Abstract
Cells are transplanted to regenerate an organs’ parenchyma, but how transplanted parenchymal cells induce stromal regeneration is elusive. Despite common use of decellularized matrix, little is known what pivotal signals must be restored for tissue or organ regeneration. We report that Alx3, a developmentally important gene, orchestrated adult parenchymal and stromal regeneration by directly transactivating Wnt3a and VEGF. Contrasting to modest parenchyma formed by native adult progenitors, Alx3-restored cells in decellularized scaffolds not only produced vascularized stroma involving VEGF signaling, but also parenchymal dentin via Wnt/β-catenin pathway. In an orthotopic large-animal model following parenchyma and stroma ablation, Wnt3a-recruited endogenous cells regenerated neurovascular stroma and differentiated into parenchymal odontoblast-like cells extending processes into newly-formed dentin with structure-mechanical equivalency to native dentin. Thus, Alx3-Wnt3a axis enables postnatal progenitors with modest innate regenerative capacity to regenerate adult tissues. Depleted signals in decellularized matrix may be reinstated by a developmentally pivotal gene or corresponding protein.
Keywords: Alx3, Wnt, mesenchyme, VEGF, MSC, BMP, decellularized matrix
Regeneration of diseased or injured tissues is attributed to unique abilities of stem cells including in vivo self-renewal and asymmetric differentiation. Stem cells are transplanted with a goal to differentiate into parenchymal cells, such as myocytes for muscle or hepatocytes for the liver1–3. Rarely are cells transplanted to regenerate an organ’s stroma. For complete tissue regeneration, neurovascular and interstitial stroma is presumed to derive endogenously from the host4. For example, regenerated myocardium was populated not only by transplanted iPS-derived cardiomyocytes, but also host endothelial cells that participate in vascular formation2. Dental pulp and dentin offer a powerful model to study parenchymal and stromal regeneration5. Parenchymal and stromal dental tissues are compartmentalized with all parenchymal tissues highly mineralized, and yet with stromal components being neurovascular, interstitial and unmineralized5.
Whether adult tissue repair recapitulates prenatal morphogenesis is often debated but incompletely understood. Hoxa9 is a key repressor for ageing satellite cells and, when attenuated, permits ageing muscle repair6. Conversely, Pitx2 promotes cardiac muscle regeneration by activating reactive oxygen species scavengers7. Delivery of follistatin-like 1 encoding protein, which declines upon myocardial infarction, rescues ischemic cardiomyocytes8. Manipulation of a proapoptotic Sept4/ARTS gene stimulates quiescent hair-follicle stem cells towards regeneration9. Hedgehog and FGF8 signals are region-specific in salamander limbs and, when transposed, enable otherwise non-regenerating blastemas to regenerate10. CTGF bridges injured spinal cord via glial cells in zebrafish, but not in mammals11. Alx3 is a highly conserved transcription factor, and regulates neural, hair and pancreatic development12. Alx3 functions as a Mitf suppressor for melanocyte differentiation and regulates rodent hair stripe patterns13. However, whether parenchymal and/or stromal tissues of an organ defect in the adult can regenerate by signals that are pivotal in development is elusive.
Decellularized matrix has yielded some success in tissue and organ regeneration14. Although cells are seeded in decellularized scaffolds, little is known what pivotal molecular signals need to be reinstated for regeneration. Here, we discovered that Alx3, a transcription factor of the homeobox gene family, was pivotal for prenatal tooth development but subsided in the adult. Adult progenitors with limited innate regenerative capacity, when restored with Alx3, not only regenerated parenchymal dentin, but also vascularized stroma with improved cell survival in vivo. Alx3 promoted parenchymal and stromal regeneration by direct transactivation of Wnt3a and VEGF promoters. We then took a translational step to infuse recombinant human Wnt3a protein, Alx3’s direct target, orthotopically in surgically ablated porcine root canals. Wnt3a not only induced regeneration of parenchymal odontoblasts with mineralized tubular dentin, but also neurovascular stroma. Thus, parenchyma and stroma regenerated not only ectopically by transplanted Alx3-restored adult progenitors, but also orthotopically by Wnt3a-responsive endogenous cells, providing a rare example that cell transplantation and endogenous repair may share common pathways, and yield similar outcome. Depleted biological molecules in decellularized matrix may be restored in an adult tissue by signals that are pivotal in its development. Alx3’s roles in prenatal dentin development and adult dentin regeneration represent another step towards our understanding of tooth regeneration.
Alx3 was robustly expressed in development but subsided in the adult
At E12.5 in the mouse tooth organ, Alx3 emerged in the frontonasal process and dental mesenchyme (Fig. 1a). Alx3 expression became increasingly robust in dental mesenchyme and peaked at E16.5 (Fig. 1b–d), synchronous with odontoblast commitment and differentiation, followed by its appearance in dental epithelium (Fig. 1e,f). Importantly, Alx3 vanished in the 8-wk adult molar (Fig. 1g,g’). Real-time qPCR confirmed increasingly robust Alx3 expression in dental mesenchyme from E14.5 to E17.5 and subsidence by 8 weeks in the adult (Fig. 1h). To confirm Alx3 roles in tissue development, we generated Alx3 knockout mice by CRISPR/Cas9 and confirmed its genotype (Supplementary Notes).
Fig. 1.
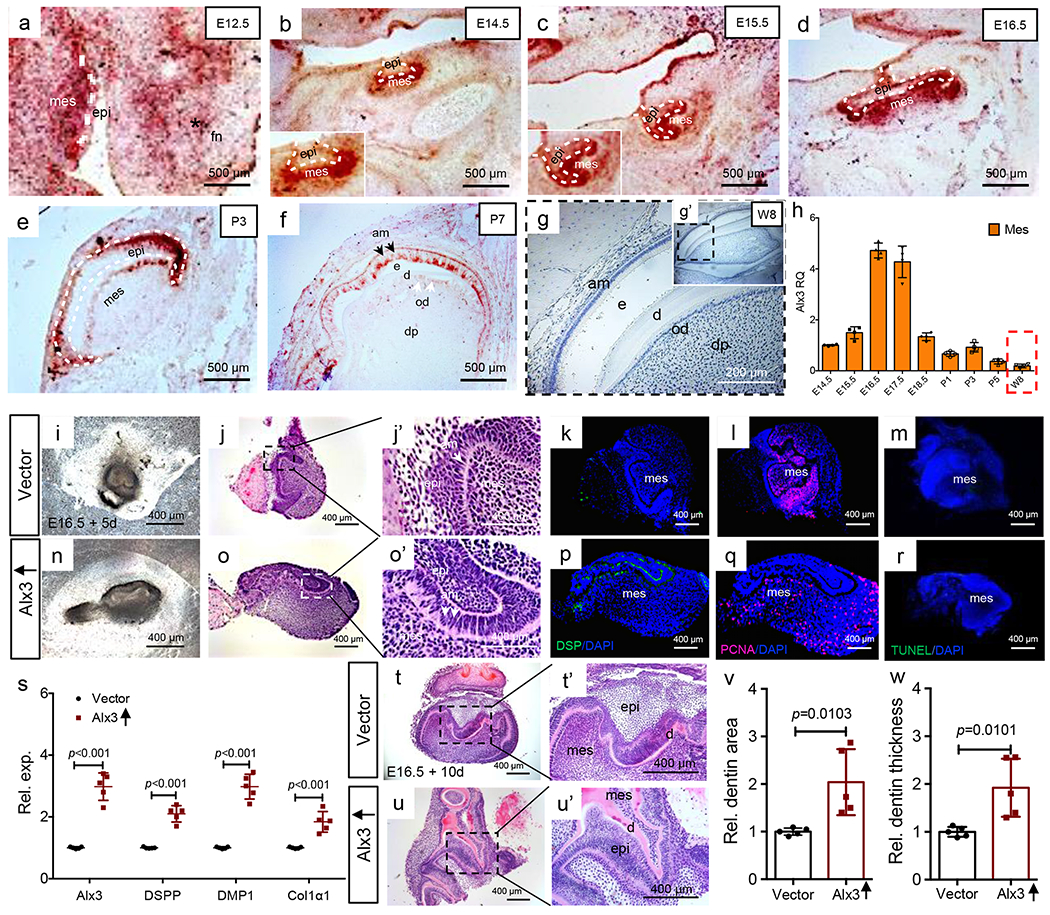
Alx3 immunomapping and tissue reconstitution, a-f: Alx3 in situ hybridization from E12.5 to postnatal day 7 tooth organs. White dash lines: boundary of epithelium and mesenchyme. Scale bar, 500 μm. 3 independent biological samples. g,g’: Alx3 immunohistochemistry of molar tooth organ of an 8-wk-old CD1 mouse and higher magnification (insert). Scale bar, 200 μm. 2 independent biological samples, h: Real-time PCR of mesenchyme Alx3 expression (n=4 independent biological samples). Presented as mean±SD, bars represent standard deviation (SD). Red dash box: Mesenchyme Alx3’s expression of 8-wk-old adult molar (w8). i-o’: Photomicrograph and HE images of reconstituted E16.5 tooth organs following 5-day culture. A total of 5 independent biological samples. k,r: Dentin sialoprotein (DSP) immunofluorescence; Proliferating cell nuclear antigen (PCNA) immunofluorescence; TUNEL staining. Scale bar, 400 μm. 3 independent biological samples, s: Real-time PCR of Alx3, DSPP (dentin sialophosphoprotein), DMP1 (dentin matrix protein-1) and Col1a1 (collagen 1 alpha 1) in reconstituted tooth organs following 5-day culture (n=5 independent biological samples; presented as mean±SD and p value; p values calculated by two-sided Student’s t-tests). t-u’: Photomicrographs and HE images of E16.5 reconstituted tooth organs following 10-day culture. 5 independent biological samples. Scale bar, 400 μm. v,w: Quantified dentin area and thickness (n=5 independent biological samples; presented as mean±SD and p value; p values calculated by two-sided Student’s t-tests). mes: mesenchyme; epi: epithelium; fn: frontonasal process; e: enamel; d: dentin; od: odontoblasts; dp: dental pulp.
Alx3 gain or loss of function directly mediated mesenchyme differentiation
We perturbed Alx3 in reconstituted E16.5 tooth organs per prior methods by us and others15,16. Alx3 was over-expressed in isolated E16.5 mouse dental mesenchyme, followed by epithelium reconstitution (Supplementary Figure 1a,b; Fig. 1i–j’,n–o’). Upon 5-day incubation, Alx3 overexpression accelerated synthesis of DSP (Fig. 1p), a hallmark of odontoblast differentiation, relative to control (Fig. 1k); yet without appreciable effects on cell proliferation (Fig. 1q) or apoptosis (Fig. 1r) relative to controls (Fig. 1l,m). Alx3 overexpression significantly promoted DSPP, DMP1 and Col1α1 mRNAs (Fig. 1s), again indicative of odontoblast differentiation. Alx3 accelerated dentinogenesis by day 10 (Fig. 1u,u’) relative to control (Fig. 1t,t’), with significantly greater dentin area and thickness than vector control (Fig. 1v,w). Conversely, Alx3 knockdown by siRNA (Supplementary Figure 1b) had virtually opposite effects, with disturbed basement membrane (Supplementary Figure 1c–h), and attenuated DSPP, collagens I and IV, and laminin (Supplementary Figure 1i,j,o), leading to reduced dentin area and thickness (Supplementary Figure 1p–s), and again without appreciable effects on cell proliferation or apoptosis (Supplementary Figure 1k–n).
