Abstract
Secondary metabolites are assembled by enzymes that often perform reactions with high selectivity and specificity. Many of these enzymes also tolerate variations in substrate structure, exhibiting promiscuity that enables various applications of a given biocatalyst. However, initial enzyme characterization studies frequently do not explore beyond the native substrates. This limited assessment of substrate scope contributes to the difficulty of identifying appropriate enzymes for specific synthetic applications. Here, we report the natural function of cyanobacterial SxtG, an amidinotransferase involved in the biosynthesis of paralytic shellfish toxins (PSTs), and demonstrate its ability to modify a breadth of non-native substrates. In addition, we report the first X-ray crystal structure of SxtG, which provides rationale for this enzyme’s substrate scope. Taken together, these data confirm the function of SxtG and exemplify its potential utility in biocatalytic synthesis.
Enzymes from secondary metabolic pathways are responsible for assembling complex molecules from simple chemical building blocks. These specialized enzymes have often evolved from proteins involved in primary metabolism to gain new function.1,2 Due to this evolutionary relationship, secondary metabolic enzymes have the potential to be inherently promiscuous in substrate preference and reactivity, rendering them attractive for biocatalytic applications.3,4 However, initial characterization studies of secondary metabolic enzymes often focus on solely on native function. Expanding investigations of the substrate scopes of novel enzymes from secondary metabolite pathways can aid in the discovery of promising new biocatalysts.5,6
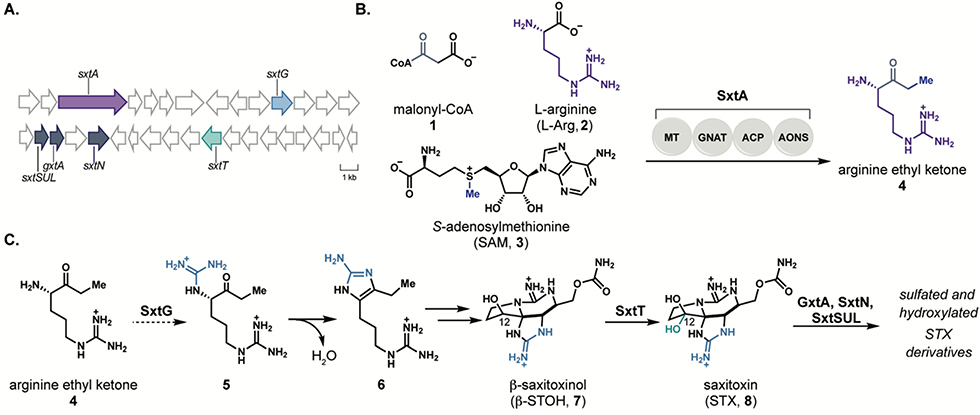
Paralytic shellfish toxins (PSTs), such as saxitoxin (STX, 8), are natural products produced by freshwater cyanobacteria and marine dinoflagellates and exhibit high affinity for voltage-gated sodium channels (VGSCs).7, 8 Gene clusters associated with PST biosynthesis have been identified in several species of cyanobacteria including Microseira wollei9, Cylindrospermopsis raciborskii10, Aphanizomenon sp. NH-511, and others12, 13; however, the majority of the gene products from these clusters remain uncharacterized (Figure 1A). We have recently demonstrated the function of enzymes involved in PST biosynthesis, including 1) the polyketide-like synthase SxtA14, which generates the first biosynthetic intermediate, arginine ethyl ketone (4, Figure 1B); 2) Rieske oxygenases SxtT and GxtA15 which perform site- and stereoselective C-H hydroxylation reactions on the tricyclic core of STX (8); and 3) sulfotransferases SxtN13 and SxtSUL16 involved in the late-stage derivatization of STX (8, Figure 1C). This work has established the initial and final steps of PST biosynthesis; however, the intervening steps remain unknown.
Figure 1.
A) Representation of the saxitoxin biosynthetic gene cluster from Microseira wollei. Genes in purple, dark blue, and green indicate enzymes that have been characterized for roles in paralytic shellfish toxin biosynthesis, and light blue indicates the enzyme SxtG. B) Reaction performed by polyketide-like synthase SxtA. C) Proposed role of SxtG to operate on the SxtA product and downstream reactions toward saxitoxin.
