.7003) | 40.3744 (37.3093–43.6055) | |
| NWC | 0.5339 (0.4876–0.5862) | 6.7376 (5.6901–7.9744) | 21.9127 (19.8790–23.9833) | 8.7922 (7.6155–10.0961) | 40.1705 (37.4994–42.9205) | |
Note: YtRr, Yangtze River region; YRr, Yellow River region; US, the United States; NC, Northern China; NWC, Northwestern inland; bold value, Maximum likelihood estimation.
Table 5.
Historical gene flow between two subpopulations sorted by Q-matrix estimated by Migrate-n.
| Pop | M (m/μ) |
||
|---|---|---|---|
| θ | 1→ | 2→ | |
| 1 | 1.6788 (1.6028–1.7596) | 11.2123 (10.5404–12.0280) | |
| 2 | 1.6074 (1.5655–1.6506) | 7.8539 (7.4344–8.2939) | |
Note: bold value, Maximum likelihood estimation.
3.4. Association analysis of yield and fiber quality traits
Linkage disequilibrium (LD) tests showed that the level of LD for G. hirsutum accessions from five ecological areas were 43, nine, zero, zero, and zero (at the significance level of p < 0.05). This finding indicates that most SSR markers did not exist linkage disequilibrium (Table S3).
For all 15 agronomic traits, including ten yield component traits and five fiber quality traits, we applied general linear model (GLM) and mixed linear model (MLM) to analyze nine environment datasets derived from the 285 accessions at four locations over two years in Jingzhou, Jiujiang, and Xinjiang as well as three years in Anyang. When we compared the results of the GLM to MLM, we found that a total of 82 maker–trait associations were detected between 17 SSR markers (p = 0.05) and 15 agronomic traits in nine environments (Table S6). The number of SSR markers associated with G. hirsutum yield traits and fiber quality traits are shown in Table 6.
Table 6.
Loci associated with more than two traits in nine environments (P < 0.05).
| Traits | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| loci | GP | PH | NFB | HF | ND | NB | LP | SB | BW | UAY | LF | UI | BS | MV | FE |
| NAU5077 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | ||||
| NAU5233 | √ | √ | √ | √ | |||||||||||
| NAU4932 | √ | √ | |||||||||||||
| NAU4956 | √ | √ | |||||||||||||
| NAU5013 | √ | √ | √ | ||||||||||||
| NAU5120 | √ | √ | |||||||||||||
| NAU5088 | √ | √ | |||||||||||||
| NAU5195 | √ | √ | √ | ||||||||||||
| NAU4860 | √ | √ | √ | √ | √ | √ | √ | √ | |||||||
| NAU5017 | √ | √ | |||||||||||||
| NAU5227 | √ | √ | √ | ||||||||||||
| NAU4951 | √ | √ | √ | √ | |||||||||||
| NAU5260 | √ | √ | |||||||||||||
| NAU5148 | √ | √ | √ | ||||||||||||
Note: GP, growth period (d); PH, plant height (cm); NFB, the number of fruit branches per plant; HF, the height of first fruit (cm); ND, the number of first fruit node; NB, the number of bolls per plant; SB, seed butter (g); LP, lint percentage (%); BW, boll weight (g); UAY, unit area yield (g·m−2); LF, the mean length of upper half fiber (mm); UI, uniformity index (%); BS, specific breaking strength (cN·tex−1); MV, micronaire value (%); FE, fiber elongation (%). √ indicates the marker associated with the traits.
Association analysis further showed that 17 SSR markers are associated with from 3.04% to 22.35% of the phenotypic variation in G. hirsutum accessions (Table S7). Interestingly, the marker NAU5077 was simultaneously associated with 11 traits, including GP, PH, NFB, ND, LP, UAY, LF, UI, BS, MV and FE. Among these traits, UI, BS and MV were closely related to fiber quality (Table 6). The marker NAU4860 was simultaneously associated with NB, SB, BW, LF, UI, BS, MV and FE, three of which are fiber quality traits. Further, NAU5233 was simultaneously associated with GP, UAY, BS and FE; NAU4951 was simultaneously associated with HF, NB, SB and MV. NAU5013 was simultaneously associated with PH, LP and SB; NAU5195 was simultaneously associated with HF, ND and LP. NAU5148 was simultaneously associated with HF, BW and UAY. NAU5260 was simultaneously associated with NB and SB. NAU4932 and NAU4956 were simultaneously associated with GP and FE. NAU5120 and NAU5088 were simultaneously associated with NFB and NB; NAU5017 was simultaneously associated with HF and UI, and NAU5227 was simultaneously associated with LP, LF and MV.