Alx3 induced biomineralization primarily via canonical Wnt signaling
To appreciate Alx3 signaling pathways, we isolated mouse E16.5 dental epithelium and mesenchyme cells (Supplementary Figure 2a–q). Alx3-overexpressed mesenchyme cells significantly upregulated Wnt3a, Wnt5a, Msx1 and DSPP in the presence of isolated dental epithelium (Fig. 2a,b). Conversely, mesenchymal Alx3 knockdown by shRNA significantly reduced Wnt3a, Wnt5a, DSPP, Msx1 and BMP4 (Fig. 2b,c). Furthermore, Alx3-overexpressed mesenchyme cells induced un-manipulated mesenchyme cells to upregulate Wnt3a, DSPP and DMP1 (Fig. 2d,e). Conversely, mesenchymal Alx3 knockdown significantly attenuated DSPP and Wnt3a in the un-manipulated mesenchyme (Fig. 2e,f).
Fig. 2.
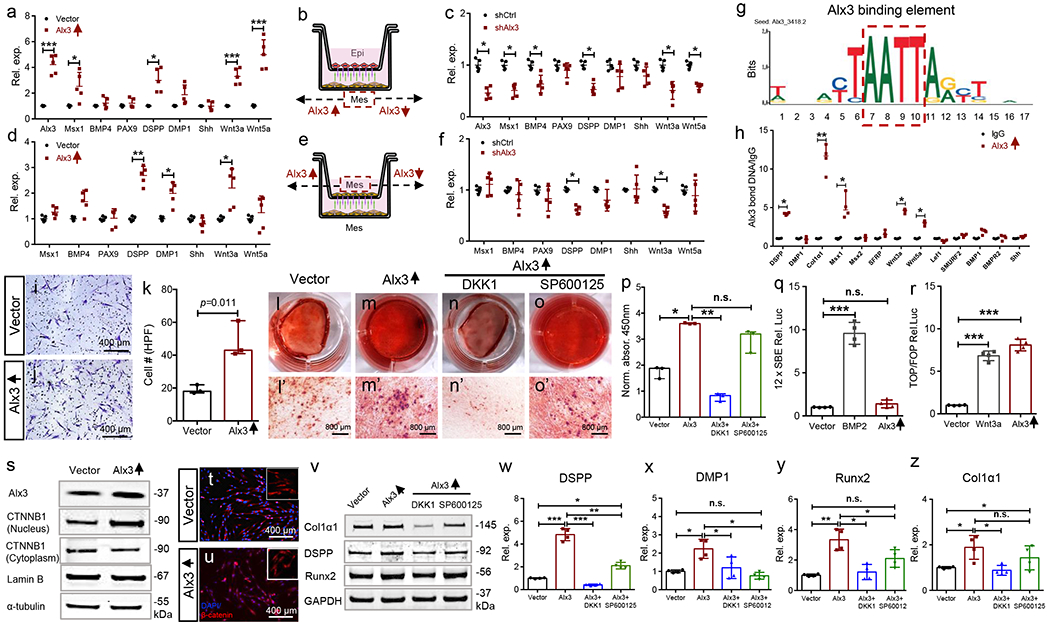
Alx3 signaling pathways, a-c: Epithelium and mesenchyme cells isolated from E16.5 tooth organs followed by mesenchymal Alx3 overexpression (a) or knockdown (c) and Transwell co-culture with isolated epithelium cells (b). d-f: E16.5 mesenchyme cells isolated, followed by mesenchymal Alx3 overexpression (d) or knockdown (f) and Transwell co-culture with un-manipulated mesenchyme cells (e) (a-f: n=5 independent biological samples; presented as mean±SD; p values calculated by multiple two-sided Student’s t-tests with Holm-Bonferroni correction; *: p<0.05; **: p<0.01; ***: p<0.001). g,h: ChIP assay of adult human dental-pulp mesenchyme stem/progenitor cells (MSCs) (n=4 independent biological samples; presented as median with range; p values calculated by Mann-Whitney tests with Bonferroni correction; *: p<0.05; **: p<0.01). Red dash box: Alx3 binding element from JASPAR CORE database, i-k: Transwell cell migration and quantification (n=3 independent biological samples; presented as median with range; p values calculated by Mann-Whitney tests with Bonferroni correction). Scale bar, 400 μm. l-p: Alizarin Red-S staining and quantification (n=3 independent biological samples; presented as median with range; p values calculated by Kruskal-Wallis tests; *: p<0.05; **: p<0.01; n.s.: not significant). Scale bar, 800 μm. q: BMP/Smad luciferase activity upon BMP2 treatment (100 ng/ml) or Alx3 overexpression (n=4 independent biological samples; presented as mean±SD; p values calculated by one-way ANOVA with Bonferroni; ***: p<0.001; n.s.: not significant), r: Wnt luciferase activity upon Wnt3a treatment (100 ng/ml) or Alx3 overexpression (n=4 independent biological samples; presented as mean±SD; p values calculated by one-way ANOVA with Bonferroni; ***: p<0.001). s: Western blot of nuclear and cytoplasmic β-catenin (CTNNB1) upon Alx3 overexpression. t,u: β-catenin transnucleation by immunofluorescence. Scale bar, 400 μm. v: Western blot of Col1α1, DSPP and Runx2; DKK1 (200 ng/ml); SP600125 (10 μW). w-z: Real-time PCR of DSPP, Runx2 and collagen1α1 (n=4 independent biological samples; presented as mean±SD; p values calculated by one-way ANOVA with Bonferroni; *: p<0.05; **: p<0.01; ***: p<0.001; n.s.: not significant).
Given its pivotal roles in dentin development and subsidence in adult tooth organs (Fig. 1g,h), we explored whether reinstating Alx3 in adult human dental-pulp mesenchymal stem/progenitor cells (MSCs) would enhance their regenerative ability. ChIP assay showed Alx3 binding to Wnt3a, Wnt5a, Msx1, DSPP and Col1a1 in MSCs (Fig. 2g,h). Alx3-restored MSCs (Supplementary Figure 2r–t) were robustly chemotactic, and induced over ~2-fold more cell migration than vector control (Fig. 2i–k), suggesting Alx3’s ability in recruiting stem/progenitor cells in vivo. Conversely, Alx3 knockdown attenuated cell migration (Supplementary Figure 3h–j). Alx3-restored cells were robustly Alizarin Red-S positive (Fig. 2m,m’) relative to control (Fig. 2l,l’), as quantified in Fig. 2p. Importantly, Alx3 failed to activate canonical BMP/Smad-responsive 12xSBE reporter (Fig. 2q), but rather upregulated Wnt luciferase activity similar to Wnt3a, a canonical Wnt ligand (Fig. 2r), suggesting that Alx3 promotes biomineralization primarily via Wnt signaling, rather than BMP. Alx3 restoration in adult MSCs increased nuclear CTNNB1 and concurrently decreased cytoplasmic CTNNB1 (Fig. 2s), and augmented β-catenin transnucleation relative to control (Fig. 2t,u). Alx3 restoration further enhanced DSPP, Runx2 and collagen1a1 mRNAs, relative to control, and these increases were robustly attenuated by DKK1, a canonical Wnt inhibitor, but only moderately by SP600125, a non-canonical Wnt inhibitor (Wnt5a/Ror2/JNK) (Fig. 2v–z) at both protein and mRNA levels, along with attenuated mineralization by DKK1 rather than SP600125 (Fig. 2n–p), again suggesting that Alx3 primarily signals via canonical Wnt to induce biomineralization. Conversely, Alx3 knockdown had virtually opposite effects on biomineralization, DSPP, Runx2 and collagen1a1, which was rescued by exogenous recombinant Wnt3a protein rather than recombinant Wnt5a (Supplementary Figure 3k–p). To further explore the Alx3-Wnt3a axis, we assayed both human and mouse Wnt3a promoter reporter luciferase activity upon Alx3 transfection (Supplementary Figure 4a–d). Importantly, Alx3 overexpression promoted Wnt3a luciferase activity in both human MSCs and mouse E16.5 dental mesenchyme cells (Supplementary Figure 4e,f), whereas Wnt3a luciferase activity was attenuated following mutation of Alx3 binding motifs on Wnt3a promoters (Supplementary Figure 4e,f), suggesting that Alx3 transactivates Wnt3a expression.
Alx3 enabled parenchyma regeneration in decellularized scaffolds
We further explored whether Alx3 enabled adult tissue regeneration in decellularized scaffolds (Fig. 3a–d). Upon 8-wk in-vivo implantation of decellularized human tooth scaffolds in the dorsum of athymic mice, un-manipulated adult MSCs produced modest interstitial tissue and little mineralized dentin (Fig. 3e,e’,g). Contrastingly, donor-matched Alx3-restored MSCs not only regenerated dental-pulp-like tissue with vascularized matrix (Fig. 3f,f), but also de novo dentin-like tissue (Fig. 3f,h), relative to vector control (Fig. 3e’,g), blank controls (Supplementary Figure 5a,c,c’e) and gel-only controls (Supplementary Figure 5b,d,d’,f). Odontoblast-like cells were present in Alx3-restored MSC samples and aligned newly-formed dentin surface (Fig. 3f,h and inserts). Quantitatively, Alx3-restored adult MSCs produced significantly more dental-pulp-like tissue than controls by measuring the percentage of dental-pulp-like tissue within the entire root-canal space (Fig. 3i), and ~6-fold more de novo dentin (Fig. 3j). Alx3 restoration further produced DSPP, especially adjacent to the newly-formed dentin (Fig. 3p) and Wnt3a (Fig. 3q) along with DMP1 (Supplementary Figure 5n) relative to controls (Fig. 3k,l and Supplementary Figure 5g–m). β-catenin-positive cells were abundant in Alx3-restored MSC samples (Fig. 3r–r”), relative to controls (Fig. 3m–m”). Human mitochondria staining identified transplanted human MSCs in both Alx3 restoration and control samples (Fig. 3o,s) and were absent in blank control or gel only samples (Supplementary Figure 5k,l), with significantly more transplanted human cells present than host mouse cells upon Alx3 restoration than control (Fig. 3t), suggesting Alx3 promotes cell survival in vivo. Nestin-positive cells were found in both control (Fig. 3u–u”) and Alx3-restoration samples (Fig. 3w–w”), although Alx3 restoration induced nestin-positive cells that aligned the newly-formed dentin surface (Fig. 3w–w”). Alx3 restoration further induced abundant PCNA staining (Supplementary Figure 5p,p’,p”) relative to control (Supplementary Figure 5o,o’,o”), suggesting that Alx3 promotes cell proliferation in vivo. Caspase 3 was sparsely present upon Alx3 restoration (Fig. 3v–x”), with quantitatively over 3-fold fewer apoptotic cells in Alx3-restored MSC samples (Fig. 3y), again suggesting that Alx3 promotes cell survival in vivo. Thus, Alx3 directly activates Wnt3a via Wnt/β-catenin signaling to promote parenchymal regeneration in adult progenitors (Fig. 6m).
Fig. 3.
