Abstract
Members of microRNA-200 (miRNA-200) family play a regulatory role in epithelial to mesenchymal transition (EMT) by suppressing Zeb1 and Zeb2 expression. Consistent with its role in suppressing EMT, Hsa-miR-200c-3p (miR-200c), a member of miR-200 family is poorly expressed in mesenchymal-like triple negative breast cancer (TNBC) cells and ectopic miR-200c expression suppresses cell migration. In this manuscript, we demonstrated that miR-200c potently inhibited TNBC cell growth and tumor development in a mechanism distinct from its ability to downregulate Zeb1 and Zeb2 expression because silencing them only marginally affected TNBC cell growth. We identified phosphodiesterase 7B (PDE7B) as a bona fide miR-200c target. Importantly, miR-200c-led inhibition in cell growth and tumor development was prevented by forcing PDE7B transgene expression while knockdown of PDE7B effectively inhibited cell growth. These results suggest that miR-200c inhibits cell growth by targeting PDE7B mRNA. To elucidate mechanism underlying miR-200c/PDE7B regulation of TNBC cell growth, we showed that cAMP concentration was lower in TNBC cells compared to estrogen receptor-positive (ER+) cells and that both miR-200c and PDE7B siRNAs were able to increase cAMP concentration in TNBC cells. High level of cellular cAMP has been shown to induce cell cycle arrest and apoptosis in TNBC cells. Our observation that ectopic expression of miR-200c triggered apoptosis indicates that it does so by elevating level of cellular cAMP. Analysis of breast tumor gene expression datasets revealed an inverse association between miR-200c and PDE7B expression. Especially, both low miR-200c and high PDE7B expression were correlated with poor survival of breast cancer patients. Our study supports a critical role of miR-200c/PDE7B relationship in TNBC tumorigenesis.
Keywords: PDE7B, miR-200c, cell growth, breast cancer
INTRODUCTION
MicroRNAs (miRNAs) are 20–22 nucleotide non-coding RNAs that downregulate the expression of target genes mostly through base-pairing with their respective 3’-untranslated regions (3’-UTRs) of mRNAs (1). Extensive evidences have firmly linked miRNAs to every event of tumor progression and development. For instance, miR-10b facilitates breast tumor invasion and metastasis by abrogating HOXD10 translation and subsequent RhoC activation (2) while miR-31 triggers apoptosis in breast cancer cells by targeting PKCε and blocks invasion through the suppression of WAVE3 (3, 4). To characterize miRNAs differentially expressed between estrogen receptor positive (ER+) breast cancer and triple negative breast cancer (TNBC) cells, we previously found that Hsa-miR-200c-3p (miR-200c), miR-205 and miR-375 are most significantly downregulated in TNBC cells (5). Consistent with the nature of TNBC cells which are mostly basal-like and thus mesenchymal-like in nature, miR-200c and miR-205 can deter EMT by targeting Zeb1 and Zeb2 (6) while miR-375 suppresses EMT by diminishing SHOX2, a potent EMT regulator (7). In addition to its well-established role in EMT, miR-200c also possesses the capability to suppress breast cancer stem cell self-renewal (8) and can sensitize breast cancer cells to microtubule-targeting chemotherapeutic agents (9). With the aid of Claudin-low TNBC mouse model, miR-200c was shown capable of suppressing breast tumor outgrowth (10, 11). However, molecular mechanism associated with miR-200c-led cell growth inhibition has not been clearly elucidated even though its ability to alter EMT status was suggested as a possible cause (10).
Cyclic nucleotide phosphodiesterases (PDEs) are enzymes regulating cellular cAMP (or cGMP) concentration by controlling the rate of hydrolysis. Based on their sequence similarity, PDEs are classified into 11 families (PDE1–PDE11) comprising of 21 PDEs and 16 of them can decomposes cAMP (12). Individual PDE has its own specific subcellular distribution, and can thus regulate the activities of compartmental downstream cAMP (or cGMP) effectors (e.g., EPAC, exchange protein directly activated by cAMP; PKA, cAMP-dependent protein kinase; PKG, cGMP-dependent protein kinase) which act at the crossroads of diverse signaling pathways and frequently altered in cancer (13). In fact, differential expression of particular PDEs are detected in various malignancies (14). For example, malignant melanoma cells overexpress PDE1C and their growth is inhibited by vinpocetine, a PDE1 inhibitor (15). PDE7B is overexpressed in chronic lymphocytic leukemia (CLL) and is required for CLL cell survival (16, 17). The findings that cellular cAMP concentration and PKA activity are often lower in cancer cells further implicate that PDEs play an important role in cancer progression (18, 19). Hence, molecules capable of inhibiting PDE activity represent as promising means to control cancer progression (20).
The objective of this study is to investigate the role of miR-200c/PDE7B relationship in breast cancer cell growth. Using multiple breast cancer cell lines, we showed that miR-200c potently inhibited both cell growth and tumor development of TNBC cells by causing apoptosis. These miR-200c-led events were apparently mediated by reducing the abundance of PDE7B because ectopically expressing PDE7B transgene prevented miR-200c from inhibiting cell growth and tumor development while PDE7B siRNA inhibited TNBC cell growth. We showed that PDE7B is a bona fide miR-200c target and the expression of miR-200c and PDE7B is inversely correlated in both established breast cancer cells as well as breast tumor specimens. To elucidate the molecular mechanism underlying miR-200c/PDE7B regulation of cell growth, we demonstrated that both miR-200c and PDE7B siRNAs greatly increased cellular cAMP concentration in TNBC cells. Since agents elevating cellular cAMP concentration can inhibit tumor cell growth, our results suggest that miR-200c/PDE7B relationship regulates TNBC cell growth by modulating cellular cAMP concentration. Finally, we analyzed publicly available breast cancer gene expression dataset and revealed that both low miR-200c and high PDE7B expression are correlated with poor overall survival of breast cancer patients. Immunohistochemistry further showed that PDE7B positivity was associated with higher tumor grade. This study demonstrates that miR-200c/PDE7B relationship is critically involved in TNBC cell growth.
RESULTS
MiR-200c inhibits TNBC cell growth in a mechanism that is not through the downregulation of EMT-associated Zeb1/2
Our previous study on miRNA expression profiles revealed that miR-200c, miR-205 and miR-375 were underexpressed in TNBC cells (5), which is consistent with their role as potent EMT-suppressors (6, 7). To determine how these miRNAs affected TNBC cell growth, we performed MTT assay on MDA-MB-231 cells that ectopically express miR-200c, miR-205 or miR-375 (21). In a growth period of 3 days, miR-200c blocked over 70% of cell growth compared to the control while miR-205 and miR-375 exerted little effect on cell growth (Fig.1A), indicating that miR-200c possesses a unique growth-inhibitory role that other EMT-suppressive miRNAs lack. To substantiate this finding, we determined growth-inhibitory effect of miR-200c on additional TNBC (BT549, Hs578T and MDA-MB-436) and ER+ lines (MCF7 and T47D). Treatment of miR-200c mimic resulted in growth inhibition ranging from 41 to 72% in TNBC lines compared to mimic control (Fig.1B). In contrast, miR-200c mimic did not significantly alter growth of MCF7 and T47D cells (Fig.1B). These results demonstrate that miR-200c specifically inhibits TNBC cell growth.
Figure 1. MiR-200c suppresses TNBC cell growth in a Zeb1/2-independent mechanism.
A. Effect of miR-200c, miR-205 and miR-375 on growth of MDA-MB-231 cells. Data are means ± SE (n = 4). B. Effect of miR-200C on growth of various breast cancer cell lines. Data are means ± SE (n = 4). C. Western blotting to analyze the abundance of MDA-MB-231, Hs578T and BT549 cells treated with Zeb1 and Zeb2 siRNA pools individually or in combination. D. Effect of Zeb1/2-knockdown on cell migration. Data are means ± SE (n = 4). *, p < 0.05 vs control. **, p < 0.01 vs control. E. Effect of Zeb1/2-knockdown on cell growth. Data are means ± SE (n = 4).
