Abstract
Background
The elderly (≥65 years) are one of the populations most at risk for respiratory tract infections (RTIs). The aim of this study was to determine whether nasal and/or oropharyngeal microbiota profiles are associated with age and RTIs.
Methods
Nasal and oropharyngeal swabs of 152 controls and 152 patients with an RTI were included. The latter group consisted of 72 patients with an upper respiratory tract infection (URTI) and 80 with a lower respiratory tract infection (LRTI). Both nasal and oropharyngeal swabs were subjected to microbiota profiling using amplicon sequencing of the 16S rRNA gene. Moraxella species were determined using quantitative real-time PCR and culture.
Results
Based on the microbiota profiles of the controls and the patients with an RTI, eight nasal and nine oropharyngeal microbiota clusters were defined. Nasal microbiota dominated by either Moraxella catarrhalis or Moraxella nonliquefaciens was significantly more prevalent in elderly compared to mid-aged adults in the control group (p = 0.002). Dominance by M. catarrhalis/nonliquefaciens was significantly less prevalent in elderly with an LRTI (p = 0.001) compared to controls with similar age.
Conclusions
Nasal microbiota dominated by M. catarrhalis/nonliquefaciens is associated with respiratory health in the elderly population.
Keywords: Elderly, Microbiota, Nasal passages, Oropharynx, Respiratory tract infection
Background
Respiratory tract infections (RTIs) remain one of the leading causes of morbidity and mortality worldwide [1, 2]. Whereas upper respiratory tract infections (URTIs) are very common but rarely life threatening, lower respiratory tract infections (LRTIs) are responsible for more severe illnesses, like pneumonia. The populations at risk are the very young (< 5 years) and the elderly (≥ 65 years).
During the first year of life, host and environmental factors, such as genetic predisposition, mode of delivery, infant feeding, exposure to antibiotics, vaccination and geographic location, affect the development of the airway microbiota [3, 4]. For the nose, this results gradually in a microbiota profile dominated by Dolosigranulum, Corynebacterium, Haemophilus, Moraxella, Staphylococcus and/or Streptococcus spp. [5]. In the first years of life, microbiota profiles dominated by Dolosigranulum and/or Corynebacterium spp. are more stable and are positively associated with lower rates of RTIs [6–9]. Less stable microbiota profiles characterized by the high abundance of the oral bacteria Haemophilus and Streptococcus spp. are associated with a higher likelihood of an RTI and their proportion is significantly higher in samples obtained during RTIs when compared to ‘healthy’ samples [6–10]. Furthermore, these microbiota profiles have been associated with an increased risk of recurrent wheeze and asthma in later childhood [9]. For microbiota profiles dominated by Moraxella spp. variable results have been reported regarding their stability and association with RTIs [6–9]. The differences in susceptibility to RTIs likely arise from a complex interplay between mucosa, innate and adaptive immunity, and airway microbiota.
In elderly, the mechanisms of the heightened susceptibility to RTIs are still poorly understood. Immunosenescence, defined as age-related deterioration of both innate and adaptive immunity, seems to impair elderly to elicit effective immune responses against pathogens [11]. In addition, immunosenescence might influence the composition of the human microbiota [12, 13]. Only few studies have addressed the upper airway microbiota in elderly [14–17], and even less in relation to RTIs [18]. The available study typically focusses on the oropharynx for LRTIs, which is suggested to be the main source of microorganisms to the lower airways in adults [19]. They observed three microbiota profiles strongly associated with pneumonia and either dominated by Lactobacillus, Rothia or Streptococcus (pseudo)pneumoniae. In contrast, three other microbiota clusters were correlated with respiratory health and were all characterized by more diverse profiles containing higher abundances of especially Prevotella, Veillonella and Leptotrichia. However, these microbiota profiles were observed in both in mid-aged adults and elderly. The aim of this study was to determine whether nasal and/or oropharyngeal microbiota profiles are associated with age and RTIs.
Methods
Source of samples
Between Augustus 2012 and Augustus 2014, respiratory swabs were collected from adult patients who were visiting the otorhinolaryngology outpatient clinic or hospitalized at the pulmonary ward of the Reiner de Graaf Gasthuis (Delft, The Netherlands). Two swabs were collected from each patient using sterile flocked swabs (Puritan Medical Products, Maine, USA). One swab was obtained from the head of the concha inferior near the anterior nares and a second swab was obtained from the oropharynx. Each swab was stored in 2 mL STGG (skim milk, tryptone, glucose, glycerol) medium. In total, swabs of 370 patients without clinical symptoms of an RTI and of 211 patients with a suspected URTI or LRTI were collected. For the current analysis, swabs collected from patients with a suspected RTI who received antibiotics in the week before visiting the outpatient clinic and swabs collected from hospitalized patients > 1 day after admission were excluded (n = 59 patients), leaving swabs of 152 patients with an RTI for further analysis. Subsequently, swabs of 152 patients without clinical symptoms (i.e. controls) were selected based on sample collection date, age and sex to match the patient group so well as possible. None of them had received antibiotics 1 week prior to sample collection.