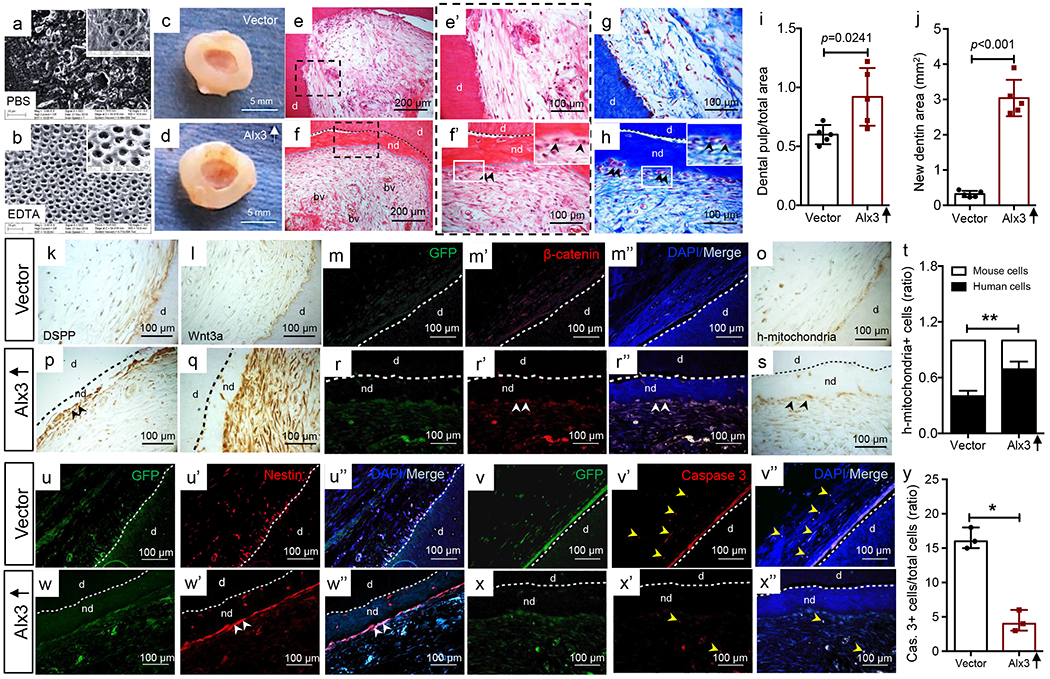
Transplanted, Alx3-restored adult human dental-pulp mesenchyme stem/progenitor cells (MSCs) regenerated both parenchyma and stroma with enhanced Wnt signaling and improved cell survival. a,b: PBS and EDTA processed, decellularized scaffolds (inserts with higher magnification); n=3 independent biological samples. c,d: Decellularized scaffolds retrieved following 8-wk in vivo implantation: vector control sample (c); Alx3 restored MSC sample (d). Scale bar, 5 mm. HE images of both vector control (e,e’) and Alx3-restored MSC samples (f,f’) samples. Black arrowheads: odontoblast-like cells that aligned newly-formed dentin surface (f’ insert); e,f: Scale bar, 200 μm; e’,f’: Scale bar, 100 μm. g,h: Masson’s Trichrome staining. Black arrowheads: odontoblast-like cells that aligned newly-formed dentin surface (h insert). Scale bar, 100 μm. c-h: n=5 independent biological samples, i: Dental pulp-like tissue area over total root-canal area (n=5 independent biological samples; presented as mean±SD and p value; p value calculated by two-side Student t-tests). j: Newly-formed dentin area (n=5 independent biological samples; presented as mean±SD and p value; p value calculated by two-side Student t-tests). k,p: DSPP immunohistochemistry (black arrowheads: DSPP). Scale bar, 100 μm. l,q: Wnt3a immunohistochemistry (black arrowheads: Wnt3a). Scale bar, 100 μm. m-r”: β-catenin-positive cells (white arrowheads), k-r”: n=3 independent biological samples. o,s: Human mitochondria staining of transplanted human cells and quantification (t) (n=5 independent biological samples; presented as mean±SD; p value calculated by two-sided Student’s t-tests; **: p<0.01). u-w”: Nestin-positive cells (white arrowheads), v-x”: Caspase-3 positive cells and quantification, u-x”: n=3 independent biological samples, (y) apoptotic cells exemplified by yellow arrowheads (n=3 independent biological samples; presented as median with range; p value calculated by Mann-Whitney tests with Bonferroni correction; *: p<0.05). Scale bar, 100 μm. d: dentin; nd: newly-formed dentin; bv: blood vessels; black dash lines: boundary between native dentin and newly-formed dentin (p,q,s); blue dash lines: boundary between newly-formed dentin and dental-pulp (f,f’,h); white dash lines: boundary between native dentin and newly-formed dentin (m-r”,u-x”).
Fig. 6.
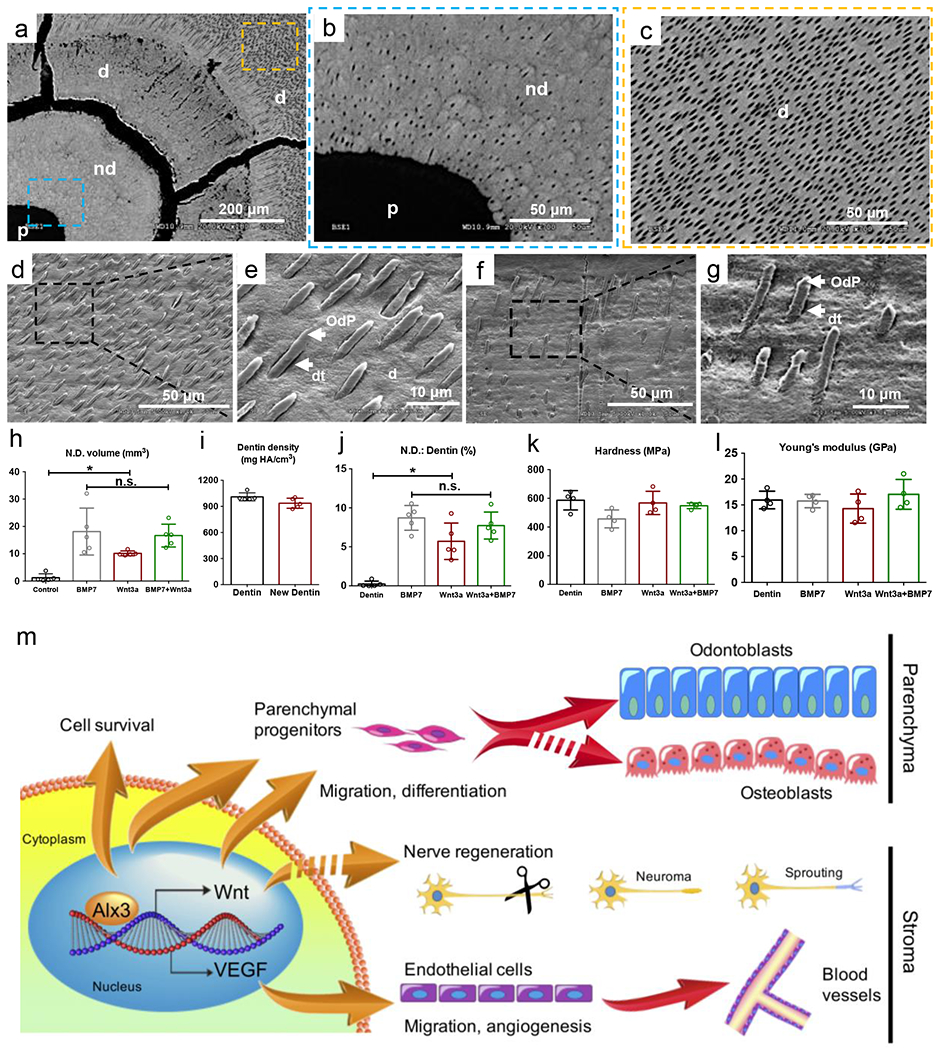
Structural and mechanical properties of regenerated dentin and overall schematic of Alx3/Wnt/VEGF cascades. a: Scanning electron microscopy of dental pulp (p), newly-formed dentin (nd) and native dentin (d) in cross-section. Scale bar: 200 μm. b,c: Dentinal tubules and resin casts in the newly-formed dentin (blue dashed box in a) and native dentin (yellow dashed box in a) by SEM. Scale bar: 50 μm. d,e: Odontoblast processes casts in the native dentin (d,e) and newly-formed dentin (f,g) of a native porcine incisor by SEM. d: dentin; dt: dentinal tubules; OdP: odontoblast process. d,f: scale bar: 50 μm. e,g: scale bar: 10 μm. h: Newly-formed dentin volume; i: Dentin mineral density, j: Ratio of newly-formed dentin (N.D.) and native dentin; k: Harness (MPa); I: Young’s modules (GPa). (h-j: n=5 independent biological tooth samples; k,l: n=4 independent biological tooth samples; Presented as mean±SD; p values calculated by one-way ANOVA with Bonferroni; *p<0.05, n.s.: not significant). m: Schematic diagram showing Alx3 regulation of Wnt3a and VEGF as its direct targets, leading to parenchymal and stromal regeneration, with multiple cellular processes including improved cell survival, parenchymal progenitor migration and differentiation, endothelial cells migration, angiogenesis and neural sprouting.
Alx3 promoted stroma regeneration via VEGF signaling
Surprisingly, we found rich vasculature in Alx3-restored MSC samples upon 4-wk in-vivo implantation (Fig. 4d,d’), along with abundant human von Willebrand factor (Fig. 4e,e’) relative to controls (Fig. 4a–b’ and Supplementary Figure 6a–j). Some regenerated blood vessels were chimeric with endothelial lining by transplanted human cells and host mouse cells (Fig. 4b’,e’). Human mitochondria were identified among cells in control and Alx3-restored MSC samples (Fig. 4c,f), indicating chimeric vessels formed by transplanted human cells and host mouse cells. Quantitatively, Alx3 restoration induced significantly more vasculature than controls (Fig. 4g). Given the surprising outcome of Alx3-induced angiogenesis, we first assayed HUVEC proliferation and migration in response to the conditioned medium of Alx3-restored MSCs. Indeed, Alx3 conditioned medium robustly augmented HUVEC proliferation and migration relative to control (Fig. 4h,i–o), suggesting that Alx3 promotes angiogenesis paracrine via MSC secretome. Conversely, HUVEC migration was attenuated by the VEGF neutralizing antibody or VEGFR2 inhibitor (Fig. 4m–o), further suggesting that Alx3 promotes angiogenesis primarily through VEGFR2. Next, we probed Alx3 binding sites on VEGF promoter and found that Alx3 transfection robustly augmented VEGF promoter luciferase reporter activity, relative to control (Fig. 4p,p’). There was significantly greater VEGF concentration in Alx3-conditioned medium than control (Supplementary Fig. 6k), further suggesting that Alx3 stimulates VEGF production. To further explore Alx3’s angiogenic effects, we assayed both VEGF and VEGF Receptor 2 (VEGFR2) expression. Indeed, Alx3 conditioned medium enhanced VEGF production, along with increased VEGFR2 phosphorylation (Fig. 4q) and quantitated in Fig. 4r, again suggesting that Alx3 exerts its angiogenic effect via VEGF signaling, especially involving VEGFR2. Tube length of HUVECs treated with Alx3 conditioned medium was significantly greater than controls and was comparable to the classic VEGF medium (Fig. 4y). Tube length decreased significantly by VEGF neutralizing antibody or inhibitor (Fig. 4w–y). PECAM1, VEGF, VE-Cadherin and FLK1 expression of HUVECs exposed to the Alx3 conditioned medium was attenuated by VEGF neutralizing antibody or VEGFR2 inhibitor, relative to controls (Fig. 4z). Additionally, tube length significantly decreased after VEGF removal by immunoprecipitation from the Alx3 conditioned medium (Supplementary Figure 6k–q). Thus, Alx3 activates VEGF signaling both directly by binding to VEGF promoter and paracrine by MSC secretome (Fig. 6m), which in totality may account for the observed cell survival and angiogenesis in vivo (Fig. 3f,f’,x”; Fig. 4d’–e’).
Fig. 4.
