Abstract
The infectivity of the human immunodeficiency virus (HIV) depends on overcoming APOBEC3 (A3) innate immunity and does so predominantly through the expression of the viral protein, Vif, which induces A3 degradation in the proteasome. Disruption of the functional interactions of Vif enables A3 mutagenesis of the HIV genome during viral replication, which can result in a broadly neutralizing antiviral effect. Vif function requires self-association along with interactions with A3 proteins, protein chaperones and factors of the ubiquitination machinery and these are described here as a potential platform for novel antiviral drug discovery. This article will examine the current state of development of Vif inhibitors that we believe to have therapeutic and functional cure potential.
Without Vif, endogenously expressed APOBEC3G confers innate immunity against HIV
Numerous studies on long-term non-progressor/elite controller (see Glossary) HIV patients [1–5], nonhuman primate [6] and mouse model systems [7–9] have shown that, in the absence of functional Vif, endogenously expressed APOBEC3G (A3G) is sufficient for robust innate immunity against retrovirus infection. This is due to A3G-dependent accumulation of catastrophic levels of dG to dA mutations in simian immunodeficiency virus (SIV) [6], HIV [1–5, 7, 8] and Moloney leukemia virus (MLV) [9] genomes. The anti-HIV DNA mutagenic activity of A3G requires that it be packaged into virions by binding to HIV Gag [10, 11] and viral and host RNAs [12–16]. Delivery of A3G into infected cells provides privileged access of A3G to nascent reverse transcripts to extensively deaminate dC to dU (i.e. hypermutations) in single-stranded stretches of nascent proviral DNA. A3G-dependent genetic damage to HIV is hypothesized to result in degradation of hypermutated proviral DNA through DNA repair pathways that prevents integration of viral genomes [17, 18] (Figure 1). Moreover, dG to dA hypermutations in the plus-strand of those viruses that integrate may negatively affect production of infectious virus through mutations in the viral untranslated regions (UTR) or mutations in the protein coding regions [17, 18]. In addition, A3G bound to DNA impairs progression of reverse transcriptase (RT) by interfering with the viral replication machinery [19–22]. It has therefore been speculated that under conditions of a spreading infection or upon activation of latent viral reservoirs, enabling A3G activity will inhibit the infectivity of virions before they are shed to spread an infection or form new reservoirs (Figure 1). To a lesser extent than that of A3G, natively expressed APOBEC3 (A3) family members A3F, A3H (haplotype specific) and A3D also are capable of inhibiting HIV infectivity through hypermutations [23, 24].
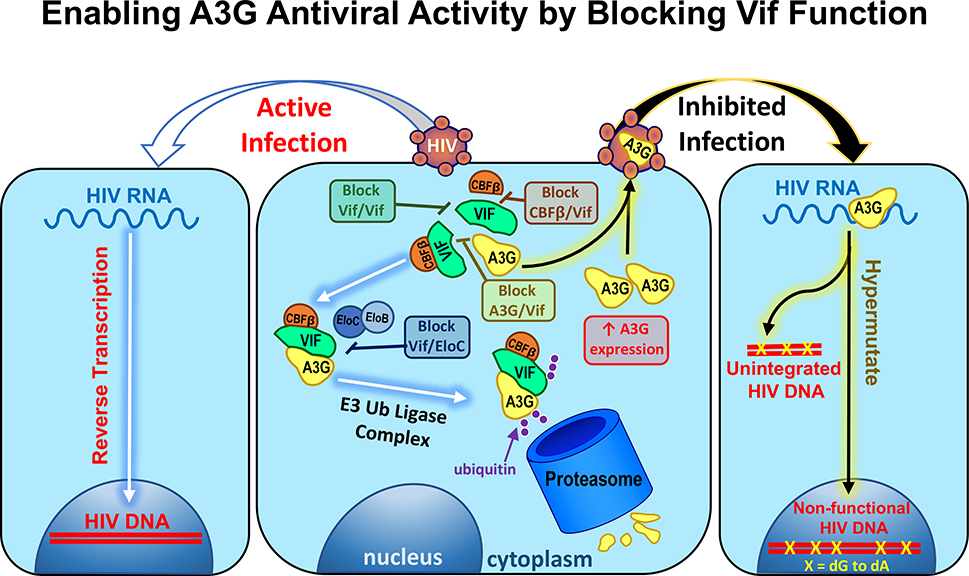
Figure 1. A schematic depiction of the potential interactions between Vif, A3G and E3 ubiquitin ligase complex in the pathway by which Vif shuttles A3G to the proteasome for degradation.
‘Block Vif/Vif’, ‘Block A3G/Vif’ and ‘Block Vif/EloC’ call out where small molecule inhibitors have been proposed to effect in the Vif-dependent A3G degradation pathway, whereas ‘Block/CBFβ’ is another possible target with no inhibitors discovered to date. If Vif is not blocked from functioning HIV particles are released from cells leading to ‘active infection’ and propagation of the virus to more cells (left, white and blue arrows). When Vif is blocked, A3G abundance is maintained and it is packaged into virions leading to ‘inhibited infection’ via A3G’s ability to hypermutate HIV DNA during reverse transcription leading to unintegrated and non-functional HIV DNA with dG to dA hypermutations (right, black and yellow arrows).
To counteract the robust anti-HIV activity of these A3 host sentinels, HIV has its own defense factor known as Vif (i.e., Viral infectivity factor) [25–27]. Vif recruits A3 proteins for polyubiquitination and proteasomal degradation through the E3 ubiquitin ligase complex [28, 29] (Figure 1). Vif also has been shown to impair translation of A3G by binding to mRNA [30, 31] and inhibiting A3G gene transcription by sequestering core binding factor-β (CBFβ) in the cytoplasm, which otherwise serves as a transcription cofactor of RUNX1 [32]. Given Vif’s multipronged attack on A3 proteins, Vif inhibitor compounds are of interest as potential first-in-class opportunities for therapy/cure initiatives but, as we will describe, have a high bar for establishing target selectivity and mechanism of action (MOA).
Specific protein-protein interactions (PPI) between Vif and multiple proteins (Figure 2) (Box 1) are known to be essential for Vif-dependent A3G degradation and were validated by the crystal structure of a Vif/EloBC/Cul5/CBFβ complex (PDB ID 4N9F) [33]. These interactions have been reviewed in detail elsewhere [34, 35]. Vif binding to A3G promotes polyubiquitination of A3G by acting as a SOCS-box type substrate receptor for an E3 ubiquitin ligase complex comprising Cullin5 (Cul5), Rbx2, ElonginB/C (EloBC), and CBFβ [29, 36–40]. Vif recruits A3G to the E3 ligase complex through direct and specific PPI formed with Vif and ElonginC (EloC) surface residues that in the recent crystal structure appear to be located on the opposite side of the Vif-CBFβ interface [33]. Vif’s interaction with CBFβ has been shown to be crucial in stabilizing Vif, preserving its intracellular abundance and enabling Vif’s functional interactions [39, 40].