Nucleic acid extraction and sequencing
Nucleic acids were extracted from 500 μL STGG medium and eluted in a final volume of 100 μL with the MagNA Pure 96 instrument using the MagNA Pure 96 DNA and Viral NA Large Volume kit and the Pathogen Universal protocol (Roche Diagnostics, Basel, Switzerland). Amplicon sequencing of the 16S ribosomal RNA (rRNA) gene was performed as described elsewhere [20]. Briefly, a fragment of ~ 464 bp of the V3-V4 region of the 16S rRNA gene was amplified and sequenced with the MiSeq desktop sequencer (Illumina, San Diego, USA).
Microbiota analysis
Sequencing data was processed following the QIIME1 pipeline. Open reference OTU clustering of high-quality sequences (≥ 100 bp in length with a quality score ≥ Q20) was conducted using UCLUST at a 97% similarity level against a pre-clustered version of the Augustus 2013 GreenGenes database. No low abundance filtering was used. See for further details Additional file 1. Operational taxonomic units (OTUs) with Alloiococcus or Propionibacterium annotation were renamed. Dolosigranulum is known to be misclassified in the GreenGenes database as Alloiococcus [21]. BLAST search confirmed that the representative sequence matched Dolosigranulum in BLAST. Propionibacterium spp. have been reclassified to the genus Cutibacterium [22].
Moraxella species determination
Moraxella species were identified using quantitative real-time PCR (qPCR) and culture. A highly specific qPCR targeting the copB gene of Moraxella catarrhalis was performed on the isolated DNA of all samples and performed as described elsewhere [20]. Culture was performed to determine which Moraxella spp. was present in the samples negative for M. catarrhalis. For culture, 200–300 μL STGG medium was inoculated on blood agar plates (Becton, Dickinson and Company, New Jersey, USA) and incubated at 35 °C in a 5% CO2 incubator. Species were identified by matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) analyzer with software version 1.6.7.1000 (Bruker corporation, Billerica, USA).
Statistical analysis
For statistical analysis the software package SPSS version 26 was used. Statistically significant differences in variables between the controls and the patients with an RTI was calculated using the Mann-Whitney U and chi-square test for continuous and categorical data, respectively. After the core members of the nasal and oropharyngeal microbiota were determined, hierarchical clusters of microbiota profiles were defined using the free python script ‘hierarchical_clustering.py’, which was written by Nathan Salomonis of the J. David Gladstone Institutes (San Francisco, CA, USA) and can be found on the following webpage: https://github.com/nsalomonis/altanalyze/blob/master/visualization_scripts/clustering.py. This script uses the Euclidean distance to measure the dissimilarity between each pair of observations. The prevalence of each microbiota cluster per age group was calculated for the controls and the patients with an RTI. Subsequently, the Fisher’s Exact test was performed to determine whether microbiota clusters were associated with age and/or RTIs. For the cluster associated with age and RTIs, Fisher’s Exact tests were performed to determine whether season of sampling, sex, smoking, young children at home, comorbidities, the use of inhaler or nasal spray were also associated with this cluster in the control group. Furthermore, statistically significant differences in the relative abundance of the genus Moraxella between groups was calculated using the Mann-Whitney U test.
Results
Study population
Nasal and oropharyngeal swabs of 152 controls and 152 patients with an RTI were selected (Table 1). The 152 controls were visiting the outpatient clinic mainly for an audiogram or hearing complaints (37%), or allergy, skin test or immunotherapy (24%). Of the 152 patients with an RTI, 72 (47%) were suffering from an URTI (i.e. a common cold, sinusitis, tonsillitis or laryngitis). The remaining 80 (53%) patients were hospitalized with a LRTI (i.e. a pneumonia, chronic obstructive pulmonary disease exacerbation, bronchitis or asthma exacerbation), which was diagnosed by the treating physician. Both groups differed significantly in age (P = 0.013).
Table 1.
