Abstract
During the early stages of the pandemic, some coronavirus disease (COVID‐19) patients were misdiagnosed as having influenza, which aroused the concern that some deaths attributed to influenza were actually COVID‐19‐related. However, little is known about whether coinfection with influenza contributes to severity of COVID‐19 pneumonia, and the optimal therapeutic strategy for these patients. We retrospectively studied 128 hospitalized patients with COVID‐19 pneumonia. All patients were positive severe acute respiratory syndrome coronavirus 2 positive by nucleic acid detection. Sixty‐four cases were coinfected with influenza A/B and the other 64 were influenza negative, matched by age, sex, and days from onset of symptoms. Among the 64 coinfected patients, 54 (84.4%) were coinfected with influenza A, and 10 (15.6%) with influenza B. The median duration of viral shedding time from admission was longer for patients with influenza coinfection (17.0 days) than for those without influenza coinfection (12.0 days) (P < .001). The multivariable Cox proportional hazards model showed that the hazards ratio of resolution in lung involvement was 1.878 (P = .020) for patients administered lopinavir/ritonavir, compared with those not administered lopinavir/ritonavir (95% confidence interval: 1.103‐3.196). Among influenza coinfected patients, those treated with lopinavir/ritonavir exhibited faster pneumonia resolution within 2 weeks after symptom onset (37% vs 1%; P = .001). There was no difference in lung involvement between influenza coinfected and noninfected groups. Lopinavir/ritonavir eliminated the difference of lung involvement between influenza coinfected and noninfected groups, indicating that lopinavir/ritonavir is associated with pneumonia resolution in COVID‐19.
Keywords: COVID‐19, influenza, lopinavir/ritonavir, pneumonia
Highlights
Coinfection with influenza contributes to severity of COVID‐19 pneumonia.
Patients with influenza coinfection had longer viral shedding time than those without influenza coinfection.
Among influenza coinfected patients, those treated with lopinavir/ritonavir exhibited faster pneumonia resolution.
Lopinavir/ritonavir is associated with pneumonia resolution in COVID‐19.
Abbreviations
- COVID‐19
coronavirus disease 2019
- CT
computerized tomography
- HR
hazard ratio
- IL‐10
interleukin‐10
- IL‐2R
interleukin‐2 receptor
- IL‐6
interleukin‐6
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
1. INTRODUCTION
Ever since December 2019, the outbreak of novel coronavirus disease (COVID‐19) has rapidly developed into a global pandemic. 1 This virus has been identified as a bat‐origin coronavirus and has been termed as severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). 2 , 3 It belongs to the family Coronaviridae, which includes two important representatives SARS‐CoV and Middle East respiratory syndrome coronavirus. 4 Akin to these previous emergent coronaviruses, SARS‐CoV‐2 is highly contagious and spread rapidly by human‐to‐human transmission. 5
The symptoms of the early stages of COVID‐19 are nonspecific, presenting with a flu‐like illness. Considering the similarity of symptoms shared with common respiratory infections, the differential diagnosis generally includes influenza and other respiratory tract diseases. 6 However, due to insufficient availability of kits and the low sensitivity of tests for throat swab specimens especially in the early days of the pandemic, some patients were classified as highly likely to be patients with COVID‐19, or were misdiagnosed as influenza patients. Thus, it was deemed concerning that some patients who seemingly died from influenza had tested positive for COVID‐19 in the posthumous diagnosis.
Anecdotal evidence suggests the presence of coinfections with other respiratory pathogens in COVID‐19 cases. 6 , 7 , 8 After an updated analysis of patients treated in an epicenter hospital designated for COVID‐19 treatment in Wuhan city, we found that the incidence of coinfection with influenza A/B in COVID‐19 was 11.8% (64 patients among 544 patients). Although old age, coexisting illness, decreased lymphocytes and increased inflammation status are known risk factors for severe COVID‐19 infections, 9 it is still not completely understood whether multiple viral infections contribute to disease severity. Therefore, we report the clinical characteristics of COVID‐19 pneumonia in patients with influenza coinfection, and compare with cases with those infected solely with SARS CoV‐2. We aimed to investigate whether coinfection with influenza in patients with COVID‐19 results in increased disease severity and whether antiviral treatment is beneficial for these patients.
