Abstract
Objective
Long non-coding RNAs (lncRNAs) are involved in carcinogenesis and could be used as diagnostic biomarkers. Our study aimed to elucidate the clinical role of serum exosomal lncRNA H19 in gastric cancer (GC).
Methods
In this prospective clinical study, we determined serum exosomal lncRNA H19 levels in 81 patients with GC and analysed the correlations between serum lncRNA H19 levels and clinical characteristics. Receiver operating characteristics (ROC) curves were constructed to determine the diagnostic performance of exosomal lncRNA H19 in GC.
Results
Serum exosomal lncRNA H19 levels were significantly upregulated in patients with GC both before and after surgery compared with healthy controls. Furthermore, serum exosomal lncRNA H19 levels were significantly decreased after compared with before surgery in patients with GC. Preoperative lncRNA H19 levels were significantly correlated with TNM stage. The area under the ROC curve (AUC) value for exosomal lncRNA H19 was 0.849, which was significantly higher than the AUC values for cancer antigens 19-9 and 72-4 and carcinoembryonic antigen, either alone or combined.
Conclusions
These results suggest that circulating exosomal lncRNA H19 may be a potential biomarker with diagnostic and prognostic value in GC.
Keywords: Gastric cancer, exosome, long non-coding RNA H19, prognosis, diagnosis, biomarker
Introduction
Gastric cancer (GC) is the fifth most common malignancy and the third leading cause of cancer deaths worldwide.1,2 Half of the world’s cases of GC occur in eastern Asia, especially in China.2 Despite advances in the diagnosis and treatment of GC, the prognosis of patients remains poor, with >70% of patients ultimately dying of cancer-related causes, mainly because their cancer is diagnosed at an advanced stage with limited treatment options, frequent metastasis, and recurrence.3,4 Early diagnosis and treatment are therefore critical to improving the prognosis of GC. The current strategy and gold standard for GC diagnosis is upper digestive endoscopy followed by pathological examination; however, this is an invasive and uncomfortable procedure.5–7 Blood-based biomarkers have the advantage of being non-invasive, and may therefore be suitable for aiding the diagnosis of GC and predicting its prognosis. However, the sensitivity and specificity of current serum biomarkers of GC, such as carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA 19-9), and carbohydrate antigen 72-4 (CA 72-4), are poor.8 There is thus a need to discover other novel biomarkers with high specificity and sensitivity for early GC screening.
Exosomes are membrane vesicles with a diameter of 50 to 100 nm that function to transfer proteins, nucleic acids, and lipids to recipient cells, and play an important role in intercellular communication within the extracellular environment.9 Numerous studies have indicated that circulating RNAs may be novel non-invasive diagnostic biomarkers in cancer patients.10 In addition, cancer cells secrete exosomes into the peripheral circulation, and exosomal RNAs can more accurately reflect changes in cancer cells during tumour progression. These exosomes not only enhance the stability of RNAs in the circulating blood by protecting RNAs from degradation by endogenous blood, but also reduce the impacts of RNAs released by circulating dysfunctional cells.10 Circulating exosomal RNAs are therefore considered to be potential novel diagnostic and prognostic biomarkers for cancers.
Long non-coding RNAs (lncRNAs) are regulatory non-coding RNAs longer than 200 nucleotides.11 Evidence has shown that lncRNAs play crucial roles in numerous biological processes, such as transcription regulation, cancer progression, and cellular differentiation.12,13 Many lncRNAs have been identified from mammalian genomes and they are considered to be vital regulatory molecules in the progression of different types of cancer. For instance, lncRNA H19 is 2.3 kb long lncRNA located on human chromosome 11p15.5, which is known as a non-coding imprinted oncofoetal RNA with a high expression level during embryogenesis, as well as in some types of tumours, including GC.14–19 Expression levels of lncRNA H19 were markedly upregulated in both tumour tissues and plasma from GC patients relative to adjacent normal gastric tissues and normal controls.20 Upregulation of lncRNA H19 expression may enhance the carcinogenesis and metastasis of GC and contribute to a poor prognosis in GC patients.21,22 However, the potential roles of exosomal lncRNA H19 in GC are not well understood. We examined the clinical value of exosomal lncRNA H19 levels to determine its suitability as a reliable non-invasive diagnostic and prognostic biomarker in patients with GC.
