Abstract
Repurposing of existing anti‐viral drugs, immunological modulators and supportive therapies represents a promising path towards rapidly developing new control strategies to mitigate the devastating public health consequences of the COVID‐19 pandemic. A comprehensive text‐mining and manual curation approach was used to comb and summarize the most pertinent information from existing clinical trials and previous efforts to develop therapies against related betacoronaviruses, particularly SARS and MERS. In contrast to drugs in current trials, which have been derived overwhelmingly from studies on taxonomically unrelated RNA viruses, a number of untested small molecule anti‐virals had previously demonstrated remarkable in vitro specificity for SARS‐CoV or MERS‐CoV, with high selectivity indices, EC50 and/or IC50. Due to the rapid containment of the prior epidemics, however, these were generally not followed up with in vivo animal studies or clinical investigations and thus largely overlooked as treatment prospects in the current COVID‐19 trials. This brief review summarizes and tabulates core information on recent or ongoing drug repurposing‐focused clinical trials, while detailing the most promising untested candidates with prior documented success against the aetiologic agents of SARS and/or MERS.
Keywords: anti‐viral agents, COVID‐19, data mining, drug repositioning, pandemics, SARS
The unprecedented public health, economic and social challenges engendered by the current COVID‐19 pandemic necessitate an urgent search for effective clinical interventions to help reduce viral load and epidemiological spread, improve prevention and control, and stem the tide of rising morbidity and mortality (Spinelli & Pellino, 2020). Due to the time lag of vaccine trials and de novo drug development based on standard drug‐target modelling, compound screens and multi‐phase clinic testing, the most rapid and practical approach towards new clinical options lies in drug repositioning of proven or promising infectious control modalities (Li & De Clercq, 2020). For SARS‐CoV‐2, the aetiologic agent of COVID‐19, this effort may be significantly assisted by previous endeavours to develop therapeutics for two prior smaller epidemics, both caused by closely related coronavirus types. Severe acute respiratory syndrome (SARS) and Middle Eastern respiratory syndrome (MERS) each originated from outbreaks of betacoronaviruses with significant sequence and structural similarity to the SARS‐CoV‐2 betacoronavirus (Zhu et al., 2020).
Thousands of COVID‐19‐related clinical trials have been proposed or launched since January 2020, at multiple global sites, with many seeking to assay for efficacy of various repurposed drugs against SARS‐CoV‐2—most available on the National Library of Medicine's registry of clinical studies, others scraped from text‐mining of the available literature. Figure 1 summarizes an early snapshot of such clinical investigations, focusing on 53 repurposing‐based trials noted to be in planning or progress as of 17 April 2020, on the basis of the tested drugs, targets or mechanisms, preliminary efficacy studies, current and prior literature on the drug's efficacy for betacoronavirus‐related disease and the specific designating information for the relevant trials themselves including their phase, scale, expected completion date and other key descriptors. The tested modalities attempt a variety of approaches to improve patient outcomes, some targeting the virus directly, others seeking to counter its deleterious physiological sequelae through immunomodulation or respiratory and circulatory support to reduce mortality and morbidity.
Figure 1.
