Abstract
The serological testing of anti‐SARS‐CoV‐2 immunoglobulin G (IgG) and/or IgM is widely used in the diagnosis of COVID‐19. However, its diagnostic efficacy remains unclear. In this study, we searched for diagnostic studies from the Web of Science, PubMed, Embase, CNKI, and Wanfang databases to calculate the pooled diagnostic accuracy measures using bivariate random‐effects model meta‐analysis. As a result, 22 from a total of 1613 articles, including 2282 patients with SARS‐CoV‐2 and 1485 healthy persons or patients without SARS‐CoV‐2, were selected for a meta‐analysis. Pooled sensitivity, specificity, and area under curve of the summary receiver operator curve (SROC) were: (a) 0.85 (95% confidence interval [CI]: 0.79‐0.90), 0.99 (95% CI: 0.98‐1.00), and 0.99 (95% CI: 0.97‐0.99) for anti‐SARS‐CoV‐2 IgG and (b) 0.74 (95% CI: 0.65‐0.81), 0.99 (95% CI: 0.97‐1.00), and 0.95 (95% CI: 0.93‐0.97) for IgM. A subgroup analysis among detection methods indicated the sensitivity of IgG and IgM using enzyme‐linked immunosorbent assay were slightly lower than those using gold immunochromatography assay (GICA) and chemiluminescence immunoassay (P > .05). These results showed that the detection of anti‐SARS‐CoV‐2 IgG and IgM had high diagnostic efficiency to assist the diagnosis of SARS‐CoV‐2 infection. And, GICA might be used as the preferred method for its accuracy and simplicity.
Keywords: antibody, COVID‐2019, diagnostic efficacy, SARS‐CoV‐2
Highlights
Detection of anti‐SARS‐CoV‐2 IgG and IgM has high diagnostic efficiency to assist the diagnosis of SARS‐CoV‐2.
1. INTRODUCTION
The coronavirus, SARS‐CoV‐2, is now widely spreading over the world and has infected millions of people. It causes a low respiratory infection (COVID‐19) pandemic. In some patients, it may lead to acute respiratory distress syndrome that contributes to most of the COVID‐19 deaths. As of 18 May 2020, nearly five million people around the world have been diagnosed with SARS‐CoV‐2, and more than 300 000 people have died from serve COVID‐19. 1 Timely diagnosis of SARS‐CoV‐2 infections and isolation of infected persons and close contacts remain priorities and challenges of epidemic prevention.
The diagnosis of SARS‐CoV‐2 infections mainly depends on the detection of SARS‐CoV‐2 nucleic acid (RNA) and SARS‐CoV‐2 immunoglobulin antibodies (IgM and/or IgG). 2 Detection of virus RNA by real‐time reverse transcriptase‐polymerase chain reaction (RT‐PCR) is considered as the golden criteria of diagnosis. However, RNA molecular detection suffers from many limitations 3 : (a) It requires expensive equipment and trained technicians in a certified laboratory, (b) it usually takes more than 2 hours to generate results, and (c) it carries the risk of false‐negatives due to low viral loads in clinical specimens. 4 Serological testing of anti‐SARS‐CoV‐2 IgG/IgM (has been used to diagnose illness, but its diagnostic efficacy remains unclear. 5 This study aims to summarize the diagnostic efficacy of the anti‐SARS‐CoV‐2 IgG/IgM test in each study, the results of which can assist in the diagnosis of SARS‐CoV‐2.
2. MATERIALS AND METHODS
2.1. Study registration
This meta‐analysis was registered on PROSPERO (ID: CRD42020184771).
2.2. Search strategy and eligibility criteria
We performed a systematic literature search in PubMed, Web of Science, Embase, CNKI (China), and Wanfang (China) databases and excluded duplicates with EndNote X9.0 software. The search terms used in PubMed were (severe acute respiratory syndrome coronavirus 2 OR Wuhan coronavirus OR Wuhan seafood market pneumonia virus OR COVID19 virus OR COVID‐19 virus OR coronavirus disease 2019 virus OR SARS‐CoV‐2 OR SARS2 OR 2019‐nCoV OR 2019 novel coronavirus) AND (antibody OR IgG OR IgM OR immunoglobulin). The searches were limited to articles published in Chinese or English in 2020. To reduce literature omissions, we checked the reference lists of the included studies.
We defined the eligibility criteria as follows: (a) numbers of true‐positives (TP), false‐positives (FP), true‐negatives (TN), and false‐negatives (FN) were available, (b) RT‐PCR test for SARS‐CoV‐2 virus nucleic acids and anti‐SARS‐CoV‐2 IgG and/or IgM test were performed. Case reports, review articles, and meta‐analysis articles were excluded.