MiR-200c is known to suppress the level of EMT-associated Zeb1 and Zeb2 (6). To determine whether miR-200c’s growth-inhibitory function was associated with downregulation of Zeb1/2, the expression of Zeb1 and Zeb2 were silenced either alone or together in Hs578T, MDA-MB-231 and BT549 cells using distinct siRNA pools (Fig.1C). Consistent with EMT-suppressing role of Zeb1/2, knockdown of Zeb1 or Zeb2 impaired cell migration (Fig.1D and S1). However, knockdown of Zeb1 or 2 only marginally decreased cell growth (Fig.1E). Our data indicate that miR-200c inhibits TNBC cell growth in a mechanism that is not through the downregulation of EMT-associated Zeb1/2.
PDE7B is a miR-200c target
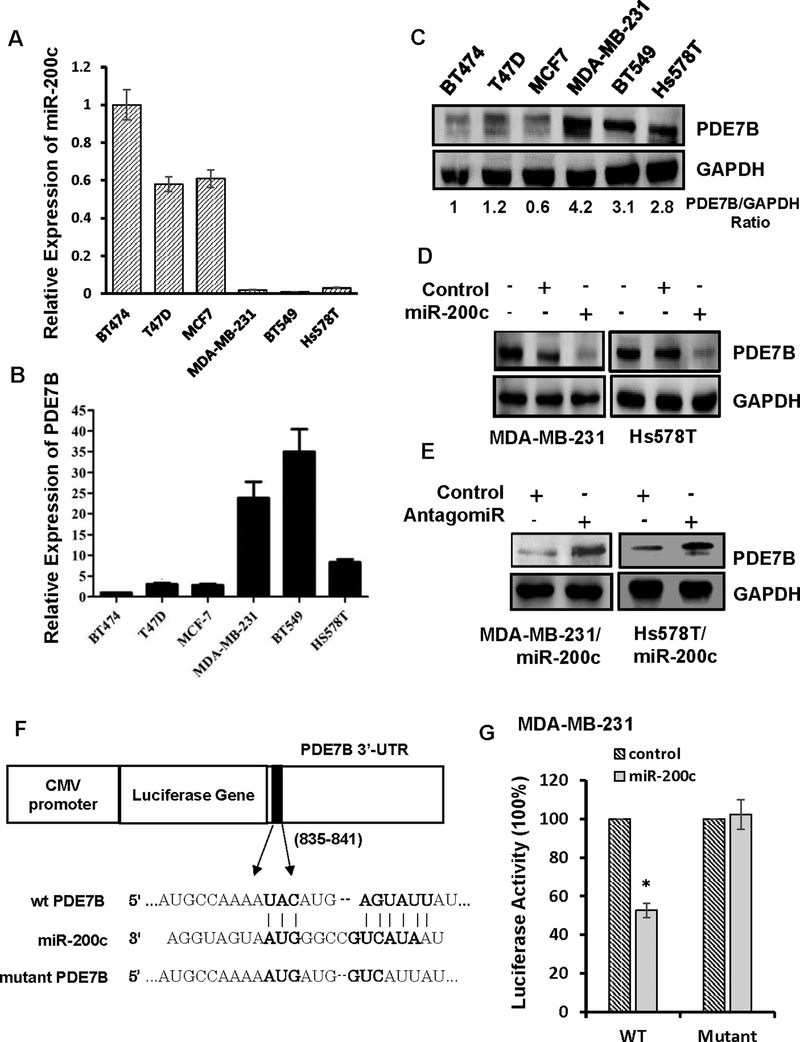
To elucidate molecular mechanism underlying miR-200c-led cell growth inhibition, we examined 28 genes we previously identified as putative miR-200c targets in breast cancer cells (5). Among them, we paid particular attention on PDE7B because cancer cell growth is often sensitive to elevated cellular cAMP concentration (22–24) and PDE7B functions to reduce cellular cAMP concentration. We first performed quantitative reverse transcription-polymerase chain reaction (qRT-PCR) to assess miR-200c and PDE7B expression in 3 ER+ and 3 TBNC lines. Little or no miR-200c was detected in TNBC lines (BT549, Hs578T and MDA-MB-231) while it was readily seen in ER+ lines (BT-474, MCF7 and T47D) (Fig.2A). In contrast, PDE7B mRNA and protein were highly expressed in TNBC lines but their abundances were much lower in ER+ lines (Fig.2B and 2C). To determine the effect of miR-200c on PDE7B expression, we treated MDA-MB-231, Hs578T and BT549 cells with control or miR-200c mimic. Western blot analysis showed that miR-200c, but not the control dramatically reduced the amount of PDE7B in all 3 lines (Fig.2D and Fig.S2). The effect of miR-200c appeared to be specific because miR-200c antagomiR greatly increased the amount of PDE7B in Hs578T and MDA-MB-231 cells with ectopic miR-200c expression (Fig.2E).
Figure 2. PDE7B is a miR-200c target and its expression is inversely correlated with miR-200c in breast cancer cells.
A. QRT-PCR of miR-200c in panel of breast cancer cell lines. Data are means ± SE (n = 4). B. QRT-PCR of PDE7B mRNA in breast cancer cell lines. Data are means ± SE (n = 4). C. Western blot of PDE7B in breast cancer cell lines. D. Effect of forced miR-200c expression on abundance of PDE7B in breast cancer cells. E. Effect of miR-200c inhibitor on abundance of PDE7B in MDA-MB-231 cells with ectopic miR-200c expression. F. Schematic diagram of miR-200c targeted site in 3’-UTR of PDE7B mRNA. G. Effect of miR-200c on activity of luciferase gene containing 3’-UTR of PDE7B in MDA-MB-231 cells. Data are means ± SE (n = 4). *, p < 0.05 vs control.
Since putative miR-200c targeting site is present in the 3’-UTR of PDE7B mRNA (Fig.2F), we investigated whether PDE7B mRNA is a miR-200c target. We linked PDE7B mRNA’s 3’-UTR to the downstream of the luciferase reporter gene in plasmid pMiR. MiR-200c, but not control mimic reduced luciferase activity in both Hs578T and MDA-MB-231 cells (Fig.2G and S3). However, introducing G/C→C/G and A/U→U/A mutation on miR-200c targeting site in 3’-UTR of PDE7B mRNA prevented miR-200c from reducing luciferase activity (Fig. 2F, 2G and S3). These results confirm PDE7B as a bona fide target of miR-200c in TNBC cells.
Growth-inhibitory capability of miR-200c is associated with reduced PDE7B abundance
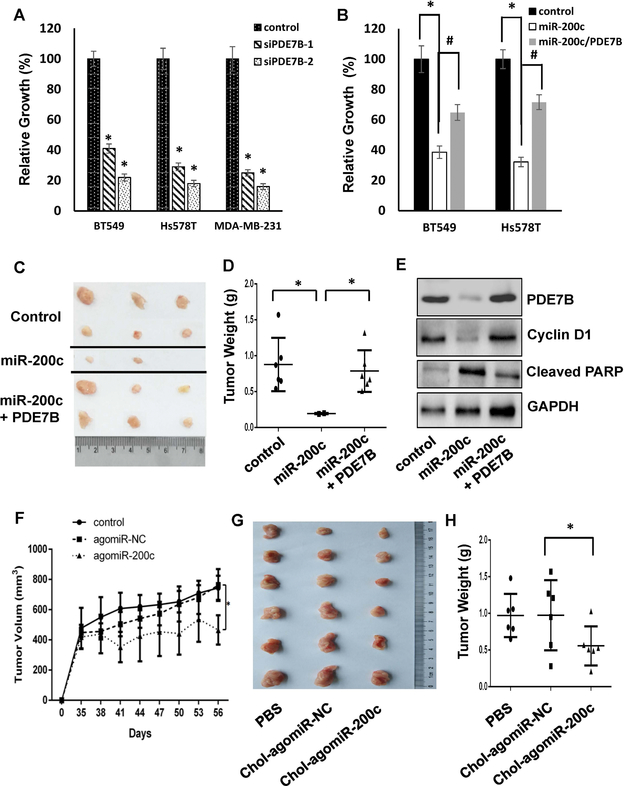
To investigate potential functional link between miR-200c and PDE7B in TNBC growth regulation, we treated BT549, Hs578T and MDA-MB-231 cells with control or two sequence-specific PDE7B siRNAs (Fig.S4). MTT assay showed that silencing PDE7B expression decreased cell growth in all 3 lines (Fig.3A). In parallel, we lentivirally introduced PDE7B transgene (only PDE7B coding region and thus not targetable by miR-200c) into BT549 and Hs578T cells followed by treatment of miR-200c mimic. Forced expression of PDE7B transgene abolished more than half of miR-200c-led growth inhibition (Fig.3B), suggesting that miR-200c inhibits TNBC cell growth at least partially by downregulating PDE7B expression.
