Abstract
Coronavirus disease 2019 (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 emerged in China in December 2019 and then rapidly spread worldwide. Why COVID‐19 patients with the same clinical condition have different outcomes remains unclear. This study aimed to examine the differences in the phenotype and functions of major populations of immune cells between COVID‐19 patients with same severity but different outcomes. Four common type adult inpatients with laboratory confirmed COVID‐19 from Beijing YouAn Hospital, Capital Medical University were included in this study. The patients were divided into two groups based on whether or not COVID‐19 polymerase chain reaction (PCR)‐negative conversion occurred within 3 weeks. Peripheral blood samples were collected to compare the differences in the phenotype and functions of major populations of immune cells between the two groups of patients. The result shows that the proportions of CD3+CD8+CD38+HLA‐DR+CD27− effector T killer cells generally declined, whereas that of CD3+CD4+CD8+ double‐positive T cells (DPTs) increased in the persistently PCR‐positive patients. In summary, considering the imbalance between effector T killer cells/CD3+CD4+CD8+ DPTs was a possible key factor for PCR‐negative conversion in patients with COVID‐19.
Keywords: COVID‐19, immune response, nucleic acid conversion
Highlights
Tregs are crucial to maintain immune cell homeostasis in numerous diseases. DPTs have a high capacity to produce cytokines that act as immunosuppressive regulators of leukocyte trafficking. In this study, the percentage of DPTs increased and the proportions of CD3+CD8+CD38+HLA‐DR+CD27‐ effector T killer cells generally declined in the RPP cases.
We therefore profiled their expression and found that the levels of functional molecules, such as HLA‐DR and CD38, were obviously higher in the NRPP group than in the RPP group. CD3+CD8+CD38+HLA‐DR+CD27‐ effector T killer cells could be helpful for reduction of viral load in COVID‐19 patients.
However, abnormalities in T‐cell profiles did not predict outcome or correlate with disease severity.
An immune imbalance may exist in RPP cases, with an increase in percentage of DPTs and reduction in effector T killer cells, following the possible recruitment of these cells to the lungs.
These observations are in line with the suggested role for T‐cell activation in the immune response to COVID‐19 infection.
1. INTRODUCTION
The novel coronavirus disease 2019 (COVID‐19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), emerged in China in December 2019, and is now a global pandemic.
There are two main challenges for the prevention and control of this disease. First, asymptomatic patients have the same infectivity and viral load as symptomatic patients. Second, it is unclear how long the virus is discharged from SARS‐CoV‐2 infected patients. 1 Zou reported that the average positive time of nucleic acid in patients with COVID‐19 was 21 days. 2 A total of 24 patients with COVID‐19 had a median time of 12 days (9‐14 days) for nucleic acid conversion, and the longest time was 34 days. A study of 191 patients with COVID‐19 in Wuhan region reported that the median nucleic acid conversion time of the patients was 20 days, and the longest time was 37 days. 3 The factors that lead to the different conversion times of patients are complex. It is reported that male gender, delayed admission and invasive mechanical ventilation are related to longer conversion times of SARS‐CoV‐2 nucleic acid. 4 The application of hormone therapy may prolong the virus clearance time. 5 , 6 Clinical observation showed that the nucleic acid shedding time of COVID‐19 critical patients was longer than that of common type. Effective antiviral therapy may improve the prognosis of COVID‐19. Gautret Philippe used hydroxychloroquine in 20 patients and found a significant reduction in viral load on day 6. 7
The duration of COVID‐19 nucleic acid positivity may be related to the host immunity. The levels of T‐cells and B‐cells were increased in patients with nucleic acid negative conversion on day 14 after the onset of the disease (P < .05). 8 Lymphocyte subsets are related to the severity of the disease, which can guide the clinical treatment. However, there is no relevant study on the immune function of patients with delayed conversion of COVID‐19 nucleic acid. The analysis of lymphocyte subsets can help to further understand the influence of immune function on the clearance of SARS‐CoV‐2.
2. METHODS
2.1. Study design and participants
This retrospective cohort study included four common type adult inpatients (≥18 years old) from Beijing YouAn Hospital, Capital Medical University, China. COVID‐19 laboratory testing involved quantitative real‐time polymerase chain reaction (PCR) for diagnosis. COVID‐19 PCR‐negative conversion was defined as two consecutive sputum, nasopharyngeal swabs, and other respiratory tract specimens with negative PCR results, when the sampling time was at least 24 hours apart.