Materials and Methods
Patients and specimens
Patients who underwent surgery for GC at The Affiliated Hospital of Jiaxing University from 2016 to 2018 were enrolled in this prospective clinical study. The inclusion criteria were drug-free patients with first-episode GC patients, aged >18 years. Patients were excluded if they had any coexisting acute or chronic infection, other active autoimmune condition, serious heart or lung disease, or a known history of other types of tumour. Peripheral blood samples were collected by venipuncture before and 14 days after surgery. The diagnosis of GC was confirmed by histopathological examination. Information on serum CEA, CA19-9, and CA72-4, as well as the results of upper digestive endoscopy, computed tomography, and magnetic resonance imaging, was also collected. No patients received chemotherapy or other anti-tumour treatments when the preoperative and postoperative blood samples were obtained. We also collected peripheral blood samples from age- and sex-matched controls who underwent routine physical examination and showed no signs of disease. The study was approved by the Ethics Committee of The Affiliated Hospital of Jiaxing University (12 December 2016). Signed consent was obtained from all the patients and controls. The study was carried out in full compliance with the relevant EQUATOR network guidelines. The clinicopathological grade of GC was staged according to the Union for International Cancer Control’s tumour-node-metastasis (TNM) classification system23 and histological grade was assessed according to the World Health Organization criteria.24
Serum collection
Peripheral blood samples (5 mL) were collected from each subject after fasting. Serum was isolated by centrifugation at 3,000 × g for 5 minutes followed by 10,000 × g for 10 minutes. The prepared serum samples were stored at –80°C for further use. The stability of exosomal lncRNA H19 in serum was determined by exposing 10 aliquots of serum samples from randomly selected patient to different conditions, including incubation at room temperature for 0, 6, 12, and 24 hours, and to repeated freeze-thaw cycles. Samples with evidence of haemolysis or lipidemia during processing were excluded.
Exosome isolation and transmission electron microscopy (TEM)
Serum (1 mL) was placed into a new sterile vessel and 250 µL of ExoQuick-TC™ Exosome Precipitation Solution (System Biosciences, Palo Alto, CA, USA) was added. The mixture was mixed by inverting and flicking the tube, followed by refrigeration for 30 minutes at 4°C and then centrifugation at 1,500 × g for 30 minutes. The exosome pellets were then resuspended in 25 µL of phosphate-buffered saline. Exosomes for TEM were spotted onto a glow-discharged copper grid, dried with a filament lamp, and then stained with 3% phosphotungstic acid solution for 5 minutes, and dried again with a filament lamp. Finally, the exosomes were examined under a JEM-1230 transmission electron microscope (JEOL, Japan).
Western blot and subcellular localisation
Total proteins were isolated from exosomes using RIPA buffer (Sigma-Aldrich, St Louis, MO, USA) and the protein concentration was measured by Bradford assay (Beyotime, China). Exosomal proteins were separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA) according to the manufacturer’s protocol. The membranes were probed using rabbit anti-CD63 (1:5000) and anti-TSG101 antibodies (1:3000) (System Biosciences) and the results were visualised using a Gel Doc XR+ system (Bio-Rad Laboratories, Hercules, CA, USA). Based on lncRNA sequence information and computational methods, the subcellular location of the H19 molecule was predicted using bioinformatics analysis tools (lncLocator: http: //www.csbio.sjtu.edu.cn/bioinf/lncLocator/index.html).