2. METHODS
2.1. Study design and participants
In this single‐center study, patient selection was carried out retrospectively at the Tongji Hospital of the Sino‐French New City District. This facility is a designated hospital for COVID‐19 treatment in Wuhan, China. Medical records for patients diagnosed with COVID‐19 pneumonia were reviewed retrospectively on admission; admission period: from 28 January to 18 February 2020. A 1:1 case‐control study was carried out involving 64 patients who tested positive for the influenza immunoglobulin M (IgM) antibody. Control cases were selected by random sampling and matched according to age (±1 years old) and sex, from patients admitted in the study period. A possible bias associated with differences in the duration of illness onset was controlled for by choosing matched pairs in which the difference was ±1 day. 10 Final follow‐up for this report was on 17 March 2020. The criteria to diagnose COVID‐19 pneumonia were pursuant to “Diagnosis and Treatment Protocols of Pneumonia caused by Novel Coronavirus (SARS‐CoV‐2) by the National Health Commission of China” (Trial Version 7). 11 Specifically, the diagnosis criteria were as follows: (a) fever or respiratory symptoms; (b) leukopenia or lymphopenia; (c) a computerized tomography (CT) scan showing radiographic abnormalities in the lung. Those who satisfied two or more diagnosis criteria (lung involvement was necessary) and showed a positive result in a high‐throughput sequencing or reverse transcription‐polymerase chain reaction (RT‐PCR) assay for SARS‐CoV‐2 were diagnosed with COVID‐19 pneumonia. 11 COVID‐19 pneumonia was classified into four types, namely, mild, moderate, severe, and critically ill, according to “Diagnosis and Treatment Protocols of Pneumonia caused by Novel Coronavirus (SARS‐CoV‐2) by the National Health Commission of China (Trial Version 7).” 11 Moderate COVID‐19 pneumonia was diagnosed based on the presence of fever, respiratory syndrome and radiological lung findings. 11 Severe COVID‐19 Pneumonia was designated when any of the following criteria were met: (a) respiratory rate >30/min, (b) oxygen saturation ≤93%, (c) a PaO2/FiO2 ratio ≤300 mm Hg, and (d) a 50% increase in chest radiological abnormalities in 24 to 48 hours. 11 Critically ill COVID‐19 pneumonia was defined when one of the following was present: (a) respiratory failure with a need for mechanical ventilation, (b) shock, or (c) organ failure with a need for intensive care unit admission. 11
COVID‐19 pneumonia was radiographically classified into four stages. 12 (a) Stage‐1 (early stage): ground glass opacities; (2) stage‐2 (progressive stage): increased crazy‐paving pattern; (c) stage‐3 (peak stage): consolidation; (d) stage‐4 (absorption stage): gradual resolution of consolidation without crazy‐paving pattern. Further, we defined a resolution of 50% as stage 4a and that of ≥50% as stage 4b.
Eligibility criteria for inclusion of patients to the study were as follows: (i) The participants were diagnosed as having COVID‐19. (ii) The patients had fulfilled an IgM test for other respiratory pathogens, including influenza A/B, respiratory syncytial virus, mycoplasma, chlamydia, and legionella.
Patients with consecutive negative results in a SARS‐CoV‐2 PCR detection test at least 24 hours apart were excluded. A total of 544 PCR positive patients were screened, and 64 of these had a positive IgM test for influenza A/B. A total of 64 counterparts of cases matched using sex, age and days from onset of symptoms, were selected. More specifically, patients without influenza infections, who were of the same sex, with a difference in age of within 1 year, 13 , 14 and the number of days from onset of symptoms within 3 days compared with patients with influenza infection, were selected to form the matched pairs. Patients with COVID‐19 coinfected with influenza A/B were grouped into the “with influenza” group, and those who tested negative for the influenza A/B IgM antibody were grouped into the “without influenza” group. Influenza A/B was defined as seasonal influenza not including avian Influenzas such as the H1N1 and H7N9 viruses. For discharge patients with COVID‐19 had to meet four strict criteria: (a) a normal temperature lasting longer than 3 days; (b) resolved respiratory symptoms; (c) substantially improved acute exudative lesions on chest CT; and (d) consecutive negative SARS‐CoV‐2 nucleic acid detection results at least 24 hours apart. 11
As the median duration of viral shedding was 20 days from illness onset, 15 we use 28 days as the cutoff value to calculate outcome and viral shedding time. As 75% of patients showed radiographic improvement at stage 4, and exhibited gradual resolution of consolidation at >14 days from onset, 12 we chose 21 days as the deadline to assess radiographic improvement.
This study was approved by the Ethics of Committees of Tongji Hospital, Tong Medical College, Huazhong University of Science and Technology (No. TJ‐IRB2020413). Informed consent for this retrospective study was waived in light of the urgency of data collection.
3. DATA COLLECTION AND MEASUREMENTS
We obtained demographic, epidemiological, clinical, laboratory, radiological characteristics, treatment, and outcome data from the Hospital Information System electronic medical records. The data were reviewed by three physicians (YJG, CY, and RZ). The onset date was defined as the day when the symptom was first noticed.
A quantitative real‐time RT‐PCR test was used to confirm SARS‐CoV‐2 positivity, as following: A Viral Nucleic Acid Kit (Tianlong Science & Technology Co, Ltd, Xi'An, China) was used to extract nucleic acids from throat swab samples and a SARS‐CoV‐2 detection kit (DAAN GENE, Guangzhou, China) was used to detect the ORF1ab gene (nCovORF1ab) and the N gene (nCoV‐NP). Respiratory Tract Profile (IgM) (Euroimmun Medizinisce Labordiagnostika AG) was used to detect IgM antibodies against. influenza A/B.