The four patients were divided into two groups based on whether or not COVID‐19 PCR‐negative conversion occurred within 3 weeks. 2 Two cases in the persistently PCR positive group (PPP group) achieved COVID‐19 PCR‐negative conversion in >3 weeks. The other two patients who achieved PCR conversion <3 weeks were classified as the non‐persistently PCR positive group (NPPP group).
We collected peripheral blood samples at two to three time points from every patient to compare the differences in lymphocyte functions between the two groups. The first time point was the time of admission, and the others were after the COVID‐19 PCR became negative. Then we analyzed the clinical characteristics, expression of infection‐related biomarkers, and lymphocyte subsets between the two groups.
2.2. Detection of viral RNA in COVID‐19
A magnetic bead‐method nucleic acid extraction kit was used along with a fully automated nucleic acid extraction instrument (Shanghai BioGerm, China). The total RNA was extracted from a 200 μL sample and fluorescence PCR (Applied Biosystems 251658240 7500 Real‐Time PCR Systems, Foster City, CA) was performed according to the manufacturer's instructions. Upper respiratory throat swab samples were taken from the patients with COVID‐19 and transported to the infectious lab in Beijing YouAn Hospital for laboratory diagnosis.
2.3. Data collection
We obtained epidemiological, demographic, clinical, laboratory, treatment, and outcome data from the electronic medical records.
2.4. 2.4. mass cytometry antibody staining
CD45 barcoding method was used as previously described. 9 Briefly, cells were pre‐stained with CD45 antibodies labeled with different metal isotopes before mixing and staining with antibody cocktail listed in the panel table below. Cells were then incubated with 0.125 μm intercalator in fixing and permeabilization buffer (Fluidigm) at 4°C overnight. Before acquisition, cells were washed three times with ice‐cold phosphate‐buffered saline and three times with deionized water, and resuspended in deionized water containing 10% EQ 4 Element Beads (Fluidigm) at a final concentration of 1 × 106 cell/mL. Data acquisition was performed on a Helios mass cytometer (Fluidigm). The original FCS data were normalized, and.fcs files of each individual were collected.
2.5. CyTOF data analysis
All.fcs files were uploaded into Cytobank, data cleaning was performed as previously reported, 9 and the population of single living cells were exported as.fcs files for further analysis. Arcsinh transform was performed to signal intensities of all channels. A viSNE analysis method was used as previously reported. 10
2.6. Statistical analysis
The categorical variables were described as frequency rates and percentages, and the continuous variables were described as mean and standard deviation, median and interquartile range (IQR) values. The Mann‐Whitney U test, χ 2 test, or Fisher's exact test was used to compare the differences between the PPP and NPPP groups, where appropriate.
3. RESULTS
3.1. Demographic and clinical characteristics of the patients with COVID‐19
By 8 March 2020, four patients with COVID‐19 admitted to the Beijing YouAn Hospital, Capital Medical University were recruited in this study. The four patients were clinically diagnosed as common cases. The median age was 58 years (IQR, 68.75‐35.25; range, 30‐70 years), and two patients were men. All four patients had a history of contact with COVID‐19 patients and were cluster onset. One patient had a history of travel or living in Hubei. Two patients had fever, cough, expectoration and fatigue. Other symptoms included shortness of breath (25%), nausea and vomiting (25%), and anorexia (25%). No patient had muscle ache and diarrhea. There were no significant differences in symptoms between the PPP and NPPP groups. Two patients (50%) had hypertension and one (25%) had diabetes and coronary heart disease (Table 1).
Table 1.
Clinical characteristics of the four patients with COVID‐19
| Clinical characteristics, symptoms | NPPP (n = 2) | PPP (n = 2) | |||
|---|---|---|---|---|---|
| Case 1 | Case 2 | Case 1 | Case 2 | ||
| Age, y | 65 | 70 | 51 | 30 | |
| Sex | F | F | M | M | |
| COVID‐19 type | Common | Common | Common | Common | |
| History of contacting patients with COVID‐19 | + | + | + | + | |
| Travel or living in Hubei | + | − | − | − | |
| Cluster onset | + | + | + | + | |
| The days of PCR‐negative conversion | 13 | 6 | 35 | 35 | |
| Symptoms | |||||
| Fever | + | − | − | + | |
| Cough | − | + | − | + | |
| Expectoration | − | + | − | + | |
| Fatigue | + | + | − | − | |
| Shortness of breath | − | + | − | − | |
| Nausea or vomiting | − | + | − | − | |
| Anorexia | − | + | − | − | |
| Muscle ache | − | − | − | − | |
| Diarrhea | − | − | − | − | |
| Concomitant diseases | |||||
| Diabetes | + | − | − | − | |
| Hypertension | + | − | + | − | |
| Coronary heart disease | + | − | − | − | |
Note: N is the total number of patients with available data.