Total exosomal RNA preparation
Total exosomal RNA was extracted and purified using an miRNeasy Micro Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. The extracted RNA was dissolved in 25 µL of diethyl pyrocarbonate-treated water and the quantity and quality of the total RNA were evaluated by NanoDrop spectrophotometry (Thermo Fisher Scientific Inc., Rockford, IL, USA).
Reverse transcription
cDNA was synthesised using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Total RNA (1 µg) was placed in a nuclease-free tube and 1 µL of random hexamer primer and nuclease-free water was added to a total volume of 12 µL. The mixture was then incubated for 5 minutes at 70°C, followed by the addition of 4 µL of 5× Reaction buffer, 1 µL of RiboLock RNase Inhibitor, 2 µL of 10 mM dNTP Mix, and 1 µL of RevertAid M-MuLV Reverse Transcriptase, and incubation for 60 minutes at 42°C followed by 10 minutes at 70°C.
Quantitative real-time polymerase chain reaction (PCR)
The primer sequences for lncRNA H19 were 5′-CGTCCGGCCTTCCTGAACA-3′ (forward) and 5′-TTGAGCTGGGTAGCACCATTTCT-3′ (reverse) and the primer sequences for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA were 5′-GGAGTCCACTGGCGTCTTC-3′ (forward) and 5′-GCTGATGATCTTGAGGCTGTTG-3′ (reverse). The primer sequences for both RNAs were designed using Primer Express 3.0 (Applied Biosystems, Foster City, CA, USA) and synthesised by BBI Life Sciences Corporation (Shanghai, China).
Real-time PCR was performed using SYBR® Premix Ex Taq™ II (Takara, Japan) on an ABI 7500 Real-Time PCR System (Applied Biosystems). The 20-µL PCR mixture included 10 µL of 2× SYBR® Premix Ex Taq™ II, 0.8 µL of 10 µM PCR forward primer, 0.8 µL of 10 µM PCR reverse primer, 0.4 µL of 50 × Rox Reference Dye II, 2 µL of cDNA, and 2 µL of ddH2O. The reaction mixture was incubated at 95°C for 10 minutes, followed by 40 reaction cycles of 95°C for 10 seconds and 60°C for 34 s. Expression levels of lncRNA H19 were normalised to GAPDH as an internal control using the 2–ΔΔCt method.25 Each sample was assayed three times, and the experimenters were blinded to the clinical and pathological diagnoses of the patients.
Electrochemiluminescence
Serum levels of CEA, CA 19-9, and CA 72-4 were measured by electrochemiluminescence immunoassay using a Cobas E601 system (Roche Diagnostics GmbH, Germany) according to the manufacturer’s protocol.
Statistical analysis
Statistical analysis was performed using SPSS Statistics for Windows, Version 22.0 (SPSS Inc., Chicago, IL, USA). Exosomal lncRNA H19 levels were compared among the three groups (healthy subjects, preoperative GC patients, and postoperative GC patients) using Student’s t-tests. Correlations between clinicopathological parameters and expression levels of exosomal lncRNA H19 in patients with GC were analysed by Mann–Whitney U tests. Data were presented as median (interquartile range). Receiver operating characteristic (ROC) curves were constructed for the GC samples and the areas under the ROC curves (AUC) were estimated to determine the feasibility of using exosomal lncRNA H19 as a biomarker for GC. ROC analysis was carried out using MedCalc software (version 18.2.1; MedCalc, Mariakerke, Belgium). The level of significance was set at P < 0.05.
Results
Patients
Eighty-one patients and 78 healthy controls were included in this study. Their mean ages (±standard deviation) were 64.23±12.50 years and 60.15±16.42 years, respectively. Detailed demographic and clinical information is provided in Table 1.
Table 1.