Two reviewers independently performed the literature search and screened the titles, abstracts, and full texts according to the eligibility criteria. Disagreements were resolved with a third reviewer or by consensus. All the eligibility studies were selected for meta‐analysis. The steps of the literature search are shown in Figure S1.
2.3. Quality assessment and risk of bias
Two reviewers assess the quality of studies enrolled in this study using QUADAS‐2, a tool for quality assessment of diagnostic accuracy studies, 6 to assess the risk of bias. We assessed statistical heterogeneity and publication bias using the I 2 statistic, Q test, and Deeks' test, respectively. Deeks' funnel plots were drawn to evaluate the risk of publication bias.
2.4. Data extraction and meta‐analysis
The two reviewers who performed the literature search also independently extracted the data from the enrolled studies using a predefined data extraction form. The variables extracted from the selected studies included author, blood collection time from symptom onset, type of anti‐SARS‐CoV‐2 (IgG or IgM), methods of antibody detection, TP, FP, TN, and FN.
2.5. Statistical analysis
We performed a meta‐analysis by the “meta4diag” package (version 2.0.8) in R soft (version 3.6.2) and “Midas” modules in the STATA statistical software (version 14.0). A bivariate random‐effects model was employed for estimating the pooled diagnostic performance measures and a 95% confidence interval (CI).
3. RESULTS
3.1. Search results
A total of 1613 articles were identified from the Web of Science, PubMed, Embase, CNKI (China), Wanfang (China), and other sources. After we removed duplicates and screened all the search records, 22 studies 3 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 meeting the predetermined inclusion and exclusion criteria were enrolled in this study for a meta‐analysis. As shown in Table 1, a total of 3767 individuals were included in this meta‐analysis, including 2282 patients with SARS‐CoV‐2 and 1485 healthy persons or patients without SARS‐CoV‐2. Their age‐bracket and sex ratio were not available in each included study.
Table 1.
The main features of the included studies for anti‐SARS‐CoV‐2 IgG/IgM in the diagnosis of COVID‐19
| No. | Author | Number (cases/controls) | Daysa | Study type | Cases | Controls | Method |
|---|---|---|---|---|---|---|---|
| 1 | Dohla et al 7 | 22/27 | 19 (IQR: 15‐24) | Prospective | PCR+ | PCR− | GICA |
| 2 | Hoffman et al 8 | 28/125 | Range 9‐29 | Retrospective | PCR+ | Healthy persons | GICA |
| 3 | Infantino et al 9 | 30/63 | 12 (range 8‐17) | Retrospective | PCR+ | Patients without SARS‐CoV‐2 and healthy persons | CLIA |
| 4 | Jin et al 10 | 27/33 | 16 (IQR: 9‐20) | Retrospective | PCR+ | PCR− | CLIA |
| 5 | Li et al 3 | 397/128 | Range 8‐33 | Retrospective | PCR+, clinical feathers | Patients without SARS‐CoV‐2 | GICA |
| 6 | Liu et al 11 | 214/100 | 15 (range 0‐55) | Retrospective | PCR+ | PCR− | ELISA |
| 7 | Pan et al 12 | 86/22 | Range 0‐34 | Retrospective | PCR+ | PCR− | GICA |
| 8 | Qu et al 13 | 41/38 | Range 3‐43 | Retrospective | PCR+ | Patients without SARS‐CoV‐2 | CLIA |
| 9 | Shen et al 14 | 97/53 | Range 0‐28 | Retrospective | PCR+ | PCR− and healthy persons | GICA |
| 10 | Spicuzza et al 15 | 23/14 | Range 3‐34 | Retrospective | PCR+ | PCR− | GICA |
| 11 | Xiang et al 16 | 90/60 | Range 13‐29 | Retrospective | PCR+ and clinical feathers | Healthy persons | ELISA |
| 12 | Bao et al 17 | 179/100 | 37 (range, 9‐62) | Retrospective | PCR+ | Healthy persons | GICA |
| 13 | Deng et al 18 | 32/44 | ⋯ | Retrospective | PCR+ | PCR− | GICA |
| 14 | Li et al 19 | 116/134 | ⋯ | Retrospective | PCR+ | PCR− | CLIA |
| 15 | Liang et al 21 | 236/59 | ⋯ | Retrospective | PCR+ | Healthy persons | GICA |
| 16 | Li et al 20 | 25/60 | Convalescence | Retrospective | PCR+ | PCR− | CLIA |
| 17 | Luo et al 22 | 101/54 | ⋯ | Retrospective | PCR+ and clinical feathers | PCR− | GICA |
| 18 | Tang et al 23 | 113/27 | 25 (range, 3‐47) | Retrospective | PCR+ and clinical feathers | Patients without SARS‐CoV‐2 | CLIA |
| 19 | Xiong et al 24 | 97/100 | ⋯ | Retrospective | PCR+ | PCR− | CLIA and ELISA |
| 20 | Xu et al 25 | 205/79 | ⋯ | Retrospective | PCR+ and clinical feathers | Patients without SARS‐CoV‐2 | CLIA |
| 21 | Zhang et al 26 | 105/138 | ⋯ | Retrospective | PCR+ and clinical feathers | PCR− and healthy persons | GICA |
| 22 | Zheng et al 27 | 25/20 | ⋯ | Retrospective | PCR+ | PCR− | CLIA and GICA |
Note: Only the first author of each study is given.