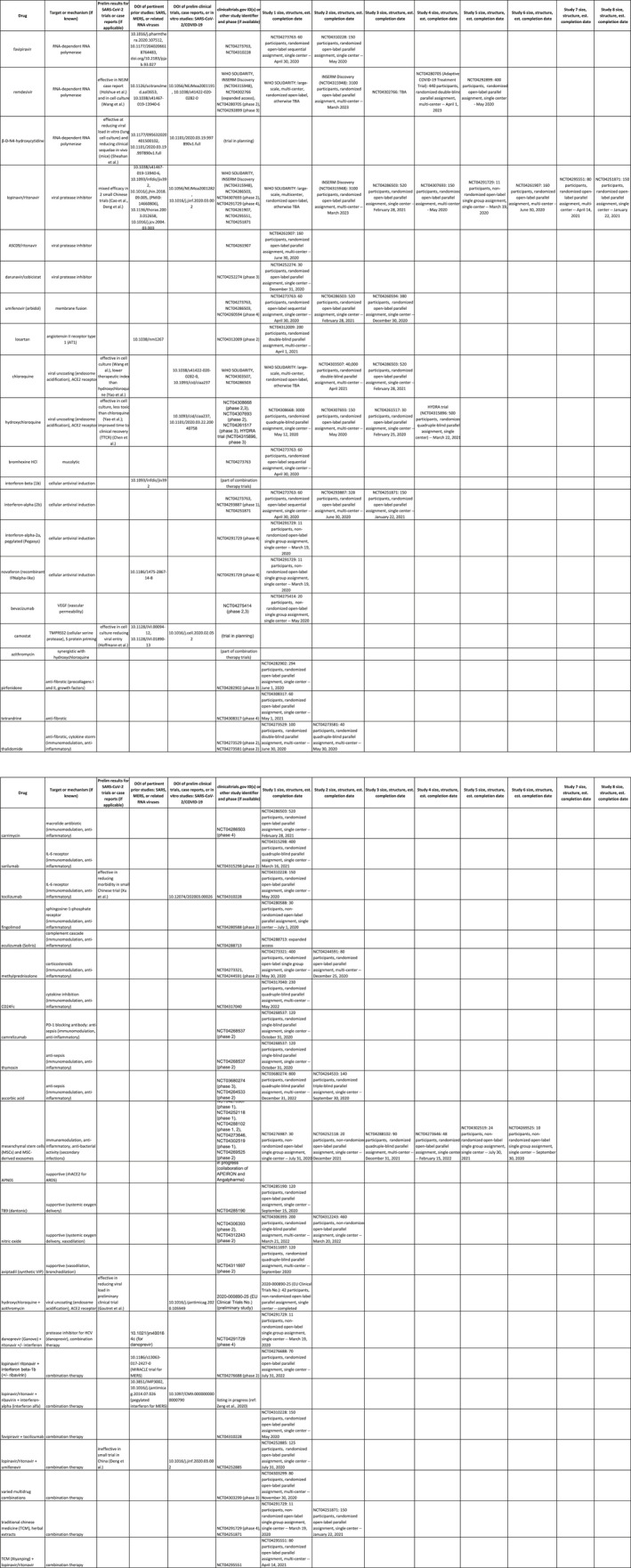
Comprehensive summary data on drug repurposing‐focused COVID‐19 clinical trials completed, in progress, or in advanced planning stages as of 17 April 2020. Trial data were organized according to drug and target or pharmacologic mechanism, along with results and pertinent literature from any preliminary COVID‐19 studies (or prior SARS and MERS studies), trial ID and phase, and data on investigational scale and structure including number of participants, blinding, single‐ vs. multi‐centre status and estimated completion date
Summary statistics for these seminal trials are provided in Figure 2. As can be seen, drugs and targets linked to unique viral components and processes—such as the viral protease and RNA‐dependent RNA polymerase—have predominated in early investigations. Several compounds had demonstrated promise in at least some preliminary studies including lopinavir/ritonavir, remdesivir, and chloroquine or hydroxychloroquine, and more than a third (for which comprehensive descriptions were provided) were already at or beyond phase 3. Nevertheless, the scale of most trials remains small, the vast majority are not multi‐centred, and evaluation of efficacy will take months or years to carry out. Only a few clinical trials are enrolling greater than 1,000 participants, including the global WHO DISCOVERY trial, the European‐based INSERM trial and the Adaptive COVID‐19 Treatment Trial. Moreover, the most commonly tested drugs have been drawn from anti‐viral studies outside the prior body of research focusing on SARS and MERS.
Figure 2.