4. Discussion
The geographic distribution of genetic variation in species is significantly associated with their evolutionary potential and future fate (Wendel et al., 1992). Generally, the domesticated crops have less genetic variability than their wildrelatives (Wendel and Cronn, 2003, Cao et al., 2014). Most cotton varieties planted in China were derived from a limited number of founder parents, such as DPL (a cotton germplasm type), Stoneville, King, Uganda, Foster, and Trice (Chen and Du, 2006). Therefore, to create association maps, it is especially critical to select samples that encompass as much genetic diversity as possible. In this study, the population panel consisted of 285 cultivars, including lines from cotton germplasm resources, historical varieties from abroad, multiple lines derived from radiation breeding programs, and some progenies of intra- and inter species. Our results showed that the level of diversity in upland cotton varieties was relatively low (PIC = 0.427), with an average number of alleles per locus of 3.2 (ranging from 2.2 and 6.6 alleles/locus). These results are similar to those detected in the variations analysis of 241 G. hirsutum cotton cultivars (Zhao et al., 2014). The average number of alleles per locus, gene diversity, and PIC in our study were less than those detected in 35 cultivars and eight inbred lines of G. hirsutum from Africa, United States, and Brazil (Lacape et al., 2007). On possible explanation for this difference is that cultivars domesticated directly in a native cotton growing area usually preserve higher levels of polymorphism than those cultivated in a non-native cotton growing areas. In our study, the average genetic diversity and PIC values were higher than those detected in previous research (Qin et al., 2015). Our results indicate that the selected markers have sufficient polymorphic information to reveal the genetic relationships between these upland cotton inbred cultivars.
Although our selection of samples emphasized genetic diversity as much as possible, when we compared the genetic diversity of five ecological groups in our study, we found that genetic differentiation among these ecological areas was still very small. This finding might be explained by frequent gene exchange and germplasm domestication events in different ecotype collections, reflecting the probable extensive exchange of parental lines by breeders. Meanwhile, the low genetic differentiation of upland cotton in five ecological regions is also probably associated with the single origin of its domestication. Over 95% of cultivated cotton crop worldwide is allotetraploid upland cotton, which was possibly initially domesticated in northern Yucatan peninsula (Stephens, 1958, Wendel et al., 2009, Coppens d'Eeckenbrugge and Lacape, 2014, Fang et al., 2017). Therefore, the low genetic differentiation of allotetraploid cotton is likely the result of frequent gene exchange with a restricted domestication source. Similarly, numerous studies have shown that within Gossypium species genetic diversity is low (Abdalla et al., 2001, Iqbal et al., 2001, Rungis et al., 2005, Lacape et al., 2007).
Additionally, population structure is important for explaining the heterogeneity of genetic architecture and is mostly affected by geographic isolation and genetic exchange isolation (Guo et al., 1997, Gutiérrez et al., 2002). We found that instead of being separated in accordance with their geographic origins, all 285 accessions could be classified into two subpopulations. This classification indicates that when upland cotton germplasm is intersperesed or crossbred, genetic exchange may occur frequently, independent of geographic restriction. These results may provide important insight into evaluating the effects of cross-breeding in molecular breeding programs.
Our results provide strong support for the validity of trait-association results. We identified nine markers that are associated with upland cotton yield traits and two markers that are associated with fiber quality within specific environments. These findings demonstrate that traits related to yield quality in G. hirsutum germplasm have potential genetic variation that may be useful for future breeding programs. We also found that yield and fiber quality traits are correlated with diverse environments. These findings suggest that agriculturalists attempting to select target traits with the same cotton cultivar but in different environments should consider using different practices (Zhang et al., 2005).