Population characteristics
| Group | Controls (n = 152) | Patients with a respiratory tract infection (n = 152) |
|---|---|---|
| Age, mean ± SD (range)* | 53 ± 19 (18–92) | 58 ± 20 (18–89) |
| Age category, n (%)** | ||
| < 65 years | 102 (67) | 81 (53) |
| ≥ 65 years | 50 (33) | 71 (47) |
| Sex, n (%) | ||
| Female | 79 (52) | 86 (57) |
| Male | 73 (48) | 66 (43) |
| Season of sampling, n (%) | ||
| Autumn | 40 (26) | 33 (22) |
| Winter | 47 (31) | 59 (39) |
| Spring | 35 (23) | 44 (29) |
| Summer | 30 (20) | 16 (11) |
| Reason for visit/hospitalisation, n (%) | ||
| Allergy/skin test/immunotherapy | 37 (24) | 5 (3) |
| Audiogram/hearing complaints | 56 (37) | 7 (5) |
| Dizzines | 9 (6) | 0 (0) |
| Infection | 0 (0) | 110 (72) |
| Follow-up | 10 (7) | 7 (5) |
| Nose spray | 0 (0) | 6 (4) |
| Other; earsa | 23 (15) | 7 (5) |
| Other; noseb | 10 (7) | 4 (3) |
| Other; throatc | 7 (5) | 2 (1) |
| Other; accompaniment | 0 (0) | 4 (3) |
| Upper airway infection, n (%) | ||
| Common cold | Not applicable | 47 (31) |
| Laryngitis | 4 (3) | |
| Sinusitis | 14 (9) | |
| Tonsillitis | 7 (5) | |
| Lower airway infection, n (%) | ||
| Asthma exacerbation | Not applicable | 2 (1) |
| Bronchitis | 3 (2) | |
| COPD exacerbation | 24 (16) | |
| Pneumonia | 51 (34) | |
Abbreviations: COPD Chronic obstructive pulmonary disease, SD Standard deviation. Statistically significant differences in variables between both groups was calculated using the Mann-Whitney U and chi-square test for continuous and categorical data, respectively. *P = 0.013. **P = 0.014
aOther; ears included cleaning of ears and inserting grommets
bOther; nose included septum deviation, frequent nosebleeds and choanal polyp
cOther; throat included complaints of long-lasting cough or difficult swallowing movement
Determination of the core microbiota of the nasal passages and oropharynx
To determine whether the nasal and/or oropharyngeal microbiota were associated with age and RTIs, first core microbiota profiles were defined using amplicon sequencing of the 16S rRNA gene. A mean of 77,414 reads per swab were obtained with sequencing, which resulted in a mean OTU of 50 for the nasal swabs and 83 for the oropharyngeal swabs.
In the nasal passages of the 152 controls and 152 patients with an RTI, the 10 most abundant genera/families were Corynebacterium (mean relative abundance of 28%), Staphylococcus (24%), Moraxella (12%), Dolosigranulum (7%), Streptococcus (5%), Haemophilus (3%), Peptoniphilus (3%), Cutibacterium (2%), Anaerococcus (2%), and Enterobacteriaceae (2%). Together these bacteria account for 88% of the classified sequences. Interestingly, three microbiota profiles were dominated (i.e. ≥ 50% relative abundance) by one of the less abundant genera Pseudomonas and Neisseria.
In the oropharynx, Prevotella (mean relative abundance of 26%), Veillonella (16%), Streptococcus (11%), Neisseria (7%), Fusobacterium (6%), Leptotrichia (5%), Haemophilus (5%), Rothia (3%), Porphyromonas (3%), and Actinobacillus (2%) were the 10 most abundant genera, accounting for 84% of the classified sequences. Four microbiota profiles were dominated by Lactobacillus or Staphylococcus.
These bacteria are the core members of the nasal and oropharyngeal microbiota of the controls and patients with an RTI. Separate analyses for both patient groups resulted in comparable core members (Additional file 2).
Microbiota clustering analysis based on nasal or oropharyngeal core members
To define clusters of microbiota profiles, hierarchical clustering was performed based on the nasal or oropharyngeal core members (Additional file 3a, b). For the nasal passages, eight microbiota clusters were defined (Additional file 4a). Cluster I was characterized by a relatively high abundance of Haemophilus, Neisseria or Streptococcus (Hae/Nei/Str), II by Moraxella (Mor), III by Staphylococcus and Corynebacterium (Sta, Cor), IV by Corynebacterium and Dolosigranulum (Cor, Dol), V by Corynebacterium (Cor), VI by Staphylococcus (Sta), VII by Moraxella and Corynebacterium (Mor, Cor), and VIII by Dolosigranulum, Haemophilus, Cutibacterium, Enterobacteriaceae or Streptococcus (Dol/Hae/Cut/Ent/Str). These microbiota clusters had a mean Shannon diversity index ranging between 2.18 and 4.50.
For the oropharynx, nine microbiota clusters were defined of which cluster I was characterized by a relatively high abundance of Prevotella and Fusobacterium (Pre, Fus), II/III by Prevotella and Veillonella (Pre, Vei), IV by Prevotella (Pre), V by Actinobacillus, Haemophilus, Staphylococcus, Rothia or Neisseria (Act/Hae/Sta/Rot/Nei), VI by Streptococcus and Veillonella (Str, Vei), VII by Lactobacillus (Lac), VIII by Streptococcus and Rothia (Str, Rot), and IX by Streptococcus, Neisseria, Actinobacillus, Lactobacillus or Staphylococcus (Str/Nei/Act/Lac/Sta) (Additional file 4b). Compared to the nasal microbiota clusters, the microbiota profiles within the oropharyngeal microbiota clusters were more variable which was illustrated by the mean Shannon diversity index ranging between 3.13 and 6.45. There was no correlation between the nasal and oropharyngeal microbiota clusters.
