Abstract
Background
Microbial biorefinery approaches are beginning to define renewable and sustainable routes to clean-burning and non-fossil fuel-derived gaseous alkanes (known as ‘bio-LPG’). The most promising strategies have used a terminal fatty acid photodecarboxylase, enabling light-driven propane production from externally fed waste butyric acid. Use of Halomonas (a robust extremophile microbial chassis) with these pathways has enabled bio-LPG production under non-sterile conditions and using waste biomass as the carbon source. Here, we describe new engineering approaches to produce next-generation pathways that use amino acids as fuel precursors for bio-LPG production (propane, butane and isobutane blends).
Results
Multiple pathways from the amino acids valine, leucine and isoleucine were designed in E. coli for the production of propane, isobutane and butane, respectively. A branched-chain keto acid decarboxylase-dependent pathway utilising fatty acid photodecarboxylase was the most effective route, generating higher alkane gas titres over alternative routes requiring coenzyme A and/or aldehyde deformylating oxygenase. Isobutane was the major gas produced in standard (mixed amino acid) medium, however valine supplementation led to primarily propane production. Transitioning pathways into Halomonas strain TQ10 enabled fermentative production of mixed alkane gases under non-sterile conditions on simple carbon sources. Chromosomal integration of inducible (~ 180 mg/g cells/day) and constitutive (~ 30 mg/g cells/day) pathways into Halomonas generated production strains shown to be stable for up to 7 days.
Conclusions
This study highlights new microbial pathways for the production of clean-burning bio-LPG fuels from amino acids. The use of stable Halomonas production strains could lead to gas production in the field under non-sterile conditions following process optimisation.
Keywords: Biofuels, Propane, Isobutane, Butane, Microbial pathway engineering, Escherichia coli, Bio-LPG
Background
Global concerns about dwindling fossil fuel supplies have led to an urgent need to transition towards more sustainable bio-economy strategies [1]. This includes implementing economically viable routes to sustainable clean-burning biofuels derived from renewable waste biomass or industrial waste streams with minimal downstream processing costs. Microbial strategies for gaseous bio-alkane production (propane and butane) are promising because of the existence of well-established global markets (20 million tonnes propane per annum) and fuels distribution infrastructures [2]. The ‘drop-in’ nature of propane can boost the calorific value of existing biogas supplies, and propane has lower energy requirements for liquefaction and/or transportation [2]. In addition, the aerosol industry generates customer-specific gaseous hydrocarbon blends (propane, butane and isobutane) for use as propellants [3].
Engineered microbial routes to bio-propane and other gaseous hydrocarbons are now well established [4–9]. These pathways are dependent on variant forms of either aldehyde deformylating oxygenase (ADO) from Prochlorococcus marinus [4–6, 9, 10] or fatty acid photodecarboxylase (CvFAP) from Chlorella variabilis [4, 8, 11]. ADO variant A134F catalyses the NADPH-dependent decarbonylation of butyraldehyde to propane in the presence of its electron transfer partner ferredoxin [4, 6, 12]. CvFAP variants G462I and G462V were shown to catalyse the blue light-dependent decarboxylation of butyric and valeric acids to propane and butane, respectively [4]. In the latter case, microbial alkane production can be achieved in vivo by direct feeding of volatile fatty acid (VFA) precursor molecules. Alternative routes require the upregulation of butyraldehyde or butyric acid, utilising pathways derived from fatty acid biosynthesis [5], reverse β-oxidation [7], valine biosynthesis [9] or the clostridial butanol pathway [6].
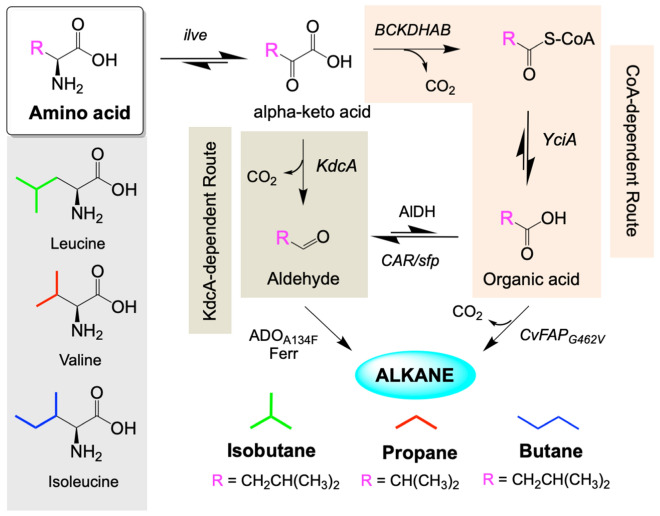
We recently explored the commercial potential of CvFAP-dependent bio-LPG production (bio-propane/butane blends) using recombinant E. coli, Halomonas and Synechocystis as microbial chassis [4]. Amino acids were explored as potential bio-alkane precursors, where valine (C3), isoleucine (n-C4) and leucine (i-C4) could be converted to propane, butane and isobutane, respectively (Fig. 1). In these pathways, amino acids undergo deamination by leucine 2-oxoglutarate transaminase (ilvE; [13]) to generate α-keto acids, followed by the action of branched-chain keto acid decarboxylase (KdcA; [14]) to generate the respective aldehyde. The action of the aldehyde dehydrogenase 3-hydroxypropionaldehyde dehydrogenase (Hpad; [15]) produced the equivalent carboxylic acid, which underwent decarboxylation to bio-alkane gas by CvFAPG462I (Fig. 1). This KdcA-dependent route successfully produced bio-LPG blends in both E. coli and Halomonas, using proteinaceous waste as the source of amino acids [4].
Fig. 1.
Overall scheme of the biocatalytic production of gaseous hydrocarbons propane, isobutane and butane from amino acids valine, leucine and isoleucine, respectively. Enzymes: ilvE = leucine 2-oxoglutarate transaminase from E. coli; BCKDHAB = human branched-chain α-keto acid dehydrogenase complex; YciA = acyl-CoA thioester hydrolase from Haemophilus influenza; CAR = carboxylic acid reductase from Mycobacterium marinum; sfp = maturation factor phosphopantetheinyl transferase from Bacillus subtilis; CvFAP = fatty acid photodecarboxylase from Chlorella variabilis; ADO = aldehyde deformylating oxygenase from Prochlorococcus marinus; Ferr = ferredoxin from Synechocystis sp. PCC6803 and KdcA = branched-chain keto acid decarboxylase from Lactococcus lactis. AlDH enzymes: αKGSDH = α-ketoglutaric semialdehyde dehydrogenase from Burkholderia ambifaria; PadA = phenylacetaldehyde dehydrogenase 17 from E. coli; or Hpad = 3-hydroxypropionaldehyde dehydrogenase puuC from E. coli
In this study, we have explored the potential of additional CvFAP- and ADO-dependent routes to gaseous bio-alkanes from the amino acids valine, isoleucine and leucine. A second coenzyme A (CoA)-dependent [16] route was designed (Fig. 1; Additional file 1: Figs. S1, S2), and the original KdcA-dependent route was refactored to identify the optimal aldehyde dehydrogenase homologue and promoter system. Optimised pathways were incorporated into E. coli and the industrial chassis Halomonas either on plasmids or as stable chromosomally integrated strains. Fermentative production of gaseous bio-alkanes was achieved, demonstrating the potential of this approach for bio-LPG production. This study illustrates a potentially viable alternative to the VFA-fed CvFAP-dependent bio-alkane route for commercial exploitation, as food waste could act as both the bio-alkane precursor and carbon source, eliminating the need to supplement cultures with potentially cytotoxic VFAs.
Results and discussion
CoA-dependent pathway design from amino acids to bio-alkanes
A CoA-dependent (VFA-producing) pathway with CvFAP was designed, based on the ADO-dependent clostridial butanol pathway from butyryl-CoA to propane [6]. Similar to the KdcA pathway, an initial ilvE-dependent amino acid deamination generates the respective α-keto acids (Fig. 1). This is followed by CoA-dependent decarboxylation of the α-keto acid, catalysed by the human branched-chain α-keto acid dehydrogenase complex (BCKDHAB) [16]. CoA is subsequently eliminated by acyl-CoA thioester hydrolase (YciA; Haemophilus influenza) [6] to generate the respective carboxylic acid (Fig. 1). This is similar to the native leucine to isovaleric acid route described in Propionibacterium freudenreichii [17]. Finally, decarboxylative alkane production is catalysed by the G462V or G462I variant of CvFAP.
To generate the ADO-dependent pathway version, the carboxylic acid precursor is converted into the respective aldehyde by carboxylic acid reductase (CAR; Mycobacterium marinum) activated by the maturation factor phosphopantetheinyl transferase (sfp; Bacillus subtilis) [6]. The terminal step is the ADO- and ferredoxin-dependent decarbonylation of the aldehydes to bio-alkanes (Fig. 1). All of the enzymes in this pathway were shown to express in E. coli in an active form (Additional file 1: Figs. S3, S4; [4, 6]). An alternative CoA-dependent route from threonine to butane was also designed (Additional file 1: Figs. S2–S4), with the initial ilvE step substituted for threonine dehydratase (ilvA) from E. coli [18] and hydrocarbon chain extension performed by the E. coli leuABCD operon [19]. However initial constructs did not produce detectable propane due to the absence of activity of the LeuABCD-catalysed steps (results not shown). Therefore, the threonine-to-butane pathway was not investigated further in this study.