The major target traits of cotton during the breeding process are quantitative; thus, phenotypic variation of each trait is directly or indirectly affected by that of other traits (Huang et al., 2018, Keerio et al., 2018, Wen et al., 2018). Numerous QTL mapping studies aimed at improving cotton fiber quality and yield traits have been previously reported (Mei et al., 2013, Adhikari et al., 2017, Wang et al., 2017a, Wang et al., 2017b, Dong et al., 2018a, Dong et al., 2018b). In this study, a total of 16 SSR markers for yield and fiber quality traits were detected. Among these markers, 14 were simultaneously associated with more than two traits, which may have resulted from gene–gene interactions or pleiotropism (Zeng et al., 2009, Lehner, 2011). For example, the SSR markers NAU5077 and NAU4860 were simultaneously associated with numerous fiber quality traits, including LF, UI, BS, MV and FE. In addition, SSR markers NAU4951, NAU5013, NAU5195, NAU5148, NAU5260, NAU5088, and NAU5017 were mainly associated with yield quality traits, including HF, NB, SB, PH, LP. These identified associations for different fiber and yield quality traits in domesticated upland cotton, along with those reported in previous studies, add toa rich cluster of yield and fiber quality QTLs (Cai et al., 2014, Adhikari et al., 2017, Ademe et al., 2017, Dong et al., 2018a, Dong et al., 2018b).
In conclusion, we used 19 SSR markers to determine the genetic structure and gene flow patterns of 285 upland cotton accessions. We then identified which markers are associated with agronomic traits in upland cotton. Our results showed that the extensive gene flow occurred in different ecological and geographic regions for crossbreeding. Specific markers identified from association analysis are potential QTLs for the selected traits inselected cotton production regions and can provide more information for marker-assisted breeding programs.
Author contributions
ZL designed the work. NZ, TZ, WL, and XM performed the experiments. WL, XZ, XP, YL, KH, WZ, KZ, DY, FZ, and ZR contributed materials/analysis tools. ZL and TZ wrote the manuscript. ZL, XM, DY, and TZ revised the manuscript.
Declaration of Competing Interest
The authors declare that they have no conflict of interests.
Acknowledgments
This research was co-supported by grants from National Key R and D Program for Crop Breeding (2016YFD0100306), National Natural Science Foundation of China (No. 31401431), the Shaanxi Science and Technology Innovation Team (2019TD-012), the Public health specialty in the Department of Traditional Chinese Medicine (Grants no. 2017-66 and 2018–43), and the Open Foundation of the Key Laboratory of Resource Biology and Biotechnology in Western China (Ministry of Education) (Grants no. ZSK2017007 and ZSK2019008).
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.03.001.
Contributor Information
Xiong-Feng Ma, Email: maxf_caas@163.com.
Dai-Gang Yang, Email: yangdaigang@caas.cn.
Zhong-Hu Li, Email: lizhonghu@nwu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abdalla A.M., Reddy O.U.K., Ei-Zik K.M., Pepper A.E. Genetic diversity and relationships of diploid and tetraploid cottons revealed using AFLP. Theor. Appl. Genet. 2001;102:222–229. [Google Scholar]
- Abdurakhmonov I.Y., Saha S., Jenkins J.N., Buriev Z.T., Shermatov S.E., Scheffler B.E., Pepper A.E., Yu J.Z., Russell J.K., Abdukarimov A. A Linkage disequilibrium based association mapping of fiber quality traits in G. hirsutum L. variety germplasm. Genetica. 2009;136:401–417. doi: 10.1007/s10709-008-9337-8. [DOI] [PubMed] [Google Scholar]