Figure 3. MiR-200c suppresses breast cancer cell proliferation by downregulating PDE7B expression.
A. Effect of PDE7B-knockdown on TNBC cell growth. Data are means ± SE (n = 4). *, p < 0.05 vs control. B. Effect of miR-200c on growth of BT549 and Hs578T cells with or without ectopic expression of PDE7B. Data are means ± SE (n = 4). *, p < 0.005 miR-200c vs control. #, p < 0.05 miR-200c vs miR200c/PDE7B. C. Image of Tumors excised from mice injected with control MDA-MB-231 cells or MDA-MB-231 cells with ectopic miR-200c alone or together with PDE7B transgene. D. Weight of tumors excised form nude mice receiving control MDA-MB-231 cells or MDA-MB-231 cells with ectopic expression of miR-200c alone or together with PDE7B. Data are means ± SE. *, p < 0.005 vs control. E. Western blot of tumor extracts to detect PDE7B, Cyclin D1, cleaved PARP and GAPDH. F. MDA-MB-231 cells (106 cells/mouse) were injected to nude mice for 5 week followed by administering PBS, 1 OD Chol-agomiR-NC or Chol-agomiR-200c once every three days for 3 weeks. Rate of tumor outgrowth was determined by measuring tumor volume once every three days. Data are means ± SD (n = 6). *, p < 0.01 vs control (PBS). G. Image of tumors excised form tumor-bearing mice treated with PBS, Chol-agomiR-NC or Chol-agomiR-200c). H. Weight of tumors excised from tumor-bearing mice treated with PBS, Chol-agomiR-NC or Chol-ago-miR-200c. Data are means ± SE. *, p < 0.05 vs Chol-agomiR-NC.
We next investigated miR-200c/PDE7B relationship in regulation of tumor outgrowth using an orthotopic breast tumor model (24–26). Empty vector-transduced MDA-MB-231 cells, MDA-MB-231 cells ectopically expressing miR-200c alone or together with PDE7B transgene were injected into mammary fat pad area of female athymic nude mice and tumors were allowed to develop for 8 weeks. Tumors were observed in all mice injected with vector-transduced cells or cells with ectopic expression of both miR-200c and PDE7B and had an average weight of 0.79± 0.45 and 0.75 ± 0.32 g/mouse respectively (Fig.3C and 3D). In contrast, tumors were only detected in 2 out 6 mice injected with cells ectopically expressing miR-200c and their weights were also much smaller (an average weight of 0.19± 0.01 g/mouse in 2 mice with tumors) (Fig.3C and 3D). Western blot of tumor extracts further showed reduction of cyclin D1 abundance and appearance of cleaved PARP in tumors developed from cells ectopically expressing miR-200c while such alteration was not observed in tumors developed from cells expressing both miR-200c and PDE7B transgene (Fig.3E and S5), indicating that miR-200c-mediated inhibited in tumor outgrowth is associated with reduced level of PDE7B and is likely due to the effect of apoptosis. Since ectopic expression of miR-200c does not distinguish between tumorigenesis and tumor growth effect, we determined how administering affected the development of established tumors. MDA-MB-231 cells were injected into nude mice and tumors were allowed to develop for 5 weeks. Cholesterol-conjugated control and miR-200c mimic (Chol-agomiR-NC and Chol-agomiR-200c) (27) were synthesized and directly injected to established tumors once every three days for 3 weeks. As indicated tumor volumes, intratumoral injection of Chol-agomiR-200c, but not Chol-agomiR-NC greatly slowed tumor outgrowth compared to PBS control group (Fig.3F). Similarly, weights of tumors developed on mice receiving Chol-agomiR-NC were similar to those on PBS-treated mice while administering Chol-agomiR-200c led to almost 50% reduction in tumor weights comparing to PBS group (Fig.3G and 3H). To ensure the reduced tumor outgrowth resulted from miR-200c-induced downregulation of PDE7B, we analyzed the abundance of miR-200c and PDE7B in two individual tumors from each group. QRT-PCR showed that level of miR-200c in mice receiving Chol-agomiR-200c was more than 40 fold higher over those receiving Chol-agomiR-NC (Fig.S6A). Western blotting further revealed that Chol-agomiR-200c led to almost 70% reduction in the abundance of PDE7B in excised tumors (Fig.S6B).
The observation that miR-200c was preferentially expressed in ER+ cells (Fig.2A) prompted us to investigate how neutralizing miR-200c affected their cell growth. MCF7 and T47D cells were treated with 100 nM control or miR-200c mimic antagomiR for 4 days followed by cell growth assessment. MTT assay showed that treatment of miR-200c antagomiR led to a moderate increase in cell growth over the control-treated cells (Fig.S7A). Similarly, ectopically expressing PDE7B increased a similar extent of cell growth in both cell lines (Fig.S7B). These results are consistent with the notion that miR-200c inhibits breast cancer cell growth by downregulating the abundance of PDE7B.
PDE7B is critical for reduced level of cellular cAMP in TNBC cells
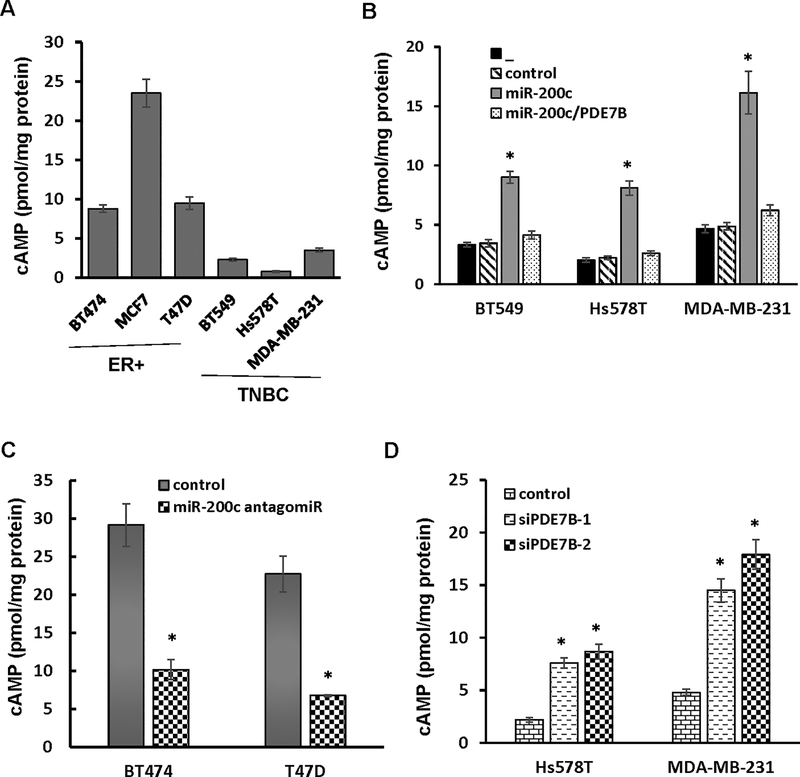
Previous studies have shown that agents elevating cellular level of cAMP can inhibit TNBC cell growth (24, 28, 29), indicating that a reduced cellular cAMP concentration is required for robust TNBC cell growth. We thus hypothesized that miR-200c inhibits TNBC cell growth by downregulating PDE7B, which in turn leads to elevation of cellular cAMP concentration. To test this hypothesis, we initially measured cellular cAMP concentration in both TNBC (low or no miR-200c/high PDE7B) and ER+ cells lines (high miR-200c/low or no PDE7B). ELISA with anti-cAMP antibody showed that level of cellular cAMP was much lower in TNBC lines than ER+ cell lines (Fig.4A), signifying that cellular cAMP concentration is positively correlated to miR-200c but negatively associated with PDE7B abundance in breast cancer cells. To determine how miR-200c affects cellular cAMP concentration in TNBC cells, we introduced control or miR-200c mimic into BT549, Hs578T and MDA-MB-231 cells. MiR-200c mimic, but not control mimic led to a remarkable increase in cellular cAMP concentration in all 3 lines (Fig.4B). However, ectopically expressing PDE7B transgene disabled miR-200c to elevate cellular cAMP concentration (Fig.4B). In parallel, we treated ER+ BT474 and T47D cells with 100 nM control or miR-200c antagomiR. Compared with cells treated with control, we observed 65 and 70% reduction in the level of cellular cAMP in BT474 and T47D cell respectively (Fig.4C). In addition, we also analyzed cellular cAMP concentration in Hs578T and MDA-MB231 cells treated with PDE7B siRNAs. Compared with scramble control, both PDE7B siRNAs greatly increased the level of cellular cAMP (Fig.4D), further supporting the notion that miR-200c elevates cellular cAMP concentration by diminishing PDE7B expression.