CoA-dependent pathway bio-alkane gas production
To test the performance of in vivo bio-alkane production from amino acids, a series of five ADO-dependent DNA expression constructs were assembled in E. coli (AFYSCIB; ADOA134F-Ferr-YciA-sfp-CAR-ilvE-BCKDHAB). These constructs varied by the presence of either one or two isopropyl β-d-thiogalactopyranoside (IPTG)-inducible (trc) or constitutive promoters (proD or trcΔlac) [20] located upstream of the ilvE and/or ADOA134F genes (Fig. 2a inset). The constitutive promoter trcΔlac was generated by removing the repressor protein gene (lacI) and the operator region from a standard trc promoter. Initial in vivo trials were performed with the single proD-controlled construct, expressed in multiple E. coli strains. Highest gaseous bio-alkane production was observed in the double deletion strain BL21(DE3)ΔΔ [4, 5], with propane making up to 79% of the total hydrocarbon gas in the culture headspace (0.11 ± 0.01 mg/L propane; Additional file 1: Fig. S5 and Table S1). This strain was designed previously to reduce the competing pathway from butyraldehyde to butanol by knocking out the E. coli aldehyde reductases Ahr and YqhD [5]. It has been shown to increase in vivo propane titres in ADO-dependent pathways [5, 6], so was selected as the E. coli chassis for the remainder of this work.
Fig. 2.
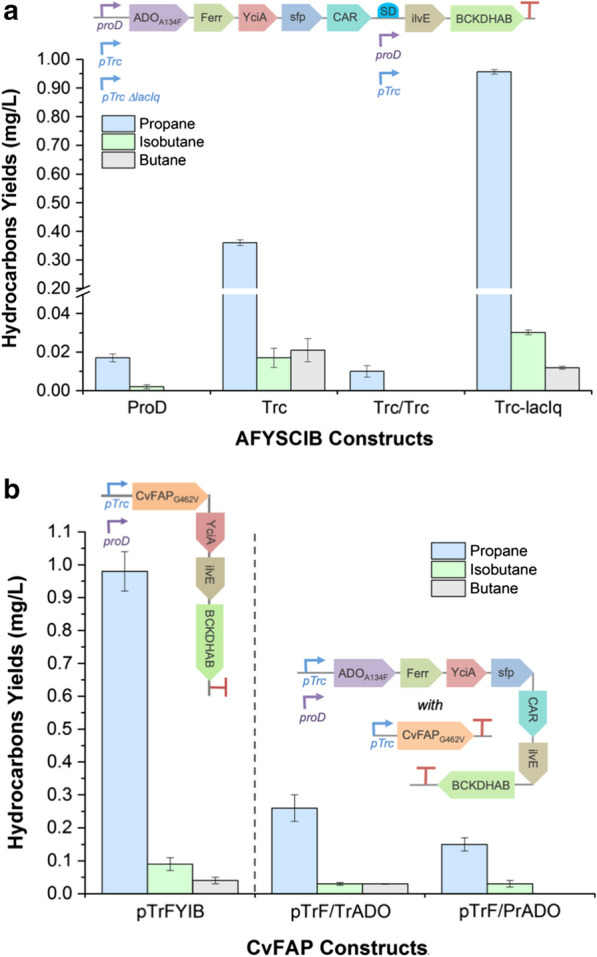
Hydrocarbon production of E. coli strain BL21(DE3)∆∆ containing engineered a ADO- and b CvFAP-dependant routes. Cultures (10 mL) were grown in LB medium containing 30 μg/mL kanamycin and 50 µg/mL chloramphenicol (pTrF/TrADO and pPrF/TrADO only) for 4–6 h at 37 °C and 180 rpm. Protein induction (100 μM IPTG) was performed for trc-containing constructs, and triplicate aliquots (1 mL) each of 3 biological replicate cultures were sealed into glass vials (3 mL) at 30 °C for 16–18 h at 200 rpm. For CvFAP-dependent constructs, the cultures were illuminated continuously with a blue LED (455 nm or 470 nm). Gaseous hydrocarbon levels were determined by manual headspace injection on an Agilent 490 Micro GC. Errors represent one standard deviation of the replicates (biological and/or technical triplicates). Constructs (inset): ProD = pPrAFYSCIB; ProD/ProD = pPrAFYSCPrIB; Trc = pTrAFYSCIB; Trc/Trc = pTrAFYSCTrIB and Trc-lacIq = pTrΔLAFYSCIB
In vivo alkane production by five AFYSCIB constructs in E. coli showed the highest bio-alkane titres when only one promoter (trc or proD) was present upstream of ADOA134F (Fig. 2a; Additional file 1: Table S2). The single trc–containing construct showed a 21-fold increase in propane titres over the equivalent proD-containing version. The best performing construct contained a single trcΔlac promoter (0.96 ± 0.01 mg propane/L culture), with yields similar to those obtained with the ADOA134F-dependent clostridial butanol pathway from glucose (3.4 mg/L [6]). Low levels of butane and/or isobutane were also detected with the single promoter constructs, with levels up to 10% of the total alkanes (up to 0.02 ± 0.01 mg butane/L culture; Fig. 2a). These data show that the CoA-dependent route can successfully generate low levels of multiple gaseous alkanes under standard E. coli cultivation conditions in the absence of additional amino acid supplementation. However, a prior study showed an ADO-dependent reverse-β-oxidation biosynthesis pathway from glycerol generated ca 22-fold higher butane titres (0.46 mg/L [7]).
Additional CoA-dependent pathways to bio-alkanes were designed to simplify the pathway and substitute the catalytically slow ADO enzyme [12] for the light-dependent CvFAPG462V variant enzyme [4]. This latter enzyme has a reported reaction quantum yield of greater than 80% [8]. Pathway modification required the elimination of the two terminal steps catalysed by CAR/sfp and ADOA134F/Ferr, as CvFAP directly acts on the carboxylic acid precursor (Fig. 1). This shortened pathway may have the added advantage of a lower overall cellular burden in E. coli as fewer recombinant genes are expressed.
We generated two CvFAP-dependent constructs (FG462VYIB; CvFAP-YciA-ilvE-BCKDHAB; Fig. 2b inset), which differed by the type of promoter upstream of CvFAPG462V (trc or proD). Gaseous alkane production was seen in E. coli only in the trc-containing construct in the presence of blue light, at titres similar to those seen with the best AFYSCIB pathway (0.98 ± 0.06 mg/L culture; Fig. 2b; Additional file 1: Table S2). Propane titres were at least tenfold higher than butane or isobutane, which differs from the CvFAP-dependent KdcA route [4] where the dominant gas produced was isobutane (4.8-fold higher than propane). This is likely due to substrate preferences of KdcA and/or differences in the amino acid concentrations present in the culture medium [21].
The requirement for blue light in CvFAP-dependent pathways necessitates adaptations to traditional in vivo bacterial cultivation strategies, as prolonged exposure to blue light (380–500 nm) is known to decrease the viability of E. coli [22] and cause the photoinactivation of CvFAP [4, 23]. Therefore, dark cycles (light absence) might be required periodically during fermentation to ensure the maintenance of sufficient cell density and the regeneration of biocatalysts. As ADO-dependent pathways do not require light, we tested E. coli strains expressing CvFAPG462V with AFYSCIB pathways for alkane gas production (Fig. 2b inset). However, in each case bio-alkane gas production was lower than what was obtained with the individual pathways alone (0.15–0.26 mg propane/L culture; Fig. 2b; Additional file 1: Table S2). This could potentially be due to the additional cellular burden of extra gene overexpression and/or the requirement of two antibiotics to maintain both plasmids.
Overall, we have demonstrated gaseous alkane production in E. coli using the CoA-dependent routes, although the levels are at best ~ 6.5-fold lower than the KdcA-dependent route with CvFAP [4]. Therefore, we focused our studies on improving the KdcA-dependent pathway, which utilises CvFAP in place of ADO. This route shows more promise as a potential industrially useful route to gaseous bio-alkanes from amino acids.
KdcA-dependent route refactoring
The existing KdcA-dependent pathway (Fig. 1) is a four-gene construct (IHKFG462I; ilvE-Hpad-KdcA-CvFAP [4]) controlled by two trc promoters upstream of ilvE and KdcA. We targeted the aldehyde dehydrogenase (AlDH) and terminal decarboxylation (CvFAP) steps in this pathway to see if further improvements in bio-alkane yields could be obtained. The existing AlDH is 3-hydroxypropionaldehyde dehydrogenase (Hpad) from E. coli, which have known activity towards isovaleraldehyde, butyraldehyde and valeraldehyde [15]. Two alternative aldehyde dehydrogenase homologues were selected, based on their known substrate specificities for short chain aldehydes. The first was α-ketoglutaric semialdehyde dehydrogenase (αKGSDH) from Azospirillum brasilense as it displays a known broad specificity [24]. The second homologue is phenylacetaldehyde dehydrogenase 17 (PadA) from E. coli, which is involved in phenylalanine degradation [25]. Each of these AlDH genes expressed in soluble active form in E. coli (Additional file 1: Figures S3, S4). In addition, both G462I and G462V variants of CvFAP were incorporated to see which supports the highest levels of bio-alkane gas production in vivo.
Six KdcA-dependent pathways were constructed under trc control, differing by the AlDH homologue and the specific CvFAP variant (Fig. 3a inset). A shortened ADO-dependent version of this pathway (IKAF; ilvE-KdcA-ADOA134F-Ferr; Fig. 1) was not investigated in this study as prior studies suggested a CvFAP-dependent route is likely to be more productive [4]. For example, the CvFAP route is much more redox efficient as ADO needs a lot of cofactor, whereas CvFAP uses light for activation. In the CvFAP-dependent version the conversion of aldehyde back to acid by Hpad or homologues should also generate a reduced cofactor.
Fig. 3.