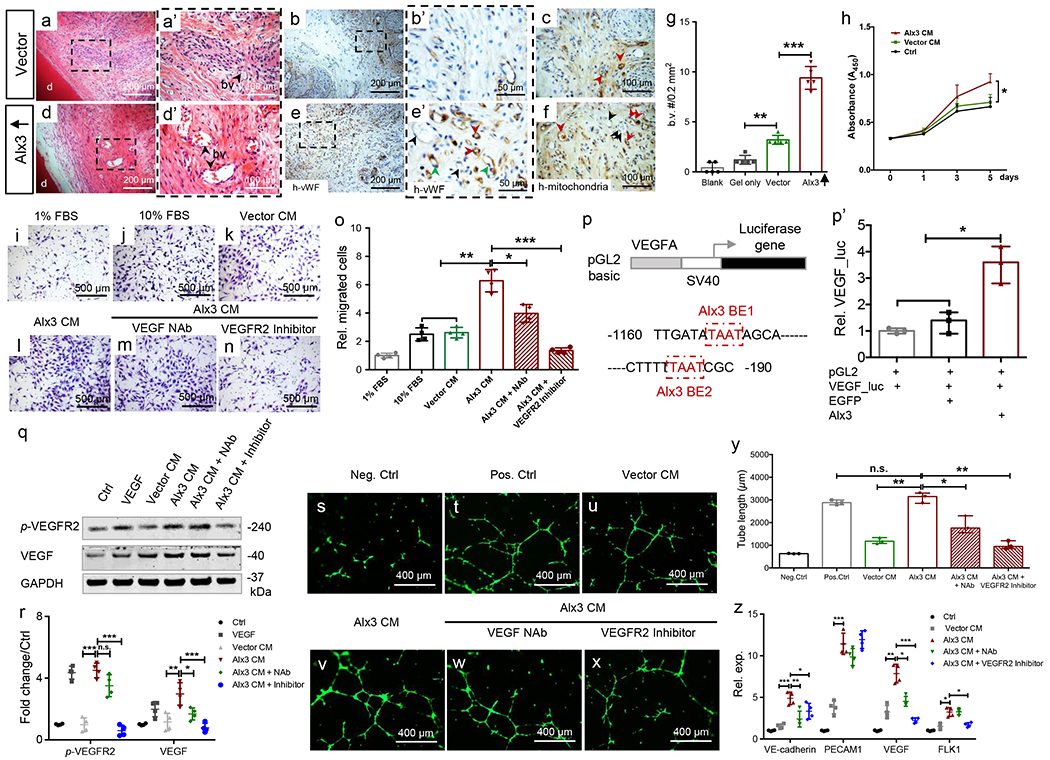
Alx3-restored adult mesenchyme stem/progenitor cells (MSCs) induced angiogenesis and VEGF signaling. a,a’,d,d’: Angiogenesis in regenerated tissues. a,d: scale bar, 200 μm. a’,d’: scale bar, 100 μm. d: native dentin. b,b’,e,e’: Human von Willebrand factor immunohistochemistry. Green arrowheads: endothelial cells of human origin (e’); black arrowheads: endothelial cells of host (mouse) origin (e’); red arrowheads: chimeric blood vessels (e’). b,e: scale bar, 200 μm. b’,e’: scale bar, 50 μm. c,f: Human mitochondria staining; red arrowheads: human cells; black arrowheads: host (mouse) cells. Scale bar, 100 μm. a-h: n=5 independent biological samples, g: Blood vessel (b.v.) quantification (n=5 independent biological samples; presented as mean±SD; p values calculated by one-way ANOVA with Bonferroni; **: p<0.01; ***: p<0.001). h: CCK8 of HUVECs treated with conditioned medium by vector control or Alx3-restored MSCs (n=4 independent biological samples; Presented as mean±SD; p values calculated by one-way ANOVA with Bonferroni; *: p<0.05). i-o: Transwell migration assay by Alx3 conditioned medium or with neutralizing antibody or VEGFR2 inhibitor (n=4 independent biological samples; presented as mean±SD; p values calculated by one-way ANOVA with Bonferroni; *: p<0.05; **; p<0.01; ***: p<0.001). Scale bar, 500 μm. p: Mouse VEGFA promoter luciferase reporter with two Alx3-binding elements in red boxes. BE: binding element, p’: VEGF luciferase assay upon Alx3 transfection (n=3 independent biological samples; presented as median with range; p values calculated by Kruskal-Wallis test; *: p<0.05). q,r: VEGF production and p-VEGFR2 activation by western blot and quantification (n=4 independent biological samples; presented as mean±SD; p values calculated by one-way ANOVA with Bonferroni; *: p<0.05; **; p<0.01; ***: p<0.001; n.s.: not significant), s-x: Tube formation of HUVECs treated by conditioned medium, Alx3 CM with or without NAb or VEGFR2 inhibitor. Scale bar, 400 μm. 3 independent biological samples, y: Tube length quantification (n=3 independent biological samples; presented as median with range; p values calculated by Kruskal-Wallis test; *: p<0.05; **; p<0.01; n.s.: not significant), z: Real-time PCR of VE-cadherin, PECAM1, VEGF and FLK1 (n=4 independent biological samples; presented as mean±SD; p values calculated by one-way ANOVA with Bonferroni; *: p<0.05; **: p<0.01; ***: p<0.001).
Recombinant Wnt3a protein induced parenchyma and stroma regeneration orthotopically in a preclinical large animal model
Since Alx3 enabled adult MSCs to produce dentin-like tissue by activation of canonical Wnt/β-catenin, we took a translational step to explore whether a canonical Wnt ligand would promote dentin regeneration. In the face, odontoblasts that form dentin and osteoblasts that form bone both drive from neural crest cells17. Genome-wide profiling of donor-matched mouse odontoblasts and osteoblasts isolated by EGFP-FACS sorting from 2.3 kb col1α1 transgenic mice showed a total of 948 differentially expressed genes (>2 folds by adjusted p values; Supplementary Figure 7). Remarkably, Alx3 was 51-fold higher in odontoblasts than donor-matched osteoblasts, whereas Wnt10a was 126-fold in favor of odontoblasts (GSE119897). Because Wnt10a currently has no recombinant protein and is a sibling isoform to Wnt3a, we tested whether Wnt3a and Wnt10a had similar or different effects on dentinogenesis. We further mapped CTNNB1, Wnt3a and Wnt10a expression from E14.5 to postnatal 8 weeks (Supplementary Figure 8a–c). Wnt3a enhanced CTNNB1 protein synthesis (Supplementary Figure 8d,e), although both Wnt3a and Wnt5a increased CTNNB1 mRNA expression (Supplementary Figure 8f) in E16.5 dental mesenchyme cells. In MSCs, Alx3 restoration increased Wnt10a, Wnt3a and Wnt5a (Supplementary Figure 8g–i), consistent with ChIP assay (Fig. 2g,h). Following 8-day culture of Wnt10a- and Wnt3a-transfected E16.5 dental mesenchyme (Supplementary Figure 8j–m’”), dentinogenesis was equivalent for Wnt3a and Wnt10a overexpression (Supplementary Figure 8p,p’ and 8q,q’) relative to control (Supplementary Figure. 8o,o’), as quantified in Supplementary Figure 8r.
In a preclinical large animal minipig model (see details in Methods), mandibular incisors were mechanically accessed to extirpate dental pulp (total of 58 teeth in 10 pigs). Following hemostasis, recombinant human Wnt3a in collagen gel was infused into clinically treated root canals. Three months later, we performed a broad array of analyses, each utilizing a maximized number of harvested tissue samples. Modest stroma was formed in surgically instrumented root canals upon collagen-gel infusion (Fig. 5a through 5a4), verifying dental-pulp ablation. BMP7 delivery induced excessive mineralization in endodontically treated root canals with embedded cells, resembling bone (Fig. 5b through 5b4). We adopted recombinant BMP7 as a control because Alx3 augments mineralization via Wnt signaling, rather than BMP pathway (Fig. 2). Wnt3a at 50 ng/mL yielded vascularized dental pulp-like tissue and mineralized dentin-like structures demarcated by dashed yellow lines (Fig. 5c through 5c4). The newly-formed dentin-like tissue (nd) consisted of dentinal tubules (dt) (Fig. 5c5) that had different orientation from those in native dentin (d) (Fig. 5c5), suggesting that Wnt3a induced de novo tubular dentin formation. Combined Wnt3a and BMP7 delivery yielded dental pulp-like and dentin-like structures also with apparent dentinal tubules (dt) (Fig. 5d5). PGP9.5 antibodies immunolocalized fiber sprouts, resembling neural filaments, upon Wnt3a delivery (Fig. 5e) or combined Wnt3a and BMP7 delivery (Fig. 5g). Neural filament-like structures upon Wnt3a delivery (S-100 in green in Fig. 5f) or combined Wnt3a and BMP7 delivery (Fig. 5h) partially overlaid with DAPI, suggesting myelinated and unmyelinated nerve sprouts, consistent with neural/glial origin of dental stem/progenitor cells18 and Wnt3a activation of neural sprouting19.
Fig. 5.
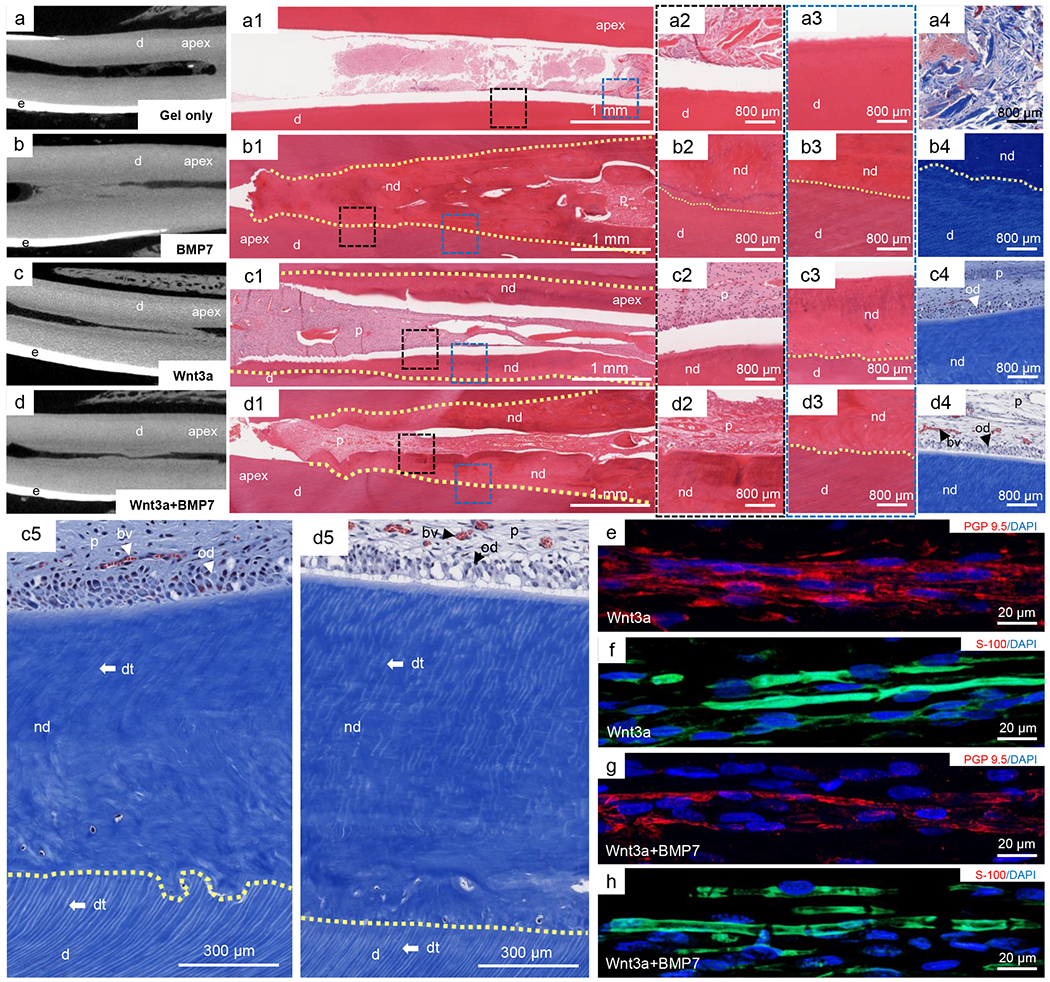
Parenchymal and stromal regeneration orthotopically in a preclinical, large animal model (total of 58 teeth in 10 minipigs), a-d: Micro-computed tomography (μ-CT) scans of porcine central incisors showing dentin (d), enamel (e) and the enclosed root canal, apex: root tip of teeth. a1-a4: HE and Masson’s Trichrome staining of collagen gel infusion alone, d: native dentin. b1-b4: HE and Masson’s Trichrome staining of BMP7 delivery in collagen gel. p: dental pulp; nd: newly-formed dentin. c1-c4: HE and Masson’s Trichrome staining of Wnt3a delivery in collagen gel. a1,b1,c1,d1: scale bar, 1 mm. a2-a4,b2-b4,c2-c4: scale bar, 800 μm. c5: High magnification of pulp-dentin boundary in Wnt3a alone group, p: dental pulp; dt: dentinal tubules; od: odontoblasts; nd: newly-formed dentin; d: native dentin. d5: High magnification of pulp-dentin boundary in Wnt3a and BMP7 combined delivery group, bv: blood vessel. Scale bar, 300 μm. e-h: Sprouts of nerve-like structures in regenerated dental pulp. e,f: PGP9.5 and S-100 immunolocalized fiber sprouts upon Wnt3a delivery. g,h: PGP9.5 and S-100 immunolocalized fiber sprouts upon combined Wnt3a and BMP7 delivery, e-h: scale bar, 20 μm.
