Abstract
Since the outbreak of severe acute respiratory syndrome (SARS) in 2003, the harm caused by coronaviruses to the world cannot be underestimated. Recently, a novel coronavirus (severe acute respiratory syndrome coronavirus‐2 [SARS‐CoV‐2]) initially found to trigger human severe respiratory illness in Wuhan City of China in 2019, has infected more than six million people worldwide by 21 June 2020, and which has been recognized as a public health emergency of international concern as well. And the virus has spread to more than 200 countries around the world. However, the effective drug has not yet been officially licensed or approved to treat SARS‐Cov‐2 and SARS‐Cov infection. NSP12‐NSP7‐NSP8 complex of SARS‐CoV‐2 or SARS‐CoV, essential for viral replication and transcription, is generally regarded as a potential target to fight against the virus. According to the NSP12‐NSP7‐NSP8 complex (PDB ID: 7BW4) structure of SARS‐CoV‐2 and the NSP12‐NSP7‐NSP8 complex (PDB ID: 6NUR) structure of SARS‐CoV, NSP12‐NSP7 interface model, and NSP12‐NSP8 interface model were established for virtual screening in the present study. Eight compounds (Nilotinib, Saquinavir, Tipranavir, Lonafarnib, Tegobuvir, Olysio, Filibuvir, and Cepharanthine) were selected for binding free energy calculations based on virtual screening and docking scores. All eight compounds can combine well with NSP12‐NSP7‐NSP8 in the crystal structure, providing drug candidates for the treatment and prevention of coronavirus disease 2019 and SARS.
Keywords: antiviral drugs, COVID‐19, drug candidates, NSP12‐NSP7‐NSP8, SARS‐CoV, SARS‐CoV‐2
Highlights
Potential inhibitors targeting RNA‐dependent RNA polymerase activity (NSP12).
The protein ‐ protein interactions (PPIs) were used as the target for virtual screening.
The binding free energy was calculated and eight potential inhibitors were obtained.
Provide drug candidates for the treatment and prevention of COVID‐19 and SARS.
1. INTRODUCTION
Since the outbreak of pneumonia caused by a novel coronavirus (severe acute respiratory syndrome coronavirus‐2 [SARS‐CoV‐2]) in Wuhan, China, in December 2019, the world has faced unprecedented challenges in treating the disease caused by this virus. 1 , 2 , 3 SARS‐CoV‐2 is closely related to the SARS virus in Guangdong, China in 2002, both of which belong to the same family of viruses. 4 , 5 , 6 China has implemented strict epidemic prevention and control measures across the country to avoid a larger‐scale epidemic. 7 However, the mortality rate of coronavirus disease 2019 (COVID‐19) caused by SARS‐CoV‐2 is much higher than that of SARS at present. 4 The virus now presents in more than 200 countries. Even now though the SARS‐CoV no longer appears on a large scale, the infectious power and harmful effects of SARS‐CoV‐2 and SARS‐CoV should not be underestimated. To date, there are no officially licensed or approved drugs against this novel coronavirus and SARS‐CoV. There is an urgent need to find new targets for the development of anti‐SARS‐CoV‐2 and anti‐SARS‐CoV agents.
ORF1a and ORF1b at the 5′‐terminus of the coronavirus (CoV) genomes encode polyprotein 1a and polyprotein 1b, the two proteins could be cleaved into 16 nonstructural proteins (NSPs), which are essential for viral replication and transcription, thus being regarded as a potential virulence factor and a target for CoV. 8 , 9 Among these NSPs, the NSP12 subunit is the essential RdRp (RNA‐dependent RNA polymerase) of the coronavirus replicative machinery, which was even able to extend a homopolymeric primer‐template substrate by a few dozen nucleotides in vitro. 10 , 11 The 3.1 Å cryo‐EM structure of the SARS‐CoV RNA polymerase NSP12 shows that it can bind with its essential cofactors NSP7 and NSP8. 12 The replication of the SARS‐coronavirus genome involves two RNA‐dependent RdRps. The first is primer‐dependent and associated with the NSP12, whereas the second is catalyzed by NSP8. NSP8 is capable of de novo initiating the replication process and has been proposed to operate as a primase. 13 In addition, NSP7, a component of the CoV replicase polyprotein, also participates in viral replication processed by binding to NSP12 as another primase. 13 The NSP12 needs to associate with NSP7 and NSP8 to activate its capability to replicate long RNA. 10 This elicits us to identify the particularly interesting compound disrupt the binding of NSP7 or NSP8 to NSP12, thus which could be used to inhibit the RdRp activity of NSP12, acting as novel antiviral agents and therapies of SARS‐CoV‐2 and SARS‐CoV.
The amino acid sequence alignment revealed that the NSP12 (PDB ID: 7BW4) of SARS‐CoV‐2 shared 96.35% similarity with the NSP12 (PDB ID: 6NUR) of SARS (Figure 1). In addition, comparative analyses of their deduced amino acid sequences revealed that NSP7 and NSP8 of SARS‐CoV‐2 shared 98.8% and 97.5% similarity with that of SARS‐CoV respectively. The similarity of the target structure may also lead to similar antiviral drugs. Therefore, the NSP12 crystal structures of SARS‐CoV‐2 and SARS‐CoV were used as the target proteins. The NSP7 and NSP8 binding pocket of NSP12 were designated as active sites for screening compounds. Computer virtual screening in known drug databases helps to quickly identify potential drug candidates for COVID‐19 prevention and treatment. What's more, previous literature has shown that a number of virtually‐screened compounds, such as ribavirin, lopinavir, and ritonavir, have proven to be effective in treating COVID‐19. 14 , 15 , 16 , 17 Here, through high‐throughput screening methods using a pool of 30 000 small molecules, several potential drug candidates were identified for preventing the binding NSP7 or NSP8 to NSP12, suggesting further assessment of the anti‐SARS‐CoV‐2 activity and anti‐SARS‐CoV activity of these compounds respectively in cell culture.