All patients underwent chest CT scanning before or after admission to the hospital, and additional chest CT scans were obtained at 5 to 14 day intervals.
4. STATISTICAL ANALYSIS
Data conforming to normal distribution were presented as mean ± SD, and as median and quartiles when in breach of normal distribution. Rate comparisons were performed by the χ 2 test. The t test, the Wilcoxon rank sum tes,t and the Fisher's exact test were used to compare paired groups. The Cox proportional hazards model was used to calculate the hazard ratio (HR) and the 95% confidence interval (CI) to identify potential risk factors for the resolution of lung involvement in COVID‐19 with influenza infection. Results are presented as 95% CI and P values. All statistical analyses were performed using SPSS version 20.0 software (SPSS).
5. RESULTS
5.1. Patient characteristics
A cohort of matched pairs of 64 cases coinfected with influenza and 64 cases without influenza but had been diagnosed as COVID‐19 pneumonia was identified in a total of 544 patients. Among the 64 coinfected patients, 54 patients (84.4%) were coinfected with influenza A, and 10 (15.6%) with influenza B. In addition to SARS‐CoV‐2, four of the patients were coinfected with influenza and mycoplasma/chlamydia, and one patient was coinfected with influenza and legionella (Table 1).
Table 1.
Clinical characteristics of patients with COVID‐19 coinfected with influenza
| Characteristics | With influenza | Without influenza | P |
|---|---|---|---|
| Number | 64/128 (50.0%) | 64/128 (50.0%) | |
| Age, y | 61.00 (48.00, 67.75) | 61.00 (49.00, 67.75) | .989a |
| Male patient, % | 28/64 (43.8%) | 27/64 (42.2%) | .858b |
| Days from onset | 8.50 (7.00, 12.00) | 9.00 (6.25, 13.00) | .916a |
| Fever, % | 56/64 (87.5%) | 53/64 (82.8%) | .456b |
| Cough, % | 44/64 (68.8%) | 38/64 (59.4%) | .269b |
| Dyspnea, % | 21/64 (32.8%) | 27/64 (42.2%) | .273b |
| Severity of pneumonia (%) | .945a | ||
| Mild/moderate | 39/64 (60.9%) | 39/64 (60.9%) | |
| Severe | 20/64 (31.3%) | 19/64 (29.7%) | |
| Critical ill | 5/64 (7.8%) | 6/64 (9.4%) | |
| Hypertension, % | 27/64 (42.2%) | 20/64 (31.3%) | .199b |
| Diabetes, % | 11/64 (17.2%) | 12/63 (19.0%)e | .785b |
| Coronary heart disease, % | 5/64 (7.8%) | 5/63 (7.9%)e | 1.000d |
| Neutrophils (109/L) | 3.31 (2.45, 5.55) | 3.24 (2.47, 4.84) | .809a |
| Lymphocytes (109/L) | 0.85 (0.62, 1.27) | 0.89 (0.64, 1.40) | .384a |
| Monocytes (109/L) | 0.41 (0.30, 0.50) | 0.43 (0.31, 0.48) | .764a |
| Eosinophils (109/L) | 0.04 (0.00, 0.10) | 0.03 (0.00, 0.10) | .975a |
| ALT, U/L | 23.00 (14.00, 39.00) | 24.00 (15.25, 38.25) | .830a |
| AST, U/L | 28.00 (20.00, 40.00) | 26.00 (21.25, 45.75) | .935a |
| LDH, U/L | 289.00 (214.00, 456.00) | 283.50 (220.25, 341.50) | .366a |
| Serum albumin, g/L | 33.56 ± 5.62 | 34.72 ± 5.60 | .243c |
| BUN, mmol/L | 4.00 (3.10, 5.90) | 4.00 (2.90, 5.00) | .826a |
| SCR, μmol/L | 66.00 (55.00, 79.00) | 70.00 (53.00, 78.75) | .862a |
| Prothrombin time, s | .207d | ||
| <16 | 58/63 (92.1%)e | 61/62 (98.4%)e | |
| ≥16 | 5/63 (7.9%)e | 1/62 (1.6%)e | |
| APTT, s | 39.11 ± 5.30 | 40.80 ± 5.74 | .090c |
| Fibrinogen, g/L | 4.34 ± 1.41 | 4.78 ± 1.39 | .081c |
| D‐dimer, mg/L | 0.97 (0.38, 2.22) | 0.57 (0.33, 1.75) | .086a |
| ≤0.5 | 20/63 (31.7%)e | 28/62 (45.2%)e | |
| 0.5‐2 | 14/63 (22.2%)e | 20/62 (22.6%)e | |
| >1 | 29/63 (46.0%)e | 39/62 (32.3%)e | |
| CRP, mg/L | 29.30 (7.50, 94.10) | 35.00 (11.00, 81.80) | .695a |