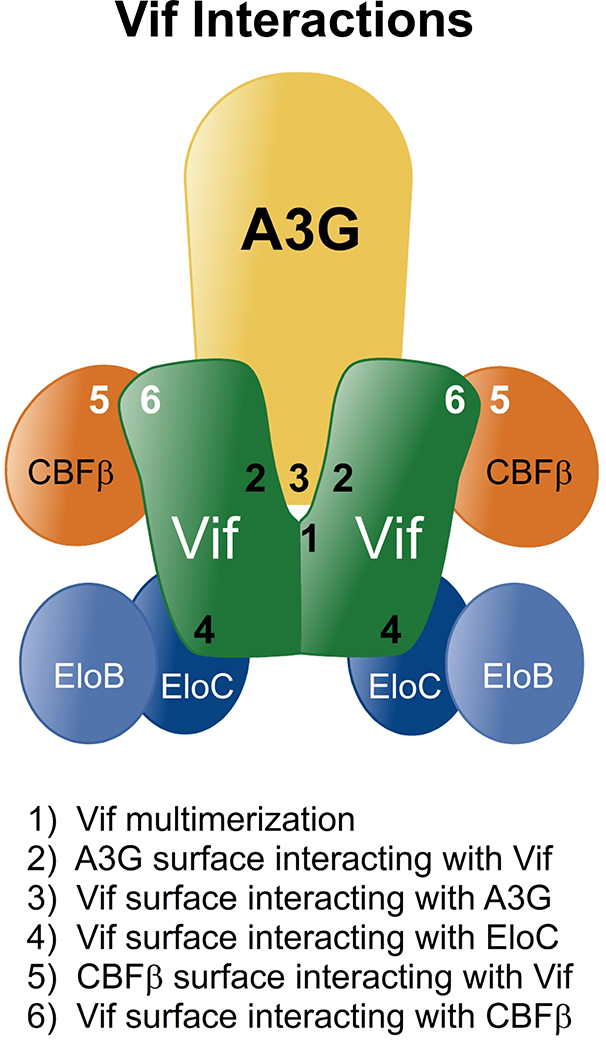
Figure 2. A conceptual model schematizing known interactions critical for Vif-dependent degradation of A3 proteins.
The interactions between Vif, A3G and E3 ubiquitin ligase complex (1 through 6) are shown for the purpose of conceptualizing the potential number of mechanisms by which small molecules might inhibit Vif function and underscore the need to use appropriate secondary assays to validate drug-target interactions and mechanism of action in lead compound development. The model should not be taken to infer that the depicted interactions co-exist at any given time in an infected cell. PPI 1–4 (black) are interactions that current small molecules are capable of disrupting. PPI 5–6 (white) are interactions that have not yet been targeted with small molecule antagonists.
Box 1. Small Molecules Have Access to Protein-Protein Interaction Interfaces.
PPI were once thought to be inaccessible as drug targets due to the amount of surface area buried at such interfaces relative to the size of small molecules that obey Lipinski’s Rule of 5 [43], but this may not be the case. The large amount of surface area involved in PPI may be dependent on a few ‘hot spot’ interactions [44–46]. Only a small subset of residues within PPI may be responsible for much of the interaction binding energy. Moreover, molecular dynamic simulations suggest that cavities and binding pockets form transiently at interfaces due to subtle movements of side chains and loops, allowing for binding of small molecules to surfaces that are not predicted in static structure models of isolated proteins or protein complexes [47, 48]. HIV entry inhibitors approved by the FDA (enfuvirtide and maraviroc) utilize allosteric binding sites to limit the conformational changes necessary for PPI between the HIV capsid protein and the CD4 receptor and CCR5/CXCR4 co-receptors [49–52]. ALLINIs are small molecules that stabilize the multimeric interface of HIV integrase with low nanomolar IC50 values in antiviral assays [53]. PPI inhibitors of human papillomavirus (E2/E1 and E6/E6AP) have been pharmacologically validated as on-target with antiviral IC50 values in the low nanomolar range [54]. Twelve small molecule therapeutics are in clinical development that target various PPI involved cancer pathways and include drugs targeting disruption of protein pairs such as MDM2-p53, tubulin α and tubulin β, BCL-2 family-BH3 domain, and IAP family-SMAC [55].
The amount of surface area that is typically buried in PPI is on average between 1200 to 2000 Å2 [45]. The complete interface between A3G and Vif and Vif multimers are not known as there are currently no co-crystal structures. However, mutational analysis of HIV-1 Vif has shown that a small pocket consisting of 5 residues in HIV Vif (40YRHHY44) is crucial for its interaction with human A3G [56]. Also, the small 161PPLP164 motif in Vif is a critical motif that affects both A3G binding and Vif multimerization [57] (detailed in Box 2). Among the known interfaces from the Vif pentameric crystal structure the Vif-EloC interface is ~715 Å2 and well below the range of typical PPI interfaces [33]. The interface between Vif and CBFβ is more than twice the typical average PPI buried surface area at ~4800 Å2 [33]. In trying to predict whether the surface area of a PPI can or cannot be disrupted by a small molecule it is important to understand how many contacts are essential for maintaining these intermolecular structures. In this regard it is of interest that HIV Vif binding to human A3G can be inhibited by mutating D128 in A3G and that natively occurring D128 prevents Vif from SIV of African green monkey (SIVagm) from binding to and degrading human A3G in HEK293T(293T) cells [58]. Overall, given the precedence of effective PPI targeting with small molecules and the evidence for A3G and Vif PPI ‘hot spot’ residues, we believe that small molecule inhibitors of Vif function targeting PPI should be aggressively pursued for therapeutic development.
The six protein interfaces that are known to affect Vif binding to A3G and lead to the reduced availability of A3G for host defense are shown in Figure 2. These are (1) the self-association region of Vif, (2) the amino acid sequences on the surface of Vif that are involved in binding to A3G, (3) the amino acid sequences on the surface of A3G that are involved in binding to Vif and (4) the amino acid sequences on Vif required to bind to EloC and Cul5 that thereby induce ubiquitination [34, 41, 42]. There are also sites on (5) CBFβ and (6) Vif involved in the interaction required to stabilize Vif in the cell [33, 39, 40]. In a small molecule targeting strategy, it is important to contemplate that there are additional opportunities for drug binding to surfaces that may be uniquely available on proteins prior to the formation of PPI or on complexes of proteins following PPI formation through induced conformational changes. The focus of this article will be on the current lead compounds reported in the literature that overcome Vif-dependent degradation of A3 proteins and enable their encapsidation in viral particles.
A Vif multimerization inhibitor enables A3 antiviral activity
Vif peptide mimetics have been reported that competitively inhibited Vif multimerization in vitro and reduced HIV replication in H9 cells, a nonpermissive human T cell line that requires Vif to counteract endogenously expressed A3G [59]. Subsequently, peptides shown to inhibit Vif multimerization in vitro [59] also prevented Vif-dependent A3G degradation and inhibited HIV infectivity in an A3G-dependent manner [60] (Box 2). This foundation motivated the development of our drug discovery program that specifically targeted Vif multimerization using cell-based fluorescence resonance energy transfer (FRET) assay in a high throughput screen (HTS). The screen resulted in the discovery and development of a small molecule, O2-16 (Figure 3, green). We evaluated the antiviral efficacy of O2-16 in PHA/IL-2 activated pooled patient peripheral mononuclear cells (PBMCs) in a 7-day infection using 16 HIV clinical isolates from 7 group M HIV subtypes as well as groups O and N. O2-16 neutralized all of the HIV isolates evaluated with an average 50% inhibitory concentration (IC50) of 170 ± 90 nM (Table 1) [61]. The compound also inhibited Vif-dependent degradation of A3G and A3F in 293T cells and enhanced recovery of A3G and A3F with viral particles in psuedotyped HIV infections, thereby enabling A3G signature hypermutations of HIV proviral DNA in the target TZM-bl cells determined by next-gen sequencing (Table 1) [61]. O2-16 had no activity against HIV Protease (PR), Integrase (IN) and RT, and a time-of-addition assay showed that the target for O2-16 was late in the viral life cycle when Vif is known to be expressed (Table 1) [61].
Box 2. The Importance of the PPLP motif in Vif for preventing A3 antiviral activity.
Predictions from molecular docking with Autodock Vina software to the Vif crystal structure suggested that the O2-16 binding site is adjacent to the highly conserved 161PPLP164 (PPLP) motif in Vif [61]. Computational docking of O2-16 considered the entire Vif surface for the lowest energy binding site for O2-16 and did not bias the search toward a specific binding region [61]. This finding is of interest as the PPLP motif was part of the most effective Vif multimerization peptide mimetics for inhibiting HIV infectivity [59, 60]. Moreover, the PPLP motif appears to be crucial for Vif self-association in both in vitro [59] and cell-based assays [57], Vif binding to A3G [57], Vif-dependent degradation of A3G [34, 62–65] and Vif binding to RNA [66]. However, the Vif crystal structure [33] does not show a direct PPLP interaction. PPLP residues were solvent-exposed, and formed a coil that is near to a Vif-Vif crystal contact comprising anti-parallel β-strands with the residues 41RHHYE45. Despite the small size of this interface, it may be part or all of a Vif dimerization interface for which PPLP may contribute in a structurally supportive role (Figure I) [34, 56]. Notably, this contact has not been evaluated for its effect on Vif dimerization and the C-terminus (174–192) of Vif was truncated prior to crystallization. The fact that these interface residues overlap with the 40YRHHY44 motif that is crucial for the interaction with A3G [34, 56] suggests the hypothesis that PPLP may have a supportive role in both Vif multimerization and A3G binding.