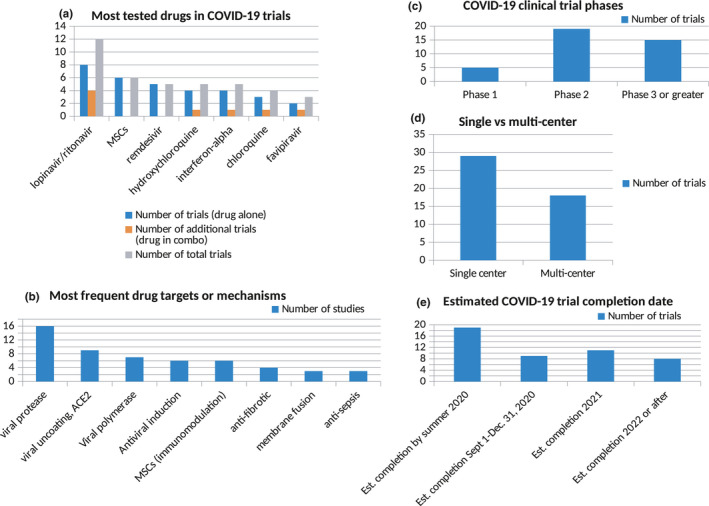
Cumulative statistics from drug repurposing‐focused COVID‐19 clinical trials, based on the aggregate number of trials (out of a total of 53 publicly reported by 17 April 2020) involving (a) a specific drug, (b) a classifying target or mechanism, (c) clinical research phase, (d) single centre vs. multi‐centre and (e) a given expected timeframe for reporting results
Since the SARS and MERS outbreaks dissipated without approaching the global impact of COVID‐19, fledgling therapeutic studies for these epidemics were generally not followed up. As a result, in vivo animal and human data for promising drug prospects, including blood concentrations and dose–response curves in animal studies, are largely unavailable. This has likely contributed to the pronounced preference, in existing COVID‐19 clinical investigations, to repurpose therapeutic candidates like favipiravir, remdesivir and lopinavir/ritonavir, all designed for taxonomically distinct viruses and viral classes (particularly HIV, Ebola and influenza) relative to SARS‐CoV‐2. These therapeutic agents nonetheless benefit from prior in vivo data which the compounds effective against SARS‐CoV and MERS‐CoV lack, making them more readily adaptable to urgent COVID‐19 clinical trials than drug candidates for fellow betacoronaviruses with higher likelihood of specific interactions with SARS‐CoV‐2 essential components.
Such factors, largely a product of practical urgencies amid a pressing pandemic and the contingent history of the SARS and MERS epidemics, further suggest that there may be substantial overlooked potential for new COVID‐19 drugs showing prior promise in vitro against other betacoronaviruses. This suggestion is reinforced by a recent study which noted significant in vitro and in vivo activity of a known nucleoside analogue with previous efficacy against SARS and MERS, β‐D‐N4‐hydroxycytidine (NHC), in reducing viral load in cell culture and tissue damage in mice secondary to SARS‐CoV‐2 infection (Sheahan et al., 2020). NHC was one of nearly a dozen drugs to have demonstrated potential in reducing the disease burden from SARS, MERS or both (De Clercq, 2006; Kumar, Jung, & Liang, 2013; Savarino, 2005), and the recent results support the notion that such repurposing may be fruitful for COVID‐19.
We have therefore systematically combed available literature, reports and commentaries to ascertain untested drugs with previous promise for SARS and MERS that merit consideration for additional COVID‐19 trials, alongside the comprehensive clinical trial data elaborated previously. We utilized an approach combining careful manual curation and algorithmic scraping using a flexible Python language‐based text‐mining tool, previously developed for research into prospective repurposable drugs for Duchenne muscular dystrophy. Concomitantly, we systematically examined the drugs with previous reported efficacy in the context of SARS and MERS, and then curated them on the basis of several factors most indicative of potential in clinical trials to repurpose them for SARS‐CoV‐2.
In assessing criteria to identify highly promising candidates for COVID‐19 drug repurposing, it has been noted that perhaps the primary predictor of eventual failure in clinical trials is non‐selectivity for the target, contributing to unacceptable toxicity (Gayvert, Madhukar, & Elemento, 2016). Therefore, in examining the as yet untested or seldom‐tested SARS and MERS drugs with potential for COVID‐19 repositioning, particular weighting was given to those exhibiting a low reported EC50 (or IC50) and high Selectivity Index (SI) from cell culture studies. Attention was likewise given to drugs which are not only selective for a viral target, especially a component indispensable for viral replication, but also substantially reduce viral load in vitro. Further consideration was given for modalities demonstrating confirmation of potential efficacy from multiple centres. With these factors as primary criteria for identifying COVID‐19 drug repurposing candidates, several especially promising potential therapeutics were identified, which is summarized in Figure 3.
Figure 3.