Figure 1.
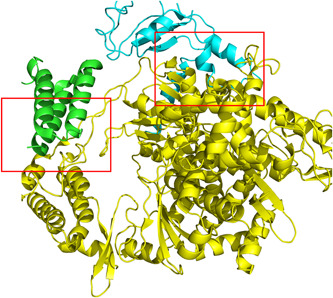
The docking boxes of the NSP12‐NSP7 interface and NSP12‐NSP8 interface. The yellow part is nsp12. The green part is nsp7. The cyan part is nsp8. The docking box on the left is NSP12‐NSP7 interface. The docking box on the right is NSP12‐NSP8 interface
2. MATERIALS AND METHODS
2.1. Construction of small molecular ligands
Each sublibrary (Food and Drug Administration [FDA], world‐not‐FDA, investigational‐only, http://zinc.docking.org/substances/subsets/) was downloaded from the zinc database. Then the model was converted to pdbqt format by prepare_receptor4.py script with assigning atomic types and atomic charges. All rotatable bonds in the molecule are set to be flexible for flexible docking.
2.2. Preparation of target proteins
The crystal structure of SARS‐CoV‐2 NSP12 (PDB ID: 7BW4) was used as the target protein of SARS‐CoV‐2. And the crystal structure of SARS NSP12 (PDB ID: 6NUR) was used as the target protein of SARS‐CoV.
2.3. Molecular docking
Vina1.1.2 was used to perform molecular docking. The docking boxes were set at the NSP12‐NSP7 interface and NSP12‐NSP8 interface (Figure 1), respectively. The search exhaustiveness was set as 32, and the number of binding modes was set as 9. Other parameters were set as default. During docking, NSP7 (or NSP8) was removed from the complex and only NSP12 was left as a receptor.
2.4. Binding free energy calculation
Each simulation system was immersed in a cubic box of TIP3P water with 10 Å distance from the solute. The Na+ or Cl− was applied to neutralize the system. General Amber force field 15 and Amber ff14SB force field were used to parameterizing the ligand and protein respectively. A total of 10 000 steps of minimization with constraints (10 kcal/mol/Å2) on heavy atoms of complex, including 5000 steps of steepest descent minimization and 5000 steps of conjugate gradient minimization, were used to optimize each system. Then each system was heated to 300 K within 0.2 ns followed by 0.1 ns equilibration in NPT ensemble. Finally, 5 ns molecular dynamics (MD) simulation on each system at 300 K was performed. The minimization, heating, and equilibrium are performed with the sander program in Amber18. The 5 ns production run was performed with pmemd.cuda. Based on the 5 ns MD simulation trajectory, binding free energy (ΔG) was calculated with MM/GBSA method according to the following equation: ΔGcal = ΔH‐TΔS = ΔEvdw + ΔEele + ΔGgb + ΔGnp‐TΔS, where ΔEele and ΔEVDW refer to electrostatic and van der Waals energy terms, respectively. ΔGgb and ΔGnp refer to polar and nonpolar solvation free energies, respectively. Conformational entropy (TΔS) was not calculated for saving time. Besides, the ligands were compared based on the same target, so it is reasonable to ignore the entropy.
3. RESULTS AND DISCUSSION
3.1. Docking results of 7496 drugs against NSP12‐NSP7 model and NSP12‐NSP8 model of SARS‐CoV‐2 and SARS‐CoV, respectively
The 7964 drugs obtained from the zinc database were screened for molecular docking. Among them, 20 top compounds showed the docking score in a range of −7.5 to −8.8 kcal/mol were selected from docking results from the NSP12‐NSP7 interface and NSP12‐NSP8 interface of SARS‐CoV‐2 (Tables 1 and 2). Similarly, 24 top compounds showed the docking score in a range of −7.5 to −8.4 kcal/mol were selected from the NSP12‐NSP7 interface and NSP12‐NSP8 interface of SARS‐CoV (Tables 3 and 4). Among the selected drugs, Tegobuvir can block both the NSP12‐NSP7 interface and the NSP12‐NSP8 interface of two coronaviruses. Cepharanthine can block both the NSP12‐NSP7 interface and the NSP12‐NSP8 interface of SARS‐CoV‐2, as well as the NSP12‐NSP8 interface of SARS‐CoV. Olysio and Filibuvir both can block the NSP12‐NSP8 interface of two coronaviruses. What is more, Lonafarnib can block the NSP12‐NSP7 interface of two coronaviruses. Further, Nilotinib, Tipranavir, and Saquinavir were all closely associated with antiviral activity.
Table 1.
Ten compounds selected from the NSP12‐NSP7 interface of SARS‐CoV‐2
| Compounds name | ID | Data | Affinity (kcal/mol) |
|---|---|---|---|
| Dihydroergotamine | ZINC3978005 | FDA | −8.8 |
| Nilotinib | ZINC6716957 | FDA | −8.4 |
| Pranlukast | ZINC1542146 | World‐not‐ FDA | −8.3 |
| Golvatinib | ZINC43195317 | Investigational‐only | −8.3 |
| Gedatolisib | ZINC49757175 | Investigational‐only | −8.1 |
| Tegobuvir | ZINC100057121 | Investigational‐only | −7.9 |
| Cepharanthine | ZINC30726863 | World‐not‐ FDA | −7.8 |
| Palovarotene | ZINC38467831 | Investigational‐only | −7.8 |
| Nafamostat | ZINC3874467 | World‐not‐ FDA | −7.6 |
| Lonafarnib | ZINC3950115 | Investigational‐only | −7.6 |
Abbreviations: FDA, Food and Drug Administration; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
Table 2.