Abbreviations: PCR+, PCR positive; PCR−, PCR negative; GICA, gold immunochromatography assay; CLIA, chemiluminescence immunoassay; ELISA, enzyme‐linked immunosorbent assay; IQR, interquartile range; IgG, immunoglobulin G; IgM, immunoglobulin M.
Days between the collection of blood samples and the symptom onset.
3.2. Quality assessments
We evaluated the quality of the 22 included studies according to QUADAS‐2 guidelines. Bias in each study was assessed as “low risk of bias,” “high risk of bias,” and “unclear risk of bias.” As shown in Figure 1, 95.45% (21 of 22) for patient selection, 36.36% (8 of 22) for index test, 13.64% (3 of 22) for flow and timing, and 4.55% (1 of 22) for reference standard showed a high risk of bias. Subjects in most of the included studies (95.45%, 21 of 22) were composed of patients with SARS‐CoV‐2 and healthy persons or patients without SARS‐CoV‐2 without “difficult to diagnose” patients, which did not avoid case‐control design and inappropriate exclusions and contributed to the high risk of bias in terms of patient selection. For the index test, 36.36% (8 of 22) studies were classified as high risk of bias mainly because they were retrospective studies, and the IgG/IgM test results were interpreted with knowledge of the RT‐PCR results not meeting the “blinding” criteria. The risk of the reference standard and flow and time bias was relatively low. Some studies did not declare aspects related to study design (ie, intervals between serologic test and RT‐PCR), which limits the ability to conclude on study quality.
Figure 1.
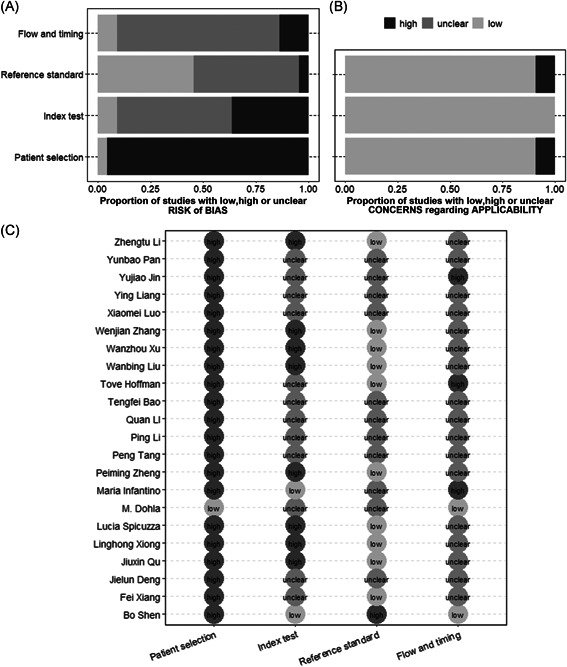
Study quality assessment using modified QUADAS‐2. (a) Risk of bias. (b) Concerns regarding applicability. (c) Details of quality assessment
3.3. Heterogeneity
The P values of the Q test were all less than .01, accompanied by I 2 > 50%. The I 2 ranging from 69.85% to 93.52% in the evaluation of anti‐SARS‐CoV‐2 IgG and/or IgM showed the heterogeneity of the statistical significance.
3.4. Diagnostic performance
The result of this bivariate random‐effects meta‐analysis is shown in Figure 2. The sensitivity and specificity was 0.85 (95% CI: 0.79‐0.90) and 0.99 (95% CI: 0.98‐1.00) for anti‐SARS‐CoV‐2 IgG, 0.74 (95% CI: 0.65‐0.81) and 0.99 (95% CI: 0.97‐1.00) for IgM, and 0.86 (95% CI: 0.79‐0.92) and 0.99 (95% CI: 0.97‐1.00) for IgG or IgM.
