Abstract
Profiling antibodies to SARS‐CoV‐2 can help to assess potential immune response after COVID‐19 disease. Luciferase IP system (LIPS) assay is a sensitive method for quantitative detection of antibodies to antigens in their native conformation. We here describe LIPS to detect antibody responses to SARS‐CoV‐2 spike (S) and nucleocapsid (N) proteins in COVID‐19 patients. The antibodies targeted both S and N fragments and gave a high assay sensitivity by identifying 26 out of 26 COVID‐19 patients with N antigen or with three protein fragments when combined into a single reaction. The assay correlated well with ELISA method and was specific to COVID‐19 as we saw no reactivity among uninfected healthy controls. Our results show that LIPS is a rapid and measurable method to screen antibody responses against SARS‐CoV‐2 antigens.
Keywords: antibodies, COVID‐19, LIPS, luciferase immunoprecipitation, SARS‐CoV‐2
Luciferase IP system (LIPS) assay is a sensitive method for quantitative detection of antibodies to antigens in their native conformation. We here describe LIPS method to detect antibody responses to SARS‐CoV‐2 spike (S) and nucleocapsid (N) proteins in COVID‐19 patients.
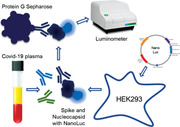
Detection of antibodies to SARS‐CoV‐2 proteins can be used to study immune responses in COVID‐19 patients [1]. Spike (S) and nucleocapsid (N) are main immunogenic proteins in SARS‐CoV‐2 [2]. Several ELISA based methods using SARS‐CoV‐2 proteins have been developed to detect the seroconversion of COVID‐19 patients, confirming the presence of antibodies in circulating blood of over 90% of COVID‐19 patients by day 14 [3].
We developed a simple and sensitive luciferase IP system (LIPS) assay to detect antibodies to S and N proteins in COVID‐19 patients. LIPS works as a liquid‐phase immunoassay, uses target antigens in their native conformation and is suitable to detect antibodies directed against linear and conformational epitopes [4].
Due to low natural background of bioluminescence, soluble crude cell lysates or culture media of the luciferase enzyme tagged recombinant proteins can be extracted from transfected cells and directly used, making it easy and fast method to produce antigens. Furthermore, the output of LIPS measurement is quantitative enabling to measure antibody levels over time during disease or in response to treatments. We used the light‐producing NanoLuc enzyme that is a small ca 20‐kDa protein and enables the secretion of fusion proteins into cell culture media, which can be used to detect antibodies to SARS‐CoV‐2 S and N proteins in COVID‐19 patients.
We carried out LIPS as previously reported [5]. We expressed S (S1 aa 1–680 and S2 aa 820–1211) and N (aa 2–419) protein fragments of SARS‐CoV‐2 in fusion with NanoLuc luciferase in HEK293 cells and confirmed the expression of the proteins by Western blotting (Supporting Information Fig. S1). After the transfection for 72 h, the cell lysates containing antigens were used in LIPS assay with COVID‐19 patients (see Supporting Information for the methods).
We tested plasma samples collected at 8–37 days after the infection from 26 COVID‐19 patients (age range 33–91 years). The COVID‐19 diagnosis was confirmed by PCR analysis for SARS‐CoV‐2 virus. In addition, we studied 26 healthy controls (age range 23–54 years) without recent infection or COVID‐19 symptoms (fever or cough) for past month. LIPS profiling detected a robust seropositive response to viral protein fragments. Using NanoLuc plasmid without the SARS‐CoV‐2 gene inserts gave no specific signal in the assay (Supporting Information Fig. S2). The statistical analysis showed highly significant reactivities to S1 (Fig. 1A), S2 (Fig. 1B), and N (Fig. 1C) antigens in the patients (Supporting Information Table S1). We found antibodies to S1 and S2 in 21 and 23 out of 26 patients, respectively, whereas all 26 patients had antibodies to the N antigen. The experimental replication of the assays gave 100% concordance of the patient positivity with high correlation coefficient in luciferase values (r = 0.98 for S1, r = 0.97 for S2, and r = 0.91 for N). Overall, the antibody responses were present to all three proteins in COVID‐19 patients (Fig. 1D), suggesting that the reactivity is not limited to a single viral epitope or antigen. Interestingly, we found no significant correlation between S1 and S2 values (Fig. 1E), but the correlation was present between S1 and N (r = 0.56; Fig. 1F) and S2 and N (r = 0.65; Fig. 1G), most likely because of the higher seropositivity for N protein.