Ten compounds selected from the NSP12‐NSP8 interface of SARS‐CoV‐2
| Compounds name | ID | Data | Affinity (kcal/mol) |
|---|---|---|---|
| Cepharanthine | ZINC30726863 | World‐not‐ FDA | −8.6 |
| Hypericin | ZINC3780340 | Investigational‐only | −8.2 |
| Avodart | ZINC3932831 | FDA | −8.1 |
| Filibuvir | ZINC100078465 | Investigational‐only | −8 |
| Olysio | ZINC164760756 | FDA | −7.8 |
| Berberine | ZINC3779067 | World‐not‐ FDA | −7.8 |
| Tegobuvir | ZINC100057121 | Investigational‐only | −7.8 |
| Grazoprevir | ZINC95551509 | FDA | −7.7 |
| Setrobuvir | ZINC100341584 | Investigational‐only | −7.7 |
| Sqv | ZINC29416466 | FDA | −7.5 |
Abbreviations: FDA, Food and Drug Administration; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
Table 3.
Thirteen compounds selected from the NSP12‐NSP7 interface of SARS‐CoV
| Compounds name | ID | Data | Affinity (kcal/mol) |
|---|---|---|---|
| Lonafarnib | ZINC3950115 | Investigational‐only | −8.4 |
| Tegobuvir | ZINC100057121 | Investigational‐only | −8.2 |
| Aromasin | ZINC3973334 | FDA | −8 |
| Troglitazone | ZINC968279 | World‐not‐ FDA | −8 |
| Hypericin | ZINC3780340 | Investigational‐only | −8 |
| Nilotinib | ZINC6716957 | FDA | −7.9 |
| Cepharanthine | ZINC30726863 | World‐not‐ FDA | −7.9 |
| Eltrombopag | ZINC11679756 | FDA | −7.8 |
| Tipranavir | ZINC100016058 | FDA | −7.8 |
| Saquinavir | ZINC26985532 | FDA | −7.8 |
| Indinavir | ZINC22448696 | FDA | −7.7 |
| Cobicistat | ZINC85537014 | FDA | −7.7 |
| Talmapimod | ZINC34001955 | Investigational‐only | −7.5 |
Abbreviations: FDA, Food and Drug Administration; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
Table 4.
Eleven compounds selected from the NSP12‐NSP8 interface of SARS‐CoV
| Compounds name | ID | Data | Affinity (kcal/mol) |
|---|---|---|---|
| Nilotinib | ZINC6716957 | FDA | −8.4 |
| Tegobuvir | ZINC100057121 | Investigational‐only | −8.4 |
| Olysio | ZINC164760756 | FDA | −8.3 |
| Cepharanthine | ZINC30726863 | World‐not‐ FDA | −8.3 |
| Rimegepant | ZINC68267814 | Investigational‐only | −8.2 |
| Eltrombopag | ZINC11679756 | FDA | −8.1 |
| Biosone | ZINC19203131 | World‐not‐ FDA | −7.9 |
| Hypericin | ZINC3780340 | Investigational‐only | −7.9 |
| Lurasidone | ZINC3927822 | FDA | −7.8 |
| Avodart | ZINC3932831 | FDA | −7.8 |
| Filibuvir | ZINC100078465 | Investigational‐only | −7.7 |
Abbreviations: FDA, Food and Drug Administration; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
3.2. Docking results of Nilotinib against SARS‐CoV‐2 NSP12‐NSP7
Nilotinib, a second‐generation small‐molecule tyrosine kinase inhibitor, is widely used in the treatment of chronic myeloid leukemia. 18 Studies have shown that Nilotinib has a potential antiviral effect. 19 Our docking results showed that Nilotinib was mainly combined with the interface between NSP12 and NSP7 of SARS‐CoV‐2 through van der Waals potential energy and hydrogen bonds, involving LYS‐411 (Figure 2A). Nilotinib could bind to the interface active pockets of the SARS‐CoV‐2 NSP12 and NSP7 (Figure 2B). Therefore, Nilotinib can be considered as a candidate drug for treating SARS‐CoV‐2 infection.
Figure 2.
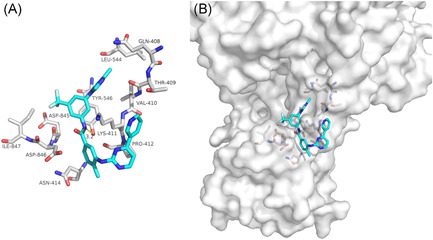
The binding model of Nilotinib against SARS‐CoV‐2 NSP12‐NSP7. A, Interactions between Nilotinib (cyan) and associated residues (off‐white) in the interface of the homology model for SARS‐CoV‐2. B, Binding models of Nilotinib (cyan) in the SARS‐CoV‐2 NSP12‐NSP7 protein interface pocket (white surface). Numbers accompanying dashed yellow lines represent the interaction distance (Å). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
3.3. Docking results of Saquinavir and Tipranavir against SARS‐CoV NSP12‐NSP7
Saquinavir, the first HIV protease inhibitor was introduced into the market in 1995. Tipranavir, a novel nonpeptide protease inhibitor approved for use in patients with resistant strains of HIV. Both of which are safe and generally well‐tolerated in HIV‐1‐infected adults. 20 , 21 Our docking results showed that five of the hydrogen bonds involving GLY‐297, PHE‐299, PHE‐325, PHE‐326, and ALA‐327 maintained upon the binding of saquinavir with interface between SARS‐CoV NSP12 and NSP7 (Figure 3A). As for Tipranavir, hydrogen bonds involving PHE‐325 maintained upon the binding of Tipranavir with interface between SARS‐CoV NSP12 and NSP7 (Figure 3C). Saquinavir and Tipranavir could bind to the interface active pockets of the SARS‐CoV NSP12 and NSP7 (Figures 3B and 3D). The previous study showed that saquinavir could bind to the SARS‐CoV‐2 RNA‐dependent RNA polymerase and inhibit the enzyme activity. 22 Our observations further confirm that saquinavir and tipranavir can bind to the NSP12‐NSP7 interface as interfacial blockers, thus making them as candidates for further in vitro evaluation of the anti‐SARS‐CoV activity.