- Ademe M.S., He S., Pan Z., Sun J.L., Wang Q.L., Qin H.D., Liu J.H., Liu H., Yang J., Xu D.Y., Yang J.L., Ma Z.Y., Zhang J.B., Li Z.K., Cai Z.M., Zhang X.L., Zhang X., Huang A.F., Yi X.D., Zhou G.Y., Li L., Zhu H.Y., Pang B.Y., Wang L.R., Jia Y.H., Du X.M. Association mapping analysis of fiber yield and quality traits in upland cotton (Gossypium hirsutum L.) Mol. Gen. Genet. 2017;292:1267–1280. doi: 10.1007/s00438-017-1346-9. [DOI] [PubMed] [Google Scholar]
- Adhikari J., Das S., Wang Z.N., Khanal S., Chandnani R., Patel J.D., Goff V.H., Auckland S., Rainville L.K., Jones D., Paterson A.H. Targeted identification of association between cotton fiber quality traits and microsatellite markers. Euphytica. 2017;213:65. [Google Scholar]
- Beerli P., Felsenstein J. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics. 1999;152:763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury P.J., Zhang Z.W., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Brown W.L. Genetic diversity and genetic vulnerability-an appraisal. Econ. Bot. 1983;37:4–12. [Google Scholar]
- Brubaker C.L., Wendel J.F. Re-evaluating the origin of domesticated cotton (Gossypium hirsutum; Malvaceae) using nuclear restriction fragment length polymorphism (RFLP) Am. J. Bot. 1994;81:1309–1326. [Google Scholar]
- Cai C.P., Ye W.X., Zhang T.Z., Guo W.Z. Association analysis of fiber quality traits and exploration of elite alleles in Upland cotton cultivars/accessions (Gossypium hirsutum L.) J. Integr. Plant Biol. 2014;56:51–62. doi: 10.1111/jipb.12124. [DOI] [PubMed] [Google Scholar]
- Campbell B.T., Saha S., Percy R., Frelichowski J., Jenkins J.N., Park W., Mayee C.D., Gotmare V., Dessauw D., Giband M., Jia X.D.Y., Constable G., Dillon S., Abdurakhmonov I.Y., Abdukarimov A., Rizaeva S.M., Abdullaev A., Barroso P.A.V., Pádua J.G., Hoffmann L.V., Podolnaya L. Status of the global cotton germplasm resources. Crop Sci. 2010;50:1161–1179. [Google Scholar]
- Cao K., Zheng Z.J., Wang L.R., Liu X., Zhu G.G., Fang W.C., Cheng S.F., Zeng P., Chen C.W., Wang X.W., Xie M., Zhong X., Wang X.L., Zhao P., Bian C., Zhu Y.L., Zhang J.H., Ma G.S., Chen C.X., Li Y.J., Hao F.G., Li Y., Huang G.D., Li Y.X., Li H.Y., Guo J., Xu X., Wang J. Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol. 2014;15:415. doi: 10.1186/s13059-014-0415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon L.R., Palmer L.J. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Chen G., Du X.M. Genetic diversity of source germplasm of upland cotton in China as determined by SSR marker analysis. Acta Genetica Sin. 2006;33:733–745. doi: 10.1016/S0379-4172(06)60106-6. [DOI] [PubMed] [Google Scholar]
- Coppens d'Eeckenbrugge G., Lacape J.M. Distribution and differentiation of wild, feral, and cultivated populations of perennial upland cotton (Gossypium hirsutum L.) in Mesoamerica and the Caribbean. PloS One. 2014;9 doi: 10.1371/journal.pone.0107458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.G., Wang J., Chen Q.J., Yu Y., Li B.C. Detection of favorable alleles for yield and yield components by association mapping in upland cotton. Genes Genome. 2018;40:725–734. doi: 10.1007/s13258-018-0678-0. [DOI] [PubMed] [Google Scholar]
- Dong C.G., Wang J., Yu Y., Li B.C. Association mapping and favorable QTL alleles for fiber quality traits in upland cotton (Gossypium hirsutum L.) J. Genet. 2018;97:1–12. [PubMed] [Google Scholar]
- Dong N., Li C.Q., Wang Q.L., Ai N.J., Hu G.H., Zhang J.B. Mixed inheritance of earliness and its related traits of short-season cotton under different ecological enviroments. Acta Gossypii Sinica. 2010;22:304–311. [Google Scholar]
- Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Earl D.A., Vonholdt B.M. Structure HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012;4:359–361. [Google Scholar]
- Esbroeck G.A.V., Bowman D.T., May O.L., Calhoun D.S. Genetic similarity indices for ancestral cotton cultivars and their impact on genetic diversity estimates of modern cultivars. Crop Sci. 1999;39:323–328. [Google Scholar]