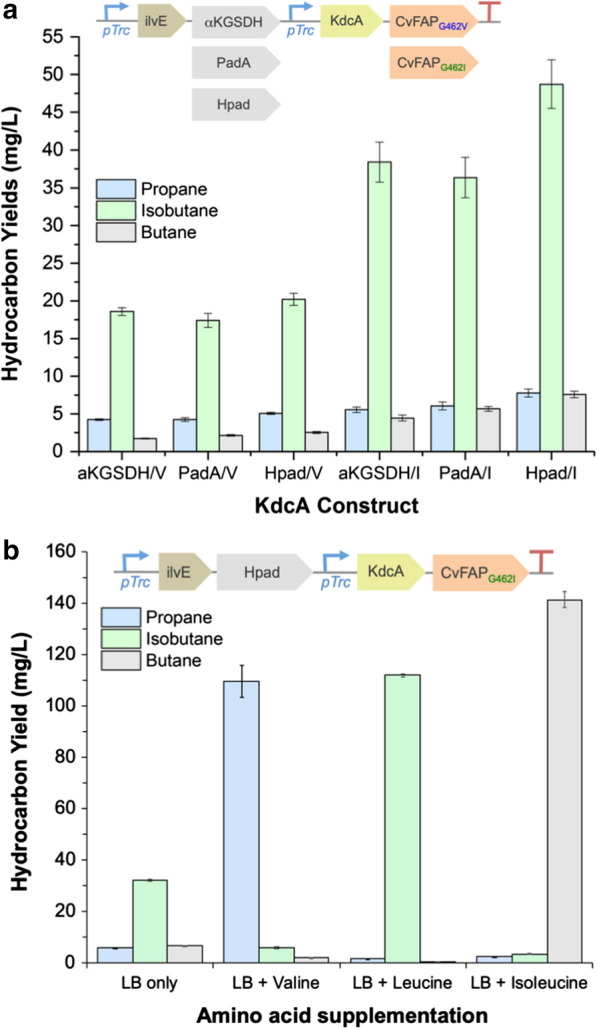
Hydrocarbons production of E. coli strain BL21(DE3)∆∆ containing engineered KdcA/CvFAPG462V-dependent routes. Gaseous alkane yields dependent on the a enzyme homologues and b amino acid supplementation for pTrIHTrKFG462I construct. Cultures (10–20 mL) were grown in LB medium containing 30 μg/mL kanamycin for 4–6 h at 37 °C and 180 rpm. Luria broth contains ca 7, 8.8 and 5.4 mM valine, isoleucine and leucine, respectively [21]. Protein induction (100 μM IPTG) was performed, followed by culture supplementation with valine, leucine or isoleucine (30 mg/mL) in part B) after 1 h at 30 °C. Triplicate samples (1 mL) each of 3 biological replicate cultures were sealed into glass vials (4 mL) and incubated at 30 °C for 16–18 h at 200 rpm, illuminated continuously with a blue LED (455 nm or 470 nm). Gaseous hydrocarbon levels were determined by manual headspace injection on an Agilent 490 Micro GC. Errors represent one standard deviation of the replicates (biological and/or technical triplicates). Data for LB + valine was obtained from previous studies [4]. Expression constructs (inset): αKGSDH/V = trc-ilvE-αKGSDH-trc-kdcA-CvFAPG462V; Hpad/V = trc-ilvE-Hpad-trc-kdcA-CvFAPG462V; PadA/V = trc-PadA-Hpad-trc-kdcA-CvFAPG462V; αKGSDH/I = trc-ilvE-αKGSDH-trc-kdcA-CvFAPG462I; Hpad/I = trc-ilvE-Hpad-trc-kdcA-CvFAPG462I and PadA/I = trc-PadA-Hpad-trc-kdcA-CvFAPG462I
As expected, dramatically higher titres of gaseous bio-alkanes were detected in all six KdcA-dependent pathways compared to the CoA-dependent routes (Fig. 3a; Additional file 1: Table S2). In each case, the major gas produced was isobutane (73–79% total bio-alkane), with propane proportions decreased to only 12–18% of the total headspace alkane. In addition, there was a 2- to 2.4-fold increase in isobutane titres with CvFAP variant G462I over G462V [4]. There was a slight improvement in bio-alkane production with Hpad, consistent with its known specificity for isovaleraldehyde, butyraldehyde and valeraldehyde [15]. The highest bio-alkane-yielding construct generated isobutane titres of 48.7 ± 3.2 mg/L in standard E. coli growth medium. This is comparable to E. coli expressing CvFAPG462I alone supplemented with isovaleric acid (87 mg/L culture; [4]). Production levels were also dependent on the plasmid backbone, as changing from a kanamycin-resistant to an ampicillin-resistant construct showed significant decreases in alkane titres (Additional file 1: Table S3).
The ratio of gases produced was similar in each KdcA-dependent pathway version, with isobutane:propane:butane ratios of ca 76:12:12 for the highest producing construct. This switch from predominantly propane to isobutane production suggests that KdcA may have a higher preference for α-ketoisocaproate over α-ketoisovalerate or α-keto-β-methylvalerate (Additional file 1: Figs. S1, S2). As each AlDH-containing construct generated similar ratios of each bio-alkane gas, the product profile might in part be influenced by the amino acid composition of the medium and their relative uptake rates into the cell.
Tuneable bio-alkane blends
Ideally, a route to bio-LPG based on microbial fermentation would be tuneable, to allow precise control of the relative ratios of propane:butane (± isobutane) to suit customer requirements. This was demonstrated successfully in E. coli expressing only CvFAP variants, where propane-to-butane ratios were dependent on the relative proportions of butyric and valeric acids fed to the cultures, respectively [4]. A similar system could be applied using the KdcA-dependent pathway by supplementing the culture with specific amino acid blends. This approach could be a suitable alternative to VFA feeding as amino acid supplementation does not have the pH and cytotoxicity effects seen when adding millimolar concentrations of organic acids to cultures. In addition, a localised anaerobic digestion plant [4] would not be required to generate VFAs from waste biomaterial.
Leucine and isoleucine supplementation of E. coli cultures expressing IHKFG462I showed proportional increases in isobutane and butane, respectively, without reaching saturation levels (Fig. 3b; Additional file 1: Fig. S6 and Table S4). These data are similar to previous studies with valine supplementation, where increasing levels of propane were generated [4]. With 30 mg/mL individual amino acid supplementation, similar yields of propane, isobutane and butane were detected (110 ± 6 [4], 112 ± 0.6 and 142 ± 3 mg/L, respectively). In each case, the predominant gas was greater than 95% of the total headspace alkane produced. Therefore, the desired country- or customer-specific requirements for propane:butane:isobutane ratios (bio-LPG or aerosol propellants [3]) during alkane gas production could be achieved by enriching the culture medium with specific amino acid blends. However, ideally the chosen microbial chassis should be modified to increase in vivo production of the required amino acid precursors, enabling the required bio-alkane blends to be generated from the locally derived waste feedstock.
Successful commercialisation of this approach would require the utilisation of cost-effective feedstocks and amino acid supply, such as proteinaceous waste (e.g. food waste). One study found the amino acid content of 39 samples of vegetal and dairy product food waste from EU industrial agro-food systems had a valine content between 6.4 and 29.4 mg/g waste [26]. Therefore, this waste could potentially be utilised as both a carbon and amino acid source.
Halomonas as industrial chassis
Economically viable scaled bio-production strategies can be difficult to achieve due to high capital and operating expenditures of typical fermentation systems. This includes energy intensive running costs such as sterilisation, aseptic growth conditions, complex monitoring systems and downstream processing. Environmental concerns include the use of vast quantities of clean water and the route of waste disposal. To address these concerns, CvFAP-dependent bio-LPG production from VFAs was recently demonstrated in the halophilic production host Halomonas [4]. This microbial chassis grows at high salinity (up to 20% w/v NaCl) and pH values (up to 12), allowing for cultivation in seawater, waste-water or recycled water without the need for costly sterilisation and aseptic conditions [27]. This allows Halomonas bioreactors to be constructed using low cost materials, such as plastics, ceramics and cement, with overall costs savings of ca 65% compared to conventional scaled E. coli cultivation [28].
Given our recent success in transitioning CvFAP-dependent VFA to propane production from E. coli to Halomonas [4], we decided to take a similar approach with the KdcA-dependent amino acid pathway. Preliminary IPTG-inducible and constitutively expressed KdcA-dependent pathways were introduced into Halomonas, using both plasmid-borne and chromosomal integration strategies [4]. Bio-propane production was successfully demonstrated in small scale in vivo batch cultivation using valine-supplemented medium [4]. However, the scalability of the process was not tested, and construct optimisation had not been performed for expression in Halomonas. To address this, we utilised the highest performing E. coli construct (IHKFG462I) to generate six Halomonas-specific plasmids, which differed only by the promoter system. An efficient IPTG-inducible MmP1 T7-like promoter (T7L) was initially employed [4], as standard pET vector-based viral systems do not function in Halomonas [29]. Following our success in re-purposing trc into two constitutive promoters for E. coli (TrΔL and ΔL; Additional file 1: Table S5), we performed equivalent truncations in T7L to generate constitutive promoters T7LΔL and HΔL, respectively. We also tested three variant Halomonas promoters (c102, c69 and c59), which are based on the major outer membrane protein porin constitutive expression system (Additional file 1: Fig. S11; [30, 31]). Preliminary studies with the constructs containing T7L and c69 promoters in standard (non-supplemented) media were reported elsewhere [4], and form the basis of comparison with the remaining four constructs under constitutive control in this study.
The best performing construct in Halomonas contained the CvFAPG462I variant under T7L control, generating primarily isobutane (9.32 ± 0.63 mg/L; Fig. 4a; [4]). This is around fourfold lower than the equivalent E. coli IHKFG462I system under double trc control (38.6 ± 2.5 mg/L isobutane). Titres of propane were also decreased around twofold (3.78 ± 0.26 mg/L) in simple medium (no amino acid supplementation). For the constitutive expression systems, the best performers were controlled by T7LΔL and HΔL promoters (Fig. 4a; Additional file 1: Table S5). Surprisingly, titres of gaseous alkanes were lower using the Halomonas-specific Pporin-like variant constitutive promoters (c59, c69 and c102) compared to the truncated T7-like ones. In the majority of these constructs, the relative proportion of propane was increased relative to isobutane, to near equivalent levels.