Figure 3.

The binding model of Saquinavir and Tipranavir against SARS‐CoV NSP12‐NSP7. A, Interactions between Saquinavir (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV. B, Binding models of Saquinavir (cyan) in the SARS‐CoV NSP12‐NSP7 protein interface pocket (white surface). C, Interactions between Tipranavir (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV. D, Binding models of Tipranavir (cyan) in the SARS‐CoV NSP12‐NSP7 protein interface pocket (white surface). Numbers accompanying dashed yellow lines represent the interaction distance (Å). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
3.4. Docking results of Lonafarnib against SARS‐CoV‐2 and SARS‐CoV NSP12‐NSP7
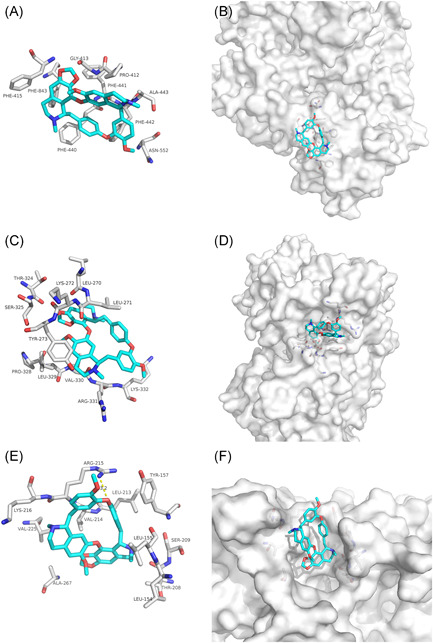
Lonafarnib as a nonpeptidomimetic inhibitor of farnesyltransferase has been used for progeria. 23 , 24 Our docking results showed that lonafarnib was mainly combined with the interface between NSP12 and NSP7 of SARS‐CoV‐2 and SARS‐CoV through van der Waals potential energy and hydrophobic accumulation, involving PHE‐843, PHE‐440, PHE‐441, PHE‐442 (Figure 4A), as well as PHE‐299, PHE‐727, PHE‐324, PHE‐325, and PHE‐326 (Figure 4C). Lonafarnib could both bind to the interface active pockets between the NSP12 and NSP7 of SARS‐CoV‐2 and SARS‐CoV (Figures 4B and 4D). Therefore, we speculate that Lonafarnib has a potential activity for the treatment of SARS‐Cov‐2 and SARS‐CoV infection.
Figure 4.
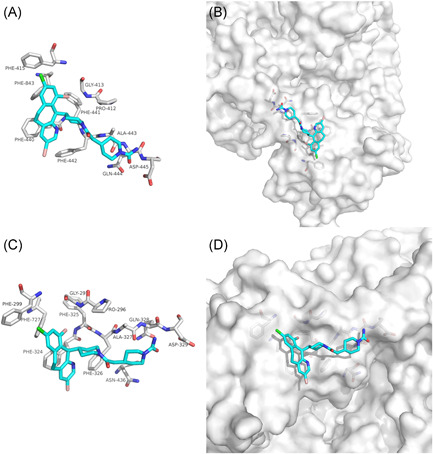
The binding model of Lonafarnib against SARS‐CoV‐2 and SARS‐CoV NSP12‐NSP7. A, Interactions between Lonafarnib (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV‐2. B, Binding models of Lonafarnib (cyan) in the SARS‐CoV‐2 NSP12‐NSP7 protein interface pocket (white surface). C, Interactions between Lonafarnib (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV. D, Binding models of Lonafarnib (cyan) in the SARS‐CoV NSP12‐NSP7 protein interface pocket (white surface). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
3.5. Docking results of Tegobuvir against SARS‐CoV‐2 and SARS‐CoV
Tegobuvir (GS‐9190) is a specific, covalent inhibitor of the hepatitis C virus (HCV) nonstructural 5B (NS5B) RdRp with demonstrated antiviral activity in patients with genotype 1 chronic HCV infection. 25 , 26 , 27 Our docking results showed that the hydrogen bonds involving ALA‐443 and LEU‐329 maintained upon the binding of Tegobuvir and SARS‐CoV‐2 NSP12 interface (Figures 5A and 5C). Tegobuvir was mainly combined with the interface between SARS‐CoV NSP12 and NSP7 through van der Waals potential energy and hydrophobic accumulation, involving PHE‐299, PHE‐727, PHE‐324, PHE‐325, and PHE‐326 (Figure 5E). Further, van der Waals forces mainly maintain the bond between Tegobuvir and the SARS‐CoV NSP12‐NSP8 interface (Figure 5G). Moreover, Tegobuvir could also bind to both of the interface active pockets of the SARS‐CoV‐2 and SARS‐CoV (Figures 5B,D and 5F,H). Thus, Tegobuvir could be as a candidate drug against SARS‐CoV‐2 and SARS‐CoV.
Figure 5.