Figure 1.
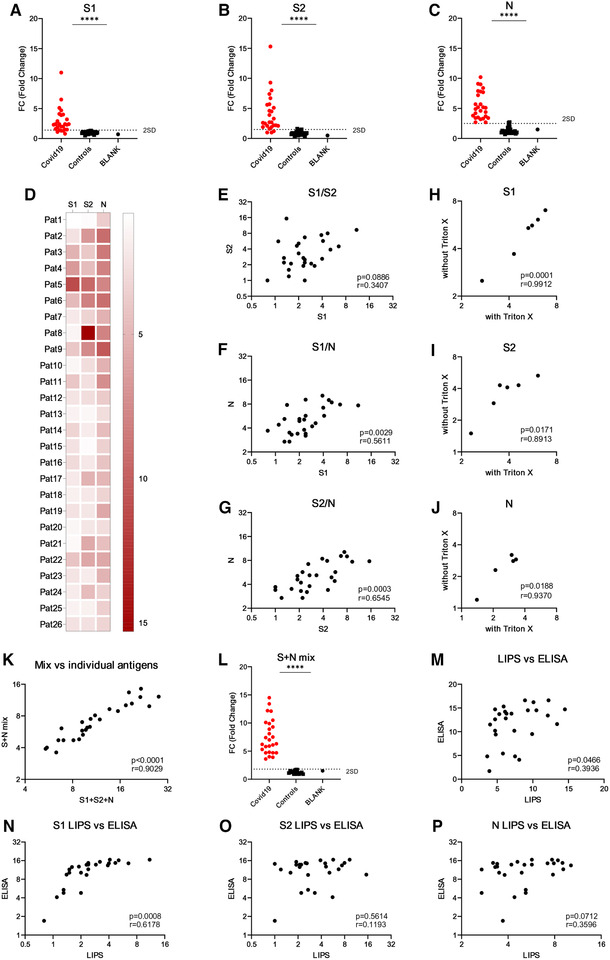
LIPS analysis of antibodies to SARS‐CoV‐2 antigens. S1, S2, and N genes were cloned in fusion with NanoLuc (Promega) gene and expressed in HEK293 cells. The cell lysates were incubated with plasma samples (in 1:40 dilution) and bound to Protein G Sepharose to capture antibody complexes with viral proteins. After the washing, luciferase substrate Nano‐Glo™ (Promega) was added and luminescence was measured in VICTOR X multilabel reader (PerkinElmer Life Sciences). Results are expressed as fold changes (FC) of luminescence units (LU) (FC = LU sample/average LU of five healthy control samples). The positive/negative discrimination level was set to the mean plus two standard deviations of the healthy control samples. The LIPS experiments were performed three times in three experimental replicates with 26 patients and 26 controls per experiment. The heatmap (D) shows average reactivities to three antigens in individual patients. The correlation of LIPS values between S1 and S2 (E), S1 and N (F), and S2 and N (G) antibody values. Triton X was added to the assays to test its impact on LIPS performance with S1 (H), S2 (I), and N (J) antigens. Mixing three antigens (S+N mix) in a single LIPS assay was correlated with the sum of the values from the three different assays (S1+S2+N) (K) and compared between patients and controls (L). The anti‐SARS‐CoV‐2 IgG ELISA (Euroimmun) was performed according to the manufacturer's instructions. Statistics were performed using unpaired Student's t‐test and Pearson correlation analysis in Graphpad Prism. ****p < 0.0001.