Figure 2.
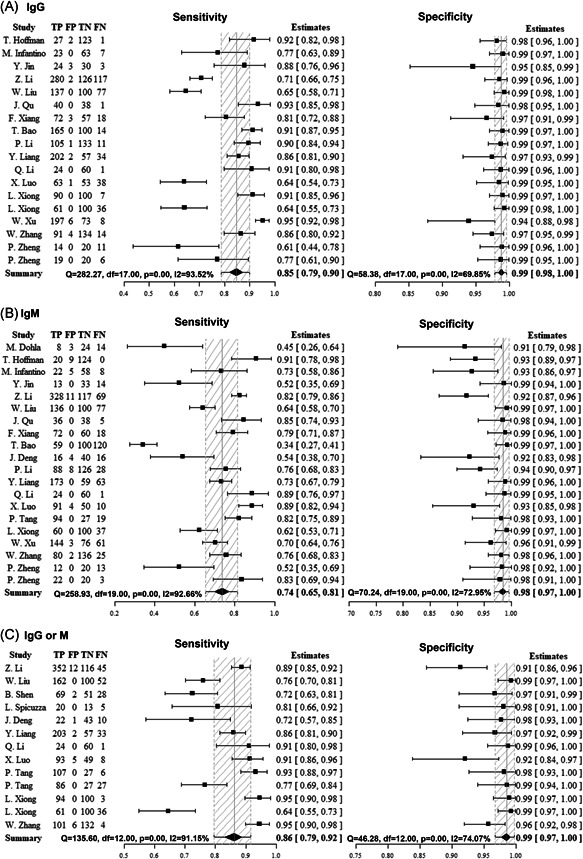
Forest plots of the pooled sensitivity and specificity for anti‐SARS‐CoV‐2 immunoglobulin G (IgG), IgM, and IgG or IgM in diagnosis of COVID‐19. (a) IgG. (b) IgM. (c) IgG or IgM. Only the first author of each study is given. Sensitivity and specificity were given with confidence intervals (CI)
Summary receiver operator characteristic (SROC) curves were generated to indicate the overall diagnostic accuracy. The area under the SROC curve (AUC) was 0.99 (95% CI: 0.97‐0.99) for anti‐SARS‐CoV‐2 IgG, 0.95 (95% CI: 0.93‐0.97) for IgM, and 0.98 (95% CI: 0.96‐0.99) for IgG or IgM (Figure 3). Pooled diagnostic odds ratio, positive likelihood ratio, and negative likelihood ratio are shown in Table 2.
Figure 3.
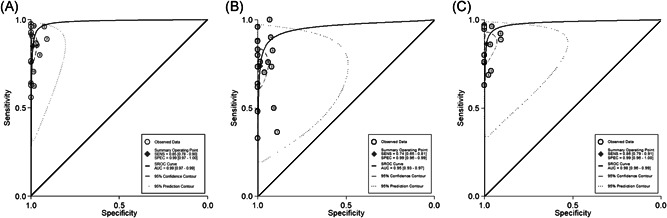
The SROC curves of the serological testing of anti‐SARS‐CoV‐2 antibodies. The regression SROC curve indicates the overall diagnostic accuracy. (a) IgG, (b) IgM, (c) IgG or IgM. AUC, area under the curve; IgG, immunoglobulin G; IgM, immunoglobulin M; SENS, sensitivity; SPEC, specificity; SROC, summary receiver operator curve
Table 2.
Summary table of the diagnostic accuracy of IgG, IgM, and IgG/IgM for SARS‐CoV‐2 infection
| Sensitivity (95% CI) | Specificity (95% CI) | DOR (95% CI) | LRpos (95% CI) | LRneg (95% CI) | AUC (95% CI) | |
|---|---|---|---|---|---|---|
| IgG | 0.85 (0.79‐0.90) | 0.99 (0.98‐1.00) | 592.62 (226.79‐1634.34) | 88.32 (38.48‐229.57) | 0.15 (0.10‐0.22 | 0.99 (0.97‐0.99) |
| IgM | 0.74 (0.65‐0.81) | 0.99 (0.97‐1.00) | 278.12 (76.02‐1029.37) | 71.41 (22.09‐259.48) | 0.27 (0.18‐0.36) | 0.95 (0.93‐0.97) |
| IgG/IgM | 0.86 (0.79‐0.92) | 0.99 (0.97‐1.00) | 777.53 (161.26‐3478.47) | 104.14 (24.99‐456.71) | 0.14 (0.08‐0.22) | 0.98 (0.96‐0.99) |
Abbreviations: AUC, area under the curve; CI, confidence interval; DOR, diagnostic odds ratio; IgG, immunoglobulin G; IgM, immunoglobulin M; LRpos, positive likelihood ratio; LRneg, negative‐positive likelihood ratio.