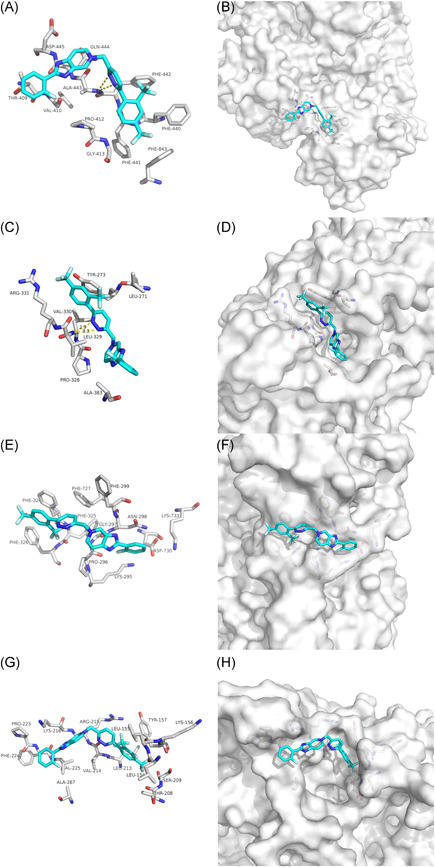
The binding model of Tegobuvir against SARS‐CoV‐2 and SARS‐CoV. A, Interactions between Tegobuvir (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV‐2. B, Binding models of Tegobuvir (cyan) in the SARS‐CoV‐2 NSP12‐NSP7 protein interface pocket (white surface). C, Interactions between Tegobuvir (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV‐2. D, Binding models of Tegobuvir (cyan) in the SARS‐CoV‐2 NSP12‐NSP8 protein interface pocket (white surface). E, Interactions between Tegobuvir (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV. F, Binding models of Tegobuvir (cyan) in the SARS‐CoV NSP12‐NSP7 protein interface pocket (white surface). G, Interactions between Tegobuvir (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV. H, Binding models of Tegobuvir (cyan) in the SARS‐CoV NSP12‐NSP8 protein interface pocket (white surface). Numbers accompanying dashed yellow lines represent the interaction distance (Å, C). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
3.6. Docking results of Olysio against SARS‐CoV‐2 and SARS‐CoV NSP12‐NSP8
Olysio, HCV NS3/4A protease inhibitor approved for the treatment of genotype 1 chronic hepatitis C in combination with pegylated interferon and ribavirin. 28 Our docking results showed that the hydrogen bonds and van der Waals forces maintained upon the binding of Olysio and SARS‐CoV‐2 NSP12‐NSP8 interface, involving VAL‐330 and TYR‐273 (Figure 6A). What is more, the hydrogen bonds involving VAL‐214 and van der Waals forces maintained upon the binding of the Olysio and SARS‐CoV NSP12‐NSP8 interface (Figure 6C). Olysio could also bind to the interface active pockets of the NSP12‐NSP8 of SARS‐CoV‐2 and SARS‐CoV (Figures 6B and 6D). Thus, based on the present results, Olysio may be considered as a candidate for further in vitro evaluation of anti‐SARS‐CoV‐2 and anti‐SARS‐CoV activity.
Figure 6.
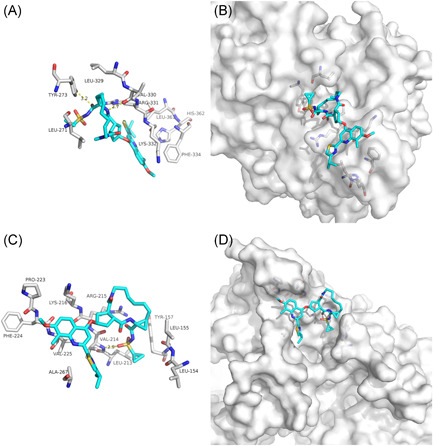
The binding model of Olysio against SARS‐CoV‐2 and SARS‐CoV NSP12‐NSP8. A, Interactions between Olysio (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV‐2. B, Binding models of Olysio (cyan) in the SARS‐CoV‐2 NSP12‐NSP8 protein interface pocket (white surface). C, Interactions between Olysio (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV‐2. D, Binding models of Olysio (cyan) in the SARS‐CoV‐2 NSP12‐NSP8 protein interface pocket (white surface). Numbers accompanying dashed yellow lines represent the interaction distance (Å, C). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
3.7. Docking results of Cepharanthine against SARS‐CoV‐2 and SARS‐CoV
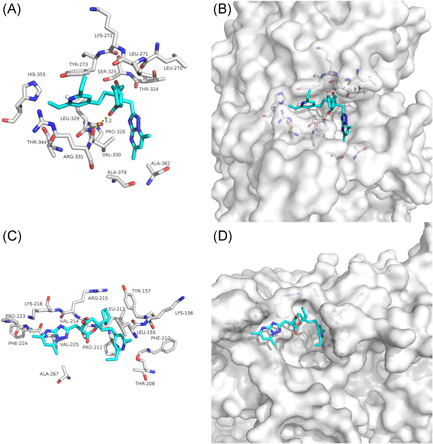
Cepharanthine, an alkaloid tetrandrine isolated from Stephania tetrandra was found to exert strong anticancer, anti‐inflammatory, and antioxidant activities. 29 In addition, it shows in vitro inhibitory effect on Herpes simplex virus type 1 (HSV‐1) infected cells. 30 Our docking results showed that Filibuvir was mainly combined with the SARS‐CoV‐2 NSP12‐NSP7 and NSP12‐NSP8 interface through van der Waals potential energy and hydrophobic accumulation, involving PHE‐843, PHE‐440, PHE‐441, PHE‐442 (Figures 7A and 7C). Further, the hydrogen bonds involving ARG‐215 maintained upon the binding of Cepharanthine and SARS‐CoV NSP12‐NSP8 interface, with additionally van der Waals forces (Figure 7E). Cepharanthine could bind to the interface active pockets of the SARS‐CoV‐2 and SARS‐CoV (Figures 7B,7D,7F). The previous study showed Cepharanthine could significantly inhibit the replication of human coronavirus strains OC43. 29 Taken together, Cepharanthine could be a potential natural antiviral compound for the prevention and treatment of SARS‐CoV‐2 and SARS‐CoV infection.
Figure 7.