Nasal and oropharyngeal microbiota clusters related to higher age in the control group
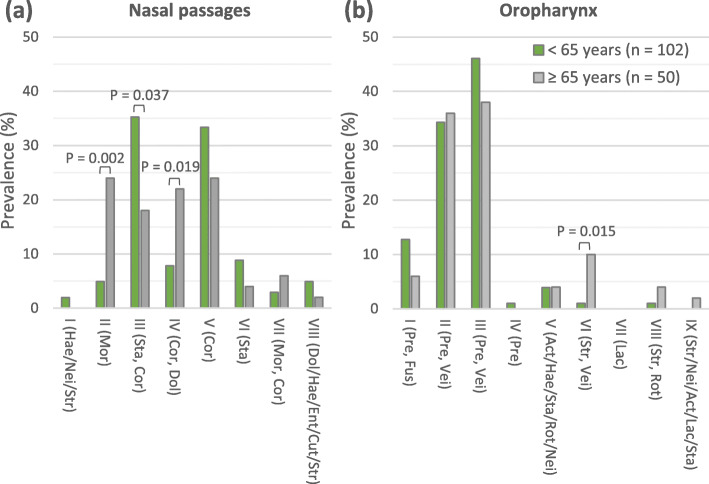
After clusters of nasal and oropharyngeal microbiota profiles were defined, their prevalence in the control group was calculated per age group (adults aged ≥65 years or < 65 years). Of the eight nasal microbiota clusters, three microbiota clusters were associated with age (Fig. 1a). Cluster II (Mor) and IV (Cor, Dol) were significantly more prevalent in adults aged ≥65 years compared to adults aged < 65 years (P ≤ 0.019), whereas cluster III (Sta, Cor) was significantly less prevalent in adults aged ≥65 years (P = 0.037). Of the nine oropharyngeal microbiota clusters, cluster VI (Str, Vei) was significantly more prevalent in adults aged ≥65 years (P = 0.015; Fig. 1b). These data showed that specific microbiota profiles of both the nasal passages and oropharynx are associated with higher age.
Fig. 1.
Prevalence of microbiota clusters among the controls aged < 65 and ≥ 65 years. a nasal microbiota clusters. b oropharyngeal microbiota clusters. Act: Actinobacillus; Cor: Corynebacterium; Cut: Cutibacterium; Dol: Dolosigranulum; Ent: Enterobacteriaceae; Fus: Fusobacterium; Hae: Haemophilus; Lac: Lactobacillus; Mor: Moraxella; Nei: Neisseria; Pre: Prevotella; Rot: Rothia; Sta: Staphylococcus; Str: Streptococcus; Vei: Veillonella. Genera separated from each other by a comma are both represented in a relatively high abundance in each microbiota profile of the relevant cluster. Genera separated from each other by a slash indicates that one of these genera is present in a relatively high abundance. All p-values are based on Fisher’s Exact test. Correction for multiple testing was not performed
Nasal and oropharyngeal microbiota clusters related to higher age and RTIs
Subsequently, the prevalence of the nasal and oropharyngeal microbiota clusters of the patients with any RTI (Fig. 2a, b), URTI (Fig. 2c, d) or LRTI (Fig. 2e, f) were compared to the control group. Nasal microbiota cluster II (Mor) was strongly associated with higher age and LRTI as it was significantly less prevalent in patients with a LRTI who passed the age of 65 years compared to controls with similar age (P = 0.001).
Fig. 2.
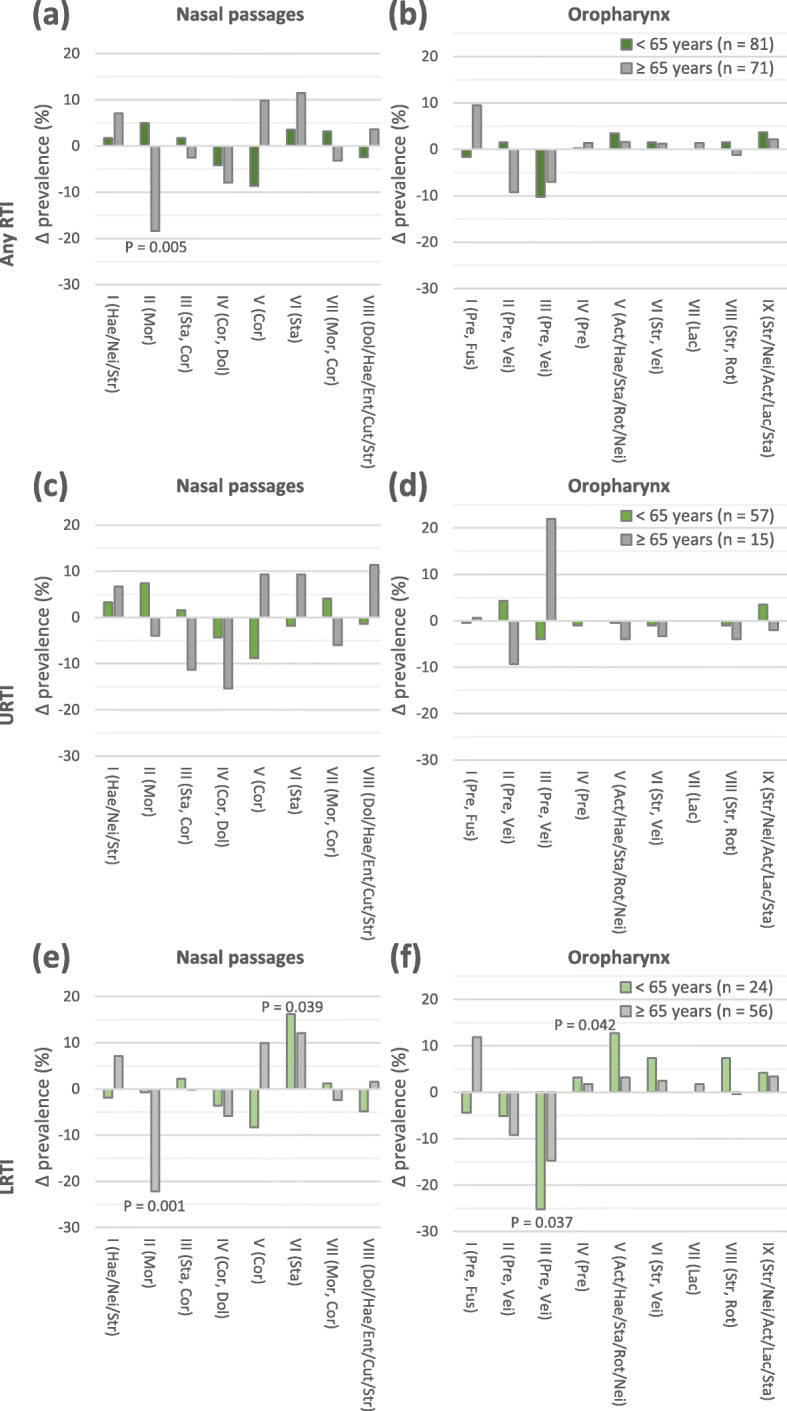