SxtG, which is annotated as an amidinotransferase, has been proposed to immediately follow SxtA in the biosynthesis of STX (8) through amidinylation of the amine of ethyl ketone 4 (Figure 1C).10 Intermediate 5 is anticipated to undergo spontaneous cyclization to deliver aminoimidazole 6, a molecule that has been previously identified in both PST-producing cyanobacteria and dinoflagellates.17–19 Amidinotransferases are well-characterized for their activity on amino acid substrates; however, reactions of amidinotransferases on other classes of substrates are less well-studied. Understanding the reactivity and substrate promiscuity of SxtG provides the opportunity to identify a biocatalyst with a complementary substrate scope to known amidinotransferases.20–22
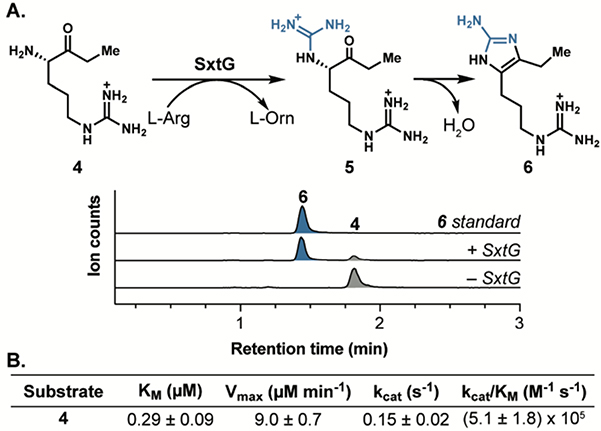
We sought to assess the native function of SxtG in the cyanobacterial biosynthesis of paralytic shellfish toxins (PSTs) using purified protein from M. wollei overexpressed in Escherichia coli (see Supporting Information). Initial SxtG activity assays were performed using L-Arg (2) as an amidino group donor and the proposed native substrate, ethyl ketone 4, as an amidino group acceptor. Liquid chromatography-mass spectrometry (LC-MS) analysis of these reactions revealed a prominent new peak at m/z 211 corresponding to aromatic product 6, resulting from amidinotransfer by SxtG and spontaneous condensation (Figure 2). The mass corresponding to the immediate amidinotransfer product 5 (m/z = 229) was only detectable in trace amounts,indicating rapid cyclization of 5 to 6 under the conditions used for the reaction and subsequent analysis. To verify the structure of the peak corresponding to m/z = 211, the reaction mixture was compared to a synthetic standard of aminoimidazole 6.17 The enzymatic product was identical in retention time and exact mass (Figure 2a). The Km of 4 was determined to be 0.29 μM and the kcat/KM was found to be 5.1 × 105 M-1 s-1 through steady-state kinetic analysis (Figure 2b, Figure S3). These data support the previously proposed hypotheses that SxtG operates on the SxtA product, ethyl ketone 4, to generate 5, which spontaneously cyclizes to afford 6.10
Figure 2.
A) Extracted ion chromatogram (EIC) LC-MS traces of the SxtG-catalyzed amidinotransfer reaction with arginine ethyl ketone (4, grey) and L-Arg (2) to generate product 6 (blue) and L-ornithine (L-Orn). B) Steady-state kinetic parameters for arginine ethyl ketone (4).