The binding model of Cepharanthine against SARS‐CoV‐2 and SARS‐CoV. A, Interactions between Cepharanthine (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV‐2. B, Binding models of Cepharanthine (cyan) in the SARS‐CoV‐2 NSP12‐NSP7 protein interface pocket (white surface). C, Interactions between Cepharanthine (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV‐2. D, Binding models of Cepharanthine (cyan) in the SARS‐CoV‐2 NSP12‐NSP8 protein interface pocket (white surface). E, Interactions between Cepharanthine (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV. F, Binding models of Cepharanthine (cyan) in the SARS‐CoV NSP12‐NSP8 protein interface pocket (white surface). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
3.8. Docking results of Filibuvir against SARS‐CoV‐2 and SARS‐CoV NSP12‐NSP8
Filibuvir is an effective oral non‐nucleoside HCV NS5B RdRp inhibitor which exhibits potent antiviral activity against subgenomic HCV replicons in cell culture assays and is a potential treatment of chronic HCV infection. 31 , 32 Studies have shown that Filibuvir was well‐tolerated and could be considered in combination with other antiviral drugs to achieve better safety and efficacy for chronic HCV. 31 Our docking results showed that the hydrogen bonds involving VAL‐330 maintained upon the binding of Cepharanthine and SARS‐CoV‐2 NSP12‐NSP8 interface, with additionally van der Waals forces (Figure 8A). Filibuvir was mainly combined with the SARS‐CoV NSP12‐NSP8 interface through van der Waals potential energy (Figure 8C). Filibuvir could bind to the interface active pockets of the SARS‐CoV‐2 and SARS‐CoV NSP12‐NSP8 (Figures 8B and 8D). Thus, Filibuvir can be considered as a candidate drug for treating SARS‐CoV‐2 and SARS‐CoV infection, providing evidence for further research.
Figure 8.
The binding model of Filibuvir against SARS‐CoV‐2 and SARS‐CoV NSP12‐NSP8. A, Interactions between Filibuvir (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV‐2. B, Binding models of Filibuvir (cyan) in the SARS‐CoV‐2 NSP12‐NSP8 protein interface pocket (white surface). C, Interactions between Filibuvir (cyan) and associated residues (off‐white) in the interface of the crystal structure for SARS‐CoV. D, Binding models of Filibuvir (cyan) in the SARS‐CoV NSP12‐NSP8 protein interface pocket (white surface). Numbers accompanying dashed yellow lines represent the interaction distance (Å). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
3.9. Binding free energy calculated by MM/GBSA
Through the simulation trajectory of 5 ns MD simulations, we calculated the binding free energy of eight drugs by MM/GBSA method. The calculated binding free energies of Nilotinib, Cepharanthine, Lonafarnib, Tegobuvir for the NSP12‐NSP7 of SARS‐CoV‐2 were −22.3412 ± 2.4994, −22.1316 ± 2.1664, −25.5364 ± 3.0488, −24.1066 ± 2.5504 kcal/mol, respectively, which highlighted Lonafarnib as the most active one (Table 5). The interaction of van der Waals forces contributed more than the electrostatic interaction for Nilotinib, Cepharanthine, and Tegobuvir, indicating that van der Waals force is the main driving force for the combination of the three drugs. What is more, the calculated binding free energies of Olysio, Cepharanthine, Tegobuvir, and Filibuvir for the NSP12‐NSP8 of SARS‐CoV‐2 were −29.2605 ± 3.6317, −29.8408 ± 3.3839, −26.7327 ± 5.0249, −28.1876 ± 2.3322 kcal/mol, respectively, indicating Tegobuvir with the strongest binding free energy (Table 6).
Table 5.
The calculated binding energies of ligand to the interface of SARS‐CoV‐2 NSP12‐NSP7
| Energya | Nilotinib | Cepharanthine | Lonafarnib | Tegobuvir |
|---|---|---|---|---|
| ΔEvdw | −36.6592 ± 3.1681 | −29.4254 ± 2.317 | −33.3535 ± 2.6564 | −33.6401 ± 2.8152 |
| ΔEele | −11.2987 ± 5.191 | −11.6919 ± 2.2657 | 0.1574 ± 3.9504 | −2.2717 ± 2.4422 |
| ΔGgb | 29.5239 ± 6.1513 | 22.2336 ± 2.157 | 11.3859 ± 3.437 | 15.8702 ± 1.9281 |
| ΔGnp | −3.9072 ± 0.4078 | −3.248 ± 0.2139 | −3.7263 ± 0.2446 | −4.065 ± 0.2181 |
| ΔGcal | −22.3412 ± 2.4994 | −22.1316 ± 2.1664 | −25.5364 ± 3.0488 | −24.1066 ± 2.5504 |
Abbreviation: SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
ΔEvdw = van der Waals energy terms; ΔEele = electrostatic energy; ΔGgb = polar solvation free energy; ΔGnp = nonpolar solvation free energy; ΔGcal = final estimated binding free energy calculated from the above terms (kcal/mol).
Table 6.
The calculated binding energies of ligand to the interface of SARS‐CoV‐2 NSP12‐NSP8
| Energya | Olysio | Tegobuvir | Filibuvir | Cepharanthine |
|---|---|---|---|---|
| ΔEvdw | −43.7511 ± 3.7818 | −39.7211 ± 4.1282 | −42.0242 ± 6.4109 | −42.6585 ± 2.7766 |
| ΔEele | −15.0669 ± 14.855 | −16.2369 ± 2.6937 | −39.6169 ± 7.4189 | 1.658 ± 2.1958 |
| ΔGgb | 34.9446 ± 12.5304 | 31.0906 ± 2.7343 | 59.7662 ± 7.7447 | 17.6229 ± 2.0283 |
| ΔGnp | −5.3871 ± 0.2575 | −4.9734 ± 0.4595 | −4.8579 ± 0.6814 | −4.8099 ± 0.2558 |
| ΔGcal | −29.2605 ± 3.6317 | −29.8408 ± 3.3839 | −26.7327 ± 5.0249 | −28.1876 ± 2.3322 |
Abbreviation: SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
ΔEvdw = van der Waals energy terms; ΔEele = electrostatic energy; ΔGgb = polar solvation free energy; ΔGnp = nonpolar solvation free energy; ΔGcal = final estimated binding free energy calculated from the above terms (kcal/mol).
