Abstract
Fusions involving neurotrophic tyrosine receptor kinase (NTRK) genes are detected in ≤2% of gliomas and can promote gliomagenesis. The remarkable therapeutic efficacy of TRK inhibitors, which are among the first Food and Drug Administration-approved targeted therapies for NTRK-fused gliomas, has generated significant clinical interest in characterizing these tumors. In this multi-institutional retrospective study of 42 gliomas with NTRK fusions, next generation DNA sequencing (n = 41), next generation RNA sequencing (n = 1), RNA-sequencing fusion panel (n = 16), methylation profile analysis (n = 18), and histologic evaluation (n = 42) were performed. All infantile NTRK-fused gliomas (n = 7) had high-grade histology and, with one exception, no other significant genetic alterations. Pediatric NTRK-fused gliomas (n = 13) typically involved NTRK2, ranged from low- to high-histologic grade, and demonstrated histologic overlap with desmoplastic infantile ganglioglioma, pilocytic astrocytoma, ganglioglioma, and glioblastoma, among other entities, but they rarely matched with high confidence to known methylation class families or with each other; alterations involving ATRX, PTEN, and CDKN2A/2B were present in a subset of cases. Adult NTRK-fused gliomas (n = 22) typically involved NTRK1 and had predominantly high-grade histology; genetic alterations involving IDH1, ATRX, TP53, PTEN, TERT promoter, RB1, CDKN2A/2B, NF1, and polysomy 7 were common. Unsupervised principal component analysis of methylation profiles demonstrated no obvious grouping by histologic grade, NTRK gene involved, or age group. KEGG pathway analysis detected methylation differences in genes involved in PI3K/AKT, MAPK, and other pathways. In summary, the study highlights the clinical, histologic, and molecular heterogeneity of NTRK-fused gliomas, particularly when stratified by age group.
Keywords: NTRK, Glioma, NGS sequencing, Methylation
Introduction
The tropomyosin receptor kinase (TRK) family of tyrosine receptor kinases is comprised of TRKA, TRKB, and TRKC, which are encoded by neurotrophic tyrosine receptor kinase (NTRK) genes NTRK1, NTRK2, and NTRK3, respectively. The three TRK proteins are structurally similar, with an extracellular region containing leucine-rich repeats, cysteine-rich clusters, and immunoglobulin-like domains, a transmembrane region, and an intracellular region including a tyrosine kinase domain [2]. Binding of neurotrophin ligands to the extracellular region triggers TRK dimerization and transphosphorylation of tyrosine residues within the activation loop of the kinase domain, which ultimately results in the upregulating of multiple pathways including mitogen-activated protein kinase (MAPK), phosphatidylinositol-3-kinase / protein kinase B (PI3K/AKT), and phospholipase C-γ (PLC-γ) signaling cascades [27]. TRK receptors are highly expressed in neural tissue, where they have a physiologic role in neuronal survival, development, proliferation, and synaptic plasticity, as well as memory and cognition [2].
Fusions involving the NTRK genes can be oncogenic drivers and typically involve the 5′ end of the fusion partner and the 3′ end of NTRK preserving the tyrosine kinase domain. Reported gene fusion partners are numerous and in many cases contain structural motifs such as coiled-coil domains and zinc finger domains that promote dimerization [10]. Thus, oncogenic NTRK fusions can result in aberrant ligand-independent TRK receptor dimerization and constitutive activation of TRK signaling pathways [3], leading to upregulated proliferation and resistance to apoptosis. NTRK fusions in which the fusion partners lack dimerization domains might alternatively promote tumorigenesis through loss of extracellular TRK regulatory domains [5].
The estimated prevalence of NTRK-fusions across all tumors is less than 1% [34, 38]. However, for certain tumors such as congenital infantile fibrosarcoma, mammary analogue secretory carcinoma, and secretory breast carcinoma, NTRK fusions occur in greater than 90% of cases and are essentially pathognomonic for those entities [48]. NTRK fusions occur in lower frequencies in a wide range of other neoplasms, including colorectal carcinoma, lung carcinoma, and papillary thyroid carcinoma, among others [48]. Approximately 0.55 to 2% of all gliomas/neuroepithelial tumors contain NTRK fusions [18, 21, 34, 38, 43, 53], though the incidence may be up to 5.3% in pediatric high grade gliomas (HGG) [34], 4% of diffuse intrinsic pontine gliomas (DIPG), and 40% of non-brainstem HGG in patients younger than 3-years-old [52].