Correlation between preoperative exosomal lncRNA H19 expression levels and clinicopathological features in 81 patients with gastric cancer.
| Characteristic | Number of cases | Exosomal lncRNA H19 level median (25%–75% interquartile range) | P-value |
|---|---|---|---|
| Age (years) | |||
| <60 | 33 | 4.781 (1.729–16.251) | 0.216 |
| ≥60 | 48 | 8.760 (4.146–16.847) | |
| Sex | |||
| Male | 51 | 9.453 (3.734–17.356) | 0.169 |
| Female | 30 | 5.291 (2.407–16.241) | |
| Lymph node metastasis | |||
| Positive | 37 | 5.160 (2.840–15.567) | 0.271 |
| Negative | 44 | 9.188 (2.908–17.084) | |
| Tumour size (cm) | |||
| <5 | 44 | 10.094 (3.332–16.260) | 0.415 |
| ≥5 | 37 | 6.052 (2.545–15.238) | |
| Tumour differentiation | |||
| Poor | 40 | 5.160 (2.096–15.555) | 0.169 |
| Moderate/well | 41 | 8.871 (4.307–17.247) | |
| TNM stage | |||
| T1/T2 | 40 | 4.991 (2.337–12.966) | 0.007 |
| T3/T4 | 41 | 12.973 (4.511–21.443) | |
Characterisation of exosomes
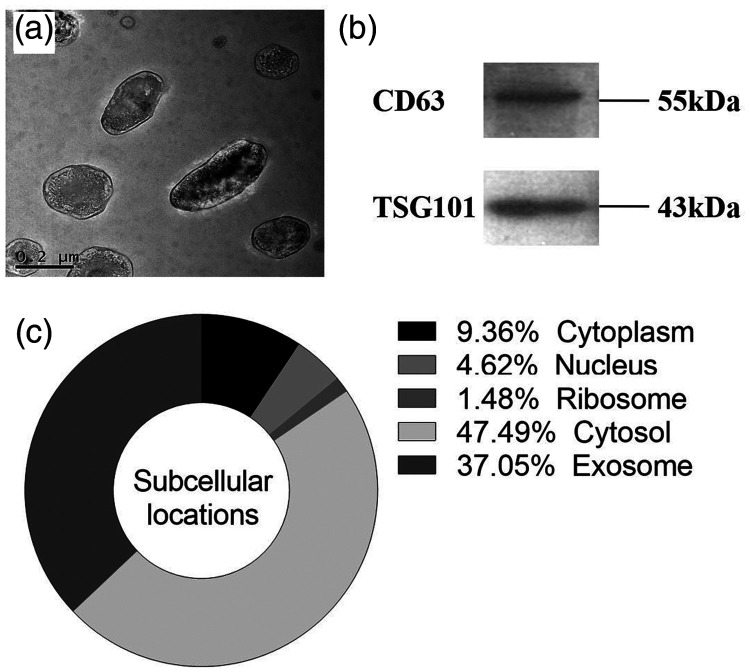
The exosomes isolated from serum were characterised by TEM and western blot. The exosomes exhibited a cup-shaped structural membrane about 50 to 150 nm in diameter on TEM (Figure 1a). The presence of exosomes in the serum samples was confirmed by western blot using anti-CD63 and anti-TSG10 antibodies specific to exosomes (Figure 1b). LncRNA H19 expression within the cell was mainly located in the cytosol and exosome (Figure 1c).
Figure 1.
Characterisation of exosomes isolated from serum. (a) Transmission electron microscopy images of exosomes in the serum of patients with gastric cancer. (b) Western blot analysis of exosomal proteins CD63 and TSG101 in exosomes. (c) Subcellular localisations of lncRNA H19 in human cells.