Abbreviations: +, positive. −, negative; COVID‐19, coronavirus disease 2019; F, female; M, male; NA, not available; NPPP, non‐persistently PCR positive; PCR, polymerase chain reaction; PPP, persistently PCR positive.
3.2. Laboratory findings on admission in patients with COVID‐19
Table 2 presents the laboratory findings in the four patients with COVID‐19. Compared to the NPPP group, the PPP group had higher levels of creatine kinase‐MB (0.61 ± 0.09 vs 0.20 ± 0.05 ng/mL, P = .047), album (ALB) (44.95 ± 1.06 vs 35.70 ± 1.41 g/L; P = .022), and Crea (86.50 ± 4.94 vs 53.50± 3 .53 μmol/L; P = .022). Meanwhile, white blood cell count (7.95±2.72 vs 3.95 ± 0.48 × 109/L), hemoglobin (152.50 ± 2.12 vs 142.00 ± 0.00 g/L), leukocyte (2.11 ± 1.16 vs 1.56 ± 1.12 × 109/L) and neutrophil (4.39 ± 1.35 vs 1.83 ± 0.53 × 109/L) counts, lactic acid (1.70 ± 0.84 vs 2.04 ± 1.27 mmol/L), platelet level (214.50 ± 28.99 vs 215.50 ± 58.68 × 109/L), alanine aminotransferase (59.00 ± 5.65 vs 40.00 ± 28.28 U/L), aspartate aminotransferase (51.50 ± 4.94 vs 62.50 ± 21.41 U/L), prothrombin time (PT) (11.85 ± 0.35 vs 13.00 ± 0.28 S), total bilirubin (15.10 ± 1.69 vs 13.10 ± 2.54 μmol/L), direct bilirubin (3.05 ± 0.49 vs 2.95 ± 0.35 μmol/L), SpO2 (97.00 ± 1.41 vs 98.00 ± 0.00%), and Troponin I (TNI) (0.01 ± 0.00 vs 0.01 ± 0.01 ng/mL) were comparable between the two groups. Compared with the NPPP group, the myocardial injury and renal function were worse in the PPP group.
Table 2.
Laboratory findings of the four patients with COVID‐19
| Laboratory findings | Normal range | NPPP (n = 2) | PPP (n = 2) | t | P |
|---|---|---|---|---|---|
| mean ± SD | mean ± SD | ||||
| WBC, ×109/L | 3.5‐9.5 | 3.95 ± 0.48 | 7.95 ± 2.72 | −1.660 | .334 |
| N, ×109/L | 1.8‐6.3 | 1.83 ± 0.53 | 4.39 ± 1.35 | −2.479 | .195 |
| L, ×109/L | 1.1‐3.2 | 1.56 ± 1.12 | 2.11 ± 1.16 | −0.480 | .679 |
| NE% | 50‐70 | 47.60 ± 19.23 | 61.90 ± 4.52 | −1.024 | .479 |
| LC% | 20‐40 | 38.25 ± 23.97 | 28.35 ± 5.44 | 0.570 | .663 |
| Hb, g/L | 120‐172 | 142.00 ± 0.00 | 152.50 ± 2.12 | −7.000 | .090 |
| PLT, ×109/L | 125‐350 | 215.50 ± 58.68 | 214.50 ± 28.99 | 0.022 | .985 |
| LA, mmol/L | 0.4‐2.0 | 2.04 ± 1.27 | 1.70 ± 0.84 | 0.311 | .789 |
| ALT, U/L | 7‐40 (F) 9‐50 (M) | 40.00 ± 28.28 | 59.00 ± 5.65 | −0.932 | .513 |
| AST, U/L | 13‐35 (F) 15‐40 (M) | 62.50 ± 21.41 | 51.50 ± 4.94 | 0.217 | .864 |
| T‐Bil, μmol/L | 3.4‐20.5 | 13.10 ± 2.54 | 15.10 ± 1.69 | −0.925 | .465 |
| D‐Bil, μmol/L | 1‐20.1 | 2.95 ± 0.35 | 3.05 ± 0.49 | −0.232 | .840 |
| ALB, g/L | 40‐55 | 35.70 ± 1.41 | 44.95 ± 1.06 | −7.400 | .022 |
| PT, S | 9.9‐12.8 | 13.00 ± 0.28 | 11.85 ± 0.35 | 3.592 | .074 |
| Crea, μmol/L | 41‐81 | 53.50 ± 3.53 | 86.50 ± 4.94 | −7.672 | .022 |
| CK‐MB, ng/mL | <3.6 | 0.20 ± 0.05 | 0.61 ± 0.09 | −5.438 | .047 |
| TNI, ng/mL | <0.056 | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.400 | .758 |
| SPO2(FIO2 = 21%) | >90% | 98.00 ± 0.00 | 97.00 ± 1.41 | 1.000 | .500 |
Note: N is the total number of patients with available data.