- Evanno G., Regnaut S., Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Lischer H.E.L. Arlequin suite v. 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fang D.D., Hinze L.L., Percy R.G., Li P., Deng D., Thyssen G. A microsatellite-based genome-wide analysis of genetic diversity and linkage disequilibrium in Upland cotton (Gossypium hirsutum L.) cultivars from major cotton-growing countries. Euphytica. 2013;191:391–401. [Google Scholar]
- Fang L., Guan X.Y., Zhang T.Z. Asymmetric evolution and domestication in allotetraploid cotton (Gossypium hirsutum L.) Crop J. 2017;5:159–165. [Google Scholar]
- Flint-Garcia S.A., Thuillet A.C., Yu J.M., Pressoir G., Romero S.M., Mitchell S.E., Doebley J., Kresovich S., Goodman M.M., Buckler E.S. Maize association population: a higher solution platform for quantitative trait locus dissection. Plant J. 2005;44:1054–1064. doi: 10.1111/j.1365-313X.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- Guo W.Z., Zhang T.Z., Pan J.J., Wang X.Y. A preliminary study on genetic diversity of Upland cotton cultivars in China. Acta Gossypii Sinica. 1997;9:19–24. [Google Scholar]
- Gutiérrez O.A., Basu S., Saha S., Jenkins J.N., Shoemaker D.B., Cheatham C.L., McCarty J.C. Genetic distance among selected cotton genotypes and its relationship with F2 performance. Crop Sci. 2002;42:1841–1847. [Google Scholar]
- Handi S.S., Katageri I.S., Adiger S., Jadhav M.P., Lekkala S.P., Lachagari V.B.R. Association mapping for seed cotton yield, yield components and fibre quality traits in upland cotton (Gossypium hirsutumL.) genotypes. Plant Breed. 2017;136:958–968. [Google Scholar]
- Holland M.M., Parson W. GeneMarker? HID: a reliable software tool for the analysis of forensic STR data. J. Forensic Sci. 2011;56:29–35. doi: 10.1111/j.1556-4029.2010.01565.x. [DOI] [PubMed] [Google Scholar]
- Huang C., Shen C., Wen T.W., Gao B., Zhu D., Li X.F., Ahmed M.M., Li D.G., Lin Z.X. SSR-based association mapping of fiber quality in upland cotton using an eight-way MAGIC population. Mol. Gen. Genet. 2018;293:793–805. doi: 10.1007/s00438-018-1419-4. [DOI] [PubMed] [Google Scholar]
- Iqbal M.J., Reddy O.U.K., El-Zik K.M., Pepper A.E. A geneticbottleneck in the ‘evolution under domestication’ of UplandcottonGossypium hirsutum L. examined using DNA fingerprinting. Theor. Appl. Genet. 2001;103:547–554. [Google Scholar]
- Iqbal M.J., Aziz N., Saeed N.A., Zafar Y., Malik K.A. Genetic diversity evaluation of some elite cotton varieties by RAPD analysis. Theor. Appl. Genet. 1997;94:139–144. doi: 10.1007/s001220050392. [DOI] [PubMed] [Google Scholar]
- Islam M.S., Thyssen G.N., Jenkins J.N., Zeng L., Delhom C.D., McCarty J.C., Deng D.D., Hinchliffe D.J., Jones D.C., Fang D.D. A MAGIC population-based genome-wideassociation study reveals functionalassociation of GhRBB1_A07 gene with superior fiber quality in cotton. BMC Genom. 2016;17:903. doi: 10.1186/s12864-016-3249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M., Rosenberg N.A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kalinowski S.T., Taper M.L., Marshall T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Keerio A.A., Shen C., Nie Y.C., Ahmed M.M., Zhang X.L., Lin Z.X. QTL mapping for fiber quality and yield traits based on introgression lines derived from Gossypium hirsutum × G. tomentosum. Int. J. Mol. Sci. 2018;19:243. doi: 10.3390/ijms19010243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacape J.M., Dessauw D., Rajab M., Noye J.L., Hau B. Microsatellite diversity in tetraploid Gossypium germplasm: assembling a highly informative genotyping set of cotton SSRs. Mol. Breed. 2007;19:45–58. [Google Scholar]