3.5. Subgroup analyses
In the selected studies, the detection methods of anti‐SARS‐CoV‐2 IgG and IgM included gold immunochromatography assay (GICA), chemiluminescence immunoassay (CLIA), enzyme‐linked immunosorbent assay (ELISA). We performed subgroup analyses among these three groups. The results showed that the sensitivity of IgG and IgM using ELISA were lower than those using GICA or CLIA. However, the meta‐regression analysis results showed that no significant differences in sensitivity and specificity were observed among these groups (P > .05) (Table 3). Additionally, the I 2 for the sensitivity of IgG, IgM, and IgG/IgM in the subgroup analyses were more than 50%. And the I 2 for the specificity of IgG using CLIA (0%) and IgG or IgM using GICA (23.78%) declined significantly.
Table 3.
The pooled sensitivity and specificity of subgroup meta‐analyses and meta‐regression
| Antidody | Test method (n) | I 2 (%) | Sensitivity (95% CI) | P value | I 2 (%) | Specificity (95% CI) | P value |
|---|---|---|---|---|---|---|---|
| IgG | GICA (7) | 87.09 | 0.83 (0.73, 0.90) | .07 | 77.27 | 0.99 (0.96, 1.00) | .71 |
| CLIA (8) | 93.16 | 0.90 (0.84, 0.95) | 0 | 0.99 (0.97, 1.00) | |||
| ELISA (3) | ⋯ | 0.69 (0.48, 0.85) | ⋯ | 0.99 (0.96, 1.00) | |||
| IgM | GICA (9) | 96.54 | 0.74 (0.60, 0.85) | .93 | 76.97 | 0.97 (0.93, 0.99) | 1.00 |
| CLIA (9) | 81.24 | 0.74 (0.60, 0.85) | 63.67 | 0.99 (0.97, 1.00) | |||
| ELISA (2) | ⋯ | 0.71 (0.40, 0.91) | ⋯ | 1.00 (1.00, 1.00) | |||
| IgG/IgM | GICA (8) | 85.30 | 0.84 (0.78, 0.90) | .06 | 23.78 | 0.95 (0.93, 0.98) | 1.00 |
| CLIA (3) | ⋯ | 0.96 (0.91, 0.98) | ⋯ | 1.00 (1.00, 1.00) | |||
| ELISA (2) | ⋯ | 0.69 (0.50, 0.85) | ⋯ | 1.00 (1.00, 1.00) |
Note: The P value was obtained comparing ELISA with GILA and CLIA.
Abbreviations: CLIA, chemiluminescence immunoassay; ELISA, enzyme‐linked immunosorbent assay; GICA, gold immunochromatography assay; IgG, immunoglobulin G; IgM, immunoglobulin M.
3.6. Influence analysis
As shown in Figure S2, we generated crosshair plots and performed influence analysis to identify outliers. Two study 3 , 11 in the meta‐analysis of IgG were identified as outliers. After excluding the outliers, the overall pooled sensitivity of IgG slightly increased from 0.85 to 0.87, specificity and AUC did not change. Moreover, the I 2 for sensitivity and specificity slightly declined from 93.52% and 69.85% to 90.53% and 66.63%, respectively. These results suggested that the outliers contributed a little heterogeneity in this meta‐analysis.
3.7. Publication bias
Deeks' funnel plot asymmetry test was used to evaluate the publication bias of the included studies. The results indicated that there was no obvious publication bias in this meta‐analysis (P > .05) (Figure S3).