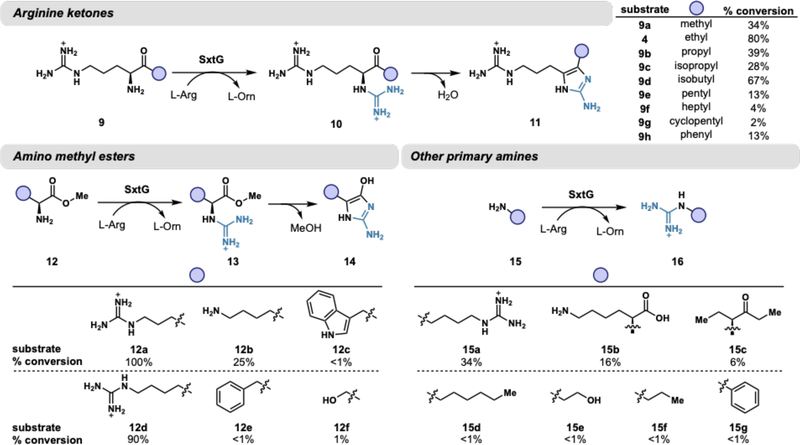
To our knowledge, amidinotransferase activity on an α-amino ketone substrate has not been evaluated with any other enzyme of this class. Optimized reaction conditions include the addition of Fe(NH4)2(So4)2, which resulted in consistently increased abundances of product 6 by LC-MS relative to controls without metal added (Figure S5). The addition of Fe(NH4)2(So4)2 was ultimately determined to not significantly influence reaction kinetics and is therefore not essential to enzymatic catalysis (Figure S4). Using these conditions, we analyzed 42 substrates including α-amino acids, α-amino methyl esters, ketone derivatives of arginine, and other primary and secondary amines for reactivity with SxtG. Of this panel, 22 substrates were productively modified by SxtG. Percent conversions for each substrate were determined by LC-MS quantification based on standard curves of chemically synthesized products (Figure 3). The putative native substrate, ethyl ketone 4, was converted in the highest yield compared with a panel of arginine ketone derivatives. Other arginine derivatives with bulky or long, greasy substituents were not optimal SxtG substrates, as exemplified by arginine heptyl ketone (9f, 4%) and arginine cyclopentyl ketone (9g, 2%). Surprisingly, SxtG exhibited a modest conversion of arginine phenyl ketone (9h, 13%) despite the substrate’s large, rigid side chain.
Figure 3.
Substrates accepted by SxtG including α-amino ketones, α-amino methyl esters, and other primary amines. Percent conversions were determined by LC-MS quantification of products. Reactions consisted of 1 μM SxtG, 1 mM substrate, 10 mM L-Arg (2), 500 μM Fe(NH4)2(So4)2, and 50 mM HEPES pH 8.5 and were incubated at 30 °C for 16 h.
α-Amino esters were accepted far more readily than their a-amino acid counterparts. For example, 90% conversion was observed with L-homoarginine methyl ester (12d) whereas L-homoarginine was not converted by SxtG (see Figure S28 for complete list of substrates not accepted by SxtG). Arginine methyl ester (12a) was completely consumed by SxtG and had comparable steady-state kinetic parameters to the putative native substrate (4) (Figure S4). Analogous to ketone substrates, all α-amino methyl ester substrates accepted by SxtG resulted in 2-aminoimidazole products, generated by the loss of methanol upon ring closure (13 to 14, Figure 3). impressively, SxtG exhibited site-selectivity with L-lysine (15b) and L-lysine methyl ester (12b), exclusively installing an amidino group onto the a-amino group rather than the terminal amine group of the side chain. This site-selectivity is orthogonal to small molecule reagents where selectivity is often governed by sterics.23 Notably, no bis-guanidinylated products were detected with either substrate. Taken together, these results suggest that SxtG is an amidinotransferase with broad substrate promiscuity, operating on a wide variety of substrates and preferring α-amino ketones and α-amino methyl esters over α-amino acids.