| PCT, ng/mL | 0.05 (0.03, 0.08) | 0.06 (0.03, 0.10) | .711a |
| Serum TNF‐α, pg/mL | 9.50 (5.40, 11.98) | 8.50 (6.08, 11.05) | .703a |
| Serum IL‐10, pg/mL | 5.00 (5.00, 8.55) | 5.00 (5.00, 9.90) | .888a |
| Serum IL‐6, pg/mL | 7.94 (3.20, 23.25) | 6.65 (2.89, 28.49) | .787a |
| Serum IL‐2R, U/mL | 543.00 (362.25, 1122.50) | 607.00 (407.00, 798.00) | .816a |
| Serum IL‐1β, pg/mL | 5.00 (5.00, 5.00) | 5.00 (5.00, 5.00) | .565a |
| Other respiratory pathogens (%) | |||
| Influenza A IgM | 54/64 (84.4%) | 0/64 (0%) | <.001 b |
| Influenza B IgM | 10/64 (15.6%) | 0/64 (0%) | .001 b |
| Respiratory syncytial virus IgM | 0/64 (0%) | 0/64 (0%) | … |
| Mycoplasma | 4/64 (6.3%) | 0/64 (0%) | .119d |
| Legionella | 1/64 (1.6%) | 0/64 (0%) | 1.000d |
| Treatment (%) | |||
| Antibiotic treatment | 42/64 (65.6%) | 48/64 (75.0%) | .246b |
| Arbidol treatment | 47/64 (73.4%) | 50/64 (78.1%) | .536b |
| Oseltamivir | 8/64 (12.5%) | 2/64 (3.1%) | .048 b |
| Lopinavir/ritonavir treatment | 27/64 (42.2%) | 10/64 (15.6%) | .001 b |
| Ribavirin treatment | 5/64 (7.8%) | 5/64 (7.8%) | 1.000b |
| Glucocorticoids treatment | 24/64 (37.5%) | 32/64 (50.0%) | .154b |
| Ventilator aid respiration | 19/64 (29.7%) | 13/64 (20.3%) | .221b |
| SARS‐CoV‐2 turned into negative in 28 d (%) | 49/64 (76.6%) | 59/64 (92.2%) | .015 b |
| Days for SARS‐CoV‐2 turned into negative | 17.00 (13.50, 20.00) | 12.00 (9.75, 13.25) | <.001 a |
| Outcome after 28 d (%) | .129a | ||
| Cured | 29/64 (45.3%) | 39/64 (60.9%) | |
| In remission | 28/64 (43.8%) | 18/64 (28.1%) | |
| Dead or deteriorated | 7/64 (10.9%) | 7/64 (10.9%) |
Note: Data are presented as a percentage or mean ± SD or median (25th‐75th percentiles). Bold values are with statistic difference.
Abbreviations: ALT, alanine transaminase; APTT, activated partial thromboplastin time; AST, aspartate transaminase; BUN, blood urea nitrogen; COVID‐19, coronavirus disease 2019; CRP, C‐reactive protein; IgM, immunoglobulin M; IL, interleukin; LDH, lactate dehydrogenase; PCT, procalcitonin; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; SCR, serum creatinine; TNF‐α, tumor necrosis factor α.
The Wilcoxon rank sum test.
The χ 2 test.
t test.
Fisher's exact test.
Missing data exists.
In the matched cohorts, the sex, age and days from onset of symptom were well‐matched between patients with influenza coinfections and those without coinfections (Table 1). There was no difference in symptoms and comorbidities between the two groups, including fever (with influenza vs without influenza; 87.5 vs 82.8%; P = .456), cough (68.8% vs 59.4%; P = .269), dyspnea (32.8% vs 42.2%; P = .273), hypertension (42.2% vs 31.3%; P = .199), diabetes (17.2% vs 19.0%; P = .785) and coronary heart disease (7.8% vs 7.9%; P = 1.000). The percentages for the severity of pneumonia on admission with influenza and the without influenza groups were similar (P = .945) in terms of mild/moderate (60.9% vs 60.9%), severe (31.3% vs 29.7%) and critically ill (7.8% vs 9.4%). Laboratory tests including hematologic, biochemical and infection‐related inflammatory cytokines were not significantly different between the two groups, as shown in Table 1. Patients who were coinfected with influenza A/B, received more antiviral therapy including lopinavir/ritonavir (42.2% vs 15.6%; P = .001) and oseltamivir (12.5% vs 3.1%; P = .048), compared with patients without influenza A/B.