4. DISCUSSION
Serological testing of anti‐SARS‐CoV‐2 IgG/IgM has been widely used to diagnose SARS‐CoV‐2 infection. However, the diagnostic efficacy of the serum antibody test reported in the earlier studies confused the clinician. The sensitivities of IgG and IgM ranged from 0.61 27 and 0.34 17 to 0.93 13 and 0.91, 8 respectively. And, there was no significant difference in the specificities of IgG and IgM among the studies. Therefore, a broad summary analysis of the diagnostic efficacy of anti‐SARS‐CoV‐2 IgG and IgM is significantly necessary to assist in the diagnosis of SARS‐CoV‐2. As of 10 May 2020, 22 studies published in Chinese or English were selected in this study. A total of 2282 patients with SARS‐CoV‐2 and 1485 controls were included in our meta‐analysis. In this unusual and urgent situation, most of the included studies were retrospective and did not meet the QUADAS guidelines well, but a summary meta‐analysis from the studies still had significantly reference value for the diagnosis of SARS‐CoV‐2. 28
This meta‐analysis results showed promising accuracy for IgG detection in diagnosing SARS‐CoV‐2, in which the pooled sensitivity was 0.85 and specificity was 0.99, with an AUC of 0.99. The pooled diagnostic performance of IgM was slightly lower than those of IgG, with a sensitivity of 0.74 and an AUC of 0.95. Additionally, combining the IgG and IgM test did not obtain a higher diagnostic accuracy than single IgG. Subgroup analysis among groups with different detection methods demonstrated that the diagnostic efficacy of the antibody test by ELISA was slightly lower than that by CLIA and GICA (P > .05). Taking the principles of accuracy and simplicity into consideration, we commented that GICA was the preferred method. A meta‐analysis of diagnostic test accuracy of the anti‐SARS‐CoV‐2 IgG/IgM test was performed in Brazil, 29 in which the pooled sensitivity of IgG and IgM (0.97 and 0.82) were all higher than those in our meta‐analysis (0.85 and 0.74). As our analysis contained more studies (22 vs 11) and patients, it is more accurate than the previous report.
Researchers have demonstrated the longitudinal change of anti‐SARS‐CoV‐2 IgG/IgM in patients with SARS‐CoV‐2. Anti‐SARS‐CoV‐2 IgM appeared in the blood and could be initially detected after 5 days (interquartile range [IQR]: 3‐6) of symptom onset, 30 and lasted for 1 month and gradually decreased. 31 And the median duration of IgG antibody detection was 14 days (IQR: 10‐18) 30 and lasted for a longer time. 31 These results suggested that the test of the serum antibodies was exceedingly helpful for the diagnosis of SARS‐CoV‐2 after the corresponding window periods. In particular, the detection efficiency of IgM was considered higher than that of the RT‐PCR method after 5.5 days of symptom onset. 30 Moreover, another study showed that a higher titer of the antibody was highly associated with a worse clinical classification. 32
However, the detection of anti‐SARS‐CoV‐2 IgG/IgM for patients in window periods had low‐diagnostic efficiency. And, the diagnostic efficiency of the serological antibody test in asymptomatic carriers was unclear. An earlier study showed that anti‐SARS‐CoV‐2 IgG was positive in only 20% (1 of 5) asymptomatic carriers, and IgM was negative in all the five carriers. 33 And, a positive IgG was detected after 18 days of diagnosis with SARS‐CoV‐2 by RT‐PCR. The sample size was relatively small, but it still indicated that the antibody test was not applicable to asymptomatic populations. Furthermore, there is no evidence for cases of SARS‐CoV‐2 reinfections in literature, but only patients with positive PCR test and IgM seroconversion several weeks after negative RT‐PCR tests. 34 As the acquired immunity and the presence of anti‐SARS‐CoV‐2 antibodies were thought to protect upon further exposure to SARS‐CoV‐2, the negative RT‐PCR test was considered as a false‐negative, which may result from reduced viral loads in convalescence, sampling errors during collection or transport. 35 And IgM seroconversion was considered deriving from an expansion of IgM+ memory B cells. There is currently no evidence to support the use of a specific antibody test to diagnose the reinfection of SARS‐CoV‐2. So, if any, I think that the diagnosis of reinfection may be based on symptoms, radiological imaging, leukocytes count, and inflammatory indexes alterations. 36
4.1. Limitations
This review has several limitations. First, none of the persons with cover, cough, and runny nose of unknown origin was enrolled in the studies. Controls in all the included studies were proven without SARS‐CoV‐2, leading to an exaggerated specificity. Second, this meta‐analysis had high heterogeneity. The bivariate random‐effects model was applied to weaken influences by heterogeneity. And also, we performed subgroup and sensitivity analysis to explore the source of heterogeneity. Finally, all the patients included in this analysis were first infected with SARS‐CoV‐2, and the diagnostic efficiency of the specific immunoglobulin for reinfection of SARS‐CoV‐2 is unclear.
5. CONCLUSIONS
This meta‐analysis showed that the detection of anti‐SARS‐CoV‐2 IgG and IgM had high diagnostic efficiency to assist the diagnosis of SARS‐CoV‐2. It was suitable for patients with symptoms for at least 5 days.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Z‐LZ and Y‐LH formulated the research questions, designed the study, developed the preliminary search strategy, and drafted the manuscript. F‐ZL and D‐TL refined the search strategy, searched, collected the articles, and then conducted quality assessment. All authors have read and approved the final version of the manuscript.