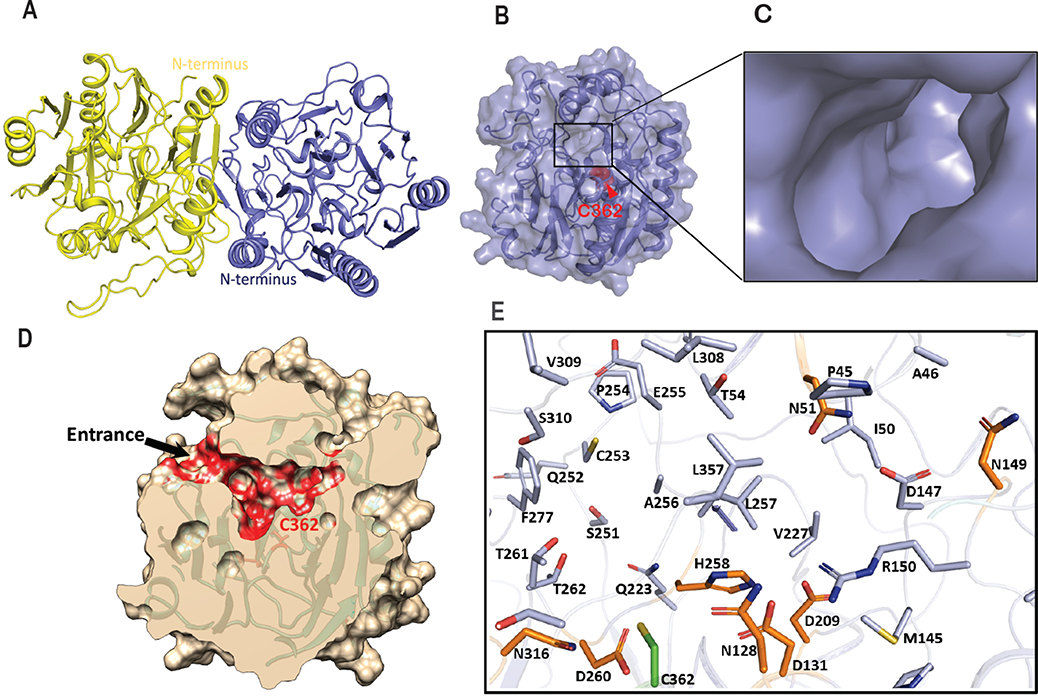
The broad substrate scope exhibited by SxtG prompted us to determine its structure with an interest in closely examining the substrate binding pocket. The structure of SxtG was solved at 1.79 Å resolution (Table S1) by molecular replacement using the available structure of L-arginine:glycine amidinotransferase (AT, 1JDW), as a search model.24 Two molecules were present per asymmetric unit, which arranged as a dimer through a noncrystallographic 2-fold symmetry axis along the hydrophobic dimer interface (Figure 4A). Formation of the SxtG dimer was verified in solution by size-exclusion chromatography (Figure S1). The final refined structure includes residues 18–377 for each monomer and exhibits five-fold pseudosymmetry with repeating ββαα-motifs surrounding the active site at the core of the protein (Figure 4B). A narrow pore visible on the surface leads to a large cavity surrounding the active site where the substrate can be accommodated (Figure 4C).
Figure 4.
Structure of SxtG from M. wollei and active site arrangement. A) Ribbon representation of SxtG dimer. Monomers are shown in yellow and blue with N-termini located on opposite sides of the dimer. B) Surface representation of SxtG with narrow opening on the surface, active site Cys362 shown in red sphere representation. C) Enlarged view of substrate entrance pore. D) Surface representation (cream color) of SxtG with active site pocket predicted by CASTp shown in red surface with an entrance on the surface leading to the active site. E) Residues at the active site of SxtG surrounding the active Cys362 (green) are shown in stick representation. Critical residues (D131, D209, H258, and D260) and four Asn (N128, N316, N51, and N149) residues at active site of SxtG are shown in orange stick representation.
Sequence alignment of SxtG with other amidinotransferases confirmed the presence of conserved catalytic residues: Cys362, His258, Asp209, Asp131 and Asp260 (Figure S29). Cys362, His258 and Asp209 of SxtG aligned exactly with the AT catalytic triad residues Cys407, His303 and Asp254 (Figure S29).24 SxtG variants C362A and D209A were generated using sitedirected mutagenesis, and each variant was inactive with 4 as a substrate (Table S3). Similarly, aspartic acid residues Asp131 and Asp260 in the SxtG active site could form hydrogen bonds with the amidino group donor L-Arg (2) and assist in stabilization of the substrate at the active site. The SxtG D260A variant was also inactive (Table S3). These data indicate that the residues determined through sequence alignment are the anticipated prototypical amidinotransferase catalytic residues.24 Further analysis comparing the structural similarity of SxtG to other structurally characterized amidinotransferases in the Protein Data Bank (PDB) was performed using the DALI server (Table S3).25 AT (Z-score = 59.4),26 inosamine-phosphate amidinotransferase StrB1 (Z-score = 46.5),21 and polyamine amidinotransferase HsvA (Z-score = 53.0)20 were identified as the closest matches. Despite SxtG sharing low sequence identity with these enzymes (<50%), the overall architecture and key amidinotransferase structural features were conserved.