6. CLINICAL OUTCOMES INMATCHED COHORTS
During the study period, the median duration (days) of viral shedding time from admission was significantly longer in patients with influenza than that those without influenza (17.0 vs 12.0 days; P < .001) (Table 1). The percentage of SARS‐CoV‐2 negative patients after 28 days was lower in the influenza coinfected group than in the control group (76.6% vs 92.2%; P = .015) (Table 1). However, there was no significant difference between the groups in the percentage of patients with the medical status of cured, partial remission, deteriorated or dead during the 28 days of observation (Table 1).
Fifty‐one patients (79.7%) in the co‐infection group completed the serial chest CT examinations within 4 weeks after illness onset, in contrast to 54 patients (84.4%) in the control group. There were no significant differences between the two groups with reference to pneumonia stages on CT scans from the second to the fourth week after illness onset (Table 2). Kaplan‐Meier survival curve analysis showed that the cumulative incidence of resolution of lung involvement was similar (P = .280 by log‐rank test) in patients with and without influenza A/B (Figure 1).
Table 2.
Comparison of initial and follow‐up CT findings in patients with COVID‐19 with or without Influenza
| Characteristics | Second wk | P | Third wk | P | Fourth wk | P | |||
|---|---|---|---|---|---|---|---|---|---|
| With (%) | Without (%) | With (%) | Without (%) | With (%) | Without (%) | ||||
| Stages of lung CT | .383 | .818 | .091 | ||||||
| Stage 1 | 1/37 (2.7) | 3/38 (7.9) | 0/44 (0) | 1/50 (2.0) | 0/51 (0) | 0/54 (0) | |||
| Stage 2 | 14/37 (37.8) | 10/38 (26.3) | 2/44 (4.5) | 5/50 (10.0) | 1/51 (2.0) | 1/54 (1.9) | |||
| Stage 3 | 16/37 (43.2) | 14/38 (36.8) | 17/44 (38.6) | 11/50 (22.0) | 5/51 (9.8) | 4/54 (7.4) | |||
| Stage 4a | 5/37 (13.5) | 8/38 (21.1) | 11/44 (25.0) | 17/50 (34.0) | 19/51 (37.3) | 12/54 (22.2) | |||
| Stage 4b | 1/37 (2.7) | 3/38 (7.9) | 14/44 (31.8) | 16/50 (32.0) | 26/51 (51.0) | 37/54 (68.5) | |||
Note: Stage 1: Ground glass opacities. Stage 2: Increased crazy‐paving pattern. Stage 3: Consolidation. Stage 4a: <50% of resolution of consolidation. Stage 4b: ≥50% of resolution of consolidation.
Abbreviations: COVID‐19, coronavirus disease 2019; CT, computed tomography.
Figure 1.
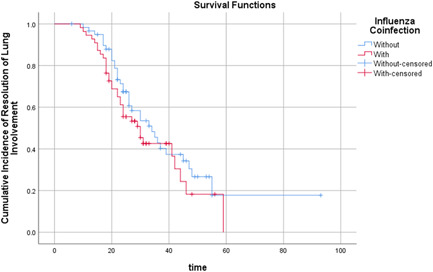
Time (d) cumulative incidence of resolution of lung involvement in patients with influenza coinfected or without
Table 3 shows the HR of the resolution of lung involvement on CT scan associated with influenza A/B coinfection and other variables. The multivariable Cox proportional hazards model showed that the HR of resolution in lung involvement was 0.869 (P = .612) for patients with influenza A/B (95% CI: 0.505‐1.495) compared with patients without influenza. The HR of resolution in lung involvement was 1.878 (P = .020) for patients who received lopinavir/ritonavir, compared with patients who did not receive lopinavir/ritonavir (95% CI 1.103‐3.196), indicating that lopinavir/ritonavir potentially improved the severity of pneumonia in COVID‐19 patients with influenza coinfection.
Table 3.
Results of Cox proportional hazards regression model
| 95.0% CI for Exp(B) | ||||||||
|---|---|---|---|---|---|---|---|---|
| B | SE | Wald | df | P | Exp(B) | Lower | Upper | |
| Influenza (with vs without) | −0.140 | 0.277 | 0.257 | 1 | .612 | 0.869 | 0.505 | 1.495 |
| Oseltamivir (used vs unused) | 0.518 | 0.414 | 1.564 | 1 | .211 | 1.678 | 0.746 | 3.778 |
| Lopinavir/ritonavir (used vs unused) | 0.630 | 0.271 | 5.386 | 1 | .020 | 1.878 | 1.103 | 3.196 |
| SARS‐CoV‐2 turned into negative in 28 d | 0.333 | 0.443 | 0.564 | 1 | .453 | 1.395 | 0.585 | 3.327 |
Note: All variables passed the proportional hazard assumption test (P > .05). Bold values are with statistic difference.