Clinical interest in NTRK-fused tumors has increased substantially due to the efficacy of Food and Drug Administration (FDA) approved TRK inhibitor therapies [4, 13, 15–17, 23, 30]. The aim of the current study is to provide insights into the clinicopathologic and molecular features of gliomas with NTRK fusions.
Materials and methods
Cohort
The surgical, consultation, and molecular pathology archives of Brigham and Women’s Hospital (BWH) (Boston, MA), Boston Children’s Hospital (BCH) (Boston, MA), Children’s Hospital of Philadelphia (CHOP) (Philadelphia, PA), Washington University School of Medicine (WashU) (St Louis, MO), Northwestern University (NWU) (Chicago, IL), and Foundation Medicine (FM) (Morrisville, NC) were reviewed for gliomas with NTRK rearrangements. In this retrospective multi-institutional study, a total of 42 cases were identified, composed of 8 cases from BWH, 7 cases from BCH, 5 cases from CHOP, 1 case from WashU, 1 case from NWU, and 20 cases from FM. The study contains 2 cases (cases 7 and 15) that have been previously reported in the literature [31, 46]. The study was conducted under BCH IRB protocol IRB-CR00027359–1 and DFCI protocol 10–417. The cases were grouped in infantile (age less than 1 year), pediatric (age ranging from 1 year to 18 years), and adult (age over 18 years). Available routine hematoxylin and eosin stained sections and immunohistochemical stains prepared from formalin-fixed, paraffin-embedded (FFPE) tissue from the 42 identified cases underwent review by neuropathologists (SA, MT, SHR, MSa, CH), with 22 of the cases undergoing central review by SA; all tumors with material for methylation were centrally-reviewed. In general, there was agreement with the initial clinical diagnosis, and a specific World Health Organization (WHO) diagnosis was sought whenever the histology allowed it. A complete set of slides was not available for the 20 cases from FM; however, these were all reviewed by one neuropathologist (SHR). A subset of the pediatric tumors had concerning histology, with occasional mitoses and pleomorphism, but, overall, these features did not reach the threshold for WHO histologic grade 3. The histologic diagnoses in cases that posed this challenge were: glioma with anaplastic features (4 cases) and anaplastic pilocytic astrocytoma (APA) (1 case). Therefore, a specific histologic grade could not be assigned for these tumors and they are referred to as having “uncertain WHO histologic grade” in the manuscript. Patient information was abstracted from the electronic medical records or from the clinical information provided on the pathology report.
Figures
The Oncoprint figure was created using R 3.6.0, RStudio 1.2.1335, and the Oncoprint function of the ComplexHeatmap 2.2.0 package. The Circos plot was generated using the online Circos Table Viewer (http://mkweb.bcgsc.ca/tableviewer). All other figures were created using GraphPad Prism (v.8) software.
Next generation sequencing (NGS)
NTRK rearrangements were detected by either DNA-based next generation sequencing (NGS) or RNA-based fusion panel performed at the time of clinical diagnosis. Given that this is a retrospective multi-institutional study, a limitation is that the NGS panels utilized are institution-specific (albeit similar in coverage of genes of interest and scope).
The NGS platforms used included the BWH hybrid capture sequencing assay (Oncopanel) (n = 12), Foundation Medicine hybrid capture sequencing assay (n = 22, including 1 case from WashU and 1 case from NWU), CHOP Comprehensive Solid Tumor Panel (n = 6), GlioSeq NGS panel (n = 1), Caris Life Sciences NGS panel (n = 1), and Integragen Genomics (next generation RNA sequencing) (n = 1) (https://www.integragen-genomics.com/bioinformatics-and-bioanalysis/mercury). In addition, RNA-based fusion testing was performed on 16 cases either as standalone targeted RNA-based anchored multiplex PCR (Archer FusionPlex) [56] or as part of a multi-assay panel (e.g. CHOP Comprehensive Solid Tumor Panel, GlioSeq NGS panel, Caris Life Sciences NGS panel).