Stability of exosomal lncRNA H19 in serum
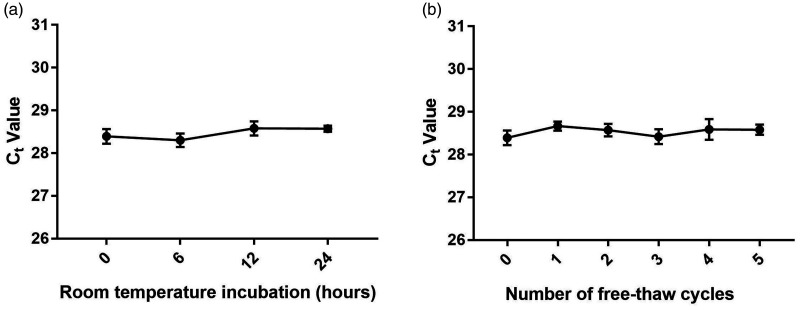
Stability is a critical prerequisite for tumour markers, and we therefore determined the stability of exosomal lncRNA H19 in serum. Expression levels of lncRNA H19 in serum exosomes were not significantly influenced by prolonged exposure to room temperature for 0, 6, 12, and 24 hours or by multiple freeze–thaw cycles (Figure 2a and 2b), indicating that exosomal lncRNA H19 was stable in serum.
Figure 2.
Stability of exosomal lncRNA H19 in serum. Expression levels of exosomal lncRNA H19 remained stable after prolonged exposure of serum samples to room temperature (a) and multiple freeze–thaw cycles (b).
Serum exosomal lncRNA H19 was upregulated in GC
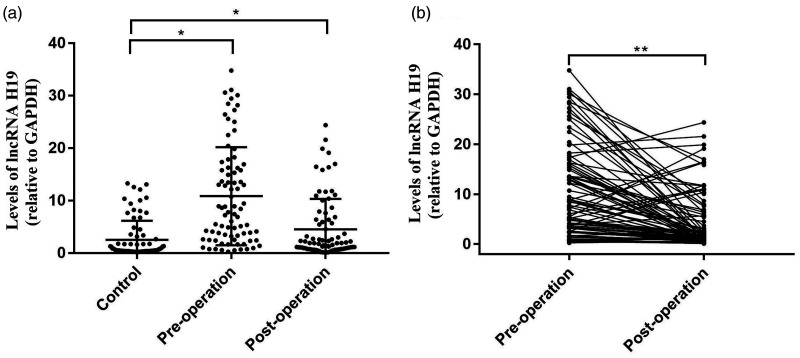
We compared serum exosomal levels of lncRNA H19 between GC patients and healthy volunteers to determine its diagnostic value for GC. Serum exosomal lncRNA H19 levels were significantly upregulated in preoperative and postoperative GC samples compared with healthy controls (P < 0.01) (Figure 3a), and lncRNA H19 levels were significantly decreased in serum exosomes from postoperative compared with preoperative GC patients (P < 0.01 (Figure 3b).
Figure 3.
Expression levels of lncRNA H19 in serum exosomes. Exosomal lncRNA H19 levels were significantly higher in patients with gastric cancer before surgery (Pre-operation) compared with levels in healthy volunteers (Control) (a) and in patients after tumour resection (Post-operation) (b). *P < 0.01 by unpaired t-test, **P < 0.01 by paired t-test.
Correlation between serum exosomal lncRNA H19 levels and TNM stage
We analysed the possible associations between lncRNA H19 expression and clinicopathological features including age, sex, lymph node metastasis, tumour size, tumour differentiation, and TNM stage, to explore the clinical value of lncRNA H19 as a GC marker. Preoperative serum levels of lncRNA H19 were significantly correlated with TNM stage in patients with GC (P = 0.007) (Table 1).