The analysis of COVID‐19 blood samples by PCR has raised concerns that the patients with severe disease may have viral RNAaemia at the acute infection phase. We therefore tested whether nonionic surfactant such as 1% Triton X, which can be used to neutralize potentially infectious plasma samples for routine laboratory handling, has an impact on the viral antigen detection in LIPS assay. The treatment of patient samples with Triton X did not affect the results as we saw high concordance of the results between samples, indicating that low concentration of mild detergent has no negative impact on assay performance (Fig. 1H‐J).
LIPS is a convenient method as it allows to pool multiple antigens to a single reaction for screening purposes. To see whether the mixing of three antigens together gives an increased signal values, we combined S1, S2, and N cell lysates and studied for COVID‐19 antibody reactivity. The combination of three viral antigens gave overall increased signals and 100% assay sensitivity with 26 patients (Fig. 1L) and correlated highly (r = 0.90) with the sum of each individual antigen values tested separately (Fig. 1K). This result shows that a relatively simple combination of antigens in the LIPS assay gives the highest performance to detect SARS‐CoV‐2 antibodies. Finally, we compared our results to the ELISA‐based test (EUROIMMUN) that measures antibodies to S protein and found a good correlation with our LIPS assay where three antigens were combined together (Fig. 1M, Supporting Information Table S1). We further compared the ELISA test for each individual antigen in LIPS method and, interestingly, found a highly significant correlation with S1 (Fig. 1N), but not with S2 (Fig. 1O) or N (Fig. 1P), suggesting that the ELISA mainly detects S1 fragment of SARS‐CoV‐2.
Previous reports have suggested earlier coronavirus infections to affect the performance of serological assays [6], however, none of our negative control samples were positive for the SARS‐CoV‐2 antibodies. We should note that our sample size of controls was relatively small and the analysis of more individuals would provide additional information on the utility of LIPS to differentiate the individuals with previous coronavirus from the positive responses to SARS‐CoV‐2.
Our results confirm the recent study [7], which used LIPS method to detect antibodies to S and N proteins in COVID‐19 patients and showed that antibodies to N protein are more sensitive for the detection of early infection. To address the safety of COVID‐19 blood samples, the study found no significant difference in antibody levels between heated and unheated plasma samples. Thus, the LIPS method is useful to study SARS‐CoV‐2 antibody levels in heat‐inactivated or Triton X treated COVID‐19 blood samples.
Taken together, we have developed sensitive and specific LIPS method combining different SARS‐CoV‐2 antigens for research screening purposes. LIPS assay with S and N antigen fragments may give useful information about the immune response in COVID‐19 patients with different clinical courses.
Conflict of interest
The authors declare no commercial or financial conflict of interests.
Abbreviations
- LIPS
luciferase IP system
- N
nucleocapsid
- S
spike
Supporting information
Supporting Information
Acknowledgments
We thank Drs Andres Männik and Mart Ustav for providing the SARS‐CoV‐2 S and N cDNA, and Liilija Verev for technical help. The study was supported by the Estonian Research Council grants PRG377 (PP) and PUT1367 (K.K.).
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202048715
References
- 1. Petherick, A. , Lancet 2020. 395: 1101–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu, W. et al., J Clin. Microbiol. 2020. 58: e00461‐20.32229605 [Google Scholar]
- 3. Amanat, F. et al., Nat. Med. 2020. 10.1038/s41591-020-0913-5. [DOI] [Google Scholar]
- 4. Burbelo, P. D. et al., J. Vis. Exp. 2009. (32): 1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fishman, D. et al., Front. Immunol. 2017. 8: 976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guo, L. et al., Clin. Infect. Dis. 2020. [Google Scholar]
- 7. Burbelo, P. D. et al., J. Infect. Dis. 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