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
The authors thank Prof Hui Chen for his support and guidance. This study was supported by the First Affiliated Hospital of Chongqing Medical University (grant number HLJJ2014‐21).
Zhang Z‐L, Hou Y‐L, Li D‐T, Li F‐Z. Diagnostic efficacy of anti‐SARS‐CoV‐2 IgG/IgM test for COVID‐19: A meta‐analysis. J Med Virol. 2021;93:366–374. 10.1002/jmv.26211
REFERENCES
- 1. IFENG News . Real‐time updates on the global epidemic of COVID‐19. https://news.ifeng.com/c/special/7uLj4F83Cqm. Accessed May 18 2020.
- 2. Tang YW, Schmitz JE, Persing DH, Stratton CW. The laboratory diagnosis of COVID‐19 infection: current issues and challenges. J Clin Microbiol. 2020;58(6):e00512–20. 10.1128/JCM.00512-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis [published online ahead of print February 27, 2020]. J Med Virol. 2020;1–7. 10.1002/jmv.25727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pang J, Wang MX, Ang IYH, et al. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019‐nCoV): a systematic review. J Clin Med. 2020;9(3):623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Farnsworth CW, Anderson NW. SARS‐CoV‐2 serology: much hype, little data [published online ahead of print April 28, 2020]. Clin Chem. 2020. 10.1093/clinchem/hvaa107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529‐536. [DOI] [PubMed] [Google Scholar]
- 7. Dohla M, Boesecke C, Schulte B, et al. Rapid point‐of‐care testing for SARS‐CoV‐2 in a community screening setting shows low sensitivity. Public Health. 2020;182:170‐172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffman T, Nissen K, Krambrich J, et al. Evaluation of a COVID‐19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS‐CoV‐2. Infect Ecol Epidemiol. 2020;10(1):1754538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Infantino M, Grossi V, Lari B, et al. Diagnostic accuracy of an automated chemiluminescent immunoassay for anti‐SARS‐CoV‐2 IgM and IgG antibodies: an Italian experience [published online ahead of print April 24, 2020]. J Med Virol. 2020;1–5. 10.1002/jmv.25932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin Y, Wang M, Zuo Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu W, Liu L, Kou G, et al. Evaluation of nucleocapsid and spike protein‐based ELISAs for detecting antibodies against SARS‐CoV‐2. J Clin Microbiol. 2020;58(6):e00461–20. 10.1128/JCM.00461-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan Y, Li X, Yang G, et al. Serological immunochromatographic approach in diagnosis with SARS‐CoV‐2 infected COVID‐19 patients. J Infect. 2020;81(1):e28–e32. 10.1016/j.jinf.2020.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qu J, Wu C, Li X, et al. Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) [published online ahead of print April 27, 2020]. Clin Infect Dis. 2020. 10.1093/cid/ciaa489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen B, Zheng Y, Zhang X, et al. Clinical evaluation of a rapid colloidal gold immunochromatography assay for SARS‐Cov‐2 IgM/IgG. Am J Transl Res. 2020;12(4):1348‐1354. [PMC free article] [PubMed] [Google Scholar]
- 15. Spicuzza L, Montineri A, Manuele R, et al. Reliability and usefulness of a rapid IgM‐IgG antibody test for the diagnosis of SARS‐CoV‐2 infection: A preliminary report [published online ahead of print April 23, 2020]. J Infect. 2020. 10.1016/j.jinf.2020.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiang F, Wang X, He X, et al. Antibody detection and dynamic characteristics in patients with COVID‐19 [published online ahead of print April 19, 2020]. Clin Infect Dis. 2020. 10.1093/cid/ciaa461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bao T, Gu J, Liu Y. Clinical application value of immunochromatography in detecting SARS‐CoV‐2 IgM and IgG antibodies [published online ahead of print May 08, 2020]. Med J Wuhan Univ. 2020. 10.14188/j.1671-8852.2020.0193 [DOI] [Google Scholar]
- 18. Deng J, Wang Y, Wang T, et al. The clinical value of GICA in the detection of serum antibodies to SARS‐CoV‐2. Int J Lab Med. 2020;41(8):964‐966+970. [Google Scholar]
- 19. Li P, Li Z, Zhao S, et al. Preliminary study of serum 2019‐nCoV IgM and IgG antibodies in the diagnosis of Novel Coronavirus pneumonia [published online ahead of print May 18, 2020]. Chin J Lab Med. 2020;43. 10.3760/cma.j.cn114452-20200302-00155 [DOI] [Google Scholar]