Fig. 4.
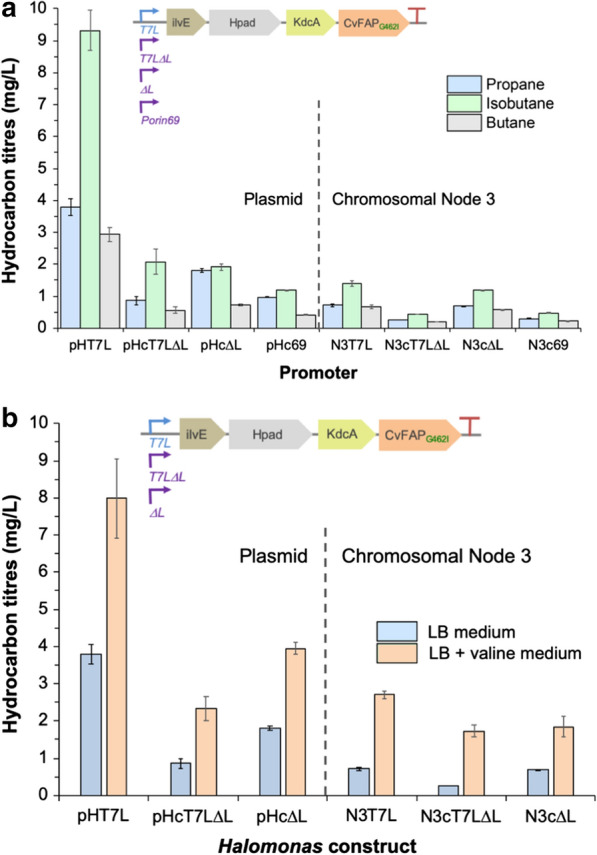
Effect of the KdcA-dependent pathway promoter on gaseous alkane production in Halomonas TQ10 in the a absence or b presence of valine culture supplementation. Pathway promoter-IHTrKFG462I was present as either a plasmid-borne or chromosomally integrated construct (single site). Cultures (20 mL) were grown in LB medium containing 30 µg/mL kanamycin for 4–6 h (OD600 ~ 0.6–0.8) at 37 °C and 180 rpm. Recombinant protein expression was induced with IPTG (0.1 mM) and culture supplementation with valine (30 mg/mL) after 1 h at 30 °C. Triplicate samples (1 mL) each of 3 biological replicate cultures were sealed into glass vials (4 mL) and incubated at 30 °C for 16–18 h at 200 rpm, illuminated continuously with a blue LED (455 nm or 470 nm). Gaseous hydrocarbon levels were determined by manual headspace injection using an Agilent 490 Micro GC, containing an Al2O3/KCl column. The errors represent one standard deviation of the data
The top three performers were further investigated by determining the increase in propane production in the presence of supplemental valine (30 mg/mL). Surprisingly, propane titres of the inducible construct only increased twofold (7.99 ± 1.07 mg/L; Fig. 4b; Additional file 1: Table S5), compared to the 17-fold increase with comparable constructs in E. coli (Fig. 3b; [4]). In each case, the relative concentrations of (iso)butane in valine-supplemented medium were dramatically decreased compared to standard media (Fig. 4b; Additional file 1: Table S5), likely due to competition for CvFAP active site. This suggests a limiting factor in this process may be the rate of amino acid uptake and/or endogenous amino acid metabolism by Halomonas compared to E. coli, and may also be contributing to the overall lower gaseous alkane titres in Halomonas compared to E. coli. Alternatively, one or more of the pathway enzymes may be unstable or poorly expressed in Halomonas. Notwithstanding, the prototype production pathway has been successfully demonstrated in Halomonas.
Chromosomal integration into Halomonas
Chromosomal integration of pathways into an industrially robust microbial chassis is preferable to maintaining the genes for these pathways on a plasmid, as it eliminates the need for expensive and/or toxic selection agents [32], and prolongs the fermentation time to maximise product yields. To investigate this, we integrated four versions of the IHKFG462I pathway into Halomonas strain TQ10 at two chromosomal loci (nodes 3 and 15; [33]), each differing only by the promoter type (T7L, T7LΔL, HΔL and c69). This was performed by utilising a novel suicide vector protocol (Additional file 1: Fig. S7), which was based on previously published methods [4, 34, 35]. Small-scale batch cultures were compared for gaseous alkane production in the presence and absence of supplemental valine (30 mg/mL).
As expected, the highest performing strain contained the inducible T7L promoter (1.39 ± 0.09 mg/L isobutane; [4]), however similar titres were obtained for the constitutive HΔL system (1.19 ± 0.01 mg/L isobutane; Fig. 4a; Additional file 1: Table S5). Overall, combined gas titres were significantly lower than for the equivalent plasmid systems, as in each case only a single copy was present in the genome. However, these differences were less marked in the presence of valine supplementation, with the HΔL construct showing only about twofold decrease in propane titres compared to the equivalent plasmid-borne system (1.84 ± 0.28 mg/L propane; Fig. 4b).
The chromosomal integration site had a significant impact on the overall gas titres, with loci 1 (node 3; [33]) showing ca a 2.5-fold increase over loci 2 (node 15; Additional file 1: Table S5). This suggests there is scope to significantly improve bio-gas titres by screening a variety of integration sites and/or multi-copy insertion of the construct into the Halomonas chromosome [33].
Fermentative amino acid to bio-alkane production in Halomonas
To investigate the scalability of this fermentative bio-alkane approach, we performed laboratory-scale cultivations of Halomonas expressing the plasmid-borne inducible IHKFG462I construct (T7L promoter) up to 48 h in a flatbed photobioreactor (400 mL). Fermentations in standard Halomonas medium (~ 0.25% amino acid blend) containing glycerol [4] showed primarily propane production, with an estimated 17 mg propane/g cells over 48 h (Additional file 1: Fig. S11). The addition of casamino acids (~ 2% amino acid blend) led to predominantly isobutane production (max 27 mg propane/g cells), with propane and butane titres around threefold lower (about 9 mg propane/g cells; Fig. 5a). This switch in bio-alkane gas predominance is consistent with the individual amino acid concentrations in each medium (Additional file 1: Fig. S12). Valine-supplemented medium (1.8%) saw propane titres rise to around 90 mg propane/g cells in 46 h (Fig. 5b), equivalent to Halomonas TQ10 expressing CvFAPG462V with supplemental butyric acid (89 mg propane/g cells; [4]). In most cases, the rate of gaseous alkane production peaked within the first 24 h, followed by a rapid decline in production (Additional file 1: Figs. S8–S10). This is attributed to plasmid instability and/or loss, or possibly CvFAP light inactivation [4, 23].
Fig. 5.
Bio-alkane production by Halomonas TQ10 expressing IHKFG462I in plasmid or chromosomally integrated constructs. Cumulative alkane production by the plasmid-borne inducible construct pHT7LIHKFG462I in LB60 medium supplemented with a casamino acids or b valine. Rate of alkane production of Halomonas TQ10 expressing chromosomally integrated c N3T7LIHKFG462I (inducible) or d N3cΔLIHKFG462I (constitutive). Photobioreactor cultivation (400 mL) was performed with medium pH 6.8 containing 0.5 mL/L antifoam and antibiotic (50 μg/mL spectinomycin or 34 μg/mL chloramphenicol for plasmid-borne and constitutive constructs, respectively). The culture was maintained at 30 °C with 60% stirring output with an airflow rate of 1.21 L/min, and ambient room lighting until mid-log phase (4–5 h). Protein expression was induced by 0.1 mM IPTG, where required, and the culture was maintained for 24–240 h with blue light exposure (1656). Culture medium feeding was employed to maintain an optical density of 0.8 and to replenish the carbon source for the chromosomal constructs. Alkane gas production was monitored by automated (a, b) or manual (c, d) headspace sampling using a Micro GC. The effect of amino acid supply on gaseous alkane titres was investigated by performing triplicate fermentations in three different culture media (Additional file 1: Figs. S8–10). Cumulative propane, isobutane and butane titres were calculated from the average production rates per hour in fermentations from freshly transformed Halomonas TQ10 in each media (a, b and Additional file 1: Fig. S11)
The longevity of the bioprocess was investigated by continuous flow fermentation of the genome integrated IHKFG462IHalomonas strains (T7L and HΔL; node 3) in valine-supplemented medium. Fermentation of the inducible (T7L) strain showed a gradual increase in propane production, peaking at around 70 h (~ 180 mg/g cells/day), followed by a slow decline to minimal titres by ~ 140 h (Fig. 5c). This suggests a gradual decrease in biocatalytic enzyme production, likely due to degradation/inactivation of IPTG and/or CvFAP photoinactivation [23] during the extended fermentation time [36]. Significant propane production in this strain was extended to nearly 5 days, however elimination of the need for IPTG could potentially extend propane production even further.
The constitutively expressed (HΔL) Halomonas strain showed a fairly consistent propane production rate from 60 to 200 h (up to 30 mg/g cells/day; Fig. 5d), followed by a rapid decline in bio-alkane production and cell viability. This loss of Halomonas viability is presumably due to the extended exposure to high-intensity blue light that is required for CvFAP activity. Cytotoxicity of blue light is a common feature of microorganisms in general [37]. Therefore, process optimisation to reduce the blue light intensity is required for efficient photocatalysis by CvFAP to extend propane production further. An alternative strategy could be employed where periodic ‘dark cycles’ are implemented, to allow cultures to regenerate to high cell density in the absence of light. For example, Halomonas TD01 (precursor of strain TQ10) is known to grow to high cell density (40 g/L cells dry weight) in 24 h with nutrient feeding [27]. This could be followed by feeding with amino acids and blue light exposure to switch on alkane gas production, until a decline in productivity is seen.