To understand how the SxtG active site accommodates an array of substrates, the active site and its adjacent substrate binding cavity were analyzed using the CASTp server to locate the interior cavities and calculate area, volume, and contributing atoms involved in the formation of each pocket.27 In total, CASTp computed 55 pockets from the structure of the SxtG monomer. Careful analysis of each pocket led us to identify the putative substrate binding pocket containing conserved amidinotransferase active site residues (i.e., Cys362, His258, Asp209, Asp131 and Asp260) (Figure 4D–4E, and Figure S32). Notably, this substrate binding pocket is significantly larger than those identified in other structurally characterized amidinotransferases, including human AT, mitochondrial AT, StrB1, and HsvA (Table S2).21 Moreover, the active site pockets of SxtG and HsvA differ from AT and StrB1 as the pockets of SxtG and HsvA are lined with polar residues (Figure 4E, Table S2). Several hydroxyl groups from Ser and Thr residues (Ser310, Ser359, Thr54, Thr261 and Thr262) surrounding the active site Cys362 in SxtG are primed to interact with the carbonyl group of a-amino ketone substrates such as 4. The importance of these residues was further tested with molecular docking using AutoDock Vina and data showed a polar interaction of Thr261 with the guanidino group of 4 (Figure S33). This interaction would anchor the amine near Cys362 and allow for the amidino transfer to the a-amino group. Unique to SxtG, the substrate binding pocket also contains four asparagine residues: Asn128 and Asn316 situated in close vicinity to Cys362, and Asn51 and Asn149 located at the distal surface of the pocket (Figure 4E). These residues at four corners of the binding pocket could facilitate substrate binding and stabilization in the active site. Given the electrostatics of the interior binding cavity, hydrophobic and/or bulky substrates, such as phenylalanine methyl ester (12e), would likely be less preferred by SxtG relative to polar substrates, such as arginine ethyl ketone 4. This is exemplified by observation of reduced percent conversions with substrates like arginine heptyl ketone (9f, 4%) and tryptophan methyl ester (12c, <1%). Furthermore, significant structural variations were observed adjacent to the substrate entrance pore upon superposition of SxtG with the HsvA (r.m.s.d 0.933), AT (r.m.s.d 0.798), and StrB1 (r.m.s.d 0.889) structures (Figure S30). Similar to HsvA, the exterior of the substrate entrance pore in SxtG is surrounded with negatively charged Glu residues, Glu58, Glu59, Glu255 and Glu276, which are not conserved in StrB1 or AT and are replaced with Lys, Asn, Gly, and Ala, respectively.20 The presence of negatively charged residues around the entrance to SxtG’s substrate binding cavity could potentially select positively charged over negatively charged or hydrophobic substrates. Taken together, the combination of an active site pocket lined with uniquely placed polar residues and a negatively charged entrance to the binding pocket likely play a significant role in determining the substrate specificity of SxtG.
Collectively, SxtG’s significantly larger substrate binding pocket allows the enzyme to accommodate a broad range of substrates. Furthermore, the highly polar environment of the pocket allows for charged substrates to be accepted more readily than hydrophobic substrates. The presence of negatively charged residues, Glu and Asp, at the substrate binding pocket and entrance pore provides a negatively charged electrostatic potential, which could facilitate the entrance of positively charged substrates, as exemplified by the substrate scope analysis. Additionally, Ser, Thr, and four Asn residues (Asn128, Asn316, Asn51 and Asn149) at the SxtG active site could participate in interactions with the guanidinium ion or carbonyl group of substrates.