Abbreviations: ALB, album; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CK‐MB, creatine kinase‐MB; COVID‐19, coronavirus disease 2019; F, female; Hb, hemoglobin; LA, lactic acid; LC, lymphocyte percentage; M, male; NE, neutrophil; NPPP, non‐persistently PCR positive; PCR, polymerase chain reaction; PLT, platelat; PPP, persistently PCR positive; PT, prothrombin time; TNI, Troponin I; WBC, white blood cell.
3.3. Complications, treatment, and outcomes of four patients with COVID‐19
All NPPP and PPP cases had pneumonia. Only one patient in the NPPP group had acute liver injury. All cases were treated with oxygen therapy. Integrated Chinese and western medicine treatment was given to the NPPP cases. All four cases were discharged after recovery.
3.4. Lymphocyte subsets analysis in patients with COVID‐19
To delineate the difference of immune response of COVID‐19 infection between PPP and NPPP patients, we performed the CD45‐barcoding methods while using mass of flight cytometry (CyTOF) as previous publication. 11 In brief, anti‐CD45 antibodies that conjugated with different metal isotopes were used to label peripheral blood mononuclear cells (PBMCs) from different patients, the labeled cells were then pooled and stained with a panel of 30 antibodies as described in Table S1. Acquired data was subject to clean‐up and debarcoding with the Cytobank and analyzed for expression of markers (Figure 1A). Furthermore, we analyzed the percentages of subsets of immune cells based on the expression of protein markers (Figure 1B). As compared to the NPPP group, the proportions of CD3+CD8+CD38+HLA‐DR+CD27− effector T killer cells generally declined, whereas CD3+CD4+CD8+ double‐positive T cells (DPTs) increased in the PPP group (Figure 1B). The expression of active markers CD38 and HLA‐DR on T cells were stronger in the NPPP group than in the PPP group (Figure 1C).
Figure 1.
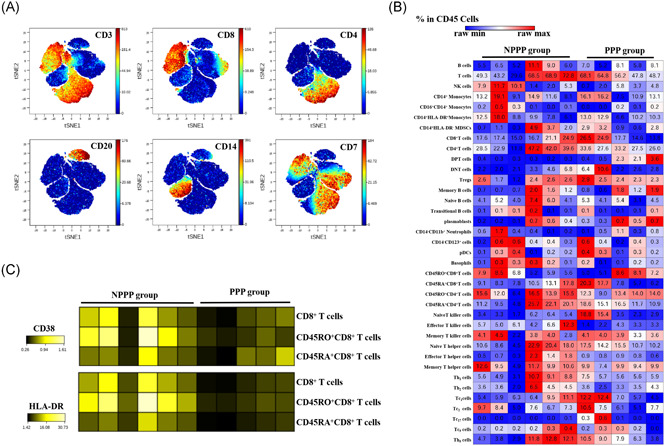
Flight cytometry (CyTOF)‐based analysis identified immune cell signatures in peripheral blood of patients with coronavirus disease 2019 (COVID‐19). A representative viSNE plot of immune cell populations from patients with persistently PCR positive (PPP) and non‐persistently PCR positive (NPPP). A, viSNE analysis from Cytobank indicated subsets of blood cells, and expression levels of relative markers were also displayed. B, Heatmap displayed percentage of different cell populations in CD45+ peripheral blood mononuclear cells (PBMCs) by raw max/min manner. C, Raw data of CD38 and HLA‐DR expression in subpopulations of blood cells were shown in heatmap
4. DISCUSSION
Cells of the innate and adaptive immune systems play important roles in defense against viral infection. In this study, we identified the signature of PBMCs from patients with COVID‐19 that were PPP group and NPPP group. By using CyTOF, we found that the frequency of DPTs cells was increased in the PPP group compared to NPPP groups (Figure 1B).
So far, DPTs had been classified as a separate T‐cell subpopulation and described in several pathological condition, such as infections, tumors, and autoimmune diseases. 12 , 13 Due to the heterogeneity exists within the DPTs sub‐population, function of DPTs remains controversial. According to previous publications, 11 DPTs have a high capacity to produce cytokines that act as immunosuppressive regulators of leukocyte trafficking. We speculate that DTPs may play an immunosuppressive role, and then inhibit the negative conversion of COVID‐19 nucleic acid. The cases of this study is limited, and further research is needed to explore the pathogenesis in the future.