Overall, propane production in the constitutive strain decreased nearly eightfold from the equivalent IPTG-inducible strain. However, stable expression levels over an extended fermentation time could potentially generate higher levels of propane overall. Therefore, this fermentative approach from amino acids to gaseous alkanes could potentially be scaled to produce bio-LPG from proteinaceous feedstocks. Process and chassis engineering to increase bio-LPG titres could provide a viable alternative approach to the simpler route from VFAs.
Conclusions
The fermentative KdcA-dependent pathway from amino acids to gaseous alkanes provides an alternative, yet complementary solution to VFA-dependent bio-LPG production. This wholly biological approach is in stark contrast to existing commercial ‘bio-LPG’ production strategies [38], which are essentially a chemically synthesised product from biologically sourced materials. Utilising Halomonas as the microbial chassis of choice, cultivated on renewable proteinaceous waste, could potentially reduce the process costs to enable successful commercialisation. Site-specific implementation of direct (VFA) or pathway driven (amino acid) strategies would be dependent on the availability and relative composition of local waste feedstocks, and the economics of waste pre-processing (hydrolysis vs anaerobic digestion). These renewable and sustainable approaches to clean-burning gaseous fuels could reduce the dependence on and environmental impact of fossil fuel combustion, and tap into the existing global market (US$19.5 billion in 2017) of amino acids.
Materials, services and equipment
All chemicals, solvents and reagents were purchased from commercial suppliers, and were of analytical grade or better. The gas calibration standard was a custom blend of 1% each of propane, butane and isobutane in nitrogen (Thames Restek, Saunderton, UK). Media components were obtained from Formedium (Norfolk, UK). The E. coli strains used for propagating all plasmids were Stellar™ (Clontech) or NEB®5α (New England Biolabs). Expression studies were carried out using E. coli strains BL21(DE3) and NiCo21(DE3) (New England Biolab). E. coli strain BL21(DE3) was modified by chromosomal deletion of two aldehyde reductase genes yqhD and yjgB (BL21(DE3)∆yqhD/∆yjgB/KanR) [5], and the kanamycin selection gene was removed (BL21(DE3)ΔΔ) as described previously [4]. Halomonas strain TQ10-MmP1 and modified pSEVA plasmids have been described previously [4].
Gene sequencing and oligonucleotide synthesis were performed by Eurofins MWG (Ebersberg, Germany). Details of all the plasmid and chromosomal constructs used in this study are found in Additional file 1: Tables S5–S7. pSBR1Ks-i-SceI is a hybrid of pSEVA and Biobrick (pBb-type) plasmids, containing oriT, I-SceI meganuclease gene under trc control, pRO1600-ColE1 double origin and both kanamycin and spectinomycin resistance markers. Plasmids pSH-N3 and pSH-N15 are based on the CRISPR/Cas9 editing Halomonas genome donor DNA pSEVA241 plasmid of Quin et al. [35], except the gRNA and antibiotic resistances were removed, and the insert contained the target DNA with a pKIKO-derived chloramphenicol resistance gene flanked by FRT sequences [39]. Node 3 and node 15 refer to Halomonas chromosomal loci for insertion of target DNA [33]. The modified pPorin-like constitutive promoter plasmids pHc102-RFP, pHc69-RFP and pHc59-RFP were kindly supplied by Duangthip Trisrivirat (Vidyasirimedhi Institute of Science and Technology; VISTEC). These plasmids are a modified pHal2-based plasmid [4] containing RFP with the T7-like promoter swapped for a modified pPorin-like constitutive promoter. The BglBrick series of vectors were obtained from Addgene (https://www.addgene.org) [40]. Gene synthesis was performed by GeneArt (ThermoFisher, Germany) or GenScript (USA). The sequences of all the oligonucleotides used in cloning and mutagenesis can be found in Additional file 1: Tables S8–S12.
The mounted high-power blue LEDs and LED drivers were from Thorlabs (Ely, U.K.), with spectra centred at 470 nm (FWHM 25 nm, 710 mW typical output). The custom-built LED blue light array had area of 396 cm2 of relatively consistent light intensity and a fixed average culture-to-LED distance of 8 cm. The photobioreactor was a thermostatic flat panel FMT 150 (500 mL; Photon Systems Instruments, Czech Republic) with integral culture monitoring (OD 680/720 nm), pH and feeding control and an LED blue light panel (465 nm; maximum PPFD = 1648 μE photons).
Single and multi-gene construct synthesis
The propane synthesis plasmids pTPC7 (YciA-sfp-CAR; [6]), ADOA134F, petF (Ferr) and pCvFAPG462V (fatty acid photodecarboxylase from Chlorella variabilis) were assembled as described previously [4, 6]. A second IPTG-inducible CvFAPG462V construct was generated by sub cloning the variant gene into pBbA1c-RFP (pTrCvFAPG462V), eliminating the RFP gene. The following genes were designed, synthesised and sub-cloned into pET21b, incorporating a C-terminal His6-tag: leucine 2-oxoglutarate transaminase from E. coli (ilvE; UniProt: P0AB80); human branched-chain α-keto acid dehydrogenase (BCKDHAB; P12694 and P21953; His6-tag on subunit B only), phenylacetaldehyde dehydrogenase 17 from E. coli (PadA; P80668), threonine dehydratase from E. coli (ilvA; P04968) and the E. coli leuABCD complex composed of 2-isopropylmalate synthase (LeuA; P09151), 2-isopropylmalate dehydrogenase (LeuB; P30125) and isopropyl malate isomerase complex (LeuC/LeuD; P0A6A6/P30126). Additional synthesised genes sub-cloned into pETM11 were α-ketoglutaric semialdehyde dehydrogenase from Azospirillum brasilense (αKGSDH; Q1JUP4) and 3-hydroxypropionaldehyde dehydrogenase from E. coli (Hpad; P23883), while branched-chain keto acid decarboxylase from Lactococcus lactis (KdcA; Q6QBS4) was sub cloned into pET28b. These latter genes contained a vector-derived N-terminal His6-tag. Genes were codon optimised to remove rare codons for optimal expression in E. coli. For LeuABCD, the native E. coli operon sequence was synthesised (no His6-tags), with gene expression controlled by a single T7 promoter (Additional file 1: Table S5).
The multi-gene construct pYSCAP (YciA-sfp-CAR-ADOA134F-Ferr) contained the genes encoding non-His6-tagged versions of acyl-CoA thioester hydrolase from Haemophilus influenza (YciA; P44886); maturation factor phosphopantetheinyl transferase from Bacillus subtilis (sfp; P39135); carboxylic acid reductase from Mycobacterium marinum (CAR; B2HN69); aldehyde deformylating oxygenase variant A134F from Prochlorococcus marinus (ADO; Q7V6D4) and ferredoxin from Synechocystis sp PCC6803 (Ferr; P27320) [5, 6]. This construct was synthesised as a complete operon with codon-optimised genes, synthetic Shine–Dalgarno (SD) sequences and the constitutive promoters R0011 (http://2015.igem.org/) and proD upstream of YciA and ADOA134F, respectively [20].
Multi-gene constructs assembly in E. coli
In most cases, the assembly of multi-gene constructs was performed by In-Fusion cloning, according to the manufacturer’s protocols [41]. Vector linearisation and insert(s) amplification were performed by polymerase chain reaction (PCR), using the CloneAmp™ HiFi PCR Premix kit (Clontech), incorporating 15–25 bp overhangs necessary for subsequent ligations. In some cases, overlap extension PCR (OEP) was performed to ligate two or more DNA fragments generated by PCR to simplify subsequent construct assembly. In this method an initial 5 PCR cycles were performed with the template DNA fragments only, followed by the addition of the forward primer of the first DNA fragment and the reverse primer of the last insert. Following In-Fusion cloning, each construct was transformed into the E. coli strain Stellar or NEB5α for plasmid recovery, and the correct assembly was confirmed by DNA sequencing. The oligonucleotide sequences and template DNA used in each PCR reaction is shown in Additional file 1: Tables S8–S12. The following sections will detail the general approaches taken for the assembly of each plasmid.
ADO-containing CoA-dependent multi-gene E. coli constructs assembly
A dual construct was assembled in pBbE2k [40] containing ADOA134F and its electron transfer partner Ferr [6], under the control of a tetracycline-inducible promoter. Both ADOA134F and Ferr genes were PCR amplified from their respective constructs in pCDFDuet-1 and pRSF-Duet1 [6]. The PCR products included the existing vector-derived 5′-SD sequence for ADOA134F and a non-native SD sequence (GGAGGACAGCTAA) for Ferr. In-Fusion cloning was performed with the linearised destination vector pBbE2k-RFP, minus the RFP gene and its SD sequence, to generate pTetADOA134FFerr (Additional file 1: Table S8).
The assembly of a butyryl-CoA to propane pathway construct pAFYSC (T7-ADOA134F-Ferr-YciA-sfp-CAR) was performed using the vector pETDuetT-1. PCR linearisation of construct TPC7 (T7-YciA-sfp-CAR) occurred between the T7 promoter and the initial SD sequence, while the two genes from pADOA134FFerr were amplified with both SD sequences. In-Fusion cloning between the two PCR products generated the IPTG-inducible pAFYSC pathway (Additional file 1: Table S8).