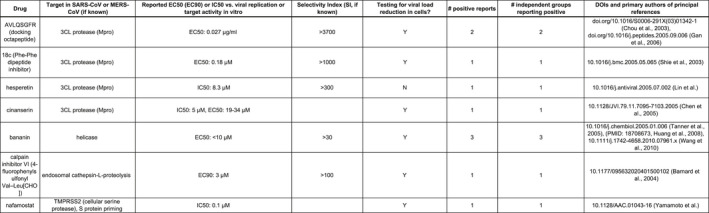
Summary data on promising untested candidates for COVID‐19 drug repurposing based on proven success in SARS and/or MERS in vitro studies, with focus on target selectivity, viral load reduction and number of positive reports with independent confirmation
Of particular promise are a docking octapeptide, AVLQSGFR (Chou, Wei, & Zhong, 2003; Gan et al., 2006), and a Phe‐Phe dipeptide inhibitor, 18c (Shie et al., 2005), with marked selectivity (> 1,000‐fold) for the betacoronaviral protease of SARS‐CoV (3C‐like protease, also known as 3Cl protease, 3CLpro or Mpro) and demonstrated capacity to reduce viral load in cell culture. Both agents exhibit not only remarkable Selectivity Index (SI) values, but also low IC50 and/or EC50 measurements that suggest viability as practical drugs in vivo. As noted previously, the viral protease is also the most frequent target of COVID‐19 clinical trials currently in progress or planning, but these candidates stand out for their proven ability to selectively target 3CLpro and to bring about a tangible reduction in viral infection capacity.
Encouraging results have also been observed for bananin (Huang, Zheng, & Sun, 2008; Tanner et al., 2005; Wang et al., 2011), a viral helicase inhibitor with > 30‐fold selectivity in cell culture studies; for calpain inhibitor VI (Barnard et al., 2004), with > 100‐fold selectivity; and for the herbal extract hesperetin (Lin et al., 2005), with an SI > 300 and a direct inhibitory effect on the 3Cl protease. Bananin (Tanner et al., 2005) and calpain inhibitor VI (Barnard et al., 2004) have likewise been found to inhibit viral load and infection in vitro. Promising seminal studies for inhibition of SARS or MERS infection have also emerged for another protease inhibitor, cinanserin (Chen et al., 2005), and for nafamostat (Yamamoto et al., 2016), a cellular serine protease inhibitor that reduces viral spike protein priming.
As indicated above, a limitation in evaluating these compounds’ potential as COVID‐19 treatments is that all reported studies thus far are in vitro, without data on therapeutic or toxic blood concentrations, ED50, or dose–response behaviour in animals—a result of the abrupt subsiding of the SARS and MERS epidemics. Yet, this very fact, in conjunction with their striking findings in cell culture studies, helps to underscore their untapped potential for the current pandemic caused by a much more persistent betacoronavirus, and the value of testing them in an in vivo context. Alongside the recent findings by Sheahan and coworkers with NHC, such results suggest that animal studies and preliminary clinical trials with these agents, or closely related chemical derivatives, may prove fruitful in expanding the arsenal of drugs to combat the relentless spread, morbidity and mortality of COVID‐19.
CONFLICT OF INTEREST
The authors certify that they have no affiliations with, or involvement in, any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other form of equity interest; and expert testimony or patent‐licensing arrangements), or non‐financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
ETHICAL APPROVAL
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required as this is a review article with no original research data.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the valuable assistance of Florian Barthélémy, PhD, in helping to format the figures and illustrations for this manuscript.
Ulm JW, Nelson SF. COVID‐19 drug repurposing: Summary statistics on current clinical trials and promising untested candidates. Transbound Emerg Dis.2021;68:313–317. 10.1111/tbed.13710
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.