Oncopanel interrogates the exons of 447 genes and 191 introns across 60 genes, and structural rearrangements are evaluated with BreaKmer analysis as previously described [20]. The Foundation Medicine NGS assay evaluates 324 genes for mutations and copy number alterations, as well as select intronic regions of a subset of genes to detect gene rearrangements. Details about the Foundation Medicine NGS assay can be found at https://www.foundationmedicine.com/genomic-testing/foundation-one-cdx. The CHOP Comprehensive Solid Tumor Panel includes sequencing and copy number analysis of 237 genes as well as targeted RNA-based anchored multiplex PCR using custom probes for over 100 genes, as previously described [49]. Caris Life Sciences performs exome sequencing on 592 genes for mutational analysis, evaluates a proportion of these genes for copy number alterations, and assesses for fusions involving targeted genes with RNA-based anchored multiplex PCR (https://www.carismolecularintelligence.com/profiling-menu/mi-profile-usa-excluding-new-york/). GlioSeq uses amplification-based DNA and RNA sequencing to evaluate for mutations, copy number alterations, and structural rearrangements involving genes relevant to primary central nervous system (CNS) tumors. A list of genes included in the GlioSeq panel can be accessed at https://mgp.upmc.com/Home/Test/GlioSeq_details. Copy number data was determined from DNA-based NGS and methylation profile plots.
DKFZ CNS tumor classification of NTRK gliomas
Genome-wide methylation profiling was performed on DNA extracted from FFPE tissue from 18 cases with available material using the Illumina EPIC Array 850 Bead-Chip (850 k) array to evaluate the DNA methylation status of over 850,000 CpG sites, as described previously [40]. The raw idat files were then analyzed by the Brain Tumor Classifier developed by Capper et al. [7], which is clinically validated at NYU. Each NTRK fusion case was compared against the CNS reference tumor cohort (82 methylation classes and 9 control tissues) using the Random Forest Classifier. The classifier generates Methylation classifier scores for each sample along with t-distributed stochastic neighbor embedding (tSNE) dimensionality reduction of queried samples against the reference cohort classes.
NTRK cohort genome-wide methylation profiling and analysis
To analyze the NTRK cohort in our study, the raw idats generated from iScan were processed and analyzed using Bioconductor R package Minfi. All the Illumina EPIC array probes were normalized using quintile normalization and corrected for background signal. Samples were then checked for their quality using mean detection p-values (p-value < 0.05). Unsupervised principal component analysis (PCA) was performed to check for biological variation within the cases. To identify the differentially methylated CpG probes, the samples were grouped based on NTRK gene involved, histologic grade, and age. Beta values were generated and probes with FDR cutoff (q < 0.05) were considered the most significantly variable probes. Heatmaps were generated in a semi-supervised manner, showing the hierarchical clustering pattern of the top 10,000 significant differentially methylated genes/probes by NTRK gene involved. KEGG pathway analysis with ClusterProfiler [54] was used to identify the signaling pathways enriched in the top 10,000 most variable genes/probes.
The types of molecular assays performed on each case are listed in Supplemental Table 1.
Results
Clinical characteristics
The cohort was comprised of 42 patients (24 males (57.1%), 18 females (42.9%)). The median patient age was 24-years-old (range < 1 to 81 years), with 7 cases arising in infants (age ≤ 1 year), 13 in pediatric patients (age ranging from > 1 to ≤18 years), and 22 in adult patients (age > 18 years). Collectively, most NTRK-fused gliomas were hemispheric (66.7%, 28/42), but also involved brainstem/spinal cord (9.5%, 4/42), cerebellum (7.1%, 3/42), optic nerve/suprasellar region/deep grey nuclei (4.8%, 2/42), septum pellucidum (2.8%, 1/42), or had a gliomatosis or widespread pattern (7.1%, 3/42); the location is not known in one case. The distribution of anatomic regions involved varied by age: while the infantile and adult NTRK-fused gliomas were typically hemispheric, the anatomic locations of pediatric NTRK-fused gliomas were more diverse: 38.5% (5/13) were hemispheric, 23.1% (3/13) were cerebellar, 23.1% (3/13) involved brainstem/spinal cord, 7.7% (1/13) were suprasellar, and 7.7% (1/13) involved the septum pellucidum. The anatomic distribution of NTRK-fused gliomas is summarized in Fig. 1.
Fig. 1.