Comparison of prevalence of microbiota clusters between controls and patients per age group. a nasal and b oropharyngeal microbiota clusters of controls and patients with any respiratory tract infection (RTI). c nasal and d oropharyngeal microbiota clusters of controls and patients with an upper respiratory tract infection (URTI). e nasal and f oropharyngeal microbiota clusters of controls and patients with a lower respiratory tract infection (LRTI). Act: Actinobacillus; Cor: Corynebacterium; Cut: Cutibacterium; Dol: Dolosigranulum; Ent: Enterobacteriaceae; Fus: Fusobacterium; Hae: Haemophilus; Lac: Lactobacillus; Mor: Moraxella; Nei: Neisseria; Pre: Prevotella; Rot: Rothia; Sta: Staphylococcus; Str: Streptococcus; Vei: Veillonella. Genera separated from each other by a comma are both represented in a relatively high abundance in each microbiota profile of the relevant cluster. Genera separated from each other by a slash indicates that one of these genera is present in a relatively high abundance. All P-values are based on Fisher’s Exact test. Correction for multiple testing was not performed
Nasal microbiota cluster VI (Sta; P = 0.039), oropharyngeal microbiota cluster III (Pre, Vei; P = 0.037) and oropharyngeal microbiota cluster V (Act/Hae/Sta/Rot/Nei; P = 0.042) were moderately associated with LRTIs in patients aged < 65 years. Interestingly, oropharyngeal microbiota cluster VII (Lac) was only present in patients with a LRTI who passed the age of 65 years. These data indicate that both the nasal and oropharyngeal microbiota differed between the controls and patients with a LRTIs.
Nasal microbiota cluster II dominated by Moraxella spp.
Of all identified microbiota clusters, nasal cluster II (Mor) was of most interest since it was associated with higher age and less prevalent in elderly with a LRTI compared to the healthy elderly population. This finding was strengthened by the significant difference in mean abundance of Moraxella spp. between the age groups of the controls (P = 0.003) and between the controls and patients with a LRTI who passed the age of 65 years (P = 0.008; Table 2). In the control group, no association with season of sampling, sex, smoking, young children at home, comorbidities, the use of inhaler or nasal spray was found.
Table 2.