REFERENCES
- Barnard, D. L. , Hubbard, V. D. , Burton, J. , Smee, D. F. , Morrey, J. D. , Otto, M. J. , & Sidwell, R. W. (2004). Inhibition of severe acute respiratory syndrome‐associated coronavirus (SARSCoV) by calpain inhibitors and beta‐D‐N4‐hydroxycytidine. Antiviral Chemistry & Chemotherapy, 15(1), 15–22. 10.1177/095632020401500102 [DOI] [PubMed] [Google Scholar]
- Chen, L. , Gui, C. , Luo, X. , Yang, Q. , Gunther, S. , Scandella, E. , … Jiang, H. (2005). Cinanserin is an inhibitor of the 3C‐like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. Journal of Virology, 79(11), 7095–7103. 10.1128/jvi.79.11.7095-7103.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, K. C. , Wei, D. Q. , & Zhong, W. Z. (2003). Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochemical and Biophysical Research Communications, 308(1), 148–151. 10.1016/s0006-291x(03)01342-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq, E. (2006). Potential antivirals and antiviral strategies against SARS coronavirus infections. Expert Review of Anti‐infective Therapy, 4(2), 291–302. 10.1586/14787210.4.2.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, Y. R. , Huang, H. , Huang, Y. D. , Rao, C. M. , Zhao, Y. , Liu, J. S. , … Wei, D. Q. (2006). Synthesis and activity of an octapeptide inhibitor designed for SARS coronavirus main proteinase. Peptides, 27(4), 622–625. 10.1016/j.peptides.2005.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayvert, K. M. , Madhukar, N. S. , & Elemento, O. (2016). A data‐driven approach to predicting successes and failures of clinical trials. Cell Chemical Biology, 23(10), 1294–1301. 10.1016/j.chembiol.2016.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. D. , Zheng, B. J. , & Sun, H. Z. (2008). Helicases as antiviral drug targets. Hong Kong Medical Journal, 14(Suppl 4), 36–38. [PubMed] [Google Scholar]
- Kumar, V. , Jung, Y. S. , & Liang, P. H. (2013). Anti‐SARS coronavirus agents: A patent review (2008 ‐ present). Expert Opinion on Therapeutic Patents, 23(10), 1337–1348. 10.1517/13543776.2013.823159 [DOI] [PubMed] [Google Scholar]
- Li, G. , & De Clercq, E. (2020). Therapeutic options for the 2019 novel coronavirus (2019‐nCoV). Nature Reviews Drug Discovery, 19(3), 149–150. England. 10.1038/d41573-020-00016-0 [DOI] [PubMed] [Google Scholar]
- Lin, C. W. , Tsai, F. J. , Tsai, C. H. , Lai, C. C. , Wan, L. , Ho, T. Y. , … Chao, P. D. (2005). Anti‐SARS coronavirus 3C‐like protease effects of Isatis indigotica root and plant‐derived phenolic compounds. Antiviral Research, 68(1), 36–42. 10.1016/j.antiviral.2005.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarino, A. (2005). Expanding the frontiers of existing antiviral drugs: Possible effects of HIV‐1 protease inhibitors against SARS and avian influenza. Journal of Clinical Virology, 34(3), 170–178. 10.1016/j.jcv.2005.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan, T. P. , Sims, A. C. , Zhou, S. , Graham, R. L. , Pruijssers, A. J. , Agostini, M. L. , … Baric, R. S. (2020). An orally bioavailable broad‐spectrum antiviral inhibits SARS‐CoV‐2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Science Translational Medicine, 12(541), eabb5883. 10.1126/scitranslmed.abb5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shie, J. J. , Fang, J. M. , Kuo, T. H. , Kuo, C. J. , Liang, P. H. , Huang, H. J. , … Wong, C. H. (2005). Inhibition of the severe acute respiratory syndrome 3CL protease by peptidomimetic alpha, beta‐unsaturated esters. Bioorganic & Medicinal Chemistry, 13(17), 5240–5252. 10.1016/j.bmc.2005.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli, A. , & Pellino, G. (2020). COVID‐19 pandemic: Perspectives on an unfolding crisis. British Journal of Surgery, 107(7), 785–787. 10.1002/bjs.11627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, J. A. , Zheng, B. J. , Zhou, J. , Watt, R. M. , Jiang, J. Q. , Wong, K. L. , … Huang, J. D. (2005). The adamantane‐derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chemistry & Biology, 12(3), 303–311. 10.1016/j.chembiol.2005.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Huang, J. D. , Wong, K. L. , Wang, P. G. , Zhang, H. J. , Tanner, J. A. , … Niccolai, N. (2011). On the mechanisms of bananin activity against severe acute respiratory syndrome coronavirus. FEBS Journal, 278(2), 383–389. 10.1111/j.1742-4658.2010.07961.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M. , Matsuyama, S. , Li, X. , Takeda, M. , Kawaguchi, Y. , Inoue, J. I. , & Matsuda, Z. (2016). Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome coronavirus s protein‐mediated membrane fusion using the split‐protein‐based cell‐cell fusion assay. Antimicrobial Agents and Chemotherapy, 60(11), 6532–6539. 10.1128/aac.01043-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N. , Zhang, D. , Wang, W. , Li, X. , Yang, B. , Song, J. , … Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.