Figure 4. Forcing PDE7B transgene expression decreases level of cAMP in breast cancer cells with ectopic miR-200c expression.
A. Level of cAMP in breast cancer cell lines. Data are means ± SE (n = 4). B. Level of cAMP in TNBC cells ectopically expressing miR-200c alone or together with PDE7B transgene. Data are means ± SE (n = 4). *, p < 0.05 vs control. “_” denotes no treatment. C. Level of cAMP in ER+ breast cancer cells treated with miR-200c antagomiR. Data are means ± SE (n = 4). *, p < 0.05 vs control. D. Level of cellular cAMP in TNBC cells with PDE7B knockdown. Data are means ± SE (n = 4). *, p < 0.05 vs control.
MiR-200c-led cells growth inhibition results from apoptosis
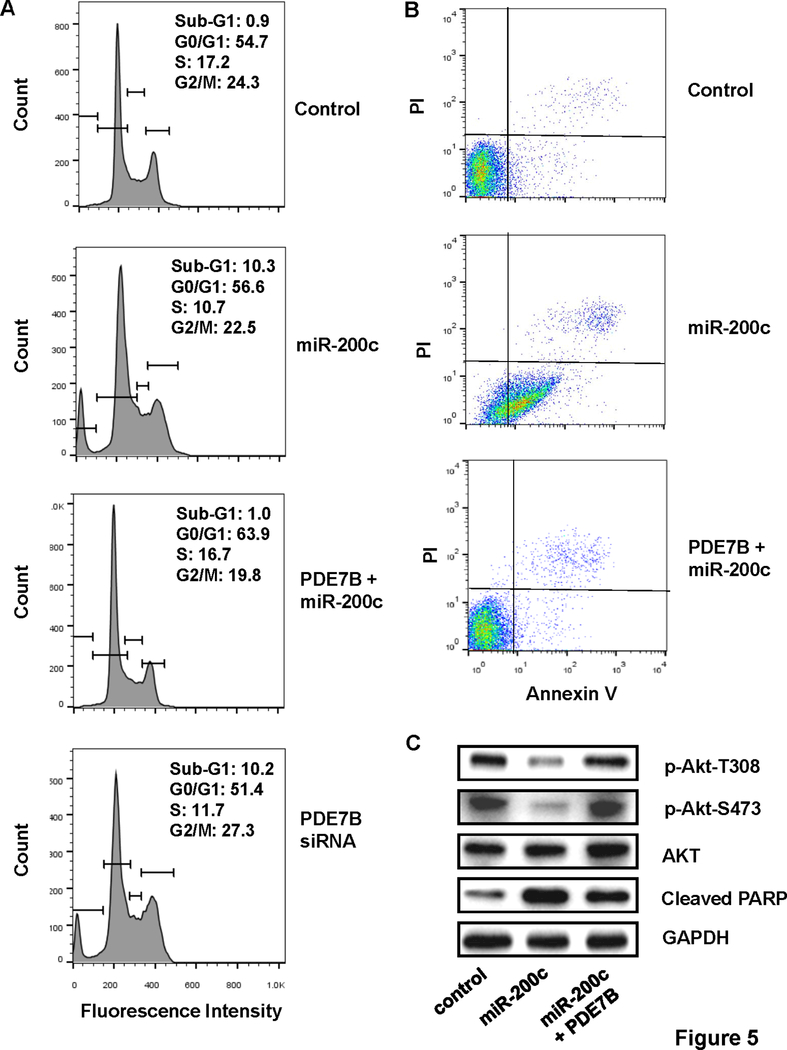
To define molecular mechanism underlying miR-200c-led cell growth inhibition, we performed flow cytometry to examine cell cycle progression on control MDA-MB-231 and MDA-MB-231 cells ectopically expressing miR-200c alone or together with PDE7B transgene. Compared with control cells, much higher percentage of MDA-MB-231 cells ectopically expressing miR-200c were present at sub-G1 stage (0.9 vs 10.3%) (Fig.5A). However, percentage of cells in sub-G1 stage was almost identical between control and cells ectopically expressing both miR-200c and PDE7B transgene (0.9 vs 1.0%) (Fig.5A). Similar to what was observed with cells ectopically expressing miR-200c, knockdown of PDE7B also led to dramatic increase in the population of cells at sub-G1 (10.2%) (Fig.5A). To determine whether the occurrence of apoptosis was the cause of high percentage of miR-200c-expressing MDA-MB-231 cells at sub-G1 stage, we performed Annexin V/propidium iodine (PI) staining-based flow cytometry. While approximately 9.3 and 7.7% of apoptotic cells (Q2 + Q3) were detected in control and cells ectopically expressing both miR-200c and PDE7B transgene, percentage of apoptotic cells increased to 77.1% in MDA-MB-231 cells with forced expression of miR-200c (Fig.5B). Western blot further revealed reduced levels of phosphor-Akt (both T308 and S473) and appearance of cleaved PARP in MDA-MB-231 cells with forced miR-200c expression but not in cells ectopically expressing both miR-200c and PDE7B (Fig.5C). These results suggest that miR-200c inhibits TNBC cell growth by inducing apoptosis most likely through the suppression of Akt activity.
Figure 5. MiR-200c-led suppression in breast cancer cell growth is associated with apoptosis.
A. Flow cytometry of cell cycle progression (control MDA-MB-231 cells, MDA-MB-231 cells ectopically expressing miR-200c alone or together with PDE7B transgene and PDE7B-knockdown MDA-MB-231 cells). B. Annexin V/PI staining-based flow cytometry of control MDA-MB-231 cells and MDA-MB-231 cells ectopically expressing miR-200c alone or together with PDE7B transgene. C. Western blot of phosphor-Akt, Akt, cleaved PARP and GAPDH in control MDA-MB-231, MDA-MB-231 cells ectopically expressing miR-200c alone or with PDE7B.