To eliminate the need for IPTG induction, a constitutive expression system was constructed using the BglBrick plasmid pBbE7k-RFP as the backbone [40]. The plasmid was linearised by reverse PCR, eliminating the T7 promoter-RFP cassette (lacIq retained), and the proD-ADOA134F insert was amplified from the multi-gene construct pYSCAP. Following In-Fusion cloning, the new ADO-containing constitutive expression vector (pPrADOA134F) was used as the backbone for the construction of a series of ADO-dependent pathways from butyryl-CoA to propane. The first constitutive pathway assembled was pPr*AFYSC (proD*-ADOA134F-Ferr-YciA-sfp-CAR), via the ligation of the pAFYSC pathway genes (SD-ADOA134F-Ferr-YciA-sfp-CAR) into the linearised proD-containing empty plasmid (pPrADOA134F minus SD-ADOA134F; Additional file 1: Table S8). This was followed by linearisation of pPr*AFYSC by reverse PCR to eliminate the now redundant lacIq repressor. The new construct (pPrAFYSC) was re-circularised by In-Fusion cloning in the absence of any insert.
The generation of ADO-dependent pathways to propane, butane and isobutane from the amino acids valine, isoleucine and leucine, respectively, requires the addition of genes ilvE and BCKDHAB to the existing AFYSC constructs (Fig. 1). Initially a constitutively controlled dual enzyme construct (pPr*IB; proD*-ilvE-BCKDHAB) was assembled in the same modified BioBrick plasmid as used for pPr*AFYSC construction. The individual genes (ilvE and BCKDHAB) were amplified by PCR from their respective synthesised constructs. This was followed by OEP to generate a dual enzyme insert, followed by ligation to the linearised empty vector (pPr*AFYSC minus AFYSC). A complete pathway from amino acid to gaseous hydrocarbon was generated by the inclusion of ilvE-BCKDHAB with AFYSC to form pPr*AFYSCIB (proD*-ADOA134F-Ferr-YciA-sfp-CAR-ilvE-BCKDHAB). This was performed by linearising pPrAFYSC after CAR, and ligating it to the ilvE-BCKDHAB insert amplified from pPr*IB. To increase the expression of ilvE and BCKDHAB, a second proD promoter was inserted downstream of CAR in pPrAFYSC by In-Fusion cloning, to generate pPrAFYSCPr (Additional file 1: Table S8). Insertion of the ilvE-BCKDHAB fragment of pPr*IB after the second proD generated pPr*AFYSCPrIB (proD*-ADOA134F-Ferr-YciA-sfp-CAR-proD-ilvE-BCKDHAB.
IPTG-inducible constructs catalysing gaseous hydrocarbon production from amino acids were generated by amplifying the seven pathway genes from pPr*AFYSCIB and ligating them into pBbE1k, linearised downstream of the trc promoter (pTrAFYSCIB; trc-ADOA134F-Ferr-YciA-sfp-CAR-ilvE-BCKDHAB). Similarly, the addition of a second trc promoter upstream of ilvE was performed by PCR coupled to In-Fusion cloning to generate pTrAFYSCTrIB (trc-ADOA134F-Ferr-YciA-sfp-CAR-trc-ilvE-BCKDHAB; Additional file 1: Table S8).
FAP-containing CoA-dependent multi-gene E. coli constructs assembly
Simplified gaseous hydrocarbon producing constructs from amino acids were generated by substituting the four genes CAR, sfp, ADOA134F and Ferr for a single gene CvFAP variant G462V or G462I. An initial construct pTrFG462VYIB was generated (trc-CvFAPG462V-YciA-ilvE-BCKDHAB) using pTrAFYSCIB as the backbone. This latter plasmid was linearised, eliminating the genes encoding AFYSC (Additional file 1: Table S9). A dual gene insert was constructed (CvFAPG462V-YciA) by PCR amplification of each individual gene, followed by OEP. This was ligated to the backbone plasmid upstream of ilvE, generating an IPTG-inducible construct. A similar constitutive construct pPrFG462VYIB (proD-CvFAPG462V-YciA-ilvE-BCKDHAB) was generated as above, except the backbone template was the proD-containing plasmid pPrAFYSCIB.
KdcA-dependent multi-gene E. coli constructs assembly
Alternative CvFAPG462V-dependent pathways were constructed by the substitution of six genes (BCKDHAB, YciA, CAR, sfp, ADOA134F and Ferr) for KdcA, an alcohol dehydrogenase and CvFAPG462V. These pathways were constructed in pBbE1k, including the insertion of a second trc promoter upstream of the latter two genes (trc-KdcA-CvFAPG462V). Each individual gene and second trc promoter were amplified by PCR, and OEP was performed between trc and KdcA DNA fragments (Additional file 1: Table S10). In-Fusion cloning was performed generating three KdcA-dependent and IPTG-inducible constructs (pTrIA*TrKFG462V, where A* = αKGSDH, PadA or Hpad; Additional file 1: Table S5). Each of these three constructs underwent site-directed mutagenesis of the CvFAP gene to produce the equivalent pathway with the variant G462I (pTrIαKTrKFG462I, pTrIΠTrKFG462I and pTrIΗTrKFG462I, respectively) as described previously [4].
Halomonas KdcA-dependent construct assembly
Six KdcA-dependent pathways were constructed in the Halomonas-compatible plasmid pHal2 [4], which varied by the type of promoter used. This was performed by multi-step In-Fusion cloning, where PCR was used to amplify the inserts, eliminating the His6-tags, and/or linearise the vectors (Additional file 1: Table S11). Each construct was propagated in the E. coli conjugative donor strain S17-1 [42]. Plasmid transformation into Halomonas was performed by conjugation according to the method described previously [4]. Plasmid content of each trans-conjugant was confirmed by DNA isolation, restriction mapping and sequencing.
The initial construct was generated under control of the IPTG-inducible MmP1 T7-like promoter (pHT7LIHKFG462I; [4]), which later underwent LacIq elimination (T7LΔL) to generate the respective constitutive construct (pHT7LΔLIHKFG462I). A second constitutively expressed construct was generated by substituting the T7-like promoter for a truncated trc promoter, which was deficient in both trc and lacI (pHΔLIHKFG462I). Three constitutive promoters were generated (c102, c69 and c59) based on the major outer membrane protein porin constitutive expression system in Halomonas (Additional file 1: Fig. S11; [30, 31]). The IHKFG462I operon was inserted downstream of each promoter, generating a further three Halomonas constructs (pHc102IHKFG462I, pHc69IHKFG462I and pHc59IHKFG462I).
Chromosomal integration of pathway genes into Halomonas
Chromosomal insertion of the KdcA-dependent pathways into Halomonas TQ10 was performed using a novel suicide vector (pSH) protocol (Additional file 1: Fig. S7) based on previously published methods [4, 34, 35]. The pSH insertion plasmids (pSH-N3 and pSH-N15) contained the biocatalytic and pKIKO-derived FRT flanked [39] chloramphenicol resistance genes surrounded by homology arms (node 3 or 15; [33]), an I-SceI restriction site and a colE1 ori (incompatible) with replication in Halomonas (Additional file 1: Fig. S10 inset). This plasmid was co-conjugated into Halomonas TQ10-MmP1 with a second spectinomycin-resistant plasmid (pSBR1Ks-i-SceI), the latter expressing the restriction enzyme I-SceI. In vivo expression of I-SceI linearised pSH plasmids facilitates chromosomal integration [34, 35]. Successful integration was seen as growth of Halomonas on chloramphenicol-selective medium, as the pSH plasmid is not replicated in Halomonas. Integration was confirmed by colony PCR, genomic sequencing and in vivo propane production after pSceI plasmid curing [34, 35].
Eight KdcA-dependent constructs (ilvE-Hpad-kdcA-CvFAPG462I) were integrated into Halomonas strain TQ10 (Additional file 1: Table S12). The constructs varied by the chromosomal loci (node 3 or 15) and the promoter type (inducible vs constitutive). The inducible system was the MmP1 T7-like promoter (T7L), while the constitutively expressed constructs were controlled by pPorin-like 69 (c69), lacIq-deficient MmP1 T7-like promoter (T7LΔL) or the truncated pTrc promoter minus trc and lacI (ΔL; Additional file 1: Table S12). The insertion of inducible T7LIHKFG462I or constitutive c69IHKFG462I constructs into pSH-N3 or pSH-N15 was performed via In-Fusion cloning using PCR linearised destination vectors (between one homology arm and upstream of the chloramphenicol gene; Additional file 1: Fig. S5) and amplified multi-gene constructs with their own promoters (N3- or N15T7LIHKFG462I; N3- or N15c69IHKFG462I). For the ΔL-containing plasmids, a similar protocol was performed as above, except the T7LIHKFG462I construct for each node was used as the template, and the only DNA eliminated/inserted was the promoter (N3- or N15cΔLIHKFG462I). To generate the constructs with the T7LΔL promoter, PCR elimination of the laciq gene was performed on the equivalent T7L-containing constructs followed by self In-Fusion cloning to re-circularise the plasmid (N3- or N15cT7LΔLIHKFG462I). Successful integration of the constructs at the correct loci was confirmed by colony PCR and genome sequencing.
Protein expression and lysate production
IPTG-dependent expression of proteins YciA, CAR, sfp, CvFAPG462V, ADOA134F and Ferr in E. coli has been demonstrated previously [4, 6]. The remaining proteins ilvE, BCKDHAB, KdcA, αKGSDH, PadA, Hpad ilvA and LeuABCD were transformed into E. coli strain BL21(DE3) for protein overexpression studies. Cultures (1 L) were grown in LB Broth Miller (Formedium) containing the required antibiotic (50 μg/mL ampicillin or 30 μg/mL kanamycin) at 37 °C with 180 rpm shaking until OD600nm = 0.6. Recombinant protein induction was performed with IPTG (0.1 mM), followed by a further 12–16 h incubation at 25 °C. Cells were harvested by centrifugation at 3320×g for 30 min at 4 °C.