The calculated binding free energies of Saquinavir, Tipranavir, Tegobuvir, and Lonafarnib for the NSP12‐NSP7 of SARS‐CoV were −23.0269 ± 4.4383, −12.1704 ± 3.2929, −24.4461 ± 3.5461, −16.8649 ± 1.8442 kcal/mol, respectively, which highlighted Tegobuvir as the most active one (Table 7). The interaction of van der Waals forces contributed more than the electrostatic interaction for Tipranavir, Tegobuvir, and Lonafarnib indicating that van der Waals force is the main driving force for the combination of the three drugs. In addition, the calculated binding free energies of Olysio, Tegobuvir, Filibuvir, and Cepharanthine for the NSP12‐NSP8 of SARS‐CoV were −28.5431 ± 3.4974, −23.8177 ± 2.8786, −30.0087 ± 2.6150, −27.9163 ± 3.1346 kcal/mol, respectively (Table 8), indicating Filibuvir with the strongest binding free energy.
Table 7.
The calculated binding energies of ligand to the interface of SARS‐CoV NSP12‐NSP7
| Energya | Saquinavir | Tipranavir | Lonafarnib | Tegobuvir |
|---|---|---|---|---|
| ΔEvdw | −32.8851 ± 4.4965 | −26.6226 ± 4.9321 | −32.2453 ± 4.5440 | −25.4293 ± 2.1237 |
| ΔEele | −68.8922 ± 8.2672 | −0.0371 ± 2.7852 | −0.3949 ± 3.4435 | −8.4345 ± 2.4795 |
| ΔGgb | 82.9788 ± 8.3596 | 18.1213 ± 3.9815 | 11.6541 ± 2.7204 | 20.1027 ± 2.4178 |
| ΔGnp | −4.2285 ± 0.5503 | −3.6321 ± 0.5170 | −3.4600 ± 0.4934 | −3.1038 ± 0.1822 |
| ΔGcal | −23.0269 ± 4.4383 | −12.1704 ± 3.2929 | −24.4461 ± 3.5461 | −16.8649 ± 1.8442 |
Abbreviation: SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
ΔEvdw = van der Waals energy terms; ΔEele = electrostatic energy; ΔGgb = polar solvation free energy; ΔGnp = nonpolar solvation free energy; ΔGcal = final estimated binding free energy calculated from the above terms (kcal/mol).
Table 8.
The calculated binding energies of ligand to the interface of SARS‐CoV NSP12‐NSP8
| Energya | Olysio | Tegobuvir | Filibuvir | Cepharanthine |
|---|---|---|---|---|
| ΔEvdw | −48.0204 ± 2.7890 | −33.3370 ± 2.6790 | −41.9872 ± 2.6531 | −39.1550 ± 3.0045 |
| ΔEele | −1.3445 ± 9.7221 | −14.0331 ± 4.1102 | −11.8905 ± 2.4355 | −164.7184 ± 10.2539 |
| ΔGgb | 26.6238 ± 7.9551 | 28.1508 ± 3.1191 | 29.0768 ± 2.5574 | 180.0938 ± 9.6691 |
| ΔGnp | −5.8020 ± 0.3219 | −4.5984 ± 0.2536 | −5.2077 ± 0.2469 | −4.1367 ± 0.2773 |
| ΔGcal | −28.5431 ± 3.4974 | −23.8177 ± 2.8786 | −30.0087 ± 2.6150 | −27.9163 ± 3.1346 |
Abbreviation: SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
ΔEvdw = van der Waals energy terms; ΔEele = electrostatic energy; ΔGgb = polar solvation free energy; ΔGnp = nonpolar solvation free energy; ΔGcal = final estimated binding free energy calculated from the above terms (kcal/mol).
4. CONCLUSION
The indiscriminate spread of the coronavirus poses a threat to human health around the world. So far, no drug has been officially approved to treat SARS‐CoV‐2 and SARS‐CoV infection. NSP12‐NSP7‐NSP8 complex is a potential target to fight against the virus. SARS‐CoV‐2 and SARS‐CoV have extremely high similarity to the target of this complex. As for a highly active NSP12 polymerase complex, viral cofactors NSP7 and NSP8 are essential, suggesting that the particularly interesting compounds could disrupt the binding of NSP7 or NSP8 to NSP12. Thus, this study carried out high‐throughput drug screening for such interfacial active pockets. Subsequently, 44 compounds were selected for further evaluation based on virtual screening and docking scores, leading eight compounds for the calculation of binding free energy. Among them, Lonafarnib, Tegobuvir, Olysio, Filibuvir, and Cepharanthine can simultaneously be used as candidate drugs for the treatment of SARS‐CoV‐2 and SARS‐CoV infection, making them possible for broad‐spectrum antiviral drugs. According to published literature, all of them have antiviral activity. Therefore, this study suggests that eight compounds can be tested in vitro for their anti‐SARS‐CoV‐2 or anti‐SARS‐CoV effect, providing more choices for clinical treatment and prevention of SARS‐CoV‐2 and SARS‐CoV infection.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
RZ, YT, JX, and HX designed the study. RZ and LC performed most studies, but a few were carried out by GY and HZ. All authors contributed thoughts and advice. YT, JX, and HX did a literature search. YT and JX collected the data. RZ. and LC analyzed and interpreted the data. RZ wrote the text, and the other authors contributed to the final text presentation. All authors have proved the submission.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (No. 31870135, No.31600116) and the “1000 Talent Plan” of Sichuan Province (No. 980, No.1060).
Ruan Z, Liu C, Guo Y, et al. SARS‐CoV‐2 and SARS‐CoV: Virtual screening of potential inhibitors targeting RNA‐dependent RNA polymerase activity (NSP12). J Med Virol. 2021;93:389–400. 10.1002/jmv.26222
Contributor Information
Xinhe Huang, Email: xinhehuang@swjtu.edu.cn.
Xu Jia, Email: jiaxu@cmc.edu.cn.
Tai Yang, Email: taiyang@cmc.edu.cn.