Scanning electron microscopy (SEM) revealed dentinal tubules in newly-formed dentin and native dentin in Wnt3a and Wnt3a+BMP7 samples, with radiolucent area of dental pulp (p), newly-formed dentin (nd) and native dentin (d) shown in Fig. 6a. Microcracks were artifacts owing to high-vacuum coating during sample preparation for SEM imaging, and separated not only the newly-formed dentin from native dentin, but also transected areas of native dentin (Fig. 6a). Additional evidence that microcracks in Fig. 6a were artifacts derived from Fig. 5c5 and Fig. 5d5 showing an intact junction between newly-formed dentin (nd) and native dentin (d). Higher magnifications showed transverse openings of dentinal tubules in the newly-formed dentin (Fig. 6b) and native dentin (Fig. 6c). Odontoblast process casts were present in the native dentin (Fig. 6d,e) and newly-formed dentin (Fig. 6f,g). Quantitatively, Wnt3a and/or BMP7 induced significantly greater volume of newly-formed dentin than carrier-only control (Fig. 6h). The optical density of regenerated dentin-like tissue by pCT analysis lacked statistically significant differences from that of native dentin (Fig. 6i). The newly-formed dentin volume accounted for ~6-9% of the native dentin (Fig. 6j). The average hardness and Young’s modulus of newly-formed dentin lacked significant differences from native dentin (Fig. 6k,l). Schematics of an Alx3-Wnt3a axis in promoting cell survival, migration and differentiation towards not only the parenchyma, but also stroma as a generalized model for tissue regeneration is depicted in Fig. 6m.
Parenchyma and stroma of adult tooth organs are regenerated by an Alx3-Wnt3a axis that is pivotal in development and reinstated in the adult. Alx3-reinstated adult progenitors differentiate into Wnt3a+, nestin+ and β-catenin+ odontoblast-like cells and produce parenchymal dentin in vivo. Alx3 activation of Wnt/β-catenin is cell autonomous, given not only direct Alx3 binding to Wnt3a promoter and attenuated Wnt3a luciferase activity following mutation of Alx3 binding motifs on Wnt3a promoter, but also DKK1 abolishment of Alx3 activation of Wnt/β-catenin. Stroma regeneration mediated by Alx3 and/or Wnt3a is a surprising outcome, and partially attributed to Alx3 activation of VEGF, by direct binding to VEGF promoter and/or paracrine by MSC secretome. Alx3-induced cell survival is consistent with its anti-apoptotic roles in neural-tube and islet development20. VEGF signaling appears to have plural effects in tissue repair as observed here: enhancing angiogenesis, promoting survival and augmenting mineralization21. Alx3 promotes chemotaxis of adult MSCs and endothelial progenitors. Similarly, Wnt3a promotes MSC migration19,22 and angiogenesis23. Proangiogenic and pro-migratory properties of molecular cues by recruiting endogenous cells and promoting cell survival in vivo are pivotal characteristics for augmenting endogenous regeneration as shown here and by others24,25. Wnt3a promotes neural sprouting19 and may account for the observed, regenerated PGP9.5 and S-100 positive filaments in vivo.
What pivotal signals must be reinstated in decellularized matrix for tissue and organ regeneration has been elusive. Alx3 is 51-fold higher in odontoblasts than donor-matched osteoblasts. Dental mesenchyme differentiates into odontoblasts that robustly produce dentin in the developing tooth organ. Adult dental-pulp mesenchyme stem/progenitors, which derive from the prenatal dental mesenchyme, have poor innate ability to regenerate dentin, as evidenced by limited dentin formation upon injury or diseases in patients5,26. Alx3 appears to enable dentinogenesis of adult progenitors by reinvigorating perhaps an otherwise suppressed circuit, which nonetheless is necessary to prevent heterotopic dentin formation in adult homeostasis. Equally, Wnt3a, as Alx3’s direct target, is pivotal for dental mesenchyme development, as shown here and by others27. Wnt3a delivered in endodontically prepared root canals recruited endogenous cells, including the likely candidate of bone-marrow MSCs, to regenerate both the parenchyma and stroma in adult teeth. Thus, depleted biological signals in decellularized matrix may be restored by molecular cues that are pivotal in the development of corresponding tissues for which defects are to be healed in the adult.
A total of ~33 million Americans will be without any teeth in the maxilla or mandible by 202028. A major cause of tooth loss is dental caries that infect dental pulp, necessitating an endodontic treatment, which in its current form, fills the root canal with prosthetic materials. A total of ~18 million root-canal treatments are performed annually in the U.S.29. Despite documented clinical success, root-canal treatments fail due to secondary infections or fractures, leading to tooth loss30. Recent work showed that Wnt ligands or analogs induced mineralization at the pulp puncture site (~1 mm diameter)31,32. However, pulp capping at pulp-puncture sites outside the root canal targets represents a different disease stage and intrinsically distinguishes from the present model and outcome of pulp and dentin regeneration along centimeter-long root-canal surfaces, which serves as an alternative to root-canal therapy. The regenerated tubular dentin with structural and mechanical equivalency to native dentin has not been previously reported; nor pulp and dentin regeneration following complete pulpectomy with an outcome of tubular dentin formation and neurovascular stroma. Translation of our findings towards therapeutic use is conceptually straightforward, especially since no cell culture or cell transplantation is needed. One could conceptualize recombinant human Wnt3a delivery in disinfected root canals in lieu of current prosthetic fillers. A randomized clinical trial is needed to appreciate the efficacy and safety of an injectable protein, such as Wnt3a, in “root-canal” patients. Alx3’s roles in the developing dental mesenchyme, as well as its transient but robust expression in dental epithelium, need to be further exploited in the context of tooth regeneration. Our observation of Alx3’s ability to induce the genesis of dentin, which forms the bulk of teeth, suggests its involvement in tooth regeneration.
Tissue engineering initially emerged as a concept with a triad of cells, biomaterials and signaling molecules33. The need for cells has been commonly practiced as ex vivo culture and cell transplantation. Cell transplantation is undoubtedly needed for regenerating missing organs2,14. Nonetheless, certain tissue and organ defects can be healed by ‘cell homing’ of endogenous progenitors25,34,35,36,37, without ex vivo cell culture and cell transplantation. The present work provides two contrasting tissue-engineering approaches: transplantation of Alx3-restored adult human progenitors and delivery of Wnt3a, as Alx3’s direct target protein, to enable endogenous regeneration without cell transplantation. Thus, cell transplantation and endogenous repair may share common pathways, and yield similar outcome. For example, transplantation of stem/progenitor cells has unveiled valuable clues for dental-pulp and dentin regeneration in ectopic and orthotopic models, including two clinical reports38–44. However, it is regulatorily and economically challenging, and clinically uncommon that a patient’s healthy adult teeth are available as donor cell sources for dental-pulp and dentin regeneration38–42,45. For severe debilitating diseases, iPS and cellular programming continue to hold great promise, whereas for tissue and organ defects, regeneration by cell homing using pivotal molecular cues and/or biomaterials may prevail.
Methods
Animals
All animal experiments including mouse and minipig use, care and surgical procedures were approved by Columbia University Institutional Animal Care and Use Committee (IACUC).
In Situ hybridization
Mouse Alx3 fragment plasmid (1,856-kb-cDNA cloned from E16 cDNA library), a generous gift from Dr. Frits Meijlink, was used to map Alx3 from E12.5 to P7. Following transformation and linearization with EcoRI or HindIII (New England BioLabs, Ipswich, MA, USA), antisense and sense RNA probes were synthesized by transcripting with Sp6 and T7 polymerases (MAXIscript® Sp6/T7 In Vitro Transcription Kit, Ambion, Austin, TX, USA), following manufacturer instructions using Digoxigenin (DIG)-11-UTP (Roche, Mannheim, Germany). The mandibular incisor and first molar tooth organs at E12.5, E14.5, E15.5, E16.5, E18.5, P1, P3, and P7 were harvested by gestation timing of CD-1 pregnant mice (Charles River, Newark, NJ, USA). The isolated tooth germs were dissected under surgical microscope in cold DEPC-treated PBS (pH=7.4), and processed for fixation with 4% paraformaldehyde for 24h at 4°C with gentle rotation. The isolated tooth germs were then exposed to 30% w/v sucrose and incubated at 4°C prior to mounting. Postnatal tooth organs were first decalcified with 0.5M EDTA (pH=8.0) and dehydrated with 30% w/v sucrose. Frozen samples at 8-pm thickness were sectioned for in situ hybridization. NBT/BCIP from DIG Nucleic Acid Detection Kit (Roche) was used to detect Alx3 expression. Alx3 transcripts were treated at 65°C with 1:5000 diluted anti-DIG-AP antibody (Roche).
Organoid reconstitution and culture
E16.5 mandibular first molar tooth organs of CD1 mice (vaginal plug=day 0.5) were harvested surgical under dissection microscope per our prior methods46. Epithelium and mesenchyme were procsssed with 50 U/ml Dispase I (BD, Franklin Lakes, NJ) for 10.5 min and mechanically separated using a 25G needle15. For overexpression, the mesenchyme was incubated with Alx3 lentivirus for 4 hrs at R.T., whereas the epithelium was immersed in organ culture medium46. For knockdown, the mesenchyme was transfected with Cy3-labeled siRNA or Alx3 siRNA by incubation with RNAiMAX-siRNA Lipofectamine duplex for 4 hrs at R.T., followed by reconstitution. At Day 5, the reconstituted tooth germs were washed with ice-cold PBS and subjected to Trizol for RNA isolation. Samples were then fixed and processed for histology and immunohistochemistry with dentin area and dentin thickness quantitated using Image J. Wnt3a and Wnt10a over-expressing lentivirus and vector-only lentivirus (control) samples were prepared and incubated with E16.5 dental mesenchyme, following reconstitution with dental epithelium. Lentiviral transfection efficiency was tested using confocal microscopy following DAPI staining.