This hypothesis is further supported by Batisse and colleagues HeLa cell-based fluorescence correlation spectroscopy (FCS), fluorescence-lifetime imaging microscopy (FLIM) and FRET analyses [57]. With these biophysical methods EGFP-Vif and mCherry-Vif were evaluated for higher order structures they form in the cell. FCS showed that there was a heterogeneous mix of monomers and homo oligomers for wild type Vif but a Vif PPLP to AALA mutant was monomeric [57]. Co-expression of wild type Vif and A3G in cells diminished Vif homo oligomerization, but monomeric Vif AALA was unable to bind to A3G [57]. These findings support a functional overlap of regions implicated in Vif multimerization (PPLP and potentially 41RHHYE45 crystal contact) and Vif binding to A3G (PPLP and 40YRHHY44) such that Vif and A3G are may be competing for the same binding regions on Vif.
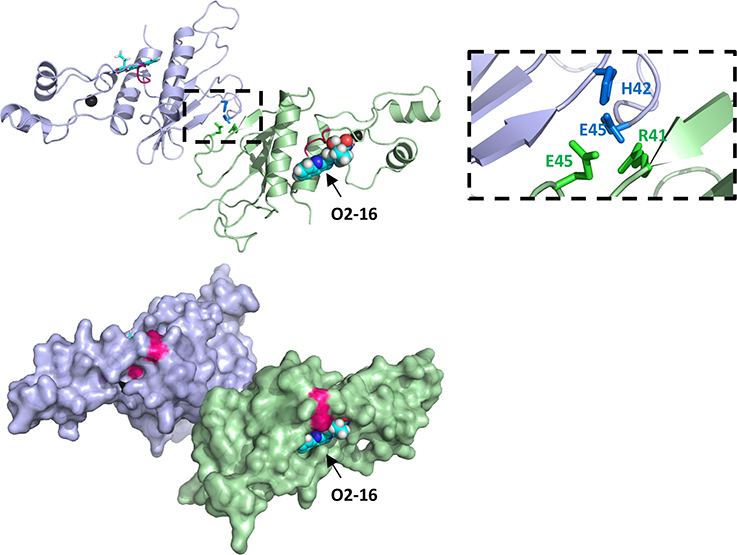
Figure I. The predicted binding site of O2-16 is adjacent to the PPLP motif.
Adjacent Vif molecules (light blue and pale green) from the asymmetric unit of the Vif/CBFβ/EloB/EloC/Cul5 pentameric complex crystal structure (PDB ID 4N9F) are shown in both cartoon and surface representation forms. The Vif-Vif interface involves an interaction between the conserved 41RHHYE45 motif and selected sidechains (sky blue and bright green) of interface residues are shown in stick format. The 161PPLP164 motif is highlighted in bright pink and is adjacent to the lowest energy binding site of O2-16 as predicted from AutoDock Vina. Zinc ions of the HCCH zinc-binding motif are shown as black spheres.
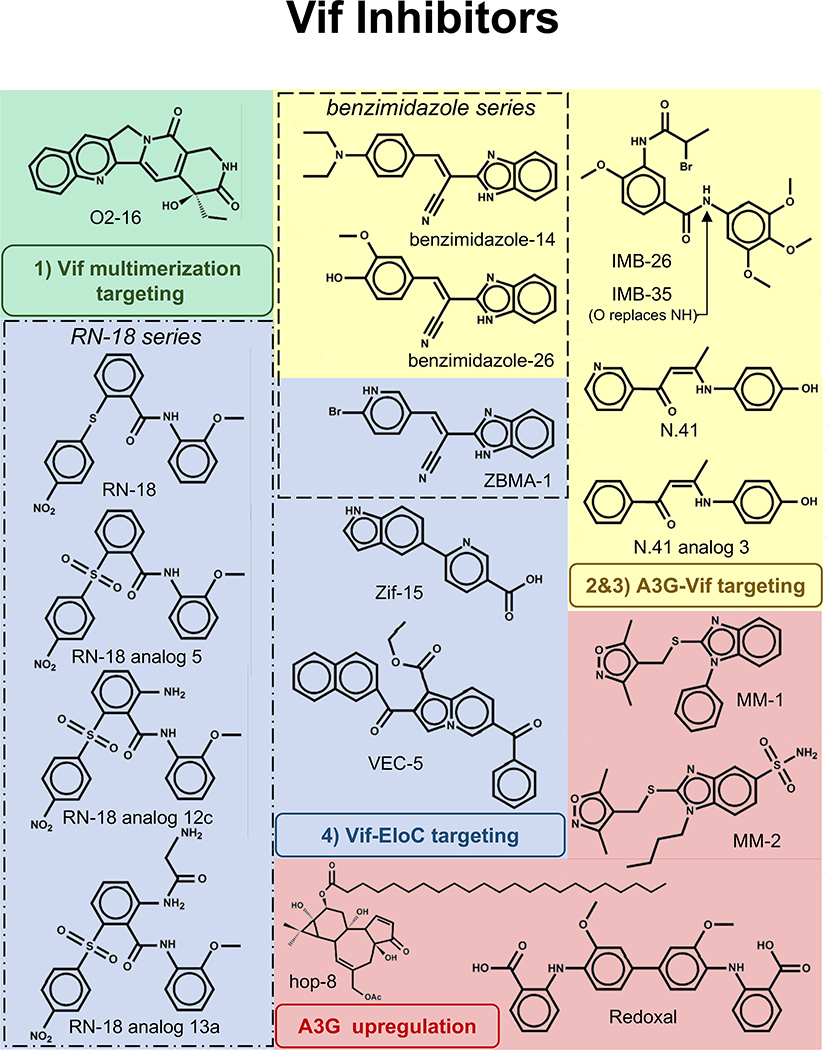
Figure 3. 2D depictions of small molecule Vif inhibitors.
Groups of compounds are color coded according to the interactions in Figure 2. These include: 1) Vif multimerization (green), 2 & 3) A3G-Vif (yellow), 4) Vif-EloC (blue) and compounds targeting A3G upregulation (red). IMB-26 and IMB-35 have similar structures but differ in that there is an O in place of NH (arrow pointing to the site of modification in IMB-35). Boxes outline the RN -18 series (dotted box) and benzimidazole series (dashed box).
Table 1.