Mean relative abundance of Moraxella spp. per population group
| Population group | All ages | < 65 years | ≥ 65 years | |||
|---|---|---|---|---|---|---|
| Mean ± SD (%) | Range (%) | Mean ± SD (%) | Range (%) | Mean ± SD (%) | Range (%) | |
| Controls and patients | 12 ± 26 | 0–100 | 9 ± 23 | 0–100 | 15 ± 29 | 0–100 |
| Controls | 13 ± 28 | 0–100 | 7 ± 20* | 0–99 | 25 ± 37 */**/*** | 0–100 |
| Patients | 10 ± 24 | 0–100 | 12 ± 27 | 0–100 | 8 ± 20** | 0–81 |
| Patients with URTI | 15 ± 29 | 0–100 | 14 ± 29 | 0–100 | 15 ± 30 | 0–81 |
| Patients with LRTI | 6 ± 18 | 0–100 | 8 ± 23 | 0–100 | 6 ± 15*** | 0–67 |
Abbreviations: LRTI Lower respiratory tract infection, SD Standard deviation, URTI Upper respiratory tract infection. Statistically significant differences between groups was calculated using the Mann-Whitney U test. *P = 0.003. **P = 0.018. ***P = 0.008
To determine whether M. catarrhalis was representing nasal cluster II (Mor), a qPCR was performed. Of all 29 nasal swabs, five (18%) were positive for M. catarrhalis. Culture data suggested that the remaining 24 (82%) of the swabs within nasal cluster II (Mor) were represented by M. nonliquefaciens.
Discussion
To the best of our knowledge, this is the largest study on the nasal and oropharyngeal microbiota and its relation to both URTIs and LRTIs in elderly. Based on the microbiota profiles of the controls and the patients with an RTI, we defined eight nasal and nine oropharyngeal microbiota clusters. One of the nasal microbiota clusters was strongly associated with age and RTIs.
The results of this study showed that nasal cluster II dominated by M. catarrhalis/nonliquefaciens, was significantly more prevalent in the healthy elderly population compared to the healthy mid-aged adults. Interestingly, M. catarrhalis/nonliquefaciens was significantly less prevalent in elderly with a LRTI compared to the healthy elderly population, suggesting an association between M. catarrhalis/nonliquefaciens and respiratory health in elderly. Previous reports have shown that Moraxella spp. become predominant community members over time in most young children [5–9]. Their microbiota as well as their immune system are in development, whereas both innate and adaptive immunity seem to deteriorate in elderly [11]. In essence, it might tolerate the same bacterial species. This might explain the significantly higher prevalence of M. catarrhalis/nonliquefaciens in the healthy elderly population. However, conflicting results have been reported regarding the role of Moraxella spp. in the pathogenesis of RTIs in young children. Some studies found that profiles dominated by M. catarrhalis/nonliquefaciens was associated with respiratory health [5–8], while others reported that Moraxella spp. were associated with high susceptibility to LRTIs [9]. Since M. catarrhalis has been considered as being a pathogen for certain disease entities (e.g. COPD exacerbation and otitis media), it is most likely that M. nonliquefaciens is actually associated with respiratory health.
Nasal and oropharyngeal microbiota clusters moderately associated with LRTIs were characterized by a relatively high abundance of a potential pathogen, such as Staphylococcus, Actinobacillus, Haemophilus, and Rothia spp. The difference in prevalence compared to the healthy population was observed in both age groups but was only significantly different in the mid-age adults. This means that no microbiota cluster was defined that could elucidate why elderly are more susceptibility to LRTIs. However, the data does indicate that both the nasal and oropharyngeal microbiota have impact on lower airway health in adults while it is generally assumed that only the oropharynx is involved in the pathogenesis of LRTI [22].
A cross-sectional study of Steenhuijsen Piters and colleagues revealed 11 (sub)clusters of oropharyngeal microbiota profiles [18]. Three clusters were associated with pneumonia which were characterized by a relative high abundance of S. (pseudo)pneumoniae, Rothia spp. or Lactobacillus spp. In contrast, three other microbiota clusters were correlated with respiratory health and contained high abundances of Prevotella, Veillonella and Leptotrichia. In our study, Streptococcus, Rothia and Lactobacillus dominated only a limited number of oropharyngeal microbiota profiles. Notably, the oropharyngeal microbiota cluster characterized by a relatively high abundance of Lactobacillus was only covered by patients with a LRTI. Furthermore, we observed a moderate association between an oropharyngeal microbiota cluster with high abundances of Prevotella and Veillonella and respiratory health in mid-aged adults.
URTIs are mainly caused by viruses and previous reports have shown that Streptococcus and Haemophilus spp. are associated with viral infections [23–26]. The interactions between viruses and the airway microbiota may affect the course of the disease and subsequent respiratory health [27]. In our study, nasal microbiota clusters characterized by a high abundance of Streptococcus or Haemophilus spp. were associated with the presence of respiratory viruses in patients with a URTI (data not shown). However, no significant difference in prevalence was observed compared to the healthy population.
A limitation of this study is that the data was collected at one timepoint. Longitudinal and more comprehensive data regarding microbiota composition and function as well as immunogenic status is required in order to elucidate the mechanism of the heightened susceptibility to RTIs in elderly. Longitudinal data is also required to confirm that nasal microbiota has impact on the lower airway health in adults. Stronger correlations might have been found when data was used from a matched case-control study, controls were healthy relatives of the patients, only patients with a confirmed pneumonia were included, nasopharynx was sampled and when specimens were collected during hospital admission, reducing antibiotic usage prior to specimen collection. Lastly, sputum collection in case of a LRTI would have been valuable to identify the causative pathogen [20].