High PDE7B expression is a poor indicator of breast malignancies
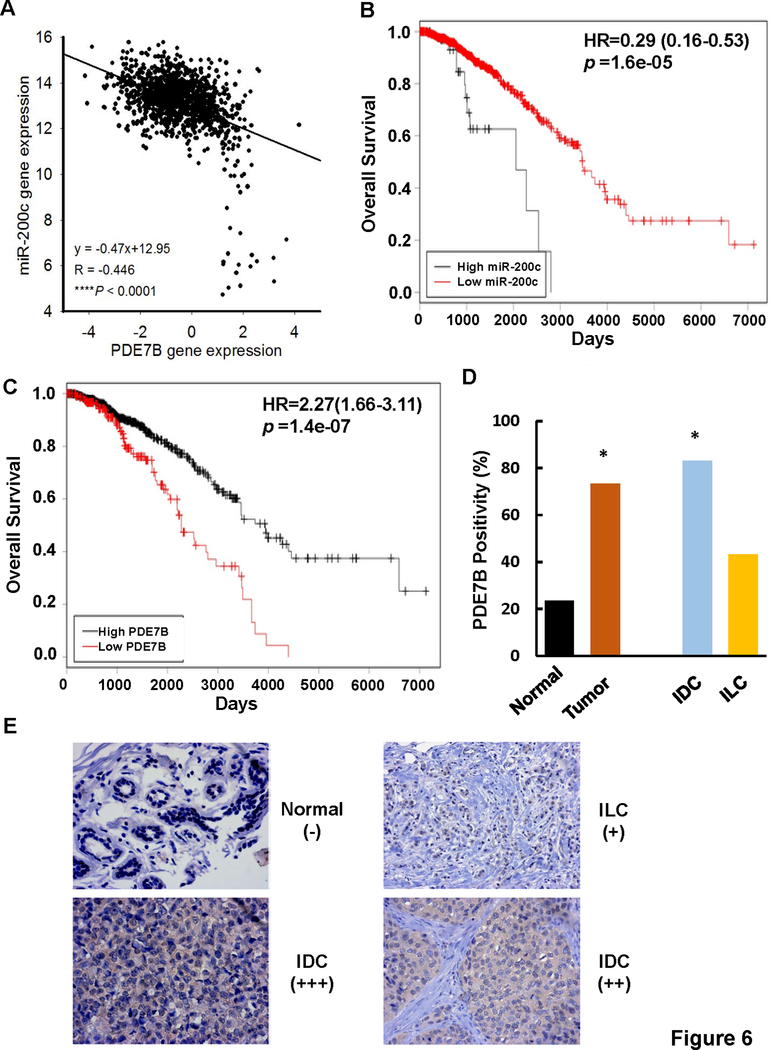
To investigate whether our laboratory studies recapitulated clinical breast tumors, we evaluated correlation between miR-200c and PDE7B expression in breast tumors using TCGA breast tumor gene expression dataset. Pearson correlation coefficient analysis showed a strong negative correlation between miR-200c and PDE7B expression (R = ‒ 0.446, P < 0.0001) in all breast cancer patients (Fig.6A), which is consistent with the observation in which their expression was inversely associated in established breast cancer cell lines (Fig.2A, 2B and 2C).
Figure 6. PDE7B expression is associated with poor prognosis of breast cancer patients.
A. Correlation between miR-200c and PDE7B expression in breast tumors. Data was analyzed using TCGA_BRCA_exp_HiseqV2_PANCAN-2015–02-24 and TCGA_BRCA_miRNA-2015–02-24 datasets; n =1192. B. miR-200c in breast cancer patient overall survival. Overall survival curve was calculated using TCGA_BRCA_miRNA-2015–02-24 dataset with the aid of Cut-off Finder; n = 1,152. C. PDE7B in breast cancer patient overall survival. Overall survival curve was calculated using TCGA_BRCA_exp_HiseqV2_PANCAN-2015–02-24 datasets with the aid of Cut-off Finder; n = 1,176. D. Percentage of breast tumor specimens positive for PDE7B expression. IDC, invasive ductal carcinoma; IFL, infiltrating lobular carcinoma. *, p < 0.001 vs Normal. E. Immunohistochemistry of PDE7B on breast tumor specimens. Scale bars, 200μm.
Abundance of miR-200c has been previously shown to negatively correlate with clinical stage, local relapse, distant metastasis and survival with limited specimens of breast cancer patients (30). The inverse association between miR-200c and PDE7B in breast cancer patients prompted us to further assess the relevance of miR-200c and PDE7B to overall survival of breast cancer patients using TCGA datasets because they possess large sample size, molecular profiles and clinical outcome information. Cutoff Finder analysis showed that both low miR-200c and high PDE7B expression correlate with poor overall survival of breast cancer patients (Fig.6B and 6C).
We extended our studies by examining PDE7B level in human tissue arrays that contained 17 normal breast and 178 breast tumor specimens (131 invasive ductal carcinomas, 44 infiltrating lobular carcinomas, 1 medullary carcinoma, 1 mucinous adenocarcinoma and 1 invasive papillary carcinoma) (Table S1). Immunohistochemistry (IHC) showed that positivity of PDE7B staining was 73.4% in breast tumor samples while 23.5% in normal breast tissues (Fig.6D, 6E and Table S1). Given the observation that the positivity of PDE7B staining in invasive ductal carcinomas was statistically higher than that in normal breast tissues (Fig.6D, 6E and Table S1), we analyzed the correlation between PDE7B positivity and various clinicopathological parameters in these tumor specimens. Although PDE7B positivity was independent of age, tumor size (T) and degree of spread to regional lymph nodes (N), it was associated with histological tumor grades (Table 1). As TNBCs are generally poorly differentiated (in grade III), positive correlation between the positivity of PDE7B staining and tumor grades is again consistent with the observation that PDE7B is preferentially expressed in TNBC cell lines.
Table 1.
Correlation between Positivity of PDE7B Staining and Clinicopathological Parameters of Invasive Ductal Carcinomas
| n=131 | PDE7B Positivity |
p value | |
|---|---|---|---|
| Negative | Positive | ||
| Age | 0.125 | ||
| < 50 | 16 | 60 | |
| ≥ 50 | 6 | 49 | |
| Histologic Grade | 0.017 | ||
| I | 1 | 7 | |
| II | 21 | 74 | |
| III | 0 | 28 | |
| T | 0.283 | ||
| 1 | 4 | 14 | |
| 2 | 13 | 79 | |
| 3 | 4 | 8 | |
| 4 | 1 | 8 | |
| N | 0.839 | ||
| 0 | 17 | 87 | |
| 1 | 5 | 20 | |
| 2 | 0 | 1 | |
| 3 | 0 | 1 | |
DISCUSSION
In our previous attempt to identify miRNAs differentially expressed between TNBC and ER+ breast cancer cells, we demonstrate that miR-200c is one of the miRNAs consistently underexpressed in TNBC cells (5). In additional to its well-established role in breast cancer cell EMT (31–33), miR-200c has also been reported to play a tumor-suppressive role by negatively regulating self-renewal of breast cancer stem cells and chemosensitivity (10, 34, 35). Importantly, nanoparticle-delivered miR-200c was shown to enhance both chemo- and radio-sensitivities in breast cancer (36–39). In fact, analyses of primary breast tumor tissues reveals that low level of miR-200c is associated with metastasis (40) and poor patient survival (30). In this study, we show that miR-200c effectively inhibits TNBC cell growth (Fig.1), thus adding an additional anti-tumorigenic function of miR-200c.
MiR-200c suppresses EMT by targeting Zeb1/2, two EMT-essential transcription factors (6, 41). Interestingly, we show that depleting Zeb1/2 exhibits little effect on TNBC cell growth (Fig.1), thus indicating that miR-200c inhibits TNBC cell growth in a mechanism that is not through the downregulation of EMT-associated Zeb1/2. In this study, we present evidence that PDE7B is a bona fide target of miR-200c (Fig.2). This finding is consistent with several available CLIP-based miRNA targetome datasets which also demonstrate PDE7B as a miR-200c target (42–44). This is clearly consistent with our observation that level of miR-200c is inversely correlated with PDE7B abundance in established breast cancer cell lines (Fig.2). Moreover, bioinformatics studies further revealed that miR-200c and PDE7B expression are inversely associated in human breast tumor specimens (Fig.6).