The proportions of CD3+CD8+CD38+HLA‐DR+CD27− effector T killer cells generally declined in the PPP cases (Figure 1B). We therefore profiled their expression and found that the levels of functional molecules, such as HLA‐DR and CD38, were obviously higher in the NPPP group than in the PPP group (Figure 1C). CD3+CD8+CD38+HLA‐DR+CD27− effector T killer cells could be helpful for reduction of viral load in patients with COVID‐19. However, abnormalities in T‐cell profiles did not predict outcome or correlate with disease severity.
This study had several limitations. First, we analyzed very few patients. Second, blood samples at different time points in this retrospective study were collected to assess the lymphocyte subpopulations, and the different time points of assessment of the lymphocyte subpopulations could have introduced bias. Third, whether the observed changes in lymphocyte subpopulation percentages in the patients are attributed to the longer conversion times of SARS‐CoV‐2 nucleic acid remains unknown. Apoptosis or infiltration of cells and serum cytokine levels were not measured, and major pathways involved in pathogenesis were not analyzed.
5. CONCLUSIONS
An immune imbalance may exist in PPP cases, with an increase in percentage of DPTs and reduction in effector T killer cells, following the possible recruitment of these cells to the lungs. These observations are in line with the suggested role for T‐cell activation in the immune response to COVID‐19 infection.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
YHB participated in the design of the study and draft the manuscript. WWJ performed the experiment and statistical analysis. TS conceived of the study and participated in its design and coordination and helped to draft the manuscript. CDX and XB participated Critical revision of the manuscript for important intellectual content and supervised this study. All authors read and approved the final manuscript.
ETHICS STATEMENT
This study and all the relevant experiments were approved by the Beijing YouAn Hospital Research Ethics Committee, and performed in accordance with the Declaration of Helsinki. All participants provided written informed consent for the collection of information, and their clinical samples were stored and used for research.
Supporting information
Supporting information
ACKNOWLEDGMENTS
This study and all the relevant experiments were approved by the Beijing YouAn Hospital Research Ethics Committee, and performed in accordance with the Declaration of Helsinki. All participants provided written informed consent for the collection of information, and their clinical samples were stored and used for research. This work was supported by grants from theKey Programs Beijing Municipal Education Commission of China (KZ202010025037).
Yu H‐b, Wang W‐j, Tang S, Chen D‐x, Xu B. Immune responses and pathogenesis in persistently PCR‐positive patients with SARS‐CoV‐2 infection. J Med Virol. 2021;93:760–765. 10.1002/jmv.26287
Contributor Information
De‐xi Chen, Email: dexichen@ccmu.edu.cn.
Bin Xu, Email: xubin1016@126.com.
REFERENCES
- 1. Lo IL, Lio CF, Cheong HH, et al. Evaluation of SARS‐CoV‐2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID‐19 in Macau. Int J Biol Sci. 2020;16:1698‐1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu K, Chen Y, Yuan J, et al. Factors associated with prolonged viral RNA shedding in patients with COVID‐19. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed]
- 5. Han Y, Jiang M, Xia D, et al. COVID‐19 in a patient with long‐term use of glucocorticoids: a study of a familial cluster. Clin Immunol. 2020;214:108413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ling Y, Xu SB, Lin YX, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J. 2020;133:1039‐1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gautret P, Lagier JC, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID‐19: results of an open‐label non‐randomized clinical trial. Int J Antimicrob Agents. 2020:105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8. Haoao X, Gao M, He L, et al. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID‐19. J Infect. 2020;433‐434:152411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Unen V, Höllt T, Pezzotti N, et al. Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat Commun. 2017;8:1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Levine JH, Simonds EF, Bendall SC, et al. Data‐driven phenotypic dissection of AML reveals progenitor‐like cells that correlate with prognosis. Cell. 2015;162:184‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang W, Su B, Pang L, et al. High‐dimensional immune profiling by mass cytometry revealed immunosuppression and dysfunction of immunity in COVID‐19 patients. Cell Mol Immunol. 2020;17:650‐652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gonzalez‐Mancera MS, Bolanos NI, Salamanca M, et al. Percentages of CD4+CD8+ double‐positive T lymphocytes in the peripheral blood of adults from a Blood Bank in Bogota, Colombia. Turk J Haematol. 2020;37:36‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parel Y, Chizzolini C. CD4+ CD8+ double positive (DP) T cells in health and disease. Autoimmun Rev. 2004;3:215‐220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