- Lehner B. Molecular mechanisms of epistasis within and between genes. Trends Genet. 2011;27:323–331. doi: 10.1016/j.tig.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Li C., Zhang J., Hu G., Fu Y., Wang Q. Association mapping and favorable allele mining for node of first fruiting/sympodial branch and its height in upland cotton (Gossypium hirsutum L.) Euphytica. 2016;210:57–68. [Google Scholar]
- Li C.Q., Guo W.Z., Ma X.L., Zhang T.Z. Tagging and mapping of QTL for yield and its components in upland cotton (Gossypium hirsutum L.) population with varied lint percentage. Acta Gossypii Sinica. 2008;20:163–169. [Google Scholar]
- Li C.Q., Xu X.J., Dong N., Ai N.J., Wang Q.L. Association mapping identifies markers related to major early-maturating traits in upland cotton (Gossypium hirsutumL.) Plant Breed. 2016;135:483–491. [Google Scholar]
- Liu G.Z., Mei H.X., Wang S., Li X.H., Zhu X.F., Zhang T.Z. Association mapping of seed oil and protein contents in upland cotton. Euphytica. 2015;205:637–645. [Google Scholar]
- May O.L., Bowman D.T., Calhoun D.S. Genetic diversity of U.S. upland cotton cultivars released between 1980 and 1990. Crop Sci. 1995;35:1570–1574. [Google Scholar]
- Mei H.X., Ai N.J., Zhang X., Ning Z.Y., Zhang T.Z. QTLs conferring FOV 7 resistance detected by linkage and association mapping in upland cotton. Euphytica. 2014;197:237–249. [Google Scholar]
- Mei H.X., Zhu X.F., Zhang T.Z. Favorable QTL alleles for yield and its components identified by association mapping in Chinese upland cotton cultivars. PloS One. 2013;8 doi: 10.1371/journal.pone.0082193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X.H., Huang H., You C.Y., Li W., Zhao W.X., Shen C., Zhang B.B., Wang H.T., Yan Z.H., Dai B.S., Wang M.J., Zhang X.L., Lin Z.X. Genome-wide SSR-based association mapping for fiber quality in nation-wide upland cotton inbreed cultivars in China. BMC Genom. 2016;17:352. doi: 10.1186/s12864-016-2662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R., Smouse P.E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H.D., Chen M., Yi X.D., Bie S., Zhang C., Zhang Y.C., Lan J.Y., Meng Y.Y., Yuan Y.L., Jiao C.H. Identification of associated SSR markers for yield component and fiber quality traits based on frame map and Upland cotton collections. PloS One. 2015;10 doi: 10.1371/journal.pone.0118073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungis D., Llewellyn D., Dennis E.S., Lyon B.R. Simple sequence repeat (SSR) markers reveal low levels of polymorphism between cotton (Gossypium hirsutum L.) cultivars. Aust. J. Agric. Res. 2005;56:301–307. [Google Scholar]
- Shappley Z.W., Jenkins J., Zhu J., McCarty J.C., Jr. Quantitative trait loci associated with agronomic and fiber traits of upland cotton. J. Cotton Sci. 1998;2:153–163. [Google Scholar]
- Shen X.L., Guo W.Z., Lu Q.X., Zhu X.F., Yuan Y.L., Zhang T.Z. Genetic mapping of quantitative trait loci for fiber quality and yield trait by RIL approach in Upland cotton. Euphytica. 2007;155:371–380. [Google Scholar]
- Shen X.L., Guo W.Z., Zhu X.F., Yuan Y.L., Yu J.Z., Kohel R.J., Zhang T.Z. Molecular mapping of QTLs for fiber qualities in three diverse lines in Upland cotton using SSR markers. Mol. Breed. 2005;15:169–181. [Google Scholar]
- Song M.Z., Yu S.X., Fan S.L., Ruan R.H., Huang Z.M. Genetic analysis of main agronomic traits in short season upland cotton. Acta Gossypii Sinica. 2005;17:94–98. [Google Scholar]
- Stephens S.G. Salt water tolerance of seeds of Gossypium species as a possible factor in seed dispersal. Am. Nat. 1958;92:83–92. [Google Scholar]
- Thornsberry J.M., Goodman M.M., Doebley J., Kresovich S., Nielsen D., Buckler E.S. Dwarf 8 polymorphisms associate with variation in flowering time. Nat. Genet. 2001;28:286–289. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- Tyagi P., Gore M.A., Bowman D.T., Campbell B.T., Udall J.A., Kuraparthy V. Genetic diversity and population structure in the US upland cotton (Gossypium hirsutum L.) Theor. Appl. Genet. 2014;127:283–295. doi: 10.1007/s00122-013-2217-3. [DOI] [PubMed] [Google Scholar]