Diagnostic value of exosomal lncRNA H19 in GC
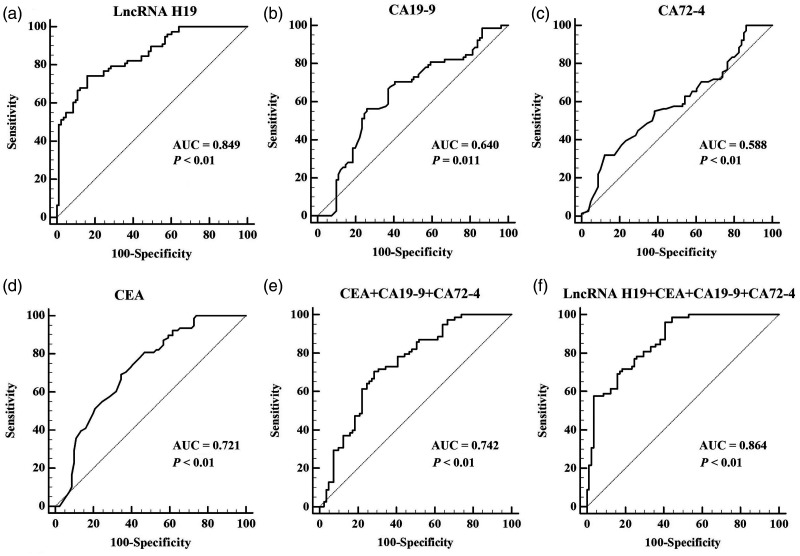
ROC curves were constructed to evaluate and compare the diagnostic abilities of preoperative exosomal lncRNA H19, CEA, CA 19-9, and CA 72-4 levels for GC. The AUC for exosomal lncRNA H19 was 0.849 (P< 0.01), the 95% confidence interval was 0.784 to 0.901, the optimal criterion value was 1.770, with a sensitivity of 74.36% and a specificity of 83.95% (Figure 4a). The AUC values for CA 19-9 (Figure 4b), CA 72-4 (Figure 4c), and CEA (Figure 4d) were 0.640, 0.588, and 0.721, respectively, which were significantly lower than that of exosomal lncRNA H19 (P = 0.011, P < 0.01, and P < 0.01, respectively).
Figure 4.
Receiver operating characteristic (ROC) curves of biomarkers. The areas under the ROC curves are shown for (a) lncRNA H19, (b) CA 19-9, (c) CA 72-4, (d) CEA, (e) the combination of CEA, CA 19-9, and CA 72-4, and (f) the combination of exosomal lncRNA H19, CEA, CA 19-9, and CA 72-4.
The AUC value for exosomal lncRNA H19 was also significantly higher than that for the combination of CEA, CA 19-9, and CA 72-4, based on pairwise comparisons (0.849 vs 0.742, P = 0.031) (Figure 4e).
We also evaluated the diagnostic value of the combination of exosomal lncRNA H19, CEA, CA 19-9, and CA 72-4 for GC. The AUC value for this combination was 0.864 (Figure 4f), which was significantly higher than those for CEA, CA 19-9, CA 72-4, and the combination of CEA, CA 19-9, and CA 72-4 (P < 0.01, P < 0.01, P < 0.01, and P < 0.01, respectively). However, there was no significant difference between the AUC for this combination and that for exosomal lncRNA H19 alone.
Discussion
Early GC is largely asymptomatic and most patients are therefore diagnosed at an advanced stage. There is thus an urgent need to find an effective indicator to allow its diagnosis at an earlier stage. Exosomes mediate local and distant cell communication by transferring specific cargos (such as proteins, lipids, and nucleic acids), which can reflect the identity of the originating cells under both physiological and pathological conditions.26 Accumulating evidence suggests that exosomes are important mediators of tumorigenesis, angiogenesis, metastasis, immune evasion, and drug resistance.27 However, the clinical role of this type of vesicle in GC diagnosis is unclear. In this study, we focussed on the potential role of exosomal molecules in GC.