This study represents the first in vitro characterization of SxtG and its involvement in PST biosynthesis. We have demonstrated that SxtG operates on the intermediate in PST biosynthesis produced by SxtA, ethyl ketone 4, to generate biosynthetic intermediate 6. Additionally, we show that SxtG has a substrate scope beyond its native substrate, accepting a variety of amines with a preference for substrates containing an α-amino carbonyl motif. In addition to a-amino ketones, SxtG can also process substrates containing bulky aromatic groups, greasy hydrocarbon chains, and simple primary amines. Analysis of the crystal structure of SxtG reveals variation in the ligand binding pocket relative to the four structurally characterized amidinotransferases, which may be responsible for its unique substrate preferences. The unusually large substrate binding pocket in SxtG accounts for its ability to accept a wide variety of substrate scaffolds. Future work will be conducted to pursue additional SxtG structures with substrates bound in the active site. Furthermore, we anticipate that SxtG’s substrate scope demonstrated here can be leveraged for the synthesis of guanidinium ion-containing compounds and natural products.
Supplementary Material
Acknowledgements
This research was supported by funds from the University of Michigan Life Sciences Institute and the National Institute of General Medical Science R35GM124880 (to A.R.H.N.) and University of Michigan Start-up funds (to M.K.). A.L.L. acknowledges the University of Michigan Rackham Graduate School and the National Institute of Neurological Disorders and Stroke F31NS111906 for support. In addition, we thank A. Frank help with molecular docking.
Footnotes
Competing Interests
The authors declare no competing interests.
Supporting Information
Complete methods, experimental details, and supplementary figures are provided in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- [1].Jensen RA (1976) Enzyme recruitment in evolution of new function, Ann. Rev. Microbiol. 30, 409–425. [DOI] [PubMed] [Google Scholar]
- [2].Maeda HA (2019) Evolutionary diversification of primary metabolism and its contribution to plant chemical diversity, Front. Plant Sci. 10, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Peracchi A (2018) The limits of enzyme specificity and the evolution of metabolism, Trends Biochem. Sci. 43, 984–996. [DOI] [PubMed] [Google Scholar]
- [4].Khersonsky O, and Tawfik DS (2010) Enzyme promiscuity: A mechanistic and evolutionary perspective, Annu. Rev. Biochem. 79, 471–505. [DOI] [PubMed] [Google Scholar]
- [5].Kohls H, Steffen-Munsberg F, and Hohne M (2014) Recent achievements in developing the biocatalytic toolbox for chiral amine synthesis, Curr. Opin. Chem. Biol. 19, 180–192. [DOI] [PubMed] [Google Scholar]
- [6].Parajuli P, Pandey RP, Nguyen THT, Dhakal D, and Sohng JK (2018) Substrate scope of O-methyltransferase from Streptomyces peucetius for biosynthesis of diverse natural products methoxides, Appl. Biochem. Biotechnol. 184, 1404–1420. [DOI] [PubMed] [Google Scholar]
- [7].Llewellyn LE (2006) Saxitoxin, a toxic marine natural product that targets a multitude of receptors, Nat. Prod. Rep. 23, 200–222. [DOI] [PubMed] [Google Scholar]
- [8].Thottumkara AP, Parsons WH, and Du Bois J (2014) Saxitoxin, Angew. Chem. Int. Ed. 53, 5760–5784. [DOI] [PubMed] [Google Scholar]
- [9].Mihali TK, Carmichael WW, and Neilan BA (2011) A putative gene cluster from a Lyngbya wollei bloom that encodes paralytic shellfish toxin biosynthesis, PLoS One 6, e14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kellmann R, Mihali TK, Jeon YJ, Pickford R, Pomati F, and Neilan BA (2008) Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria, Appl. Environ. Microbiol. 74, 4044–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mihali TK, Kellmann R, and Neilan BA (2009) Characterisation of the paralytic shellfish toxin biosynthesis gene clusters in Anabaena circinalis AWQC131C and Aphanizomenon sp. NH-5, BMC Biochem 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Soto-Liebe K, Murillo AA, Krock B, Stucken K, Fuentes-Valdes JJ, Trefault N, Cembella A, and Vasquez M (2010) Reassessment of the toxin profile of Cylindrospermopsis raciborskii T3 and function of putative sulfotransferases in synthesis of sulfated and sulfonated PSP toxins, Toxicon 56, 1350–1361. [DOI] [PubMed] [Google Scholar]