- Ulloa M., Meredith W.R., Jr. Genetic linkage map and QTL analysis of agronomic and fiber quality traits in an intraspecifc population. J. Cotton Sci. 2000;4:161–170. [Google Scholar]
- Wang B.H., Draye X., Zhuang Z.M., Zhang Z.S., Liu M., Lubbers E.L., Jones D., May O.L., Paterson A.H., Chee P.W. QTL analysis of cotton fiber length in advanced backcross populations derived from a cross between Gossypium hirsutum and G. mustelinum. Theor. Appl. Genet. 2017;130:1297–1308. doi: 10.1007/s00122-017-2889-1. [DOI] [PubMed] [Google Scholar]
- Wang B.H., Guo W.Z., Zhu X.F., Wu Y.T., Huang N.T., Zhang T.Z. QTL mapping of fiber quality in an elite hybrid derived-RIL population of upland cotton. Euphytica. 2006;152:367–378. [Google Scholar]
- Wang K.B., Wang Z.W., Li F.G., Ye W.W., Wang J.Y., Song G.L., Yue Z., Cong L., Shang H.H., Zhu S.L., Zou C.S., Li Q., Yuan Y.L., Lu C.R., Wei H.L., Gou C.Y., Zheng Z.Q., Yin Y., Zhang X.Y., Liu K., Wang B., Song C., Shi N., Kohel R.J., Percy R.G., Yu J.Z., Zhu Y.X., Wang J., Yu S.X. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012;44:1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- Wang L., Wu S.M., Zhu Y., Fan Q., Zhang Z.N., Hu G., Peng Q.Z., Wu J.H. Functional characterization of a novel jasmonate ZIM-domain interactor (NINJA) from upland cotton (Gossypium hirsutum) Plant Physiol. Biochem. 2017;112:152–160. doi: 10.1016/j.plaphy.2017.01.005. [DOI] [PubMed] [Google Scholar]
- Wen T.W., Wu M., Shen C., Gao B., Zhu D., Zhang X.L., You C.Y., Lin Z.X. Linkage and association mapping reveals the genetic basis of brown fiber (Gossypium hirsutum) Plant Biotechnol. J. 2018;16:1654–1666. doi: 10.1111/pbi.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F., Brubaker C., Alvarez I., Cronn R., Stewart J.M. Evolution and natural history of the cotton genus. In: Paterson A.H., editor. Genetics and Genomics of Cotton. Springer.; New York: 2009. pp. 3–22. [Google Scholar]
- Wendel J.F., Brubaker C.L., Percival A.E. Genetic diversity in Gossypium hirsutum and the origin of upland cotton. Am. J. Bot. 1992;79:1291–1310. [Google Scholar]
- Wendel J.F., Cronn R.C. Polyploidy and the evolutionary history of cotton. Adv. Agron. 2003;78:139–186. [Google Scholar]
- Xu Q.H., Zhang X.L., Nie Y.C. Genetic diversity evaluation of cultivars (G. hirsutum L.) from the Changjiang river valley and Yellow river valley by RAPD markers. Acta Genetica Sin. 2001;28:683–690. [PubMed] [Google Scholar]
- Yeh F., Boyle T.J.B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1996;129:157. [Google Scholar]
- Yu J.M., Pressoir G., Briggs W.H., Bi I.V., Yamasaki M., Doebley J.F., McMullen M.D., Gaut B.S., Nielsen D.M., Holland J.B., Kresovich S., Buckler E.S. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zeng L.H., Meredith W.R., Jr., Gutiérrez O.A., Boykin D.L. Identification of associations between SSR markers and fiber traits in an exotic germplasm derived from multiple crosses among Gossypium tetraploid species. Theor. Appl. Genet. 2009;119:93–103. doi: 10.1007/s00122-009-1020-7. [DOI] [PubMed] [Google Scholar]
- Zhang J.F., Lu Y.Z., Cantrell R., Hughs S.E. Molecular marker diversity and field performance in commercial cotton cultivars evaluated in the Southwestern USA. Crop Sci. 2005;45:1483–1490. [Google Scholar]
- Zhao Y.L., Wang H.M., Chen W., Li Y.H. Genetic structure, linkage disequilibrium and association mapping of verticillium wilt-resistance in elite cotton (Gossypium hirsutum L.) germplasm population. PloS One. 2014;9 doi: 10.1371/journal.pone.0086308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C.S., Gore M., Buckler E.S., Yu J.M. Status and prospects of association mapping in plants. Plant Genome. 2008;1:5–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.