Conclusions
We showed that nasal microbiota dominated by M. catarrhalis/nonliquefaciens is associated with respiratory health in the elderly population. Further research is required to determine which species is associated with respiratory health and whether it is a positive association. In case of a positive association, efforts should be made to uphold these bacteria to promote respiratory health in the elderly population.
Supplementary information
Additional file 1. Qiime1 version 1.9.1 Scripts and settings.
Additional file 2 Core microbiota members of the (a) nasal passages or (b) oropharynx based on the profiles of controls and/or patients with a respiratory tract infection (RTI).
Additional file 3 Hierarchical clustering of (a) the nasal and (b) the oropharyngeal microbiota profiles of the 152 controls and 152 patients with a respiratory tract infection based on the core members.
Additional file 4 Characteristics of (a) nasal and (b) oropharyngeal microbiota clusters based on the core members of the 152 controls and 152 patients with a respiratory tract infection.
Acknowledgements
The authors are grateful to Ingrid Poot, Anne Hout and Michiel Weber (DDL Diagnostic Laboratory) for their technical assistance and Maurine A. Leverstein-van Hall (Alrijne Hospital, Leiden, the Netherlands) for providing the culture data.
Abbreviations
- Act
Actinobacillus
- COPD
Chronic obstructive pulmonary disease
- Cor
Corynebacterium
- Cut
Cutibacterium
- Dol
Dolosigranulum
- Ent
Enterobacteriaceae
- Fus
Fusobacterium
- Hae
Haemophilus
- Lac
Lactobacillus
- LRTIs
Lower respiratory tract infections
- Nei
Neisseria
- MALDI-TOF MS
Matrix-assisted laser desorption ionization time of flight mass spectrometry
- Mor
Moraxella
- OTUs
Operational taxonomic units
- Pre
Prevotella
- qPCR
Quantitative real-time PCR
- Rot
Rothia
- rRNA
Ribosomal RNA
- RTIs
Respiratory tract infections
- Sta
Staphylococcus
- STGG
Skim milk, tryptone, glucose, glycerol
- Str
Streptococcus
- URTIs
Upper respiratory tract infections
- Vei
Veillonella
Authors’ contributions
EM, HH, MK were involved in the design of the study. HH and JK recruited the patients. EM collected the swabs, performed the molecular tests, performed data analyses and drafted the manuscript with support from EK and CW. All authors discussed the results and contributed to the final manuscript. The authors read and approved the final manuscript.
Funding
There was no external funding involved.
Availability of data and materials
The datasets generated datasets are available in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) repository with the accession number PRJNA596902.
Ethics approval and consent to participate
Local medical ethical review board approval was obtained for this study (METC Zuidwest Holland, The Hague, The Netherlands; reference 2012–181). Written, informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
LD and WQ are shareholders of DDL Diagnostic Laboratory. The other authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12931-020-01443-8.
References
- 1.Collaborators GBDCoD Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: a systematic analysis for the global burden of Disease study 2017. Lancet. 2018;392(10159):1736–1788. doi: 10.1016/S0140-6736(18)32203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Disease GBD, Injury I, Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the global burden of Disease study 2016. Lancet. 2017;390(10100):1211–1259. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Unger SA, Bogaert D. The respiratory microbiome and respiratory infections. J Inf Secur. 2017;74(Suppl 1):S84–SS8. doi: 10.1016/S0163-4453(17)30196-2. [DOI] [PubMed] [Google Scholar]
- 4.Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15(5):259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch A, Levin E, van Houten MA, Hasrat R, Kalkman G, Biesbroek G, et al. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMed. 2016;9:336–345. doi: 10.1016/j.ebiom.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190(11):1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 7.Bosch A, de Steenhuijsen Piters WAA, van Houten MA, Chu M, Biesbroek G, Kool J, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Am J Respir Crit Care Med. 2017;196(12):1582–1590. doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- 8.Man WH, van Houten MA, Merelle ME, Vlieger AM, Chu M, Jansen NJG, et al. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir Med. 2019;7(5):417–426. doi: 10.1016/S2213-2600(18)30449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17(5):704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Man WH, Clerc M, de Steenhuijsen Piters WAA, van Houten MA, Chu M, Kool J, et al. Loss of microbial topography between Oral and nasopharyngeal microbiota and development of respiratory infections early in life. Am J Respir Crit Care Med. 2019;200(6):760–770. doi: 10.1164/rccm.201810-1993OC. [DOI] [PubMed] [Google Scholar]