Immunohistochemistry and immunofluorescence
Tissue samples were embedded in paraffin and sectioned at 5-μm thickness, followed by hematoxylineosin staining. Unstained sections were prepared for immunohistochemistry. Immunohistochemical staining was performed using HRP-DAB Cell & Tissue Staining Kit (R&D, Minneapolis, MN, USA) per our prior methods47. Briefly, tissue sections were deparaffinized, blocked and incubated with anti-Alx3 (sc-22070, 1:200, Santa Cruz, Dallas, TX, USA), anti-DSPP (ab122321, 1:400, abcam, Cambridge, MA, USA), anti-vWF (ab68545, 1:400, abcam), anti-mitochondria (ab92824, 1:400, abcam), anti-Wnt3a (09-162, 1:400, Millipore, Darmstadt, Germany), anti-DMP1 (ab103203, 1:400, abcam) overnight at 4°C, followed by washing and incubation with HRP conjugated secondary antibodies.
For immunofluorescent staining, sections or cells were incubated with anti-CTNNB1 (ab32572, 1:500, abcam), anti-CD146 (ab75769, 1:400, abcam), anti-Collagen IV (ab6586, 1:400, abcam), anti-GFP (ab6556, 1:500, abcam), anti-PCNA (ab29, 1:500, abcam), anti-Ki67 (ab15580, 1:400, abcam), anti-Vimentin, ab92547, 1:400, abcam), anti-Caspase3 (AB3623, 1:400, Millipore), anti-nestin (MAB353, 1:400, Millipore), anti-ameloblastin (sc-50534, 1:200, Santa Cruz), anti-amelogenin (sc-32892, 1:200, Santa Cruz), anti-CK14 (sc-17104, 1:200, Santa Cruz), anti-DSP (sc-33586, 1:200, Santa Cruz), anti-S100 (S2644, 1:400, Sigma), anti-PGP 9.5 (ab8189, 1:400, abcam). Secondary antibodies, Alexa Fluor® 488 Goat anti-rabbit IgG (H+L) Antibody (1:1000, Invitrogen, Carlsbad, CA, USA), Alexa Fluor® 594 Goat anti-rabbit IgG (H+L) Antibody (1:1000, Invitrogen), Alexa Fluor® 594 Goat anti-mouse IgG (H+L) Antibody (1:1000, Invitrogen) or Alexa Fluor® 647 donkey anti-goat IgG (H+L) Antibody (1:1000, Invitrogen) were applied for 60 min. Samples were sealed with Vecta shield mounting medium containing DAPI. Images were taken by using Nikon A1 Confocal or Leica DMI6000B.
Cell culture
Following IRB approval and informed consent, freshly extracted human mandibular third molar teeth from two anonymous female donors (17 and 18 years of age) were collected to isolate mesenchymal stem/progenitor cells (MSCs) from dental pulp tissue per our prior methods46. All pulp tissue was removed and immediately treated with 3 mg/mL type I collagenase and 4 mg/mL dispase in L-DMEM for 1 h at 37°C. Following enzymatic digestion, single-cell suspension was made by filtering through a cell strainer and plated in 25 cm2 flasks (L-DMEM with 10% FBS, 2 mM L-glutamine, 1% Antibiotic-Antimycotic) at 37°C with 5% CO2.
Dental epithelium and mesenchyme cells were isolated from E16.5 tooth germs under dissection microscope with enzymatic digestion. Dental epithelium cells were co-cultured with lentiviral transfected dental mesenchyme cells in Transwell. Cells were cultured in DMEM/F12 medium with 10% fetal bovine serum and 1% Antibiotic-Antimycotic for 4 days. RNA was extracted separately from epithelium and mesenchyme cells.
Human Umbilical Vein Endothelial Cells (HUVECs) were purchased from Thermo Scientific (Cat#: C0035C, Rockford, IL, USA) and cultured in Medium 200 supplemented with Large Vessel Endothelial Supplement (LVES). Cells were passaged upon 80% confluence, with medium change every other day. Only low passage (≤3) cells were used.
TUNEL
Apoptotic cells were immunodetected by commercial DeadEnd™ Fluorometric TUNEL System (Promega, Madison, WI, USA) per manufacturer instructions. Paraffin sections were fixed by 4% paraformaldehyde once deparaffinized and rehydrated, followed by permeabilization and accessibility of DNA strand breaks by Terminal deoxynucleotidyl transferase (TdT). Labeled dUTPs were visualized using immunofluorescence microscopy.
Lentivirus transfection
Alx3’s reading frame was cloned into pWPI vector with GenScript, control pWPI (Plasmid #12254, Addgene, Cambridge, MA, USA) or pWPI-Alx3 (6.25 μg), pMD2.G (Plasmid #12259, Addgene) (0.625 μg) and psPAX2 (Plasmid #12260, Addgene) (3.125μg), per our prior methods46. Vectors were transfected into 293T cells at 80% confluency using Calcium Phosphate Transfection Kit (Invitrogen). Virus supernatant was collected 2 days following transfection and filtered with a 0.45-μm membrane, and further purified with Lenti-X Maxi Purification Kit (Clontech, Mountain View, CA, USA) per manufacturer protocol. Cells (< passage 3) were cultivated up to 30-50% confluence and transfected with lentivirus in 8 μg/mL polybrene (Santa Cruz). Transfected cells were GFP-sorted and further passaged 3-5 times to verify GFP labeling. For Alx3 knockdown, cells (< passage 3) were cultivated up to 30-50% confluence and infected with shAlx3 particles (sc-38645-V) in 5 μg/mL polybrene with infected cells selected with 5 μg/mL puromycin dihydrochloride (Santa Cruz) and further passaged 3-5 times, with shRNA lentiviral particles (sc-108080) as a control.
Chromatin Immunoprecipitation (ChIP)
ChIP assay was performed with Imprint Chromatin Immunoprecipitation Kit (Sigma, St. Louis, MO, USA). Briefly, stripwells were incubated with 1-μg Alx3 polyclonal antibody (sc-22068, Santa Cruz) or 1-μg normal Mouse IgG as a control in 100-μl Antibody Buffer for 2 hrs at R.T. on an orbital shaker. Formaldehyde cross-linked cells (1×107 cells) were quenched with ice-cold 1.25 M glycine, followed by resuspension and lysing. Nuclear pellets were resuspended in 400 μl Shearing Buffer containing Protease/Phosphatase Inhibitor Cocktail, and sonicated to shear cross-linked DNAto an optimal fragment of 200bp to 1000bp. Sonicated chromatins were diluted with equal volume of Dilution Buffer, with 5% diluted samples set aside on ice as Input. A total of 100-μl sonicated samples were added to preincubated beads in previously prepared stripwells and incubated at R.T. for 2 hrs on an orbital shaker. Following wash (4x) with IP-wash buffer, bound DNA-protein complexes were eluted from the beads by crosslinking reversal through incubating with Reversing Solution at 4°C overnight. Finally, the lysates were mixed with Binding Solution and collected following flow through and centrifugation. Final eluted DNAs were subjected to qPCR with ChIP PCR primers. Primer sequences are listed in Supplementary Table 1 including Alx3 binding element (JASPAR CORE database).
Western Blot
Total proteins were extracted by RIPA Buffer (Thermo Scientific) with Protease/Phosphatase Inhibitor Cocktail (Cell Signaling Technology, Danvers, MA, USA). Nucleus proteins were extracted with NE-PER® Nuclear and Cytoplasmic Extraction kit (Thermo Scientific) per manufacturer protocols. Cell lysate proteins (12 pg) were separated by 4-12% SDS-PAGE, transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA), and detected with anti-Alx3 (ab64985, 1:500, abcam), anti-CTNNB1 (ab32572, 1:500, abcam), anti-DSPP (sc-73632, 1:200, Santa Cruz), anti-Runx2 (ab76956, 1:400, abcam), anti-DMP1 (sc-6551, 1:200, Santa Cruz), anti-Axin2 (ab32197, 1:400, abcam) anti-VEGF (sc-7269, 1:200, Santa Cruz), anti-pVEGFR2 (2474S, 1:500, CST), anti-Wnt3a (09-162, 1:400, Millipore), anti-Wnt5a (ab72583, 1:400, abcam), anti-Wnt10a (ab106522, 1:400, abcam), anti-Collagen I (ab34710, 1:400, abcam), anti-tubulin (sc-398103, 1:200, Santa Cruz), anti-lamin B (sc-6216, 1:200, Santa Cruz), anti-Gapdh (sc-25778, 1:500, Santa Cruz) primary antibodies. Signals were detected by Odyssey® Imaging System (LI-COR) using IRDye® 800CW Secondary Antibodies (1:10000, LI-COR, Lincoln, NB, USA).
Quantitative RT-PCR
RNA was extracted using TRIzol (Invitrogen). Complementary DNA (cDNA) was synthesized with random hexamer primers and iScript™ cDNA Synthesis Kit (Bio-Rad). mRNA was measured by real-time qPCR following SYBR or Taqman protocol. Relative mRNA levels of target genes were normalized to GAPDH, with primer sequences listed in Supplementary Table 2.
Transient transfection and luciferase assay
Cells were electroporated following Neon™ protocol. Briefly, 1×106 MSCs were resuspended and mixed with 4-μg Alx3 plasmid, 20-ng reporter plasmid (pGL4) and 4-μg Topflash/Fopflash plasmid (Millipore) or 4-μg constructed SBE-OC-pGL4 reporter plasmid, with a SV-40 Renilla plasmid co-transfected. Cells treated with rhWnt3a (100 ng/ml) or rhBMP2 (100 ng/ml) were positive controls. Following 36-48 hrs, cells were lysed and assayed for reporter activity. Luciferase was measured using Dual Luciferase reporter assay (Promega). Relative luciferase was calculated by normalizing Firefly activity to Renilla activity.
We then amplified the 2.2-kb region of Wnt3a promoters of Homo sapiens (−2008 to +100, relative to transcription start site) and Mus musculus (−2008 to +100) followed by separate cloning into the Kpnl-Xhol and Kpnl-Hindlll sites of the promoterless vector (pGL4 Basic) containing firefly luciferase gene as a reporter (Promega). Additional luciferase constructs were generated from wild type by mutating Alx3 binding motifs (TAAT or ATTAto GCCG), and were confirmed by sequencing. 1.6 kB VEGFJuc plasmid was a gift from Patricia D’Amore (Addgene plasmid # 29667). Generally, a total of 1×105 MSCs or HUVECs were transfected with 1-μg Wnt3a-pGL4 or Wnt3a-m-pGL4 or VEGFJuc plasmid, along with 20-ng reporter plasmid (pGL4) and 1-μg Alx3 plasmid or EGFP plasmid by Lipofectamine 2000 per manufacturer protocol. After 48 hrs, cells were lysed and assayed for reporter activity.
Cell differentiation
Lentiviral transfected or shRNA transfected MSCs were cultivated in DMEM with 10% FBS, 5 mM β-glycerophosphate, 50-mM L-ascorbic acid 2-phosphate, 10-nM dexamethasone and 1% Antibiotic-Antimycotic per our prior methods46. In lentiviral transfected samples, 200 ng/ml rhDKKI (R&D Systems) or 10μM SP600125 (Sigma-Aldrich) were used to assay canonical and non-canonical Wnt pathways. shRNA transfected cells were exposed to 100 ng/ml Wnt3a (R&D Systems) or 200 ng/ml Wnt5a (R&D Systems). Mineral nodules were detected with 1% Alizarin Red-S (Sigma-Aldrich). Total RNA and proteins were detected from at least biological triplicates forqRT-PCR and western blot.