Recent studies indicated that lncRNAs, which are contained in exosomes, exert important regulatory functions in GC development. Li et al.28 completed a pioneering investigation of the expression levels of long intergenic non-protein-coding RNA 152 (LINC00152) in patients with GC and revealed that both plasma and plasma exosomal levels of LINC00152 were significantly elevated in GC patients compared with healthy controls. Similarly, three other exosomal lncRNAs, ZFAS1, HOTTIP, and lncUEGC1, were also remarkably upregulated in GC patients compared with healthy controls.10,11,29 These studies indicated that exosomal lncRNAs may be predictive biomarkers of GC. The role of exosomal lncRNA H19 has been studied in several types of cancer, including hepatocellular carcinoma, bladder cancer, lung cancer, and colorectal cancer.30–33 CD90+ liver cancer cells modulated the endothelial cell phenotype via the release of exosomes containing H19 lncRNA, thus identifying lncRNA H19 as a putative therapeutic target in hepatocellular carcinoma.30 Serum exosomal H19 levels were significantly upregulated in patients with bladder cancer compared with healthy people.31 Furthermore, H19 promoted gefitinib resistance by packaging into exosomes in non-small cell lung cancer (NSCLC) cells, indicating that it might be a promising therapeutic target in patients with epidermal growth factor receptor-positive NSCLC.32 Carcinoma-associated fibroblasts also promoted stemness and chemoresistance by transferring exosomal lncRNA H19 in colorectal cancer.33 These findings confirm the importance of exosomal lncRNA H19 in tumours; however, its role in GC is not well studied and warrants further investigation. In this study, we explored the expression patterns of serum exosomal lncRNA H19 in GC patients and healthy volunteers, and showed that serum levels were significantly upregulated in patients with GC, both pre- and postoperatively, compared with healthy controls. Furthermore, serum exosomal lncRNA H19 levels in GC patients were markedly decreased after surgery compared with before surgery. These results indicate that exosomal lncRNA H19 may be a potential biomarker of GC, and that surgery may affect serum levels of lncRNA H19 in GC patients.
We also investigated the diagnostic value of serum exosomal lncRNA H19 in GC patients. TEM examination and western blotting analysis showed that exosomes were successfully obtained from serum, and stability analysis showed that serum H19 was stable in exosomes, confirming the suitability of serum exosomal lncRNA H19 as a potentially suitable diagnostic biomarker of GC. We also analysed the correlations between exosomal lncRNA H19 expression levels and clinicopathological features in GC patients and showed that preoperative exosomal lncRNA H19 levels were significantly correlated with TNM stage. This correlation indicates that the expression levels of exosomal lncRNA H19 may help to assess tumour stage in patients with GC. The preoperative diagnostic capacities of exosomal lncRNA H19, CEA, CA 19-9, and CA 72-4 for GC were also evaluated and compared by ROC curve analysis. The AUC of exosomal lncRNA H19 was significantly higher than that for any of the other markers, either alone or combined, and was similar to that for the combination of exosomal lncRNA H19, CEA, CA 19-9, and CA 72-4. These results indicate that the diagnostic ability of exosomal lncRNA H19 was superior to CEA, CA 19-9, and CA 72-4, suggesting that it might be an appropriate diagnostic marker for GC.
This study was limited by the lack of a sample size calculation, and the limited number of samples may thus affect the significance of the results.
In summary, the results of this study demonstrate that measuring serum exosomal lncRNA H19 may provide a reliable, non-invasive method for determining the diagnosis and prognosis in patients with GC. This study may thus offer new insights into future diagnostic and prognostic methods for GC.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Science and Technology Project of Jiaxing City [2018AD32059; 2020AD30058] and by the 2019 Jiaxing Key Discipline of Medicine – Clinical Laboratory Diagnostics [Innovation Subject 2019-cx-03].