Proposed Mechanism of Action Detail and antiviral activities for Vif Inhibitors
| Compound (publication date, reference) |
Proposed Target |
A3G Deg. |
A3F Deg. |
A3G Dependent Antiviral Activity |
More A3G in Virus |
A3G hyper mutati ons |
Modeling predictions |
reported μM IC50 (cell type tested) |
Multiple Clades Tested |
Drug Resistant Strains Tested |
Counters creens to other HIV Targets |
|---|---|---|---|---|---|---|---|---|---|---|---|
| O2-16 (December 2016, 61) | Vif Multimerization | + | + | + | + | + | Binds Vif near PPLP | 0.17 (PBMC) | 16 isolates from all HIV clades | 7 mult-/single-drug resistant strains | PR, RT, IN and time-of-addition |
| N4.1analogs (February 2015, 67) | A3G-Vif interaction | + | + | + | + | ND | ND | 4.2 (PBMC) | 4 isolates | ND | + |
| IMB-26/35 (April 2010, 68) | A3G-Vif interaction | + | ND | + | + | + | ND | 0.017 (H9)* | ND | ND | (PR, RT, IN)* |
| Benzimidazole series | |||||||||||
| analogs 14/26 (April 2015, 69) | A3G-Vif interaction | + | ND | + | ND | ND | ND | 0.004/0.05 8 (H9) | ND | ND | ND |
| ZBMA-1 (June 2015, 70) | Vif-EloC interaction | + | ND | + | ND | ND | Binds EloC near Asp 111 | 1.01 (PBMC) | ND | ND | ND |
| RN-18 series | |||||||||||
| analog 12c/13a (January 2017, 73) | Vif-EloC interaction | + | ND | ND | ND | ND | Binds BC box region of Vif | 1.54/0.25 (H9) | 4 isolates | ND | ND |
| analog 5 (May 2012, 72) | ND | + | ND | + | ND | ND | ND | 1.1 (H9) | ND | ND | ND |
| RN-18 (October 2008, 71) | ND | + | + | + | ND | ND | ND | 6.0 (H9) | ND | ND | ND |
| Other compounds | |||||||||||
| Zif-15 (July 2017, 75) | Vif-EloC interaction | + | ND | ND | + | ND | Binds BC box region of Vif | ND | ND | ND | ND |
| VEC-5 (February 2012, 74) | Vif-EloC interaction | + | + | + | ND | ND | Binds Vif binding region of EloC | 24.5 (CEM) | ND | ND | + |
| hop-8 (September 2017, 76) | A3G expression | + | ND | +/− | + | ND | ND | 0.11 (PBMC) | 3 isolates | 3 drug resistant strains, ΔVif and HIV-2 | PR, RT, IN and ENV |
| Redoxal (October 2015, 78) | A3G expression | + | ND | + | ND | ND | ND | 1.4 (PBMC) | 3 isolates | ND | + |
| MM-1/2 (July 2014, 77) | A3G expression | + | ND | + | ND | ND | ND | ND | ND | ND | ND |
Footnote: ND = not determined
claimed in report with data not shown
The PPI between A3G and Vif has potential as an antiretroviral drug discovery target
Three groups have reported compounds that affect A3G binding to Vif (Figure 1, block A3G/Vif). A target-based HTS approach was developed based on in vitro FRET assay with Vif and A3G peptides corresponding to the known Vif and A3G binding regions to identify inhibitors for Vif and A3G binding [67]. The lead compound series from the resulting screen was N.41 and its analogs (Figure 3, yellow). Through structure-activity relationship (SAR) Pery and colleagues reported that the compounds were predicted to inhibit direct contact between Vif and A3G [67]. While both Vif-dependent A3G and A3F degradation were inhibited by N.41 (Table 1), A3C, which is known to have an alternative Vif binding region, was not [67]. They also established that these compounds had A3G-dependent antiviral activity (Table 1), led to an increased A3G recovery within viral particles and had low micromolar IC50 against four different HIV isolates in PBMC (Table 1) [67]. They suggested that the compounds bound to A3G (Figure 2, #2) based on the activities of different compound analogs. Analog 4 in these studies affected A3G degradation in the region known to bind to Vif, but analog 4 also inhibited viral packaging of A3G. This suggested that there may be an overlap of A3G residues affected by analog 4 binding (i.e. 128DPD130) [67]. Notably, despite strong MOA and antiviral data, the N.41 series of compounds are enanimones that are unstable and will require structural optimization to progress as lead drug candidates [67].
The two other groups that identified potential Vif inhibitors used a fluorescent A3G degradation screen coupled with lower throughput biophysical methods and co-immunoprecipitation (co-IP) experiments to establish A3G-Vif binding as the target. Cen and colleagues identified IMB-26/35 (Figure 3, yellow) and used surface plasmon resonance (SPR) and co-IP to show that the compounds bound to A3G and prevented binding to Vif (Figure 2, #2). However, both experiments were single data point analyses so more in-depth analysis will be required for confirmation that the IMB-26/35 antiviral mechanism relied on direct A3G binding [68]. They convincingly showed that the antiviral activity of IMB-26/35 i) was A3G-dependent determined by antiviral activity being present in A3G expressing H9 cells but not in A3G lacking SupT1 cells, ii) promoted increased recovery of A3G in viral particles revealed through western blot analyses, iii) and caused A3G signature hypermutations in the HIV genome observed through sequencing analysis of infected TZM-bl cells (Table 1) [68]. Moreover, their compound had low cytotoxicity in mice based on their being no effect on weight or hepatic and renal function when mice were given high doses of the compound [68]. They also tested IMB-26/35 against a single HIV isolate (BH10) and reported low nanomolar (17 nM) IC50 (Table 1).
Benzimidazole analogs 14 and 26 (Figure 3, yellow) have been identified as promising compounds that had a similar experimental profile to IMB-26/35 [69]. These compounds also appeared to prevent A3G degradation and A3G-dependent antiviral activity (Table 1) with low cytotoxicity in mice [69]. Low nanomolar IC50 in H9 cells was also reported using one viral isolate (Table 1) [69]. This study also used SPR and co-IP in transfected 293T cells to show that benzimidazole analogs 14 and 26 bound to A3G or Vif to block the interaction (Figure 2, #2 or #3), but which protein the compound interacted with could not be distinguished from the studies [69].
Vif-EloC inhibitors are antiviral and prevent A3 degradation
ZBMA-1 has been reported as a compound that inhibited Vif binding to EloC (Figure 1, block Vif/EloC); [69, 70]. Notably, ZBMA-1 is also in the benzimidazole series of compounds (Figure 3, dashed box) and was discovered using the same experimental design as used in the discovery of benzimidazole analogs 14 and 26 (Tables 1) [69, 70]. The two major differences are that: i) the reported IC50 for ZBMA-1 was markedly higher (low micromolar range (1 μM) as opposed to low nanomolar for analogs 14 (4 nM) and 26 (58 nM)) (Table 1) [69, 70], and ii) ZBMA-1 inhibited Vif binding to EloC and did not affecting Vif binding to A3G [70]. Computational modeling has docked ZBMA-1 onto the Vif crystal structure with EloBC, CBFβ, and Cul5 [33, 70] and predicted hydrogen bonding between Asp111 of EloC and the NH on the pentameric ring of ZBMA-1 (Figure 3, blue). Mutation of Asp111 of EloC to either Leu or Ala prevented ZBMA-1 inhibition of Vif-EloC interaction in co-IP studies in 293T cells [70]. Though not tested for activity on Vif-EloC complexes, benzimidazole analogs 14 and 26 also have an NH on the pentameric ring (Figure 3, dashed box) and therefore could be active on the same target [70]. Given the proposed MOA for ZBMA-1, a follow up binding study for the more efficacious analogs 14 and 26 would be illuminating.
Other compounds that have been identified by high throughput screening are in the RN-18 series (Figure 3, dotted box). RN-18 was the first reported small molecule Vif inhibitor in the literature [71]. In the original study, RN-18 was identified as the lead inhibitor from a 293T cell-based screen for Vif-dependent degradation of fluorescently labeled A3G, similar to the screens that identified IMB-26/35 and the benzimidazole series [71]. A subsequent study used SAR and identified analog 5, which showed a modest improvement in IC50 compared to RN-18 [72] (Table 1). Both reports showed A3G-dependent antiviral activity by comparing infectivity in non-permissive (i.e. A3G expressing) cells (H9 or CEM) and in permissive (i.e. A3G lacking) cells (MT-4 or CEM-SS), but they did not confirm MOA, test multiple clades for antiviral activity, or counterscreen for activity with other HIV targets (Table 1) [71, 72].
RN-18 has been further developed through medicinal chemistry and SAR studies on 33 different analogs [73]. Docking studies suggested that the RN-18 analogs bind to Vif in a pocket within the Vif-EloC interface in the Vif BC-box region (a.a. 139–158) [73]. RN-18 analogs 12c/13a had mid nanomolar to low micromolar IC50 values (0.25–1.54 μM) for antiviral activity in H9 cells when tested against four different isolates of HIV (Table 1) [73]. Notably, there was broad inter-lab variability of the IC50 range reported for the parental RN-18 compound (6 μM-263 μM). It is unclear why there is such a broad difference since both labs used H9 cells and the NL4-3 isolate for HIV infectivity [71–73]. The major difference seems to be in infectivity end points; a 48 h infection was used in the first two studies [71, 72] while a 7-day infection was used in the last study [73]. Perhaps RN-18 stability was reduced in the longer study and caused the reduced efficacy. A follow up study on RN-18 stability compared to the newer analogs could shine light on the discrepancy.