This inverse correlation between miR-200c and PDE7B suggest that miR-200c specifically inhibits TNBC cell growth by diminishing PDE7B expression. This notion is supported by our findings that depleting PDE7B resulted in cell growth suppression (Fig.3A) and forced PDE7B expression greatly prevented miR-200c-led cell growth suppression and tumor outgrowth (Fig.3B, 3C, 3D and 3E). However, we observed that forced PDE7B did not fully rescue miR-200c-led growth suppression (Fig.3B). MiR-200c has been reported to target K-Ras and CRKL (45, 46); it is thus highly likely that in addition to PDE7B, miR-200c may also inhibit TNBC cell growth by suppressing other growth-relevant genes.
PDE7B has been shown to play a critical role in CLL because PDE7B is not only overexpressed in CLL but also required for their survival (16). Analyses of clinical specimens further demonstrate that PDE7B is an unfavorable characteristic in CLL and Mantle cell lymphoma (17, 47). Recently, PDE7B was reported to be overexpressed in glioblastoma and serve as a significant cell growth mediator (48). In this study, we found that PDE7B is preferentially expressed in TNBC cells (Fig.2) and depletion of PDE7B led to TNBC growth inhibition (Fig.3), suggesting that PDE7B is involved in TNBC tumorigenesis. The role of PDE7B in breast cancer development is apparently also supported by the finding that high expression of PDE7B correlates with poor breast cancer patient survival (Fig.6). We propose a notion that concurrent loss of miR-200c and gain of PDE7B expression may potentially be an important event in breast tumor progression.
As a PDE isotype, PDE7B functions to reduce cellular cAMP concentration through hydrolysis (12). We show that abundance of PDE7B is inversely correlated with cellular cAMP concentration in breast cancer cells (Fig.4), indicating that low level of cellular cAMP in TNBC cells may be maintained by PDE7B (Fig.4). This notion is supported by our observation that both miR-200c and PDE7B siRNAs are able to increase cellular cAMP concentration in TNBC cells (Fig.4). It has been well established that agents raising cellular cAMP concentration can suppress growth of various cancer types including breast, colon and medullary thyroid cancer cells (22–24). In this study, we found that miR-200c raised cellular cAMP concentration by downregulating PDE7B expression (Fig.4). Cell cycle arrest and apoptosis have been implicated as the mechanisms responsible for cAMP-induced cell growth inhibition (49, 50). We showed that ectopic miR-200c expression resulted in spontaneous apoptosis in TNBC cells in both in vitro and in vivo models (Fig.3 and 5), suggesting cAMP-induced apoptosis as the most likely mechanism responsible for miR-200c-led inhibition in both cell growth and tumor development. A recent study by Weinberg’s group reported that increase in cellular cAMP concentration and subsequently PKC activation induce a mesenchymal-to-epithelial transition in breast cancer cells, which promotes their differentiation and loss of tumor-initiating ability (51). We showed that exogenously administering synthesized miR-200c mimic effectively deterred tumor development (Fig.3). We reason that miR-200c’s tumor-suppressive effect can also be contributed by the ability of cAMP/PKA signaling axis to trigger MET and cause loss of tumor-initiating ability of breast cancer cells.
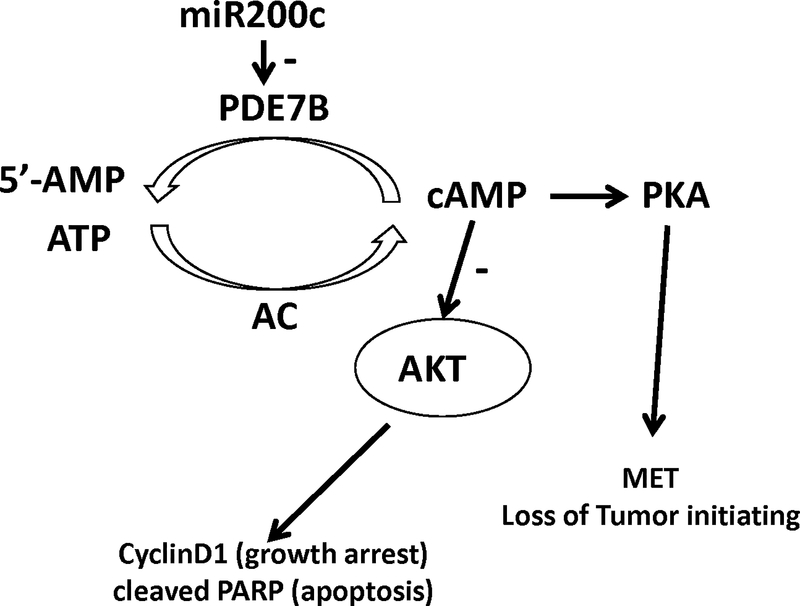
In this study, we establish the importance of miR-200c/PDE7B relationship in regulation of TNBC cell growth using established lines. The detection of positive correlation between PDE7B and overall survival but negative correlation between miR-200c and overall survival (Fig.6) are strongly consistent with our experimental findings and support a critical role of miR-200c/PDE7B relationship in TNBC tumor cell growth and progression. We propose a model (Fig.7) in which balance of miR-200c and PDE7B controls cellular cAMP concentration. Increase in cellular cAMP concentration suppresses Akt activity, leading to cell growth arrest and apoptosis. Meanwhile, elevated level of cellular cAMP also induces MET and causes the loss of tumor initiating ability in breast cancer cells. This study suggests that an anti-TNBC therapeutic strategy may be developed by modulating miR-200c/PDE7B balance.
Figure 7. Model of PDE7B/miR-200c regulation of breast cancer cell growth.
Balance of miR-200c and PDE7B controls cellular cAMP concentration in breast cancer cells. Elevated cellular cAMP concentration both suppresses Akt activity and activates PKA, leading to cell growth arrest/apoptosis and loss of tumor initiating ability.
MATERIALS AND METHODS
Cell culture and other reagents.
All cell lines were obtained from American Tissue Culture Collection (Manassas, VA) and verified for authenticity. Cells were cultured in DMEM containing 10% fetal bovine serum. Detailed information on antibodies used for Western blot were described in Supplemental Information. Micro-RNA, control mimics and antagomiR were purchased from Invitrogen (Carlsbad, CA). Cholesterol-conjugated miRNAs (Chol-agomiR-NC and Chol-agomiR-200c) were purchased from GenePharma (Shanghai, China). The specificity of miR-200c mimic (agomiR-200c) were confirmed by several recent publications (39, 52)
Cell migration.
Transwells (Corning, Tewksbury, MA) were used to analyze cell migration as previously described (21). Briefly, the undersurface of the upper chamber was coated with 10 μg/ml of collagen I. Cells were suspended in a density of 106cells/ml and 100μl of cell suspension was added into the upper chamber of each Transwell. Cells were allowed to migrate for 4 h, cells on the undersurface of the upper chamber were then stained and counted under a phase-contrast microscope.
Cell growth.
MTT assay was performed to measure cell growth as previously described (53). Briefly, cells were suspended in a density of 105cells/ml and 0.5 ml of cell suspension was added into each well of a 24-well plates. Cells were cultured for 1 to 3 days followed by adding MTT solution to cells for 2 h. After removing culture medium, MTT formazan crystals were dissolved in DMSO and measured with a microplate reader at 560 nm.
qRT–PCR.
Total RNA extracted from cells using Trizol (Invitrogen) was used to analyze the levels of PDE7B and β-actin mRNA. The levels of miR-200c and U6 snRNA were measured using miR-200c and U6 snRNA TaqMan microRNA Assay Kits (Invitrogen). Level of PDE7B mRNA was standardized using β-actin mRNA as the internal standardization while miR-200c was standardized by U6 snRNA.