Abbreviations: CI, confidence interval; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
We compared the clinical outcomes of patients with lopinavir/ritonavir treatment with those without lopinavir/ritonavir treatment in the influenza coinfected group. We found faster pneumonia resolution in lopinavir/ritonavir treated patients, which occurred within 2 weeks after symptom onset. Among the influenza coinfected patients, 37% (10 of 27) of patients with lopinavir/ritonavir treatment achieved absorption in the second week, compared to 3.1% (1 of 37) of patients in the control group (P = 0.001) (Table 4). Kaplan‐Meier analysis showed that the time cumulative incidence of resolution of lung involvement was higher (P = .002 by log‐rank test) in patients with lopinavir/ritonavir treatment (Figure 2).
Table 4.
Clinical characteristics of COVID‐19 patients with lopinavir/ritonavir group and without lopinavir/ritonavir (control group)
| Parameters | Total patients (n = 128) | Patients with Influenza (n = 64) | ||||
|---|---|---|---|---|---|---|
| Lopinavir/ritonavir (n = 37) | Control (n = 91) | P | Lopinavir/ritonavir (n = 27) | Control (n = 37) | P | |
| Age | .239a | .324a | ||||
| Median, y | 56.0 | 61.0 | 56.0 | 61.0 | ||
| IQR | 43.5‐68.0 | 50.0‐68.0 | 44.0‐67.0 | 50.5‐68.0 | ||
| Range, y | 10.0‐77.0 | 9.0‐83.0 | 10.0‐77.0 | 31.0‐83.0 | ||
| Male, n (%) | 15 (40.5) | 40 (44.0) | .844b | 10 (37.0) | 18 (48.6) | .447b |
| Influenza A IgM (+), n (%) | 30 (81.1) | 34 (37.4) | <.000 b | 20 (74.0) | 34 (91.9) | .081b |
| Influenza B IgM (+), n (%) | 7 (18.9) | 3 (3.3) | .003 b | 7 (26.0) | 3 (8.1) | .081b |
| Mycoplasma (+), n (%) | 0 (0) | 4 (4.4) | .323b | 0 (0) | 4 (10.8) | .132b |
| Respiratory syncytial virus A/B IgM (+), n (%) | 0 (0) | 0 (0) | 1.000b | 0 (0) | 0 (0) | 1.000b |
| Legionella (+), n (%) | 0 (0) | 1 (1.1) | 1.000b | 0 (0) | 1 (2.7) | 1.000b |
| Lymphocyte count, median (IQR), 109/L | 0.8 (0.5‐1.1) | 1.0 (0.6‐1.4) | .092a | 0.78 (0.46‐1.06) | 1.03 (0.67‐1.33) | .037 a |
| Lactate dehydrogenase, median (IQR), U/L | 285.0 (247.5‐438.0) | 285.5 (210.8‐372.5) | .332a | 274.5 (209.3‐462.7) | 329.0 (261.0‐456.0) | .359a |
| D‐dimer, median (IQR), μg/mL FEU | 0.8 (0.4‐2.3) | 0.7 (0.4‐2.0) | .904a | 0.80 (0.38‐2.80) | 1.0 (0.41‐2.14) | .862a |
| IL‐6, median (IQR), pg/mL | 7.9 (6.0‐21.8) | 7.5 (2.8‐28.8) | .510a | 8.1(2.7‐29.1) | 7.9 (6.0‐21.7) | .690a |
| SARS‐CoV‐2 turned into negative in 28 d, % | 34 (91.9) | 74 (81.3) | .430b | 23 (85.2) | 26 (70.3) | .253b |
| Days for SARS‐CoV‐2 turned into negative | 13.0 (10.0‐16.0) | 16.5 (12.25‐23.75) | .003 a | 16.0 (13.0‐19.5) | 19.0 (14.0‐26.25) | .036 a |
| Absorption stage of chest CT | Lopinavir/ritonavir (n = 37) | Control group (n = 91) | P b | Lopinavir/ritonavir (n = 27) | Control group (n = 37) | P b |
|---|---|---|---|---|---|---|
| Wk 1, no. (%) | 3 (8.8) | 3 (3.7) | .359 | 2 (7.4) | 1 (3.1) | .588 |
| Wk 2, no. (%) | 11 (32.4) | 11 (13.8) | .036 | 10 (37.0) | 1 (3.1) | .001 |
| Wk 3, no. (%) | 22 (64.7) | 44 (55.0) | .095 | 16 (59.3) | 15 (46.9) | .269 |
| Wk 4, no. (%) | 32 (94.1) | 67 (81.7) | .147 | 25 (92.6) | 26 (81.3) | .147 |
Note: Values are expressed as mean (standard deviation), median (25th‐75th percentile) or n (%). *P < .05. Bold values are with statistic difference.
Abbreviations: COVID‐19, coronavirus disease 2019; CT, computed tomography; FEU, fibrinogen equivalent unit; IgM, immunoglobulin M; IL, interleukin; IQR, interquartile range.
Whitney U test.
Fisher's exact test.
Figure 2.