Virtual screens probing the Vif-EloC binding pocket (i.e. the region around Vif’s BC box, 144SLQ[Y/F]LAL150) have identified two other potential Vif-EloC targeting compounds (Figure 2, #4). Prior to the Vif pentameric crystal structure [33], homology modeling and a virtual screen was used to identify VEC-5 (Figure 3, blue) [74]. VEC-5 antiviral activity was only mid micromolar (25 μM) in CEM cells (Table 1), but the MOA was confirmed with multiple methods (e.g. A3G-dependent antiviral activity, co-IPs, viral packaging and A3G degradation assays in 293T cells) (Table 1) [74]. Recently, Zif-15 (Figure 3, blue) was identified from a structure-based virtual screen using the Vif crystal structure to model the binding region between Vif and EloC [75]. However, MOA confirmation was limited to Vif-dependent A3G degradation and antiviral mechanistic details were lacking for Zif-15 (Table 1) [75]. ZBMA-1 and VEC-5 were predicted to bind to EloC whereas RN-18 analogs and Zif-15 were predicted to bind in the consensus BC-box region of Vif that directly binds to EloC (Table 1). Overall, none of the Vif-EloC targeting compounds have particularly low antiviral IC50 values. This could be due to the lack of specificity for Vif relative to other BC-box containing proteins in cells that may be competing for the compound binding. The clarity from the co-crystal structure on the Vif-EloC interaction and the fact that modeling could predict hits on this target make it worthwhile to pursue, yet counterscreens to other BC-box containing proteins would be a crucial part of any SAR efforts to develop leads (Box 3) to eliminate compounds that generally target the E3 ubiquitin ligase system and focus on leads that are more on-target for Vif specifically.
Box 3. Preclinical Hurdles to Lead Development.
In preclinical drug development, there are multiple hurdles to overcome even after compounds are identified with a verifiable MOA and low or sub nanomolar antiviral activity against multiple clades. The first hurdle would be absorption, distribution, metabolism, excretion and toxicity (ADMET) along with pharmacokinetics (PK) studies in mice or rats. Many promising compounds can be thwarted because they are quickly metabolized in plasma. Establishing a strong in vivo half-life and maximum plasma concentration are required for a preclinical lead.
Compounds that impair the function of essential viral proteins are likely, under appropriate conditions, to select for drug resistance engendering HIV mutations. To date, all antiretroviral (ARV) therapies targeting HIV proteins have demonstrated this characteristic as part of their preclinical development. Drug resistant Vif mutations (no longer bind to compounds but persist in Vif-dependent A3G degradation) have proven difficult to identify [83]. This may be anticipated given that it is the abundance and function of multiple host A3 proteins and not the viral Vif protein that is ultimately the determining factor for antiviral Vif inhibitors. Vif mutations within critical residues in PPI, that also impair drug binding, may also prevent inhibition of A3 by Vif and thereby reduce viral infectivity. A drug-resistant mutation in Vif may have to be a gain of function mutation that perhaps enables additional residues to support Vif PPI and functionality.
Solid ADMET, PK, in vivo efficacy, and drug resistance data will require large scale synthesis of the lead compound and testing a full panel of HIV clades and drug resistant strains in an acute infection of PBMC treated over a dose range. Drug combination studies in PBMC with various ARV therapies (e.g. NNRTI, NRTI, PI, INI) to evaluate antagonistic or synergistic antiviral effects as recommended by the FDA guidelines are also key datasets required before clinical trials. Therefore, although many avenues exist to clinical trials for a Vif inhibitor as a novel ARV with therapeutic and curative potential, the process is very much in the early stages of preclinical development.
Since 2008 there has been an emergence of compounds in the literature identified as affecting the A3 and Vif antiviral pathway. All current modalities inhibiting Vif function seek to protect A3 proteins from Vif-dependent degradation and thereby neutralize HIV through catastrophic hypermutation of the viral genome. This approach of enabling innate immunity to inhibit HIV is unique and might conceptually afford this therapeutic approach a lower probability that drug-resistance will emerge.
A3G upregulators are antiviral but have off-target effects
Three compounds were identified that upregulated A3G expression by western blot analysis of A3G protein [76–78]. However, these compounds did not inhibit A3G binding to Vif in 293T cell-based transfection models [76–78]. Conceptually these compounds may enhance A3 innate immunity through mass action that overwhelms the ability of Vif to clear A3 from the cell. Higher expression of A3G mRNA has been correlated with reduced viremia and increased CD4+ cell counts in patients known to be long term nonprogressors and elite controllers [1–5]. All three compounds increased expression of A3G protein and increased recovery of A3G with viral particles despite the presence of Vif in 293T cells (Table 1) [76–78]. However, all compounds had off-target issues that mitigate enthusiasm for their further development. Compounds MM-1 and MM-2 (Figure 3, red) have been identified as A3G upregulators, but their effective dose was too close to their toxic dose in 293T cells and there was limited analysis of MOA (Table 1), thus further SAR will be necessary [77]. Redoxal (Figure 3, red), a pyrimidine biosynthesis inhibitor, has been identified as a stabilizer of A3G expression [78]. The antiviral activity of Redoxal was in the low micromolar range against three HIV isolates (Table 1), though the mechanism by which reducing nucleoside pools affected A3G stabilization was not determined [78].
A phorbol ester, hop-8 (Figure 3, red), also has been identified as a potent inhibitor of replication of wild-type HIV-1 and HIV-2 strains and drug-resistant strains tested in C8166 (human T cell leukemia) cells and PBMC [76]. Hop-8 had an IC50 value ranging from 0.1 μM to 8 μM [76] and produced low cytotoxicity (Table 1). HIV proteins PR, IN, RT and envelope (ENV) were ruled out as being targeted by hop-8 in tests using commercially available in vitro kits. Although hop-8 antiviral activity positively correlated with increasing A3G expression, its antiviral activity was not strictly Vif-dependent as evidenced by hop-8 antiviral activity in C8166 cells (which have low expression of A3G) infected with HIV lacking Vif expression [76]. The authors speculated that phorbol ester mediated downregulation of CD4 at the cell surface may have contributed to the antiviral effect [76]. The authors also pointed out that the therapeutic use of hop-8 may be limited by the high tumorigenicity of phorbol esters [76].
Overall, there are two potentially major drawbacks to compounds that induce overexpression of A3 proteins. First it is not clear whether A3G expression can be selectively increased without collateral effects on off-target protein expression. Secondly, uncontrolled A3 activity through increased expression may pose a risk for undesirable effects on mRNA editing, mRNA translation and genotoxic hypermutations. A3G has been shown to counteract micro-RNA (miRNA) mediated repression of protein translation [79], thus increased A3G expression could have downstream effects related to epigenetic regulatory pathways in the cell. Although under physiological conditions, A3G genotoxicity is mitigated by the cytoplasmic retention of A3G and A3F [80], unanticipated upregulation of the expression of other members of the family that are naturally localized in the cell nucleus such as A3A or A3B could pose an increased risk for genotoxicity and cancer [81]. Any future development of these compounds (Box 3) must vet them with appropriate counterscreens for off-target mRNA expression and genotoxicity.
Although A3 are expressed in all mammals and SIV produces Vif, there are species-specific structural requirements for HIV Vif binding to human A3 and therefore the PPI and drug binding requirements for an HIV therapeutic may not be testable in all animal models. Therefore, the second hurdle is to establish whether in vitro efficacy translates to an in vivo model. Viruses lacking functional Vif are unable to propagate in multiple humanized mouse models, making this perhaps an ideal in vivo model to test an inhibitor of HIV Vif that is A3-dependent [7, 8, 82]. Notably, human T-cells produced in hu-NOG mice expressed A3 with comparable abundance to that observed for endogenously expressed A3G/A3F in humans and HIV infection was absolutely dependent on the expression of functional HIV Vif [7, 82].