Luciferase reporter gene constructs and luciferase assay.
PDE7B 3’-UTR luciferase reporter gene plasmid was constructed by inserting the entire human PDE7B 3’-UTR sequence into the pMiR vector (Invitrogen). PDE7B 3’-UTR containing mutation at miR-200c targeting site was generated through site-directed mutagenesis on wild-type plasmid using QuickChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). To determine the effect of PDE7B 3’-UTR on luciferase activity, cells were transfected with empty pMiR or pMiR containing wild-type or mutant PDE7B 3’UTR for 2 days and then lysed for measurement of luciferase activity. The trransfection efficiency was normalized by including pTK-RLuc plasmid (1:200 ratio) in all transfection experiments and Renella luciferase activity was determined for standardization. Dual-Luciferase® Reporter Assay System (Promega, Madison, WI) was used to measure both firefly and Renella luciferase activities.
Measurement of cAMP.
Cyclic AMP Assay Kit (Cayman Chemical, Ann Arbor, MI) was employed to quantitate cAMP according to manufacturer’s protocol. To determine effect of miR-200c and PDE7B knockdown on cellular cAMP concentration, cells were transfected with control, miR-200c mimic or PDE7B siRNAs for 4 days prior to the analysis of cAMP.
Flow cytometry to analyze cell cycle and apoptosis.
Cell cycle was analyzed as previously described (24). Briefly, cells were detached with trypsin, washed and fixed in 100% ethanol. Fixed cells were suspended in PBS containing 20μg/mL of propidium iodide and analyzed by flow cytometry using FACSCanto II flow cytometer (BD Biosciences, Bedford, MA). Generated data were analyzed using the BD FACSDiva Software. Extent of apoptosis was determined using Annexin V-FITC detection kit (BD Biosciences) as previously described (54) and the percentage of apoptotic cells was calculated using ModFit LT 3.0 software (BD Biosciences).
Tumorigenesis study.
Vertebrate animal procedures were approved by the Ethical Committee of Shanghai University of Traditional Chinese Medicine and performed as previously described (25, 26). Briefly, 1×106 cells were implanted into 4th mammary fat pad of 6-week-old female BALB/c nude mice (Academia Sinica, Shanghai, China). After 8 week, mice were sacrificed, tumors were excised and weighed. Parts of excised tumors were also homogenized for western blotting. To determine the effect of miR-200c on the development of established tumors, nude mice were first injected with 1×106 MDA-MB-231 cells. After 5 weeks, total of 1 OD Chol-agomiR-NC or Chol-agomiR-200c was injected at 3 different spots on each established tumor. Treatment was repeated once every 3 days and rate of tumor outgrowth was determined by measuring tumor volumes (V) which were calculated using formula of V = 0.5 × Dmax x (Dmin)2, where Dmax is the maximal tumor diameter and Dmin is the corresponding perpendicular diameter. After 3 weeks of treatment, mice were sacrificed and tumors were excised for weight measurement and miR-200c/PDE7B quantitation.
Immunohistochemistry.
Intensity of PDE7B in breast tumors and normal breast tissues were evaluated by IHC using anti-PDE7B polyclonal antibody on commercial tissue array (BR2086, Alena Biotechnology Ltd, Xi´an, China) which contains 17 normal breast tissues and 178 breast tumor specimens. Intensity of PDE7B immunostaining was graded based on the percentage of cells exhibiting PDE7B staining and average 1,000 cells counted for each sample. “-” was negative PDE7B staining; “+”, “++” and “+++” were <25%, 25~50% and >50% cells positive for PDE7B staining respectively.
Statistical analysis.
Statistical analyses of cell growth, migration, tumor outgrowth and luciferase activities were done by ANOVA. Adjusted χ2 test was used to compare covariates between PDE7B staining and clinicopathological parameters. Correlation between miR-200 or PDE7B expression and overall survival of breast cancer patients was analyzed using breast cancer gene expression dataset (TCGA_BRCA_miRNA-2015–02-24 or TCGA_BRCA_exp_HiseqV2_PANCAN-2015–02-24 respectively) with the aid of Cutoff Finder (55). The cutoff values of high and low expression groups for miR-200c and PDE7B are 11.14 and 0.5712 respectively, and p < .05 was considered as significant. Correlation between PDE7B expression and clinical features were investigated using Pearson’s chi-square tests. The correlation between miR-200c and PDE7B expression in human breast cancer patients was analyzed by Pearson correlation coefficient using datasets TCGA_BRCA_exp_HiSeqV2_PANCAN-2015–02-24 and TCGA_BRCA_miRNA-2015–02-24). p < 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from NSF of China 81773946, 81573673, 81001666 (DZ), NIH CA187152 (SH) and Florida Breast Cancer Foundation (SH).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 2.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–8. [DOI] [PubMed] [Google Scholar]
- 3.Korner C, Keklikoglou I, Bender C, Worner A, Munstermann E, Wiemann S. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon). J Biol Chem. 2013;288(12):8750–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sossey-Alaoui K, Downs-Kelly E, Das M, Izem L, Tubbs R, Plow EF. WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. Int J Cancer. 2011;129(6):1331–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo D, Wilson JM, Harvel N, Liu J, Pei L, Huang S, et al. A systematic evaluation of miRNA:mRNA interactions involved in the migration and invasion of breast cancer cells. J Transl Med. 2013;11:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008;10(5):593–601. [DOI] [PubMed] [Google Scholar]
- 7.Hong S, Noh H, Teng Y, Shao J, Rehmani H, Ding HF, et al. SHOX2 Is a Direct miR-375 Target and a Novel Epithelial-to-Mesenchymal Transition Inducer in Breast Cancer Cells. Neoplasia. 2014;16(4):279–90 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Molecular cancer therapeutics. 2009;8(5):1055–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knezevic J, Pfefferle AD, Petrovic I, Greene SB, Perou CM, Rosen JM. Expression of miR-200c in claudin-low breast cancer alters stem cell functionality, enhances chemosensitivity and reduces metastatic potential. Oncogene. 2015;34(49):5997–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones R, Watson K, Bruce A, Nersesian S, Kitz J, Moorehead R. Re-expression of miR-200c suppresses proliferation, colony formation and in vivo tumor growth of murine claudin-low mammary tumor cells. Oncotarget. 2017;8(14):23727–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev. 2006;58(3):488–520. [DOI] [PubMed] [Google Scholar]
- 13.Maurice DH, Ke H, Ahmad F, Wang Y, Chung J, Manganiello VC. Advances in targeting cyclic nucleotide phosphodiesterases. Nature reviews Drug discovery. 2014;13(4):290–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy I, Horvath A, Azevedo M, de Alexandre RB, Stratakis CA. Phosphodiesterase function and endocrine cells: links to human disease and roles in tumor development and treatment. Current opinion in pharmacology. 2011;11(6):689–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu K, Murata T, Watanabe Y, Sato C, Morita H, Tagawa T. Characterization of phosphodiesterase 1 in human malignant melanoma cell lines. Anticancer Res. 2009;29(4):1119–22. [PubMed] [Google Scholar]
- 16.Zhang L, Murray F, Zahno A, Kanter JR, Chou D, Suda R, et al. Cyclic nucleotide phosphodiesterase profiling reveals increased expression of phosphodiesterase 7B in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2008;105(49):19532–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Murray F, Rassenti LZ, Pu M, Kelly C, Kanter JR, et al. Cyclic nucleotide phosphodiesterase 7B mRNA: an unfavorable characteristic in chronic lymphocytic leukemia. Int J Cancer. 2011;129(5):1162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drees M, Zimmermann R, Eisenbrand G. 3’,5’-Cyclic nucleotide phosphodiesterase in tumor cells as potential target for tumor growth inhibition. Cancer Res. 1993;53(13):3058–61. [PubMed] [Google Scholar]