Cell migration assays
Cell migration was quantitated using 8-pm pore size Transwell@ (Corning, New York, NY, USA). Following 12-hr starvation, 2×105MSCs with Alx3 knockdown or over-expression in culture medium with 1% fetal bovine serum were plated in Transwell inserts, with 2% FBS in 750-μl DMEM as induction medium in the lower chamber. A total of 1×105 HUVECs in 500-μl L-DMEM + 1% FBS medium were serum-starved for 12 hrs and then plated in the Transwell insert, whereas Alx3 conditioned medium (CM) with L-DMEM + 10% FBS medium, vector control conditioned medium, Alx3 CM with VEGF neutralizing antibody or VEGFR2 inhibitor were added in the lower chamber. Following 16-hr incubation at 37°C and 5% CO2, unmigrated HUVECs were removed, and migrated HUVECs were washed with PBS, fixed with 4% paraformaldehyde, stained by 1% crystal violet staining and counted using Image J.
Cell proliferation assays
HUVECs were plated at a density of 3×103 cells per well. Viable cells were measured by Cell Counting Kit-8 (Sigma-Aldrich) at 450-nm absorbance, upon exposure to 10% FBS control medium, vector control conditioned medium or Alx3 conditioned medium.
Conditioned medium and angiogenesis assay
FACS-sorted, vector control or Alx3-restored MSCs were plated and then transferred to L-DMEM + 1% FBS medium at ~60% confluence. Conditioned medium was collected 2 days later, centrifuged at 1200 rpm for 5 mins to remove debris, further filtered with a 0.22-μm membrane and stored in Protease/Phosphatase Inhibitor Cocktail (Cell Signaling Technology) at −80°C until use48. Protein concentrations in the conditioned medium were determined by Pierce™ BCA Protein Assay Kit (Thermo Scientific).
In vitro angiogenesis/tube assays were performed using Angiogenesis Kit (Life Technology). Briefly, 5×104 HUVECs were seeded into Geltrex® Matrix (100 μl) coated 24-well plates containing L-DMEM + 1% FBS as negative control, or with rhVEGF (20 ng/mL) as positive control, or vector control conditioned medium, Alx3 conditioned medium and VEGF immunoprecipitated Alx3 conditioned medium or Alx3 conditioned medium combined with VEGF neutralizing antibody (sc7269, 1 ug/ml, Santa Cruz) or VEGF receptor 2 kinase inhibitor (5 nM, ab145888, abcam). HUVECs were incubated at 37°C for 16 hrs, stained with Calcein AM (2 mg/ml, Trevigen, Gaithersburg, MD, USA) to detect tube formation using Leica DFC300 FX fluorescence microscope. Quantification of tube length was performed using Image J in randomly selected 4 fields per sample49.
Total proteins were extracted from HUVECs upon treatment with 1% FBS control medium, medium with 20 ng/ml rhVEGF, vector control conditioned medium, Alx3 conditioned medium or Alx3 CM with VEGF neutralizing antibody or VEGFR2 inhibitor exposures for 30 mins. RNAs were extracted from the same groups at Day 2.
Immunoprecipitation (IP) and Enzyme-linked immunosorbent assay (ELISA)
VEGF was quantitatively measured in vector control conditioned medium, Alx3 conditioned medium and VEGF-depleted Alx3 conditioned medium using commercial ELISA Kit (DVE00, R&D) per manufacturer protocol. Briefly, 100-μl conditioned samples were incubated in microplate stripes at R.T. for 2 hrs, followed by 2-hr incubation with 200-μl conjugate and incubation of 200 μl substrate for 30 mins, followed by measuring 450-nm absorbance. Each experiment was conducted in at least biological triplicates. Alx3 conditioned medium was incubated with VEGF (sc-7269, 1 μg per 400 μg total protein, Santa Cruz) at 4°C overnight, with normal goat IgG serum as a negative control. A total of 100-μl agarose bead slurry was incubated for 1 hr with gentle agitation to obtain supernatants.
Preparation of human tooth scaffolds
Following IRB approval, healthy human premolars and molars were collected immediately following medically necessary tooth extractions. Periodontal tissue was scraped off, followed by PBS washing plus 2% Antibiotic-Antimycotic. The roots of disinfected teeth were sectioned with IsoMet® low-speed saw at 2-mm thickness50. Human tooth scaffolds were decellularized and treated with graded EDTA, followed by copious PBS washing plus 2% Antibiotic-Antimycotic ultrasonically50. All decellularized scaffolds were sterilized with ethylene oxide and incubated in DMEM at 37°C with 5% CO2 before use.
Scanning electron microscopy
Briefly, PBS- and EDTA-treated tooth scaffolds were washed with PBS, fixed with 2% glutaraldehyde, dehydrated and dried in a CO2 critical-point dryer. Dentinal tubules in tooth scaffolds were visualized by scanning electron microscope (SEM) (Hitachi S3500N, Dartronics, Pert Amboy, NJ, USA). Mandibular incisors were dissected and fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1-M cacodylate buffer solution (pH 7.4) at room temperature for 24 hrs followed by sample immersion in 0.1-M cacodylate, gradual dehydration in ascending ethanol concentrations and embedding in methylmethacrylate (MMA) resin (Fisher Scientific Company, Waltham, MA, USA). Microcloths with Metadi Supreme polycrystalline diamond suspensions of decreasing sizes (Buehler Lake Bluf, IL, USA) were used to polish sample surface. To expose MMA casts of dentine tubules for SEM, dentin surface was acid-etched with 37% phosphoric acid for 2-10s to remove the mineral, washed with 5.25% sodium hypochlorite for 5 min to remove the non-mineralized collagen, and coated with gold and palladium for imaging by high-vacuum SEM.
In vivo cell inoculation
Vector control or Alx3-restored adult MSCs (1×106 cells) were suspended at 37°C and 5% CO2 for 24 hrs in 1 mg/mL collagen gel (BD Bioscience) and inoculated into sterilized tooth scaffolds, followed by incubation at 37°C with 5% CO2 for 30 min for gelation. Cell-inoculated scaffolds, along with empty and gel only scaffolds, were implanted subcutaneously in the dorsum of athymic mice (6-8 wks, Harlan, East Millstone, NJ, USA) under anesthesia with isoflurane, after IACUC approval. Implants were retrieved after 4 and 8 weeks following euthanasia, and processed with 4% paraformaldehyde. Samples were demineralized in 0.5M EDTA (pH8) for 3 months. Paraffin sections at 4 μm thickness were processed, followed by hematoxylin and eosin, Masson’s Trichrome and immunohistochemical staining.
Tissue harvest and processing
Fourteen-day-old 2.3kb col1α1 promoter-EGFP transgenic mice were obtained from Jackson Laboratories (Sacramento, CA, USA) and sacrificed immediately for cell isolation per our prior methods47,51. Mandible and calvaria samples were harvested and fixed in 4% paraformaldehyde for 24 hrs at 4°C, followed by demineralization in 0.5M EDTA (pH 7.4) for 3 wks. For frozen sections, samples were embedded in OCT and sectioned at 10-μm thickness. Randomly selected sections were stained with hematoxylin and eosin with adjacent sections processed for immunohistochemistry.
Isolation of donor-matched odontoblasts and osteoblasts
Dental pulp of mandibular incisors from 14-day-old 2.3kb col1α1 promoter-EGFP transgenic mice was harvested per our prior methods47, and treated with 0.05% trypsin-EDTA (Invitrogen) and 1.5 U/ml collagenase P (Roche) in PBS at 37°C for 30 min on a shaker. Single-cell suspension was obtained by passing cells through narrowed Pasteur pipettes and a 70-μm strainer47,51. Donor-matched calvaria samples were isolated after removing cranial sutures and four sequential 15-min enzyme digestions at 37°C in 0.05% trypsin-EDTA and 1.5 U/ml collagenase P in PBS. GFP-positive cells from dental pulp and calvaria were isolated by FACS per our previous methods47,51.
In vivo subrenal capsule transplantation
Donor-matched odontoblasts and osteoblasts from 14-day-old 2.3kb col1α1 promoter-EGFP transgenic mice were loaded in collagen gel and transplanted in subrenal capsule in athymic mice (2-month-old, Harlan, Indianapolis, IN) for 4 wks. A longitudinal incision (~1.5 cm) was made on the central dorsal surface, following by blunt dissection. Another incision was made along the kidney’s longitudinal axis, followed by manual displacement of the kidney to relieve subrenal capsule and creation of a pocket for cell-loaded collagen gel graft. The subrenal pocket was sutured with 2 stitches, followed by skin closure with 2 clips. The implanted cell grafts were harvested in 4 wks and processed for paraffin embedding/sections and frozen sections.
RNA Microarray
Total RNA was extracted from GFP positive cells using a mirVana miRNA Isolation Kit (Ambion, Invitrogen, Grand Island, NY). For Gene Set Enrichment Analysis (GSEA) analysis, a tooth specific geneset was first built from the literatures and covered known genes expressed and/or functionally important in tooth development. The differentially expressed genes between tooth and bone from the microarray data were run on GSEA software for Tooth Specific Geneset enrichment analysis52. Positive enrichment scores indicate tooth-specific geneset enriched as compared to bone phenotype.
Endodontic procedures in preclinical large animal model and protein delivery
Minipigs (micro-Yucatan minipig, aged 24 months and weighing 110-120 kg; Sinclair Bio Resources, Windham, Maine) were fasted for 10 hrs before general anesthesia. Following sedation with Telazol, general anesthesia was induced by 2-5 mg/kg propofol intravenously and maintained by isoflurane and oxygen (1-5% with 2L O2/min). Once sedated, minipigs received prophylactic cefazolin (22 mg/kg; IV). Bupivacaine (0.5%) was injected intramucosally for mental nerve block. Six mandibular incisors in each minipig underwent endodontic or root-canal procedures. A preoperative digital radiograph was taken to check root-canal anatomy. Following rubber dam isolation, six anterior teeth were mechanically accessed with sterile high speed round burs. Dental pulp was extirpated with hand files and irrigated with sterile saline, followed by drying of the canal with sterile paper points. Recombinant human Wnt3a (50 ng/mL) and BMP7 (1.25 mg/mL) (R&D Systems, Minneapolis, MN) were mixed with 1-mg/mL neutralized rat tail collagen gel solution (BD Biosciences, Bedford, MA) and backfill-injected into the surgically prepared root canal. Protein-free collagen gel served as a control. Following gelation, the access cavity was sealed with Cavit™ and composite resin. All teeth were harvested 3 months later following minpig euthanasia.
Radiographs and micro-computed tomography (μCT)
The renal capsule transplantation sample was imaged with a Faxitron MX-20 radiography system (Faxitron, Buffalo Grove, IL). The fixed minipig mandibular incisors were scanned by μCT (Scanco, Bmttisellen, Switzerland). The scout views were used to locate the region of interest. Sequential transaxial images were acquired using 20-μm isotropic voxel size with 70 kV peak tube voltage, 114 μA current and 200 ms integration time. A 600 mg HA/cm threshold was adopted to distinguish mineralized tissue from soft tissue. Quantitative μCT data analysis was conducted in two different regions for each sample. Tissue volume and density were calculated by running the Scanco script for 3D segmentation evaluation. Typical 2D and 3D pCT cross-section images were acquired.
Nanoindentation
Newly-formed and original, native dentin were exposed following embedding with epoxy resin. The samples were mechanically polished up to 2000 grit and indented with a Nanoindenter (CSM instruments, Ashland, VA) with a 142.35° curvature diamond triangular pyramid shaped head, 50-nm radius and load set at 1 mN with 10 s loading, 10 s hold and 10 s unloading. Each sample was tested for 20 randomly selected areas.