Concluding Remarks
There have been some reports that question whether A3G expressed in the HIV-positive patient population will have sufficient antiviral activity to inhibit viral replication. Studies have shown that conditional selection pressure can cause A3G to promote the emergence of drug resistance [84, 85]. If Vif inhibitors were only partially effective, residual Vif function might continue to induce degradation of A3G below an abundance enabling hypermutation activity to the necessary level to completely inactivate HIV but sufficient to drive the emergence of strains resistant to other ARVs [86]. However, it has been reported from in vitro 293T cell-based studies that a single A3G molecule per HIV virion was sufficient to induce hypermutation frequencies comparable to inactivating hypermutation levels found in patients [87]. The level of hypermutation observed in vitro and in vivo correlated to a level of ~10-fold greater than the level calculated in silico to be necessary for mutation frequencies to be sub-lethal [87].
We do not know what level of A3G mutagenic activity is necessary and sufficient to extinguish the virus. An estimate for the in vivo mutation frequency of error-prone RT is 1.8 × 10−6/bp/cycle [88], which may only need to be increased 3- to 4-fold before the mutational events reach a saturation and distribution to neutralize HIV [89]. Whether the necessary increase in mutations over the existing RT mutation rate will be sufficient for lethal viral mutations and whether A3G can catalyze this level of mutation in the presence of a Vif inhibitor remains to be determined empirically. It is our position that the development of highly effective Vif inhibitors that enable catastrophic hypermutation of HIV by preserving native expression of A3 be aggressively pursued. Such compounds may enable innate immunity and therein be a novel strategy to add to the current arsenal of ARV. We maintain that Vif inhibitors could have a significant role in ‘cure’ strategies for eradicating latent viral reservoirs.
Currently O2-16 is the most advanced published lead in this novel drug category. O2-16 is broadly neutralizing of HIV of numerous tested HIV clades and drug-resistant strains (Table 1) and at present has the most extensive validation of MOA (Table 1). The IC50 of O2-16 is only 170 nM and therefore will need optimization to low or sub nanomolar range before the preclinical hurdles in Box 3 are addressed. ZBMA-1 and RN-18 analogs 12c/13a will also need further SAR to reduce IC50 values as well. Other compounds like IMB-26/35 and benzimidazole analog 14 have demonstrated low nanomolar activity against HIV and are promising candidates for development. However, both scaffolds need to be tested against multiple clades and drug resistant isolates. Furthermore, the benzimidazole series has multiple candidate Vif interactions (Figures 2 & 3) and confirmation of the PPI affected needs clarification. We think that the breadth of leads on multiple targets suggest that low nanomolar antivirals targeting the Vif/A3 pathway are achievable. Barring success in overcoming preclinical hurdles described in Box 3, in 3–10 years we anticipate at least one if not multiple Vif inhibitors in clinical trials. If such compounds reach clinical trials the data will reveal whether Vif inhibitors have therapeutic potential in treatment, prevention and cure for HIV.
Box 4. Clinicians Corner.
Although some researchers argue there is no pressing need for new targets for HIV antiviral drug development, history teaches us that the eventual emergence of drug resistance to microbicides is a certainty, although unpredictable in its timing.
For those living with HIV, one cannot ignore the opportunity for a novel therapeutic and functional cure afforded by fully enabling innate immunity through A3 proteins.
Disabling Vif places antiviral agents, A3 proteins, within viral particles before they leave an infected cell. This renders virions incapable of productive infections and we predict, will reduce the ability of HIV to establish latent viral reservoirs. Due to the MOA of this novel antiviral approach, Vif inhibitors may reduce the reliance on, or the doses required for current treatment ARV regiments.
We predict this to be true based on experimental cell and animal model systems, but proof of concept in patients will depend on the outcome of Vif inhibitors in clinical testing, which we estimate to be initiated within the next 3–10 years.
Open Questions.
Are there small molecule compounds that will inhibit the interaction between Vif and CBFβ and not prevent the function of CBFβ in gene transcription?
Are there small molecule compounds that will selectively inhibit the interaction between Vif and EloC without affecting the proteosomal degradation of other proteins with BC box motifs?
What are the macromolecular binding requirements of the benzimidazole series of compounds and is there a chemical reactivity that explains for differences in the effects of ZBMA-1 and analogs 14 and 26 on Vif?
Will Vif inhibitors enable A3 hypermutation sufficient to neutralize all strains and clades of HIV in patients?
Can low nanomolar inhibitors of Vif PPI be developed with drug-like properties (e.g. ADMET, PK, in vivo efficacy) and sufficient efficacy for viral suppression with once-a-day dosing?
Can Vif inhibitors be part of a solution addressing the long-standing need for novel approaches to cure HIV?
Highlights.
Both model systems and clinical data suggest that the interactions between HIV Vif and APOBEC3 innate immunity are critical for HIV infection and have antiretroviral therapeutic potential.
There are multiple protein-protein interactions required for HIV Vif to block APOBEC3 antiviral activity.
Diverse small molecule Vif inhibitors targeting different interactions establish proof-of-concept for antiviral sites for drug development.
Vif inhibitors could have the potential to enhance HIV treatment options and might be part of future prevention and cure initiatives.
Acknowledgments
This work was supported by PHS grants R21NS067671, R21AI095007, R21AI122845, R01GM110038, R01GM110568 and a Bill and Melinda Gates Fund Grand Challenges Explorations Grant 51715 awarded to HCS.
Glossary
- Long-term non-progressors
A subset of HIV-infected persons having prolonged elevation in CD4 cell counts for many years in the absence of treatment.
- Elite controllers
A subset of HIV-infected persons who can spontaneously control plasma viral load without treatment.
- Hypermutation
a form of gene editing in which a multitude of nucleotides on the same strand of DNA are changed.
- APOBEC3 (A3)
A family of seven deaminase enzymes in humans that act as cellular sentinels targeting foreign (e.g. viruses) and endogenous (e.g. retroelements) DNA to protect the genome through innate immunity.
- Viral infectivity factor (Vif)
An HIV accessory protein that is essential for viral replication in cells expressing A3 proteins whose mechanism of action is to prevent A3 assembly with nascent viral particles through targeted degradation of A3 by the proteasome.
- E3 ubiquitin ligase complex
A highly diverse class of ubiquitin conjugating enzymes (ligases) that are assembled on a Cullin protein scaffold, which binds to a RING-box protein at its N-terminus (e.g. Rbx2), an adaptor protein (e.g. EloBC) and a substrate receptor at its C-terminus (in this case, Vif).
- Core binding factor-β (CBFβ)
A transcription co-factor that interacts with the transcription factor RUNX-1 to regulate gene expression leading to the differentiation of hematopoietic stem cells into mature blood cells, but also serves as a molecular chaperone for Vif ensuring its proper protein folding within the cell.
- ALLINIs
Allosteric inhibitors of HIV integrase that disrupt both integrase multimerization and its interaction with co-factor LEDGF/p75.
- Fluorescence resonance energy transfer (FRET)
A process involving an energy donor and acceptor chromophore in which the excitation of the donor’s energetic state is transferred to an acceptor within an appropriate proximity (a FRET pair). Proteins tagged with FRET pairs can be used to assess their interactions through FRET signal intensity.
- High throughput screen (HTS)
A cell-based or in vitro method by which an optimized automation process with robotic liquid handlers and plate readers allows for rapid testing of a large library of small molecules for relevant interactions or effects.
- Fluorescence correlation spectroscopy (FCS)
A correlation analysis of fluctuation of the fluorescence intensity in a defined space (e.g. cytoplasm). The analysis gives the average number of fluorescent molecules and average diffusion time, when the molecule is passing through the space to determine both its concentration and size.
- Fluorescence-lifetime imaging microscopy (FLIM)
An imaging technique based on the differences in the exponential decay rate of the fluorescent molecules. In FRET assays energy transfer from the donor molecule to the acceptor molecule will decrease the lifetime of the donor. FLIM-based FRET measurements are insensitive to the concentration of fluorophores and can thus filter out artifacts introduced by variations in the concentration and emission intensity across the sample.