Cells were resuspended in lysis buffer (5 mL/g pellet; 50 mM Tris pH 7.0 containing 1 mM MgCl2, 1 mM β-mercaptoethanol, 10% glycerol, 2X protease inhibitors, 50 μg/mL DNAse and 50 μg/mL lysozyme) and freeze-thawed in liquid nitrogen. Cells were lysed by sonication, and clarified using centrifugation (48,000×g). Protein content was determined using 12% SDS-PAGE gels (Mini-Protean TGX Stain-Free Precast Gels, Bio-Rad). Protein gels were imaged using a BioRad Gel Doc EZ Imager and relative protein band intensity was determined using the BioRad ImageLab software. Identification of His6-tagged proteins was performed by Western blots using the Trans-Blot® Turbo™ Transfer system (PVDF membranes; BioRad) and the Western Breeze Chemiluminescent Immunodetection kit (alkaline phosphatase; Life Technologies) with mouse (His tag monoclonal antibody) and alkaline phosphatase-containing (Anti-C-My) primary and secondary antibodies, respectively.
Gaseous hydrocarbon production
In vivo gaseous hydrocarbon production by recombinant E. coli was performed using the following general protocol: cultures (10 mL) were incubated for 4–6 h (OD600 ~ 1.6–2) at 37 °C and 180 rpm in LB or TB medium containing 50 µg/mL ampicillin, 30 µg/mL kanamycin or 50 µg/mL chloramphenicol, dependent on the antibiotic resistance (Additional file 1: Table S5). Supplemental valine, leucine or isoleucine (up to 30 mg/L) were included in the medium where required. Protein induction (0.1 mM IPTG) was performed for trc or T7-containing constructs, and triplicate samples (1 mL) each of 3 biological replicate cultures were sealed into glass vials (3 mL) and incubated at 30 °C for 16–18 h at 200 rpm, illuminated continuously with a blue LED (455 nm or 470 nm).
For propane production in Halomonas, LB60 medium (1% tryptone, 0.5% yeast extract, 6% NaCl) pH 9.0 was used containing spectinomycin (50 μg/mL). Cultures were agitated (180 rpm) at 37 °C for 5 h incubation (OD ~ 1.6–2) prior to induction. IPTG (0.1 mM) was added (where necessary), and the remaining methodology was performed as described above for E. coli cultures.
Photobioreactor cultivation
General photobioreactor cultivation (400 mL) was performed with high salt glycerol medium (30–32% seawater or Instant Ocean, NaCl to 6%, 0.1% glycerol and 0.5% yeast extract) pH 6.8 containing 0.5 mL/L antifoam and antibiotic (50 μg/mL spectinomycin or 34 μg/mL chloramphenicol for plasmid-borne and constitutive constructs, respectively). Alternative growth media were based on LB60 pH 6.8, which were supplemented with 1.5% valine (LB60Val) or casamino acids (LB60Cas). Cultivation was performed in batch mode, pre-equilibrated at 30 °C with 60% stirring output. An overnight starter culture (10–15 mL) of Halomonas TQ10 expressing pHT7LIHKFG462I was added, to achieve a starting OD680 nm of ~ 0.2, and the culture was maintained at 30 °C with an airflow rate of 1.21 L/min, automated pH maintenance, culture optical density monitoring and ambient room lighting until mid-log phase (4–5 h). Protein induction by IPTG (0.1 mM) was performed for T7L-promoter systems with continual monitoring for 2–10 days with blue light exposure (1656 or 600 μE for plasmid-borne and chromosomal systems, respectively). Alkane gas production was monitored at 20-min intervals by automated headspace sampling using a Micro GC, while aqueous amino acid and glycerol depletion were quantified by HPLC.
Fermentations of Halomonas TQ10 containing chromosomally integrated N3T7LIHKFG462I or N3cΔLIHKFG462I was performed as above with LB60Val pH 6.8, except culture medium feeding was employed to maintain an optical density of 0.8 and to replenish the carbon source. Cultures were maintained for about 240 h, with alkane gas production monitored at 2–3 times daily by manual sampling using a Micro GC.
Analytical techniques
Propane levels were determined by manual headspace injection or automated (fermentation off gas monitoring) using an Agilent 490 Micro GC, containing an Al2O3/KCl column and a thermal conductivity detector (TCD). Aqueous culture metabolites (VFAs and glycerol) were analysed by HPLC using an Agilent 1260 Infinity HPLC with a 1260 ALS autosampler, TCC SL column heater, a 1260 refractive index detector (RID) with an Agilent Hi-Plex H column (300 × 7.7 mm; 5 mM H2SO4). The running conditions for both the Micro GC and HPLC were the same as described previously [4]. For amino acid quantitation, analysis was performed according to the method of Bartolomeo and Maisano [43]. Each analyte concentration was calculated by comparing the peak areas to a standard curve generated under the same running conditions. Error bars indicate one standard deviation of the data obtained for the replicates (biological and/or technical triplicates).
Supplementary information
Additional file 1. Additional Figures and Tables with additional data and methods.
Acknowledgements
This is a contribution from the EPSRC/BBSRC Future Biomanufacturing Research Hub and the BBSRC/EPSRC Synthetic Biology Research Centre SYNBIOCHEM. M.A. was funded by a Ph.D. scholarship from the Newton-Mosharafa fund.
Abbreviations
- ADO
Aldehyde-deformylating oxygenase
- BCKDHAB
Human 2-oxoisovalerate dehydrogenase α-and β-subunits
- CAR
Carboxylic acid reductase
- CvFAP
Chlorella variabilis fatty acid photodecarboxylase
- Ferr
Ferredoxin
- Hpad
3-Hydroxypropionaldehyde dehydrogenase
- ilvA
l-Threonine dehydratase biosynthetic
- ilvE
Branched-chain amino acid aminotransferase
- IPTG
Isopropyl β-d-1-thiogalactopyranoside
- KdcA
Branched-chain keto acid decarboxylase
- LB
Luria broth
- LeuABCD
Isopropyl malate synthase, dehydrogenase and isomerases complex
- LPG
Liquefied petroleum gas
- PadA
Phenylacetaldehyde dehydrogenase
- sfp
Maturation factor phosphopantetheinyl transferase
- YciA
Acyl-CoA thioester hydrolase
- αKGSDH
α-Ketoglutaric semialdehyde dehydrogenase
- NADPH
Reduced nicotinamide dinucleotide phosphate
- bio-LPG
Biologically sourced gaseous alkane gases
- IPTG
Isopropyl β-d-thiogalactopyranoside
- PCR
Polymerase chain reaction
- DNA constructs
AFYSCIB: ADOA134F-Ferr-YciA-sfp-CAR-ilvE-BCKDHAB
- FG462VYIB
CvFAPG462V-YciA-ilvE-BCKDHAB
- IHKFG462I
ilvE-Hpad-KdcA-CvFAPG462I
- IPTG
Isopropyl β-d-thiogalactopyranoside
Authors’ contributions
MA designed and performed the experimental work, analysed the results, interpreted the data and produced the initial draft of the manuscript. HST participated in the design of the study, analysing the results, interpreting the data and drafting the manuscript. RH, MF and PK technically supported MA during the experimental work. HST, MHS and NSS coordinated the project. NSS directed the project and secured funding. All authors read and approved the final manuscript.
Funding
The work was supported by C3 Biotechnologies Ltd, the UK Engineering and Physical Sciences Research Council (EP/S01778X/1; EP/J020192/1), and the Biotechnology and Biological Sciences Research Council (BB/M017702/1; BB/L010798/1). MA was funded by a PhD scholarship from the Newton-Mosharafa fund, a science and innovation partnership between the UK and Egypt.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
A patent application (PCT/EP2019/060013) entitled ‘Hydrocarbon production’ is pending in relation to the production of hydrocarbon gases in engineered microbial strains. M.S. and N.S.S. are founding directors of C3 Biotech Ltd.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohamed Amer, Email: mohamed.amer@manchester.ac.uk.
Robin Hoeven, Email: robin.hoeven@manchester.ac.uk.
Paul Kelly, Email: paul.kelly-6@manchester.ac.uk.
Matthew Faulkner, Email: matthew.faulkner@manchester.ac.uk.
Michael H. Smith, Email: mikesmith@c3biotechnologies.com
Helen S. Toogood, Email: helen.toogood@manchester.ac.uk
Nigel S. Scrutton, Email: Nigel.scrutton@manchester.ac.uk
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13068-020-01766-0.
References
- 1.Sanz-Hernández A, Esteban E, Garrido P. Transition to a bioeconomy: perspectives from social sciences. J Clean Prod. 2019;224:107–119. doi: 10.1016/j.jclepro.2019.03.168. [DOI] [Google Scholar]
- 2.de Jong E, Jungmeier G. Biorefinery concepts in comparison to petrochemical refineries. In: Pandey A, Höfer R, Taherzadeh M, Nampoothiri KM, Larroche C, editors. Industrial biorefineries & white biotechnology. Amsterdam: Elsevier B.V; 2015. pp. 3–33. [Google Scholar]
- 3.BOC. Industrial gases UK. 2020. https://www.boconline.co.uk/en/products-and-supply/packaged-chemicals/propellants/propellants.html. Accessed 12 Apr 2020.