REFERENCES
- 1. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Habibzadeh P, Stoneman EK. The novel coronavirus: a bird's eye view. Int J Occup Environ Med. 2020;11(2):65‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang ZW, Liang MX, Fu YZ, Han K, Lyu XZ. [2019‐nCoV: new challenges from coronavirus]. Zhonghua Yu Fang Yi Xue za Zhi. 2020;54:E001. [DOI] [PubMed] [Google Scholar]
- 4. Rabaan AA, Al‐Ahmed SH, Haque S, et al. SARS‐CoV‐2, SARS‐CoV, and MERS‐COV: a comparative overview. Infez Med. 2020;28(2):174‐184. [PubMed] [Google Scholar]
- 5. Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS‐CoV‐2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B. 2020;10(5):766‐788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu J, Zhao S, Teng T, et al. Systematic comparison of two animal‐to‐human transmitted human coronaviruses: SARS‐CoV‐2 and SARS‐CoV. Viruses. 2020;12(2):244. 10.3390/v12020244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang F, Liu N, Wu JY, Hu LL, Su GS, Zheng NS. [Pulmonary rehabilitation guidelines in the principle of 4S for patients infected with 2019 novel coronavirus (2019‐nCoV)]. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43:E004. [DOI] [PubMed] [Google Scholar]
- 8. Snijder EJ, Decroly E, Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. 2016;96:59‐126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327‐347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Subissi L, Posthuma CC, Collet A, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci USA. 2014;111(37):E3900‐E3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao Y, Yan L, Huang Y, et al. Structure of the RNA‐dependent RNA polymerase From COVID‐19 Virus. Science. 2020;368(6492):779‐782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kirchdoerfer RN, Ward AB. Structure of the SARS‐CoV nsp12 polymerase bound to nsp7 and nsp8 co‐factors. Nat Commun. 2019;10(1):2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. te Velthuis AJ, van den Worm SH, Snijder EJ. The SARS‐coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40(4):1737‐1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang H, Yan B, Yue L, He M, Liu Y, Li H. Virtual screening, ADME/Tox predictions and the drug repurposing concept for future use of old drugs against the COVID‐19. Life Sci. 2020;27:117963‐203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pant S, Singh M, Ravichandiran V, Murty USN, Srivastava HK. Peptide‐like and small‐molecule inhibitors against Covid‐19. J Biomol Struct Dyn. 2020:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choy KT, Wong AYL, Kaewpreedee P, et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS‐CoV‐2 replication in vitro. Antiviral Res. 2020;178:104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jean S‐S, Lee P‐I, Hsueh P‐R. Treatment options for COVID‐19: the reality and challenges. J Microbiol Immunol Infect. 2020;53(3):436‐443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hussain T, Zhao D, Shah SZA, et al. Nilotinib: a tyrosine kinase inhibitor mediates resistance to intracellular mycobacterium via regulating autophagy. Cells. 2019;8(5):506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ananthula HK, Parker S, Touchette E, et al. Preclinical pharmacokinetic evaluation to facilitate repurposing of tyrosine kinase inhibitors nilotinib and imatinib as antiviral agents. BMC Pharmacol Toxicol. 2018;19(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. la Porte CJL. Saquinavir, the pioneer antiretroviral protease inhibitor. Expert Opin Drug Metab Toxicol. 2009;5(10):1313‐1322. [DOI] [PubMed] [Google Scholar]
- 21. Kandula VR, Khanlou H, Farthing C. Tipranavir: a novel second‐generation nonpeptidic protease inhibitor. Expert Rev Anti Infect Ther. 2005;3(1):9‐21. [DOI] [PubMed] [Google Scholar]
- 22. Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (2019‐nCoV), Wuhan, China through a drug‐target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784‐790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wong NS, Morse MA. Lonafarnib for cancer and progeria. Expert Opin Investig Drug. 2012;21(7):1043‐1055. [DOI] [PubMed] [Google Scholar]
- 24. Asselah T, Loureiro D, Tout I, et al. Future treatments for hepatitis delta virus infection. Liver Int. 2020;40(suppl 1):54‐60. [DOI] [PubMed] [Google Scholar]
- 25. Erdağ TK, Kurtoğlu G. The 100 most cited Turkish papers in the otorhinolaryngology journals of web of science. Turk Arch Otorhinolaryngol. 2015;120:112‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zeuzem S, Buggisch P, Agarwal K, et al. The protease inhibitor, GS‐9256, and non‐nucleoside polymerase inhibitor tegobuvir alone, with ribavirin, or pegylated interferon plus ribavirin in hepatitis C. Hepatology. 2012;55(3):749‐758. [DOI] [PubMed] [Google Scholar]
- 27. Hebner CM, Han B, Brendza KM, et al. The HCV non‐nucleoside inhibitor Tegobuvir utilizes a novel mechanism of action to inhibit NS5B polymerase function. PLoS One. 2012;7(6):e39163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Izquierdo L, Helle H, François F, et al. Simeprevir approved for hepatitis C virus infection. Pharmgenomics Pers Med. 2014;71(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weber C Jr, Opatz T. Bisbenzylisoquinoline Alkaloids. Alkaloids Chem Biol. 2019;81:1‐114. [DOI] [PubMed] [Google Scholar]
- 30. Liu X, Wang Y, Zhang M, Li G, Cen Y. [Study on the inhibitory effect of cepharanthine on herpes simplex type‐1 virus (HSV‐1) in vitro]. Zhong Yao Cai. 2004;27(2):107‐110. [PubMed] [Google Scholar]
- 31. Rodriguez‐Torres M, Yoshida EM, Marcellin P, et al. A phase 2 study of filibuvir in combination with pegylated IFN alfa and ribavirin for chronic HCV. Ann Hepatol. 2014;13(4):364‐375. [PubMed] [Google Scholar]
- 32. Beaulieu PL. Filibuvir, a non‐nucleoside NS5B polymerase inhibitor for the potential oral treatment of chronic HCV infection. IDrugs. 2010;13(12):938‐948. [PubMed] [Google Scholar]