- Enanimones
Any compound having a carbonyl group adjacent to the double bond of an enamine. An enamine is an unsaturated compound produced by condensing an aldehyde or ketone with a secondary amine. They have been shown to have instability in aqueous and acidic environments.
- Structure-activity relationship (SAR)
The analysis of the relationship between chemical structure and its impact on a desired biological activity which seeks to determine the underlying chemical groups responsible for evoking a specific biological effect.
- Surface plasmon resonance (SPR)
A biophysical measurement of binding between a ligand (e.g. small molecule) flowed over a substrate covalently linked to a metal chip. Ligand binding causes electro-magnetic surface changes that are measured by a sensor that detects light reflected off the metal chip.
- hu-NOG mice
A humanized mouse model that uses cord blood hCD34+ stem cells to reconstitute a human immune system in immune compromised mice. This model has been shown to contain human T cells that endogenously expressed human A3G/A3F at levels comparable to that observed in humans and where HIV infection is absolutely dependent on functional Vif for sustaining an HIV infection.
Footnotes
Conflict of Interest Statement
H.C.S. is a professor at the University of Rochester School of Medicine and Dentistry. He is also the founder and CEO of the University of Rochester spin-out company OyaGen, Inc. The company has a financial interest in the development of antiviral and anticancer drugs based on APOBEC technology. R.P.B. and J.D.S. are employees of OyaGen, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kikuchi T, et al. Anti-APOBEC3G Activity of HIV-1 Vif Protein Is Attenuated in Elite Controllers. J Virol. 2015;89(9):4992–5001. doi: 10.1128/JVI.03464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Pasquale M, et al. Lower HIV provirus levels are associated with more APOBEC3G protein in blood resting memory CD4+ T lymphocytes of controllers in vivo. PLoS One. 2013;8(10):e76002. doi: 10.1371/journal.pone.0076002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kourteva Y, et al. APOBEC3G expression and hypermutation are inversely associated with human immunodeficiency virus type 1 (HIV-1) burden in vivo. Virology. 2012;430(1):1–9. doi: 10.1016/j.virol.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rangel HR, et al. Deletion, insertion and stop codon mutations in vif genes of HIV-1 infecting slow progressor patients. J Infect Dev Ctries. 2009;3(7):531–8. doi: 10.3855/jidc.471. [DOI] [PubMed] [Google Scholar]
- 5.Jin X, et al. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J Virol. 2005;79(17):11513–6. doi: 10.1128/JVI.79.17.11513-11516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krupp A, et al. APOBEC3G polymorphism as a selective barrier to cross-species transmission and emergence of pathogenic SIV and AIDS in a primate host. PLoS Pathog. 2013;9(10):e1003641. doi: 10.1371/journal.ppat.1003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato K, et al. APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. PLoS Pathog. 2014;10(10):e1004453. doi: 10.1371/journal.ppat.1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krisko JF, et al. HIV restriction by APOBEC3 in humanized mice. PLoS pathogens. 2013;9(3):e1003242. doi: 10.1371/journal.ppat.1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stavrou S, et al. Different modes of retrovirus restriction by human APOBEC3A and APOBEC3G in vivo. PLoS Pathog. 2014;10(5):e1004145. doi: 10.1371/journal.ppat.1004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. The Journal of biological chemistry. 2004;279(33):34083–6. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- 11.Cen S, et al. The interaction between HIV-1 Gag and APOBEC3G. J Biol Chem. 2004;279(32):33177–84. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- 12.Apolonia L, et al. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog. 2015;11(1):e1004609. doi: 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan MA, et al. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svarovskaia ES, et al. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;279(34):35822–8. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 15.Wang T, et al. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81(23):13112–24. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.York A, et al. The RNA Binding Specificity of Human APOBEC3 Proteins Resembles That of HIV-1 Nucleocapsid. PLoS Pathog. 2016;12(8):e1005833. doi: 10.1371/journal.ppat.1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangeat B, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424(6944):99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 18.Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113(6):803–9. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 19.Guo F, et al. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J Virol. 2006;80(23):11710–22. doi: 10.1128/JVI.01038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li XY, et al. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J Biol Chem. 2007;44:32065–74. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- 21.Ara A, et al. Mechanism of Enhanced HIV Restriction by Virion Coencapsidated Cytidine Deaminases APOBEC3F and APOBEC3G. J Virol. 2017;91(3) doi: 10.1128/JVI.02230-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollpeter D, et al. Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G. Nat Microbiol. 2018;3(2):220–233. doi: 10.1038/s41564-017-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaipan C, et al. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J Virol. 2013;87(1):444–53. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li MM, et al. The range of human APOBEC3H sensitivity to lentiviral Vif proteins. J Virol. 2010;84(1):88–95. doi: 10.1128/JVI.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher AG, et al. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237(4817):888–93. doi: 10.1126/science.3497453. [DOI] [PubMed] [Google Scholar]
- 26.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 27.Strebel K, et al. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328(6132):728–30. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 28.Stopak K, et al. HIV-1 Vif Blocks the Antiviral Activity of APOBEC3G by Impairing both Its Translation and Intracellular Stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302(5647):1056–60. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 30.Mercenne G, et al. HIV-1 Vif binds to APOBEC3G mRNA and inhibits its translation. Nucleic Acids Res. 2010;38(2):633–46. doi: 10.1093/nar/gkp1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerrero S, et al. Translational regulation of APOBEC3G mRNA by Vif requires its 5′UTR and contributes to restoring HIV-1 infectivity. Sci Rep. 2016;6:39507. doi: 10.1038/srep39507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson BD, Harris RS. Transcriptional regulation of APOBEC3 antiviral immunity through the CBF-beta/RUNX axis. Sci Adv. 2015;1(8):e1500296. doi: 10.1126/sciadv.1500296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y, et al. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505(7482):229–33. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 34.Salter JD, et al. Structural insights for HIV-1 therapeutic strategies targeting Vif. Trends Biochem Sci. 2014;39(9):373–80. doi: 10.1016/j.tibs.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barraud P, et al. Advances in the structural understanding of Vif proteins. Curr HIV Res. 2008;6(2):91–9. doi: 10.2174/157016208783885056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehle A, et al. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J Biol Chem. 2006;281(25):17259–65. doi: 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- 37.Mehle A, et al. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18(23):2861–6. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hultquist JF, et al. Vif Proteins of Human and Simian Immunodeficiency Viruses Require Cellular CBFbeta To Degrade APOBEC3 Restriction Factors. J Virol. 2012;86(5):2874–7. doi: 10.1128/JVI.06950-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jager S, et al. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012;481(7381):371–5. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, et al. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012;481(7381):376–9. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 41.Aydin H, et al. Structure-guided analysis of the human APOBEC3-HIV restrictome. Structure. 2014;22(5):668–84. doi: 10.1016/j.str.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 42.Letko M, et al. Identification of the HIV-1 Vif and Human APOBEC3G Protein Interface. Cell Rep. 2015;13(9):1789–99. doi: 10.1016/j.celrep.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipinski CA, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 44.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267(5196):383–6. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 45.Moreira IS, et al. Hot spots--a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68(4):803–12. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 46.Keskin O, et al. Hot regions in protein--protein interactions: the organization and contribution of structurally conserved hot spot residues. J Mol Biol. 2005;345(5):1281–94. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 47.Brown SP, Hajduk PJ. Effects of conformational dynamics on predicted protein druggability. ChemMedChem. 2006;1(1):70–2. doi: 10.1002/cmdc.200500013. [DOI] [PubMed] [Google Scholar]