- 4.Amer M, Wojcik EZ, Sun C, Hoeven R, Hughes JMX, Faulkner M, Yunus IS, Tait S, Johannissen LO, Hardman SJO, et al. Low carbon strategies for sustainable bio-alkane gas production and renewable energy. Energy Environ Sci. 2020 doi: 10.1039/d0ee00095g. [DOI] [Google Scholar]
- 5.Kallio P, Pásztor A, Thiel K, Akhtar MK, Jones PR. An engineered pathway for the biosynthesis of renewable propane. Nat Commun. 2014;5:4731. doi: 10.1038/ncomms5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menon N, Pásztor A, Menon BRK, Kallio P, Fisher K, Akhtar MK, Leys D, Jones PR, Scrutton NS. A microbial platform for renewable propane synthesis based on a fermentative butanol pathway. Biotechnol Biofuels. 2015;8:61. doi: 10.1186/s13068-015-0231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheppard MJ, Kunjapur AM, Prather KLJJ. Modular and selective biosynthesis of gasoline-range alkanes. Metab Eng. 2016;33:28–40. doi: 10.1016/j.ymben.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Sorigué D, Légeret B, Cuiné S, Blangy S, Moulin S, Billon E, Richaud P, Brugière S, Couté Y, Nurizzo D, et al. An algal photoenzyme converts fatty acids to hydrocarbons. Science. 2017;907:903–907. doi: 10.1126/science.aan6349. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Liang Y, Wu W, Tan X, Lu X. Microbial synthesis of propane by engineering valine pathway and aldehyde-deformylating oxygenase. Biotechnol Biofuels. 2016;9:80. doi: 10.1186/s13068-016-0496-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schirmer A, Rude M, Li X, Popova E, del Cardayre S. Microbial biosynthesis of alkanes. Science. 2010;329:559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 11.Huijbers MME, Zhang W, Tonin F, Hollmann F. Light-driven enzymatic decarboxylation of fatty acids. Angew Chem Int Ed Engl. 2018;57:13648–13651. doi: 10.1002/anie.201807119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khara B, Menon N, Levy C, Mansell D, Das D, Marsh ENG, Leys D, Scrutton NS. Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase. ChemBioChem. 2013;14:1204–1208. doi: 10.1002/cbic.201300307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X, Wang X, Engel PC. The specificity and kinetic mechanism of branched-chain amino acid aminotransferase from Escherichia coli studied with a new improved coupled assay procedure and the enzyme’s potential for biocatalysis. FEBS J. 2013;281:391–400. doi: 10.1111/febs.12609. [DOI] [PubMed] [Google Scholar]
- 14.Gocke D, Nguyen CL, Pohl M, Stillger T, Walter L, Müller M. Branched-chain keto acid decarboxylase from Lactococcus lactis (KdcA), a valuable thiamine diphosphate-dependent enzyme for asymmetric C–C bond formation. Adv Synth Catal. 2007;349:1425–1435. doi: 10.1002/adsc.200700057. [DOI] [Google Scholar]
- 15.Jo J-EE, Mohan Raj S, Rathnasingh C, Selvakumar E, Jung W-CC, Park S. Cloning, expression, and characterization of an aldehyde dehydrogenase from Escherichia coli K-12 that utilizes 3-hydroxypropionaldehyde as a substrate. Appl Microbiol Biotechnol. 2008;81:51–60. doi: 10.1007/s00253-008-1608-x. [DOI] [PubMed] [Google Scholar]
- 16.Ævarsson A, Chuang JL, Wynn RM, Turley S, Chuang DT, Hol WG. Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease. Structure. 2000;8:277–291. doi: 10.1016/S0969-2126(00)00105-2. [DOI] [PubMed] [Google Scholar]
- 17.Thierry A, Maillard MB, Yvon M. Conversion of l-leucine to isovaleric acid by Propionibacterium freudenreichii TL 34 and ITGP23. Appl Environ Microbiol. 2002;68:608–615. doi: 10.1128/AEM.68.2.608-615.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong X, Quinn PJ, Wang X. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for the production of l-threonine. Biotechnol Adv. 2010;29:11–23. doi: 10.1016/j.biotechadv.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Yu P, Chen X, Li P. Enhancing microbial production of biofuels by expanding microbial metabolic pathways. Biotechnol Appl Biochem. 2017;64:606–619. doi: 10.1002/bab.1529. [DOI] [PubMed] [Google Scholar]
- 20.Davis JH, Rubin AJ, Sauer RT. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 2011;39:1131–1141. doi: 10.1093/nar/gkq810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giannakis S, Rtimi S, Darakas E, Escalas-Cañellasbd A, Pulgarin C. Light wavelength-dependent E. coli survival changes after simulated solar disinfection of secondary effluent. Photochem Photobiol Sci. 2015;14:2238–2250. doi: 10.1039/C5PP00110B. [DOI] [PubMed] [Google Scholar]
- 23.Lakavath B, Hedison TM, Heyes DJ, Shanmugam M, Sakuma M, Hoeven R, Tilakaratna V, Scrutton NS. Radical-based photoinactivation of fatty acid photodecarboxylases. Anal Biochem. 2020; Accepted. [DOI] [PubMed]
- 24.Watanabe S, Yamada M, Ohtsu I, Makino K. α-Ketoglutaric semialdehyde dehydrogenase isozymes involved in metabolic pathways of d-glucarate, d-galactarate, and hydroxy-l-proline: molecular and metabolic convergent evolution. J Biol Chem. 2007;282:6685–6695. doi: 10.1074/jbc.M611057200. [DOI] [PubMed] [Google Scholar]
- 25.Ferrández A, Prieto MA, García JL, Díaz E. Molecular characterization of PadA, a phenylacetaldehyde dehydrogenase from Escherichia coli. FEBS Lett. 1997;406:23–27. doi: 10.1016/S0014-5793(97)00228-7. [DOI] [PubMed] [Google Scholar]
- 26.Prandi B, Faccini A, Lambertini F, Bencivenni M, Jorba M, van Droogenbroek B, Bruggeman G, Schöber J, Petrusan J, Elst K, et al. Food wastes from agrifood industry as possible sources of proteins: a detailed molecular view on the composition of the nitrogen fraction, amino acid profile and racemisation degree of 39 food waste streams. Food Chem. 2019;286:567–575. doi: 10.1016/j.foodchem.2019.01.166. [DOI] [PubMed] [Google Scholar]
- 27.Tan D, Xue Y-S, Aibaidula G, Chen G-Q. Unsterile and continuous production of polyhydroxybutyrate by Halomonas TQ10. Bioresour Technol. 2011;102:8130–8136. doi: 10.1016/j.biortech.2011.05.068. [DOI] [PubMed] [Google Scholar]
- 28.Ye J, Huang W, Wang D, Chen F, Yin J, Li T, Zhang H, Chen GQ. Pilot scale-up of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) production by Halomonas bluephagenesis via cell growth adapted optimization process. Biotechnol J. 2018;13:1800074-10. doi: 10.1002/biot.201800074. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H, Zhang HM, Chen X, Li T, Wu Q, Ouyang Q, Chen G-Q. Novel T7-like expression systems used for Halomonas. Metab Eng. 2017;39:128–140. doi: 10.1016/j.ymben.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Li T, Li T, Ji W, Wang Q, Zhang H, Chen G-Q, Lou C, Ouyang Q. Engineering of core promoter regions enables the construction of constitutive and inducible promoters in Halomonas sp. Biotechnol J. 2016;11:219–227. doi: 10.1002/biot.201400828. [DOI] [PubMed] [Google Scholar]
- 31.Yin J, Fu X-Z, Wu Q, Chen J-C, Chen G-Q. Development of an enhanced chromosomal expression system based on porin synthesis operon for halophile Halomonas sp. Appl Microbiol Biotechnol. 2014;98:8987–8997. doi: 10.1007/s00253-014-5959-1. [DOI] [PubMed] [Google Scholar]
- 32.Friehs K. Plasmid copy number and plasmid stability. Adv Biochem Eng Biotechnol. 2004;86:47–82. doi: 10.1007/b12440. [DOI] [PubMed] [Google Scholar]
- 33.Yin J, Wang H, Fu X-Z, Gao X, Wu Q, Chen G-Q. Effects of chromosomal gene copy number and locations on polyhydroxyalkanoate synthesis by Escherichia coli and Halomonas sp. Appl Microbiol Biotechnol. 2015;99:5523–5534. doi: 10.1007/s00253-015-6510-8. [DOI] [PubMed] [Google Scholar]
- 34.Fu XZ, Tan D, Aibaidula G, Wu Q, Chen JC, Chen GQ. Development of Halomonas TQ10 as a host for open production of chemicals. Metab Eng. 2014;23:78–91. doi: 10.1016/j.ymben.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Qin Q, Ling C, Zhao Y, Yang T, Yin J, Guo Y, Chen GQ. CRISPR/Cas9 editing genome of extremophile Halomonas spp. Metab Eng. 2018;47:219–229. doi: 10.1016/j.ymben.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 36.Politi N, Pasotti L, Zucca S, Casanova M, Micoli G, De Angelis MGC, Magni P. Half-life measurements of chemical inducers for recombinant gene expression. J Biol Eng. 2014;8:5. doi: 10.1186/1754-1611-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai T. The antimicrobial effect of blue light: what are behind? Virulence. 2017;8:649–652. doi: 10.1080/21505594.2016.1276691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherrard A. Calor introduces BioLPG to the UK and sets target to be fully renewable by 2040. 2018. https://bioenergyinternational.com/biogas/calor-introduces-biolpg-uk-sets-target-fully-renewable-2040. Accessed 12 Jan 2020.
- 39.Sabri S, Steen JA, Bongers M, Nielsen LK, Vickers CE. Knock-in/Knock-out (KIKO) vectors for rapid integration of large DNA sequences, including whole metabolic pathways, onto the Escherichia coli chromosome at well-characterised loci. Microb Cell Fact. 2013;12:60. doi: 10.1186/1475-2859-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J Biol Eng. 2011;5:15–17. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raman M, Martin K. One solution for cloning and mutagenesis: in-fusion HD cloning plus. Nat Methods. 2014;11:972. doi: 10.1038/nmeth.f.373. [DOI] [Google Scholar]
- 42.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnol Adv. 1983;1:784–791. [Google Scholar]
- 43.Bartolomeo MP, Maisano F. Validation of a reversed-phase HPLC method for quantitative amino acid analysis. J Biomol Tech. 2006;17:131–137. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional Figures and Tables with additional data and methods.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.