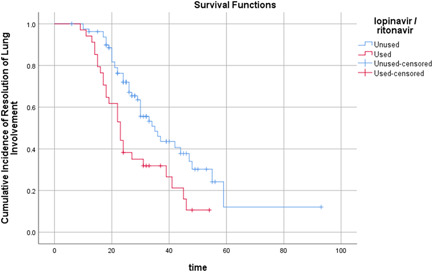
Time (d) cumulative incidence of resolution of lung involvement in patients with lopinavir/ritonavir treatment or without
7. DISCUSSION
The common clinical manifestations of COVID‐19, including fever, cough, dyspnea, and ground‐glass opacity and consolidation with bilateral lung involvement, 7 , 9 are similar to the characteristics presented in influenza A and other respiratory viruses infections. 16 These clinical features may also cause complications and influence mortality. Because of the insufficient sensitivity of tests to detect SARS‐CoV‐2 from upper respiratory specimens, coinfection cases can easily be misdiagnosed as seasonal influenza, 17 which poses several challenges to the diagnosis and treatment of COVID‐19.
In a recent small sample descriptive study of five patients coinfected with both SARS‐CoV‐2 and the influenza virus, it was reported that the coinfected patients presented with similar clinical symptoms and radiological characteristics as did patients infected with only SARS‐CoV‐2. 18 However, the study did not analyze the risk related to the prognosis of COVID‐19 pneumonia.
A previous study has demonstrated that 53% of COVID‐19 pneumonia patients at stage 2 showed increased crazy‐paving pattern on lung CT after 5 to 8 days. 12 Additionally, in 75% of these patients, time period to reach stage 4 was >14 days; this clinical pattern was defined as gradual resolution of consolidation without crazy‐paving pattern. 12 The absorption stage extended beyond 26 days (last few days of follow‐up) from the onset of initial symptoms. 12 In this study, patients coinfected with influenza showed similar radiological progression as that common in COVID‐19. There was no difference between the two groups in pneumonia stages from the second to the fourth week after illness onset, suggesting that, under rational medication use, coinfection with influenza is not associated with increased severity of COVID‐19 pneumonia.
The duration of virus replication is an important factor in assessing the risk of transmission and in guiding clinical decisions. Prolonged viral shedding time was associated with a fatal outcome. 19 Therefore, the viral shedding duration had strategic value for antiviral treatment. Multiple antiviral medications were administered to patients with influenza coinfections, including oseltamivir (“with influenza” vs “without influenza” groups: 12.5% vs 3.1%) and lopinavir/ritonavir (42.2% vs 15.6%). We observed longer viral shedding duration in patients receiving several antiviral therapies compared with those receiving fewer antiviral therapy. Lopinavir‐ritonavir treatment was more effective in pneumonia resolution in influenza coinfected patients, and 37% (10 of 27) of patients administered lopinavir/ritonavir treatment achieved resolution of consolidation in the second week, compared to 3.1% (1 of 37) of patients in the control group (P = .001).
Lopinavir is a human immunodeficiency virus protease inhibitor, which is usually combined with ritonavir. Ritonavir inhibits cytochrome P450 and increases the half‐life of lopinavir. 20 By using a pre‐trained Molecule Transformer‐Drug Target Interaction deep‐learning model, lopinavir was predicted to have an inhibitory potency against the SARS‐CoV‐2 3C‐like proteinase, with a K d of 204.05 nM. 21 However, this drug was not efficacious in very ill patients with COVID‐19, which may be due to delayed treatment. 22 Patients with severe COVID‐19 who received lopinavir‐ritonavir showed a lowering of overall mortality (lopinavir‐ritonavir group:19% vs standard‐care group:25%), the risk of severe adverse events (20% vs. 32%), and the risk of respiratory failure or acute respiratory distress syndrome (13% vs 27%). 22 Lopinavir/ritonavir lowered the body temperature and restored normal physiological mechanisms with no evident toxicity or side effects. 23 COVID‐19 messenger RNA clearance time correlated positively with the length of hospital stay in patients treated with lopinavir/ritonavir. 24 These recent studies have identified that lopinavir/ritonavir was beneficial for viral clearance and for the treatment of COVID‐19, 23 , 24 , 25 which is consistent with our findings.
This study has several limitations. First, due to the retrospective study design, not all laboratory and radiologic examinations were performed according to the clinical course of the disease. Therefore, their role may be underestimated in outcome judgment. Second, the viral shedding data were not precise due to limitations in frequency of respiratory specimen collection, qualitative viral RNA detection, and the relatively low positive rate of SARS‐CoV‐2 RNA detection in throat swabs. 26 Third, we only chose SARS‐CoV‐2 RNA positive patients, so the fatality ratio in our study may not reflect the true mortality rate of co‐infection patients. Fourth, this is a retrospective study using a limited sample size and with limited variable adjustments which may affect our findings. Additional robust scientific studies with proper controls are needed. Last, we applied pair‐matched analyses, but not all confounders were reported; this could have introduced biased into our results.