- 11.Krone CL, van de Groep K, Trzcinski K, Sanders EA, Bogaert D. Immunosenescence and pneumococcal disease: an imbalance in host-pathogen interactions. Lancet Respir Med. 2014;2(2):141–153. doi: 10.1016/S2213-2600(13)70165-6. [DOI] [PubMed] [Google Scholar]
- 12.Bomar L, Brugger SD, Lemon KP. Bacterial microbiota of the nasal passages across the span of human life. Curr Opin Microbiol. 2018;41:8–14. doi: 10.1016/j.mib.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bischoff SC. Microbiota and aging. Curr Opin Clin Nutr Metab Care. 2016;19(1):26–30. doi: 10.1097/MCO.0000000000000242. [DOI] [PubMed] [Google Scholar]
- 14.Whelan FJ, Verschoor CP, Stearns JC, Rossi L, Luinstra K, Loeb M, et al. The loss of topography in the microbial communities of the upper respiratory tract in the elderly. Ann Am Thorac Soc. 2014;11(4):513–521. doi: 10.1513/AnnalsATS.201310-351OC. [DOI] [PubMed] [Google Scholar]
- 15.Roghmann MC, Lydecker AD, Hittle L, DeBoy RT, Nowak RG, Johnson JK, et al. Comparison of the microbiota of older adults living in nursing homes and the community. mSphere. 2017;2(5):e00210–e00217. doi: 10.1128/mSphere.00210-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira PAB, Aho VTE, Paulin L, Pekkonen E, Auvinen P, Scheperjans F. Oral and nasal microbiota in Parkinson’s disease. Parkinsonism Relat Disord. 2017;38:61–67. doi: 10.1016/j.parkreldis.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 17.Liu CM, Price LB, Hungate BA, Abraham AG, Larsen LA, Christensen K, et al. Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv. 2015;1(5):e1400216. doi: 10.1126/sciadv.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Steenhuijsen Piters WA, Huijskens EG, Wyllie AL, Biesbroek G, van den Bergh MR, Veenhoven RH, et al. Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J. 2016;10(1):97–108. doi: 10.1038/ismej.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson RL, de Koff EM, Bogaert D. Characterising the respiratory microbiome. Eur Respir J. 2019;53(2):1801711. doi: 10.1183/13993003.01711-2018. [DOI] [PubMed] [Google Scholar]
- 20.van den Munckhof EHA, de Koning MNC, Quint WGV, van Doorn LJ, Leverstein-van Hall MA. Evaluation of a stepwise approach using microbiota analysis, species-specific qPCRs and culture for the diagnosis of lower respiratory tract infections. Eur J Clin Microbiol Infect Dis. 2019;38(4):747–754. doi: 10.1007/s10096-019-03511-4. [DOI] [PubMed] [Google Scholar]
- 21.Walker RE, Walker CG, Camargo CA, Jr, Bartley J, Flint D, Thompson JMD, et al. Nasal microbial composition and chronic otitis media with effusion: a case-control study. PLoS One. 2019;14(2):e0212473. doi: 10.1371/journal.pone.0212473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scholz CFP, Kilian M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. Nov., Cutibacterium gen. Nov. and Pseudopropionibacterium gen. Nov. Int J Syst Evol Microbiol. 2016;66(11):4422–4432. doi: 10.1099/ijsem.0.001367. [DOI] [PubMed] [Google Scholar]
- 23.Mansbach JM, Hasegawa K, Henke DM, Ajami NJ, Petrosino JF, Shaw CA, et al. Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol. 2016;137(6):1909–1913. doi: 10.1016/j.jaci.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal microbiota, host Transcriptome, and Disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194(9):1104–1115. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edouard S, Million M, Bachar D, Dubourg G, Michelle C, Ninove L, et al. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur J Clin Microbiol Infect Dis. 2018;37(9):1725–1733. doi: 10.1007/s10096-018-3305-8. [DOI] [PubMed] [Google Scholar]
- 26.de Steenhuijsen Piters WAA, Jochems SP, Mitsi E, Rylance J, Pojar S, Nikolaou E, et al. Interaction between the nasal microbiota and S. pneumoniae in the context of live-attenuated influenza vaccine. Nat Commun. 2019;10(1):2981. doi: 10.1038/s41467-019-10814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly MS, Surette MG, Smieja M, Pernica JM, Rossi L, Luinstra K, et al. The nasopharyngeal microbiota of children with respiratory infections in Botswana. Pediatr Infect Dis J. 2017;36(9):e211–e2e8. doi: 10.1097/INF.0000000000001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Qiime1 version 1.9.1 Scripts and settings.
Additional file 2 Core microbiota members of the (a) nasal passages or (b) oropharynx based on the profiles of controls and/or patients with a respiratory tract infection (RTI).
Additional file 3 Hierarchical clustering of (a) the nasal and (b) the oropharyngeal microbiota profiles of the 152 controls and 152 patients with a respiratory tract infection based on the core members.
Additional file 4 Characteristics of (a) nasal and (b) oropharyngeal microbiota clusters based on the core members of the 152 controls and 152 patients with a respiratory tract infection.
Data Availability Statement
The datasets generated datasets are available in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) repository with the accession number PRJNA596902.