- 20. Li Q, Liu D, Qiao Z, et al. Value of SARS‐CoV‐2 IgM/IgG antibody detection in diagnosis of new coronavirus pneumonia. Int J Lab Med. 2020. http://kns.cnki.net/kcms/detail/50.1176.r.20200304.1041.006.html [Google Scholar]
- 21. Liang Y, Zeng S, Liu T, et al. Clinical application value of novel coronavirus antibody detection in the diagnosis of COVID‐19 [published online ahead of print March 17, 2020]. Med J Wuhan Univ. 2020. 10.14188/j.1671-8852.2020.0167 [DOI] [Google Scholar]
- 22. Luo X, Wang J, Zhang Y, et al. Detection of specific SARS‐CoV‐2 IgM and IgG antibodies in COVID‐19 and its clinical application. J Southwest Univ (Nat Sci Ed.). 2020;42(3):30‐34. [Google Scholar]
- 23. Tang P, Zhao Z, Liu Y, et al. Comparison between chemiluminescence and colloidal gold method in the detection of 2019 novel coronavirus specific antibodies and the value in the COVID‐19 diagnosis and treatment. Med J Wuhan Univ. 2020. 10.14188/j.1671-8852.2020.0131 [DOI] [Google Scholar]
- 24. Xiong L, Huang Y, Meng J, et al. Comparision of chemiluminescence and ELISA kits for SARS‐CoV‐2 IgM/IgG detection. J Trop Med. 2020. http://kns.cnki.net/kcms/detail/44.1503.R.20200429.1523.002.html [Google Scholar]
- 25. Xu W, Li J, He X, et al. The diagnostic value of joint detection of serum IgMand IgG antibodies to 2019‐nCoV in 2019‐nCoV infection. Chin J Lab Med. 2020;43(3):230‐233. [Google Scholar]
- 26. Zhang W, Lv X, Huang C, et al. Clinical evaluation and application of detection for SARS‐ CoV‐2 IgM and IgG antibodies with colloidal gold immunochromatography assay. Chinese. J Virol. 2020. 10.13242/j.cnki.bingduxuebao.003676 [DOI] [Google Scholar]
- 27. Zheng P, Ciu F, Zhang F, et al. Clinical evaluation of different detection methods of SARS‐CoV‐2 IgM/IgG antibody in the COVID‐19 diagnosis [published online ahead of print April 1, 2020]. Lab Med. 2020. http://kns.cnki.net/kcms/detail/31.1915.R.20200331.1804.002.html [Google Scholar]
- 28. Di Giambenedetto S, Ciccullo A, Posteraro B, Lombardi F, Borghetti A, Sanguinetti M. Still much to learn about the diagnostic role of SARS‐CoV‐2 antibody detection [published online ahead of print May 2, 2020]. Clin Infect Dis. 2020. 10.1093/cid/ciaa532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Castro R, Luz PM, Wakimoto MD, Veloso VG, Grinsztejn B, Perazzo H. COVID‐19: a meta‐analysis of diagnostic test accuracy of commercial assays registered in Brazil. Braz J Infect Dis. 2020;24(2):180–187. 10.1016/j.bjid.2020.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID‐19) [published online ahead of print March 21, 2020]. Clin Infect Dis. 2020. 10.1093/cid/ciaa310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang G, Nie S, Zhang Z, Zhang Z. Longitudinal change of SARS‐Cov2 antibodies in patients with COVID‐19 [published online ahead of print May 2, 2020]. J Infect Dis. 2020. 10.1093/infdis/jiaa229 [DOI] [Google Scholar]
- 32. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS‐CoV‐2 in patients of novel coronavirus disease 2019 [published online ahead of print March 28, 2020]. Clin Infect Dis. 2020. 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yongchen Z, Shen H, Wang X, et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID‐19 patients. Emerg Microbes Infect. 2020;9(1):833‐836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bentivegna E, Sentimentale A, Luciani M, Speranza ML, Guerritore L, Martelletti P. New IgM seroconversion and positive RT‐PCR test after exposure to the virus in recovered COVID‐19 patient [published online ahead of print June 11, 2020]. J Med Virol. 2020. 10.1002/jmv.26160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roy S. COVID‐19 reinfection: myth or truth? [published online ahead of print May 29, 2020]. SN Compr Clin Med. 2020:1‐4. 10.1007/s42399-020-00335-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang ZL, Hou YL, Li DT, Li FZ. Laboratory findings of COVID‐19: a systematic review and meta‐analysis [published online ahead of print May 23, 2020]. Scand J Clin Lab Invest. 2020:1‐7. 10.1080/00365513.2020.1768587 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information
Supporting information