In conclusion, although patients coinfected with influenza had prolonged viral shedding times, coinfection with influenza was not associated with increased disease severity of COVID‐19 pneumonia. It is possible that treatment with lopinavir/ritonavir hastened the resolution of COVID‐19, and eliminated the difference of lung involvement between the two groups.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
YY, WL, and RZ designed the study. CY, YG, PG, and JS collected the data. CY, ZZ, and YG prepared the figures and tables. CY and ZZ contributed with regard to analytical tools. CY, ZZ, and RZ wrote the paper. RZ, WL, and YY conceived the project and supervised and coordinated all the work.
ETHICS STATEMENT
The study protocol was approved by the Medical Ethics Committee of Tongji Hospital (No.TJ‐IRB2020413).
ACKNOWLEDGMENTS
The authors greatly appreciate the entire hospital staff for their efforts in recruiting and treating patients and thank all patients involved in this study. This study was financially supported by the National Natural Science Foundation of China (grant no: 81770681, 81974086, 8177068481974087, and 81700597).
Yu C, Zhang Z, Guo Y, et al. Lopinavir/ritonavir is associated with pneumonia resolution in COVID‐19 patients with influenza coinfection: A retrospective matched‐pair cohort study. J Med Virol. 2021;93:472–480. 10.1002/jmv.26260
Chong Yu and Zhiguo Zhang contributed equally to this work.
Contributor Information
Ying Yao, Email: yaoyingkk@126.com.
Wenhui Liao, Email: liaowh126@126.com.
Rui Zeng, Email: zengr126@126.com.
REFERENCES
- 1. Velavan TP, Meyer CG. The COVID‐19 epidemic. Trop Med Int Health. 2020;25(3):278‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bordi L, Nicastri E, Scorzolini L, et al. Differential diagnosis of illness in patients under investigation for the novel coronavirus (SARS‐CoV‐2), Italy, February 2020. Euro Surveill. 2020;25:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rusakow LS, Guarín M. A cystic fibrosis mutation associated with mild lung disease. N Engl J Med. 1995;333(24):1644. [DOI] [PubMed] [Google Scholar]
- 11.National Health Commission of the People's Republic of China. Diagnosis and treatment protocols of pneumonia caused by a novel coronavirus (trial version 7). 2020. http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989/files/ce3e6945832a438eaae415350a8ce964.pdf
- 12. Pan F, Ye T, Sun P, et al. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID‐19) pneumonia. Radiology. 2020:200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kernan WN, Viscoli CM, Brass LM, et al. Phenylpropanolamine and the risk of hemorrhagic stroke. N Engl J Med. 2000;343(25):1826‐1832. [DOI] [PubMed] [Google Scholar]
- 14. Johansson K, Cnattingius S, Näslund I, et al. Outcomes of pregnancy after bariatric surgery. N Engl J Med. 2015;372(9):814‐824. [DOI] [PubMed] [Google Scholar]
- 15. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020. [DOI] [PMC free article] [PubMed]
- 16. Sullivan SJ, Jacobson RM, Dowdle WR, Poland GA. 2009 H1N1 influenza. Mayo Clin Proc. 2010;85(1):64‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu X, Cai Y, Huang X, et al. Co‐infection with SARS‐CoV‐2 and influenza A virus in patient with pneumonia, China. Emerg Infect Dis. 2020;26:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding Q, Lu P, Fan Y, Xia Y, Liu M. The clinical characteristics of pneumonia patients co‐infected with 2019 novel coronavirus and influenza virus in Wuhan, China. J Med Virol. 2020. [DOI] [PMC free article] [PubMed]
- 19. Wang Y, Guo Q, Yan Z, et al. Factors associated with prolonged viral shedding in patients with avian influenza A(H7N9) virus infection. J Infect Dis. 2018;217(11):1708‐1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mangum EM, Graham KK. Lopinavir‐ritonavir: a new protease inhibitor. Pharmacotherapy. 2001;21(11):1352‐1363. [DOI] [PubMed] [Google Scholar]
- 21. Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS‐CoV‐2) through a drug‐target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784‐790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cao B, Wang Y, Wen D, et al., A trial of lopinavir‐ritonavir in adults hospitalized with severe COVID‐19. N Engl J Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ye XT, Luo YL, Xia SC, et al. Clinical efficacy of lopinavir/ritonavir in the treatment of coronavirus disease 2019. Eur Rev Med Pharmacol Sci. 2020;24(6):3390‐3396. [DOI] [PubMed] [Google Scholar]
- 24. Yuan J, Zou R, Zeng L, et al. The correlation between viral clearance and biochemical outcomes of 94 COVID‐19 infected discharged patients. Inflamm Res. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Z, Chen X, Lu Y, Chen F, Zhang W. Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Biosci Trends. 2020;14(1):64‐68. [DOI] [PubMed] [Google Scholar]
- 26. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]