- 19.Cho-Chung YS, Nesterova MV. Tumor reversion: protein kinase A isozyme switching. Ann N Y Acad Sci. 2005;1058:76–86. [DOI] [PubMed] [Google Scholar]
- 20.Savai R, Pullamsetti SS, Banat GA, Weissmann N, Ghofrani HA, Grimminger F, et al. Targeting cancer with phosphodiesterase inhibitors. Expert Opin Investig Drugs. 2010;19(1):117–31. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Zhang M, Chen H, Dong Z, Ganapathy V, Thangaraju M, et al. Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer Res. 2010;70(20):7894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dicitore A, Grassi ES, Caraglia M, Borghi MO, Gaudenzi G, Hofland LJ, et al. The cAMP analogs have potent anti-proliferative effects on medullary thyroid cancer cell lines. Endocrine. 2016;51(1):101–12. [DOI] [PubMed] [Google Scholar]
- 23.McEwan DG, Brunton VG, Baillie GS, Leslie NR, Houslay MD, Frame MC. Chemoresistant KM12C colon cancer cells are addicted to low cyclic AMP levels in a phosphodiesterase 4-regulated compartment via effects on phosphoinositide 3-kinase. Cancer Res. 2007;67(11):5248–57. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Li Y, Zhu JY, Fang D, Ding HF, Dong Z, et al. Triple negative breast cancer development can be selectively suppressed by sustaining an elevated level of cellular cyclic AMP through simultaneously blocking its efflux and decomposition. Oncotarget. 2016;7(52):87232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Zhu G, Li Y, Padia RN, Dong Z, Pan ZK, et al. Extracellular signal-regulated kinase signaling pathway regulates breast cancer cell migration by maintaining slug expression. Cancer Res. 2009;69(24):9228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Q, Lu YY, Noh H, Hong S, Dong Z, Ding HF, et al. Interleukin enhancer-binding factor 3 promotes breast tumor progression by regulating sustained urokinase-type plasminogen activator expression. Oncogene. 2013;32(34):3933–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nature biotechnology. 2007;25(10):1149–57. [DOI] [PubMed] [Google Scholar]
- 28.Naviglio S, Di Gesto D, Romano M, Sorrentino A, Illiano F, Sorvillo L, et al. Leptin enhances growth inhibition by cAMP elevating agents through apoptosis of MDA-MB-231 breast cancer cells. Cancer Biol Ther. 2009;8(12):1183–90. [DOI] [PubMed] [Google Scholar]
- 29.Naviglio S, Di Gesto D, Illiano F, Chiosi E, Giordano A, Illiano G, et al. Leptin potentiates antiproliferative action of cAMP elevation via protein kinase A down-regulation in breast cancer cells. J Cell Physiol. 2010;225(3):801–9. [DOI] [PubMed] [Google Scholar]
- 30.Song C, Liu LZ, Pei XQ, Liu X, Yang L, Ye F, et al. miR-200c inhibits breast cancer proliferation by targeting KRAS. Oncotarget. 2015;6(33):34968–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67(17):7972–6. [DOI] [PubMed] [Google Scholar]
- 32.Radisky DC. miR-200c at the nexus of epithelial-mesenchymal transition, resistance to apoptosis, and the breast cancer stem cell phenotype. Breast Cancer Res. 2011;13(3):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howe EN, Cochrane DR, Richer JK. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011;13(2):R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng ZM, Qiu J, Chen XW, Liao RX, Liao XY, Zhang LP, et al. Essential role of miR-200c in regulating self-renewal of breast cancer stem cells and their counterparts of mammary epithelium. BMC cancer. 2015;15:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim YY, Wright JA, Attema JL, Gregory PA, Bert AG, Smith E, et al. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J Cell Sci. 2013;126(Pt 10):2256–66. [DOI] [PubMed] [Google Scholar]
- 36.Lin J, Liu C, Gao F, Mitchel RE, Zhao L, Yang Y, et al. miR-200c enhances radiosensitivity of human breast cancer cells. J Cell Biochem. 2013;114(3):606–15. [DOI] [PubMed] [Google Scholar]
- 37.Sun Q, Liu T, Yuan Y, Guo Z, Xie G, Du S, et al. MiR-200c inhibits autophagy and enhances radiosensitivity in breast cancer cells by targeting UBQLN1. Int J Cancer. 2015;136(5):1003–12. [DOI] [PubMed] [Google Scholar]
- 38.Kopp F, Oak PS, Wagner E, Roidl A. miR-200c sensitizes breast cancer cells to doxorubicin treatment by decreasing TrkB and Bmi1 expression. PLoS One. 2012;7(11):e50469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Meng T, Yuan M, Wen L, Cheng B, Liu N, et al. MicroRNA-200c delivered by solid lipid nanoparticles enhances the effect of paclitaxel on breast cancer stem cell. International journal of nanomedicine. 2016;11:6713–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Damiano V, Brisotto G, Borgna S, di Gennaro A, Armellin M, Perin T, et al. Epigenetic silencing of miR-200c in breast cancer is associated with aggressiveness and is modulated by ZEB1. Genes, chromosomes & cancer. 2017;56(2):147–58. [DOI] [PubMed] [Google Scholar]
- 41.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & development. 2008;22(7):894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic acids research. 2014;42(Database issue):D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic acids research. 2018;46(D1):D239–D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi Y, Zhao Y, Li C, Zhang L, Huang H, Li Y, et al. RAID v2.0: an updated resource of RNA-associated interactions across organisms. Nucleic acids research. 2017;45(D1):D115–D8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kopp F, Wagner E, Roidl A. The proto-oncogene KRAS is targeted by miR-200c. Oncotarget. 2014;5(1):185–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura M, Sasaki Y, Kobashi K, Takeda K, Nakagaki T, Idogawa M, et al. CRKL oncogene is downregulated by p53 through miR-200s. Cancer science. 2015;106(8):1033–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang C, Dong HJ, Zou ZJ, Fan L, Wang L, Zhang R, et al. High expression of cyclic nucleotide phosphodiesterase 7B mRNA predicts poor prognosis in mantle cell lymphoma. Leuk Res. 2013;37(5):536–40. [DOI] [PubMed] [Google Scholar]
- 48.Brooks MD, Jackson E, Warrington NM, Luo J, Forys JT, Taylor S, et al. PDE7B is a novel, prognostically significant mediator of glioblastoma growth whose expression is regulated by endothelial cells. PLoS One. 2014;9(9):e107397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho-Chung YS, Clair T, Bodwin JS, Berghoffer B. Growth arrest and morphological change of human breast cancer cells by dibutyryl cyclic AMP and L-arginine. Science (New York, NY. 1981;214(4516):77–9. [DOI] [PubMed] [Google Scholar]
- 50.Kim SN, Ahn YH, Kim SG, Park SD, Cho-Chung YS, Hong SH. 8-Cl-cAMP induces cell cycle-specific apoptosis in human cancer cells. International journal of cancer Journal international du cancer. 2001;93(1):33–41. [DOI] [PubMed] [Google Scholar]
- 51.Pattabiraman DR, Bierie B, Kober KI, Thiru P, Krall JA, Zill C, et al. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science (New York, NY. 2016;351(6277):aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng Q, Zhang D, Yang YU, Cui X, Sun J, Liang C, et al. MicroRNA-200c impairs uterine receptivity formation by targeting FUT4 and alpha1,3-fucosylation. Cell death and differentiation. 2017;24(12):2161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong S, Noh H, Chen H, Padia R, Pan ZK, Su SB, et al. Signaling by p38 MAPK Stimulates Nuclear Localization of the Microprocessor Component p68 for Processing of Selected Primary MicroRNAs. Sci Signal. 2013;6(266):ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L, Fang D, Chen H, Lu Y, Dong Z, Ding HF, et al. Cyclin-dependent kinase 2 is an ideal target for ovary tumors with elevated cyclin E1 expression. Oncotarget. 2015;6(25):20801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7(12):e51862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.