- [13].Cullen A, DAgostino PM, Mazmouz R, Pickford R, Wood S, and Neilan BA (2018) Insertions within the saxitoxin biosynthetic gene cluster result in differential toxin profiles, ACS Chem. Biol. 13, 3107–3114. [DOI] [PubMed] [Google Scholar]
- [14].Chun SW, Hinze ME, Skiba MA, and Narayan ARH (2018) Chemistry of a unique polyketide-like synthase, J. Am. Chem. Soc. 140, 2430–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lukowski AL, Ellinwood DC, Hinze ME, DeLuca RJ, Du Bois J, Hall S, and Narayan ARH (2018) C-H hydroxylation in paralytic shellfish toxin biosynthesis, J. Am. Chem. Soc. 140, 11863–11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lukowski AL,, Denomme N, Hinze ME, Hall S, Isom LL, and Narayan ARH (2019) Biocatalytic detoxification of paralytic shellfish toxins, ACS Chem. Biol. 14, 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tsuchiya S, Cho Y, Konoki K, Nagasawa K, Oshima Y, and Yotsu-Yamashita M (2014) Synthesis and identification of proposed biosynthetic intermediates of saxitoxin in the cyanobacterium Anabaena circinalis (TA04) and the dinoflagellate Alexandrium tamarense (Axat-2), Org. Biomol. Chem. 12, 3016–3020. [DOI] [PubMed] [Google Scholar]
- [18].Tsuchiya S, Cho Y, Konoki K, Nagasawa K, Oshima Y, and Yotsu-Yamashita M (2015) Synthesis of a tricyclic bisguanidine compound structurally related to saxitoxin and its identification in paralytic shellfish toxin-producing microorganisms, Chem.-Eur. J. 21, 7835–7840.. [DOI] [PubMed] [Google Scholar]
- [19].Tsuchiya S, Cho Y, Yoshioka R, Konoki K, Nagasawa K, Oshima Y, and Yotsu-Yamashita M (2017) Synthesis and identification of key biosynthetic intermediates for the formation of the tricyclic skeleton of saxitoxin, Angew. Chem. Int. Ed. 56, 5237–5331. [DOI] [PubMed] [Google Scholar]
- [20].Shanker S, Schaefer GK, Barnhart BK, Wallace-Kneale VL, Chang D, Coyle TJ, Metzler DA, Huang J, and Lawton JA (2017) The virulence-associated protein HsvA from the fire blight pathogen Erwinia amylovora is a polyamine amidinotransferase, J. Biol. Chem. 292, 21366–21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fritsche E, Bergner A, Humm A, Piepersberg W, and Huber R (1998) Crystal structure of L-arginine:inosamine-phosphate amidinotransferase StrB1 from Streptomyces griseus: An enzyme involved in streptomycin biosynthesis, Biochemistry 37, 17664–17672. [DOI] [PubMed] [Google Scholar]
- [22].Humm A, Fritsche E, Mann K, Gohl M, and Huber R (1997) Recombinant expression and isolation of human L-arginine:glycine amidinotransferase and identification of its activesite cysteine residue, Biochem. J. 322, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huang Z, and Dong G (2017) Site-selectivity control in organic reactions: A quest to differentiate reactivity among the same kind of functional groups, Acc. Chem. Res. 50, 465–471.. [DOI] [PubMed] [Google Scholar]
- [24].Humm A, Fritsche E, Steinbacher S, and Huber R (1997) Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: A mitochondrial enzyme involved in creatine biosynthesis, Embo J. 16, 3373–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Holm L, and Rosenstrdm P (2010) Dali server: Conservation mapping in 3D, Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fritsche E, Humm A, and Huber R (1999) The ligand-induced structural changes of human L-arginine:glycine amidinotransferase-A mutational and crystallographic study, J. Biol. Chem. 274, 3026–3032. [DOI] [PubMed] [Google Scholar]
- [27].Tian W, Chen C, Lei X, Zhao J, and Liang J (2018) CASTp 3.0: Computer atlas of surface topography of proteins, Nucleic Acids Res. 46, W363–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.