ORCID iD
Pingyang Shao https://orcid.org/0000-0002-9378-0952
References
- 1.Cheng J, Zhuo H, Xu M, et al. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med 2018; 16: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–E386. [DOI] [PubMed] [Google Scholar]
- 3.Pasechnikov V, Chukov S, Fedorov E, et al. Gastric cancer: prevention, screening and early diagnosis. World J Gastroenterol 2014; 20: 13842–13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015; 385: 977–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park JY, Von Karsa L, Herrero R. Prevention strategies for gastric cancer: a global perspective. Clin Endosc 2014; 47: 478–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy I, Gralnek IM. Complications of diagnostic colonoscopy, upper endoscopy, and enteroscopy. Best Pract Res Clin Gastroenterol 2016; 30: 705–718. [DOI] [PubMed] [Google Scholar]
- 7.Gupta N, Bansal A, Wani SB, et al. Endoscopy for upper GI cancer screening in the general population: a cost-utility analysis. Gastrointest Endosc 2011; 74: 610–624. [DOI] [PubMed] [Google Scholar]
- 8.Shimada H, Noie T, Ohashi M, et al. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer 2014; 17: 26–33. [DOI] [PubMed] [Google Scholar]
- 9.Shen M, Ren X. New insights into the biological impacts of immune cell-derived exosomes within the tumor environment. Cancer Lett 2018; 431: 115–122. [DOI] [PubMed] [Google Scholar]
- 10.Lin LY, Yang L, Zeng Q, et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer 2018; 17: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao R, Zhang Y, Zhang X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer 2018; 17: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tye CE, Gordon JA, Martin-Buley LA, et al. Could lncRNAs be the missing links in control of mesenchymal stem cell differentiation? J Cell Physiol 2015; 230: 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet 2010; 19: R152–R161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou J, Wang L, Wu Q, et al. Long noncoding RNA H19 upregulates vascular endothelial growth factor A to enhance mesenchymal stem cells survival and angiogenic capacity by inhibiting miR-199a-5p. Stem Cell Res Ther 2018; 9: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays 2010; 32: 473–480. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Zhou Y, Huang T, et al. The interplay of lncrna-h19 and its binding partners in physiological process and gastric carcinogenesis. Int J Mol Sci 2017; 18: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi D, Wang M, Yu F. Knockdown of lncRNA-H19 inhibits cell viability, migration and invasion while promotes apoptosis via microRNA-143/RUNX2 axis in retinoblastoma. Biomed Pharmacother 2019; 109: 798–805. [DOI] [PubMed] [Google Scholar]
- 18.Ding D, Li C, Zhao T, et al. LncRNA H19/miR-29b-3p/PGRN axis promoted epithelial-mesenchymal transition of colorectal cancer cells by acting on Wnt signaling. Mol Cells 2018; 41: 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 2013; 52: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan J, Zhang Y, She Q, et al. Long noncoding RNA H19/miR-675 axis promotes gastric cancer via FADD/caspase 8/caspase 3 signaling pathway. Cell Physiol Biochem 2017; 42: 2364–2376. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014; 5: 2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen JS, Wang YF, Zhang XQ, et al. H19 serves as a diagnostic biomarker and up-regulation of H19 expression contributes to poor prognosis in patients with gastric cancer. Neoplasma 2016; 63: 223–230. [DOI] [PubMed] [Google Scholar]
- 23.Webber C, Gospodarowicz M, Sobin LH, et al. Improving the TNM classification: findings from a 10-year continuous literature review. Int J Cancer 2014; 135: 371–378. [DOI] [PubMed] [Google Scholar]
- 24.Solcia E, Capella C. Endocrine tumours of the gastrointestinal tract In: E Solcia, G Klöppel, LH Sobin. (eds) Histological Typing of Endocrine Tumours. 2nd ed Berlin: Springer, 2000, pp.61–68. [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 26.Fu M, Gu J, Jiang P, et al. Exosomes in gastric cancer: roles, mechanisms, and applications. Mol Cancer 2019; 18: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012; 18: 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Shao Y, Zhang X, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol 2015; 36: 2007–2012. [DOI] [PubMed] [Google Scholar]
- 29.Pan L, Liang W, Fu M, et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol 2017; 143: 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conigliaro A, Costa V, Lo Dico A, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer 2015; 14: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Yang K, Yuan W, et al. Determination of serum exosomal H19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med Sci Monit 2018; 24: 9307–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei Y, Guo W, Chen B, et al. Tumor-released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non-small cell lung cancer. Oncol Rep 2018; 40: 3438–3446. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Ren J, Ding L, Zhang D, et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018; 8: 3932–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]