- 48.Eyrisch S, Helms V. Transient pockets on protein surfaces involved in protein-protein interaction. J Med Chem. 2007;50(15):3457–64. doi: 10.1021/jm070095g. [DOI] [PubMed] [Google Scholar]
- 49.Veljkovic N, et al. Preclinical discovery and development of maraviroc for the treatment of HIV. Expert Opin Drug Discov. 2015;10(6):671–84. doi: 10.1517/17460441.2015.1041497. [DOI] [PubMed] [Google Scholar]
- 50.Zhan P, et al. Targeting protein-protein interactions: a promising avenue of anti-HIV drug discovery. Curr Med Chem. 2010;17(29):3393–409. doi: 10.2174/092986710793176357. [DOI] [PubMed] [Google Scholar]
- 51.Labrecque J, et al. HIV-1 entry inhibition by small-molecule CCR5 antagonists: a combined molecular modeling and mutant study using a high-throughput assay. Virology. 2011;413(2):231–43. doi: 10.1016/j.virol.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 52.Zhou G, Chu S. Discovery of small molecule fusion inhibitors targeting HIV-1 gp41. Curr Pharm Des. 2013;19(10):1818–26. doi: 10.2174/1381612811319100006. [DOI] [PubMed] [Google Scholar]
- 53.Sharma A, et al. A new class of multimerization selective inhibitors of HIV-1 integrase. PLoS Pathog. 2014;10(5):e1004171. doi: 10.1371/journal.ppat.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D’Abramo CM, Archambault J. Small molecule inhibitors of human papillomavirus protein - protein interactions. Open Virol J. 2011;5:80–95. doi: 10.2174/1874357901105010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nero TL, et al. Oncogenic protein interfaces: small molecules, big challenges. Nat Rev Cancer. 2014;14(4):248–62. doi: 10.1038/nrc3690. [DOI] [PubMed] [Google Scholar]
- 56.Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol. 2007;81(15):8201–10. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Batisse J, et al. APOBEC3G impairs the multimerization of the HIV-1 Vif protein in living cells. J Virol. 2013;87(11):6492–506. doi: 10.1128/JVI.03494-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu H, et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc Natl Acad Sci U S A. 2004;101(15):5652–7. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang B, et al. Potent suppression of viral infectivity by the peptides that inhibit multimerization of human immunodeficiency virus type 1 (HIV-1) Vif proteins. J Biol Chem. 2003;278(8):6596–602. doi: 10.1074/jbc.M210164200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller JH, et al. The dimerization domain of HIV-1 viral infectivity factor Vif is required to block virion incorporation of APOBEC3G. Retrovirology. 2007;4:81. doi: 10.1186/1742-4690-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett RP, et al. An analog of camptothecin inactive against Topoisomerase I is broadly neutralizing of HIV-1 through inhibition of Vif-dependent APOBEC3G degradation. Antiviral Res. 2016;136:51–59. doi: 10.1016/j.antiviral.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bergeron JR, et al. The SOCS-box of HIV-1 Vif interacts with ElonginBC by induced-folding to recruit its Cul5-containing ubiquitin ligase complex. PLoS Pathog. 2010;6(6):e1000925. doi: 10.1371/journal.ppat.1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donahue JP, et al. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology. 2008;377(1):49–53. doi: 10.1016/j.virol.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolfe LS, et al. Dissection of the HIV Vif interaction with human E3 ubiquitin ligase. J Virol. 2010;84(14):7135–9. doi: 10.1128/JVI.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang S, et al. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J Biol Chem. 2001;276(7):4889–93. doi: 10.1074/jbc.M004895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernacchi S, et al. Importance of the proline-rich multimerization domain on the oligomerization and nucleic acid binding properties of HIV-1 Vif. Nucleic Acids Res. 2011;39(6):2404–15. doi: 10.1093/nar/gkq979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pery E, et al. Identification of a novel HIV-1 inhibitor targeting Vif-dependent degradation of human APOBEC3G protein. J Biol Chem. 2015;290(16):10504–17. doi: 10.1074/jbc.M114.626903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cen S, et al. Small molecular compounds inhibit HIV-1 replication through specifically stabilizing APOBEC3G. J Biol Chem. 2010;285(22):16546–52. doi: 10.1074/jbc.M109.085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pan T, et al. Development of benzimidazole derivatives to inhibit HIV-1 replication through protecting APOBEC3G protein. Eur J Med Chem. 2015;95:500–13. doi: 10.1016/j.ejmech.2015.03.050. [DOI] [PubMed] [Google Scholar]
- 70.Zhang S, et al. Identification of an HIV-1 replication inhibitor which rescues host restriction factor APOBEC3G in Vif-APOBEC3G complex. Antiviral Res. 2015;122:20–7. doi: 10.1016/j.antiviral.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 71.Nathans R, et al. Small-molecule inhibition of HIV-1 Vif. Nat Biotechnol. 2008;26(10):1187–92. doi: 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohammed I, et al. SAR and Lead Optimization of an HIV-1 Vif-APOBEC3G Axis Inhibitor. ACS Med Chem Lett. 2012;3(6):465–469. doi: 10.1021/ml300037k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou M, et al. Synthesis, biological evaluation and molecular docking study of N-(2-methoxyphenyl)-6-((4-nitrophenyl)sulfonyl)benzamide derivatives as potent HIV-1 Vif antagonists. Eur J Med Chem. 2017;129:310–324. doi: 10.1016/j.ejmech.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 74.Zuo T, et al. Small-molecule inhibition of human immunodeficiency virus type 1 replication by targeting the interaction between Vif and ElonginC. J Virol. 2012;86(10):5497–507. doi: 10.1128/JVI.06957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pu C, et al. Design, synthesis and biological evaluation of indole derivatives as Vif inhibitors. Bioorg Med Chem Lett. 2017;27(17):4150–4155. doi: 10.1016/j.bmcl.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 76.Chen H, et al. Anti-HIV Activities and Mechanism of 12-O-Tricosanoylphorbol-20-acetate, a Novel Phorbol Ester from Ostodes katharinae. Molecules. 2017;22(9) doi: 10.3390/molecules22091498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsui M, et al. Small molecules that inhibit Vif-induced degradation of APOBEC3G. Virol J. 2014;11:122. doi: 10.1186/1743-422X-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pery E, et al. Redoxal, an inhibitor of de novo pyrimidine biosynthesis, augments APOBEC3G antiviral activity against human immunodeficiency virus type 1. Virology. 2015;484:276–87. doi: 10.1016/j.virol.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu C, et al. APOBEC3G inhibits microRNA-mediated repression of translation by interfering with the interaction between Argonaute-2 and MOV10. J Biol Chem. 2012;287(35):29373–83. doi: 10.1074/jbc.M112.354001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Salter JD, et al. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem Sci. 2016;41(7):578–594. doi: 10.1016/j.tibs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Henderson S, Fenton T. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol Med. 2015;21(5):274–84. doi: 10.1016/j.molmed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 82.Sato K, et al. Remarkable lethal G-to-A mutations in vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice. J Virol. 2010;84(18):9546–56. doi: 10.1128/JVI.00823-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Albin JS, et al. Long-term restriction by APOBEC3F selects human immunodeficiency virus type 1 variants with restored Vif function. J Virol. 2010;84(19):10209–19. doi: 10.1128/JVI.00632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim EY, Bhattacharya T, Kunstman K, Swantek P, Koning FA, Malim MH, Wollinsky SM. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J Virol. 2010;84(19):10402–5. doi: 10.1128/JVI.01223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sadler HA, Stenglein MD, Harris RS, Mansky LM. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J Virol. 2010;84(14):7396–404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fourati S, Malet I, Binka M, Boukobza S, Wirden M, Sayon S, Simon A, Katlama C, Simon V, Calvez V, Marcelin AG. Partially active HIV-1 Vif alleles facilitate viral escape from specific antiretrovirals. AIDS. 2010;24(15):2313–21. doi: 10.1097/QAD.0b013e32833e515a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Armitage AE, et al. APOBEC3G-induced hypermutation of human immunodeficiency virus type-1 is typically a discrete “all or nothing” phenomenon. PLoS Genet. 2012;8(3):e1002550. doi: 10.1371/journal.pgen.1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mansky LMaT, HM Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69(8):5087–94. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Drake JWaH, JJ Mutation rates among RNA viruses. Proc Natl Acad Sci USA. 1999;96(24):13910–3. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]