Statistical analysis
Power analysis was calculated for all quantitative pig data with a type-one error rate of 0.05 and 80% power to claim non-inferiority of the treated groups (Wnt3a and/or BMP7) to the control. Two-sided Student’s t-tests were used for paired comparisons. One-way analysis of variance (ANOVA) was used with post-hoc Bonferroni adjustment for comparison between three or more groups. For smaller sample sizes or samples with skewed distribution, Mann–Whitney U or Kruskal-Wallis tests were performed. A p value <0.05 was considered as statistically significant. All statistical analyses were performed in SAS version 9.4. All in vitro experiments were performed in at least three independent biological samples.
Supplementary Material
Acknowledgements
We thank Q. Guo, P. Ralph-Birkett, and Y. Tse for administrative and technical assistance. We thank Dr. F. Meijlink and J. Deschamps for Alx3 fragment plasmid as a generous gift, Dr. M.S. Shelanski for suggestions of two antibodies, Dr. J.E. Nor for sharing his insight on VEGF signaling, Dr. Y. Zhang for technical help with Wnt3a luciferase promoter constructs and Dr. A.F. Fouad for citation of Survey of Dental Services Rendered. The work is supported by NIH grants R01DE025643, R01DE023112, R01AR065023, R01DE025969 and R01DE026297, and an Osteo Science Foundation grant to J.J. Mao. Scholarly effort of several co-authors was supported by National Natural Science Foundation of China grants 81570939, 81741106 and Beijing Municipal Administration of Hospitals’ Youth Programme (QML20161503) to J. Zhou, National Natural Science Foundation of China grants 81271110 to Z. Wang, 81170932 to J. Ling, 81371136 to X. Zhou, National Science and Technology Support Program 2014DFA31990 to Z. Wang, Program of International Science and Technology Cooperation 2014DFA31990 to L. Ye.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests in this article. J.J.M. has co-founded Innovative Elements and Xinkewo with goals to develop regenerative products.
Data Availability Statement
Data supporting the findings of this study are available within the article from corresponding authors upon reasonable request.
References
- 1.Sacco A, Doyonnas R, Kraft R, Vitorovic S & Blau HM Self-renewal and expansion of single transplanted muscle stem cells. Nature 456, 502–506, doi: 10.1038/nature07384 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiba Y et al. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 538, 388–391, doi: 10.1038/nature19815 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Fox IJ et al. Stem cell therapy. Use of differentiated pluripotent stem cells as replacement therapy fortreating disease. Science 345, 1247391, doi: 10.1126/science.1247391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmons PJ, Przepiorka D, Thomas ED & Torok-Storb B Host origin of marrow stromal cells following allogeneic bone marrow transplantation. Nature 328, 429–432, doi: 10.1038/328429a0 (1987). [DOI] [PubMed] [Google Scholar]
- 5.Mao JJ & Prockop DJ Stem cells in the face: tooth regeneration and beyond. Cell Stem Cell 11,291–301, doi: 10.1016/j.stem.2012.08.010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schworer S et al. Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature 540, 428–432, doi: 10.1038/nature20603 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao G et al. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature 534, 119–123, doi: 10.1038/nature17959 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei K et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature 525, 479–485, doi: 10.1038/nature15372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs Y et al. Sept4/ARTS regulates stem cell apoptosis and skin regeneration. Science 341, 286–289, doi: 10.1126/science.1233029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nacu E, Gromberg E, Oliveira CR, Drechsel D & Tanaka EM FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature 533, 407–410, doi: 10.1038/nature17972 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Mokalled ΜH et al. Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science 354, 630–634, doi: 10.1126/science.aaf2679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ten Berge D et al. Mouse Alx3: an aristaless-like homeobox gene expressed during embryogenesis in ectomesenchyme and lateral plate mesoderm. Dev Biol 199, 11–25, doi: 10.1006/dbio.1998.8921 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Mallarino R et al. Developmental mechanisms of stripe patterns in rodents. Nature 539, 518–523, doi: 10.1038/nature20109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song JJ et al. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat Med 19, 646–651, doi: 10.1038/nm.3154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakao K et al. The development of a bioengineered organ germ method. Nat Methods 4, 227–230, doi: 10.1038/nmeth1012 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Jiang N et al. Exosomes mediate epithelium-mesenchyme crosstalk in organ development. ACS Nano 11,7736–7746, doi: 10.1021/acsnano.7b01087 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thesleff I, Tummers M. Tooth organogenesis and regeneration. StemBook; Cambridge (MA); 2008. [PubMed] [Google Scholar]
- 18.Kaukua N et al. Glial origin of mesenchymal stem cells in a tooth model system. Nature 513, 551–554, doi: 10.1038/nature13536 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Endo Y et al. Wnt-3a and Dickkopf-1 stimulate neurite outgrowth in Ewing tumor cells via a Frizzled3- and c-Jun N-terminal kinase-dependent mechanism. Mol Cell Biol 28, 2368–2379, doi: 10.1128/MCB.01780-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakhwani S, Garcia-Sanz P & Vallejo M Alx3-deficient mice exhibit folic acid-resistant craniofacial midline and neural tube closure defects. Dev Biol 344, 869–880, doi: 10.1016/j.ydbio.2010.06.002 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Hu K & Olsen BR Osteoblast-derived VEGF regulates osteoblast differentiation and bone formation during bone repair. J Clin Invest 126, 509–526, doi: 10.1172/JCI82585 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song J et al. Smadl transcription factor integrates BMP2 and Wnt3a signals in migrating cardiac progenitor cells. Proceedings of the National Academy of Sciences of the United States of America 111, 7337–7342, doi: 10.1073/pnas.1321764111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivkin E et al. The linear ubiquitin-specificdeubiquitinase gumby regulates angiogenesis. Nature 498, 318–324, doi: 10.1038/nature12296 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carmeliet P & Jain RK Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298–307, doi: 10.1038/nature10144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee CH et al. Regeneration of the articular surface of the rabbit synovial joint by cell homing: a proof of concept study. Lancet 376, 440–448, doi: 10.1016/S0140-6736(10)60668-X (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diogenes A, Ruparel NB, Shiloah Y & Hargreaves KM Regenerative endodontics: Away forward. J Am Dent Assoc 147, 372–380, doi: 10.1016/j.adaj.2016.01.009 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Zhang YD, Chen Z, Song YQ, Liu C & Chen YP Making a tooth: growth factors, transcription factors, and stem cells. Cell Res 15, 301–316, doi: 10.1038/sj.cr.7290299 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Oral Health in America: A Report of the Surgeon General (Executive Summary). Rockville, MD: US Department of Health and Human Services, National Institute of Dental and Craniofacial Research, National Institutes of Health; 2010. [Google Scholar]
- 29.Survey of Dental Services Rendered, <http://www.aae.org/about-aae/news-room/endodontic-treatment-statistics.aspx> (2007).
- 30.Ng YL, Mann V, Rahbaran S, Lewsey J & Gulabivala K Outcome of primary root canal treatment: systematic review of the literature - part 1. Effects of study characteristics on probability of success. International endodontic journal 40, 921–939, doi: 10.1111/j.1365-2591.2007.01322.x (2007). [DOI] [PubMed] [Google Scholar]
- 31.Hunter DJ et al. Wnt Acts as a Prosurvival Signal to Enhance Dentin Regeneration. J Bone Miner Res 30, 1150–1159, doi: 10.1002/jbmr.2444 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Arany PR et al. Photoactivation of endogenous latent transforming growth factor-betal directs dental stem cell differentiation for regeneration. Science translational medicine 6, 238ra269, doi: 10.1126/scitranslmed.3008234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langer R & Vacanti JP Tissue engineering. Science 260, 920–926 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Lee CH et al. Protein-releasing polymeric scaffolds induce fibrochondrocytic differentiation of endogenous cells for knee meniscus regeneration in sheep. Sci Trans Med 6, 266ra171, doi: 10.1126/scitranslmed.3009696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franz WM, Zaruba M, Theiss H & David R Stem-cell homing and tissue regeneration in ischaemic cardiomyopathy. Lancet 362, 675–676, doi: 10.1016/S0140-6736(03)14240-7 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Askari AT et al. Effect of stromal-cell-derived factor 1 on stem-cell homing and tissue regeneration in ischaemic cardiomyopathy. Lancet 362, 697–703, doi: 10.1016/S0140-6736(03)14232-8 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Kavanagh DP & Kalia N Hematopoietic stem cell homing to injured tissues. Stem Cell Rev 7, 672–682, doi: 10.1007/s12015-011-9240-z (2011). [DOI] [PubMed] [Google Scholar]
- 38.Bottino MC et al. Bioactive nanofibrous scaffolds for regenerative endodontics. J Dent Res 92, 963–969, doi: 10.1177/0022034513505770 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang GT et al. Stem/progenitor cell-mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an in vivo model. Tissue engineering. Part A 16, 605–615, doi: 10.1089/ten.TEA.2009.0518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iohara K et al. Complete pulp regeneration after pulpectomy by transplantation of CD105+ stem cells with stromal cell-derived factor-1. Tissue engineering. Part A 17, 1911–1920, doi: 10.1089/ten.TEA.2010.0615 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Iohara K et al. A novel combinatorial therapy with pulp stem cells and granulocyte colony-stimulating factor for total pulp regeneration. Stem cells translational medicine 2, 521–533, doi: 10.5966/sctm.2012-0132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conde MC et al. Stem cell-based pulp tissue engineering: variables enrolled in translation from the bench to the bedside, a systematic review of literature. Int Endod J 49, 543–550, doi: 10.1111/iej.12489 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Nakashima M et al. Pulp regeneration by transplantation of dental pulp stem cells in pulpitis: a pilot clinical study. Stem Cell Res Ther 8, 61, doi: 10.1186/s13287-017-0506-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xuan K et al. Deciduous autologous tooth stem cells regenerate dental pulp after implantation into injured teeth. Sci Transl Med 10, doi: 10.1126/scitranslmed.aaf3227 (2018). [DOI] [PubMed] [Google Scholar]
- 45.He L et al. Regenerative Endodontics by Cell Homing. Dent Clin North Am 61, 143–159, doi: 10.1016/j.cden.2016.08.010 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Zhou C et al. Lhx8 mediated Wnt and TGFbeta pathways in tooth development and regeneration. Biomaterials 63, 35–46, doi: 10.1016/j.biomaterials.2015.06.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang N et al. Postnatal epithelium and mesenchyme stem/progenitor cells in bioengineered amelogenesis and dentinogenesis. Biomaterials 35, 2172–2180, doi: 10.1016/j.biomaterials.2013.11.061 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawamura R, Hayashi Y, Murakami H & Nakashima M EDTA soluble chemical components and the conditioned medium from mobilized dental pulp stem cells contain an inductive microenvironment, promoting cell proliferation, migration, and odontoblastic differentiation. Stem Cell Res Ther 7, 77, doi: 10.1186/s13287-016-0334-z (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khoo CP, Micklem K & Watt SM A comparison of methods for quantifying angiogenesis in the Matrigel assay in vitro. Tissue Eng Part C Methods 17, 895–906, doi: 10.1089/ten.TEC.2011.0150 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li R et al. Human treated dentin matrix as a natural scaffold for complete human dentin tissue regeneration. Biomaterials 32, 4525–4538, doi: 10.1016/j.biomaterials.2011.03.008 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Zheng Y et al. Mesenchymal Dental Pulp Cells Attenuate Dentin Resorption in Homeostasis. Journal of dental research, doi: 10.1177/0022034515575347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