Anatomic distribution of NTRK-fused gliomas: NTRK-fused gliomas are primarily hemispheric, particularly in infant and adult patients. NTRK-fused gliomas in pediatric patients are more diverse in anatomic location. *Includes case involving septum pellicidum
Survival and progression-free survival data were available in 21 cases (4 infantile, 9 pediatric, and 8 adult patients). For 2 infantile cases, a diagnosis was made at autopsy. Excluding these 2 cases, the median follow-up period after diagnostic procedure was 23 months (range 4–189 months). During this time, 57.9% (11/19) of cases showed tumor recurrence/progression. Cases with recurrence (n = 11) were mostly either high histologic grade (WHO grade 3 or 4) (54.5%, 6/11) or of uncertain WHO grade (36.4%, 4/11), and only one case was of low histologic grade (9.1%, 1/11). Both infantile NTRK-fused gliomas demonstrated progression/recurrence (100%, 2/2); in the pediatric and adult age groups, tumor progression/recurrence occurred in 55.6% (5/9) and 50.0% (4/8) of cases, respectively. Death occurred in 28.6% (6/21) of cases (2 infantile and 4 adult patients), all with high-grade histology. Patient clinical characteristics are summarized in Table 1.
Table 1.
Clinicopathologic characteristics
| Patient characteristics | Total, n (%) |
|---|---|
| Total Number of Patients | 42 (100%) |
| Demographics | |
| Sex | |
| Male | 24 (57.1%) |
| Female | 18 (42.9%) |
| Age, median (range), years | 24 (<1-81) |
| Infantile (≤1), n (%) | 7 (16.7%) |
| Pediatric (>1 to ≤18), n (%) | 13 (31.0%) |
| Adult (>18), n (%) | 22 (52.4%) |
| Radiology | |
| Location | |
| Hemispheric | 28 (66.7%) |
| Brainstem/spinal cord | 4 (9.5%) |
| Cerebellar | 3 (7.1%) |
| Gliomatosis pattern/widespread | 3 (7.1%) |
| Optic nerve/suprasellar/deep grey nuclei | 2 (4.8%) |
| Other/unknown | 2 (4.8%) |
|
Survival Data (n=21: 4 infantile, 9 pediatric, 8 adult) (cases without any available follow-up data excluded) | |
| Deaths | 6 (28.6% of all cases; all HG) |
| Infantile | 2 (50%) |
| Pediatric | 0 (0%) |
| Adult | 4 (50%) |
| Tumor Recurrence/Progression | 11 (57.9%; 6 HG, 1 LG, 4 of certain WHO grade)a |
| Infantile | 2 (100%)a |
| Pediatric | 5 (55.6%) |
| Adult | 4 (50%) |
| Follow-up, median (range), months | 23 (4-189)a |
aExcludes 2 infantile tumors diagnosed at autopsy
HG High histologic grade, LG Low histologic grade
Targeted therapy with larotrectinib was administered in 3 patients: 2 pediatric patients (one of whom showed a decrease in tumor burden (Fig. 2a-b), and one of whom has shown stable disease), and 1 infantile patient whose course of larotrectinib was terminated due to elevated liver function tests.
Fig. 2.
Representative T1 postcontrast MRI images of a pediatric NTRK-fused glioma treated with targeted TRK inhibitor therapy. (a) Recurrent/residual tumor along the patient’s resection cavity (arrow) demonstrated (b) radiologic response to larotrectinib (arrow)
NTRK fusions
NTRK rearrangements involved NTRK1 in 47.6% of cases (20/42), NTRK2 in 33.3% of cases (14/42), and NTRK3 in 19.0% (8/42) of cases (Fig. 3). The frequencies of NTRK genes involved varied by patient age. All NTRK genes were involved in infantile NTRK-fused gliomas in approximately equal proportions. In comparison, the majority of pediatric NTRK-fused gliomas involved NTRK2 (69.2%, 9/13). Most adult NTRK-fused gliomas involved NTRK1 (68.2%, 15/22), with NTRK3 (18.2%, 4/22) and NTRK2 (13.6%, 3/22) comprising subsets of cases. Overall, there were 29 unique fusion partners. Several rearrangements were recurrent, including BCAN-NTRK1 (n = 4), TPM3-NTRK1 (n = 4), ETV6-NTRK3 (n = 4), ARHGEF2-NTRK1 (n = 2), LMNA-NTRK1 (n = 2), BCR-NTRK2 (n = 2), and TRIM24-NTRK2 (n = 2). In our cohort, no fusion partner was shared by more than one NTRK gene. Furthermore, intrachromosomal rearrangements comprised the vast majority of NTRK1 fusions (95.0%, 19/20), whereas interchromosomal rearrangements comprised most fusions involving NTRK2 (85.7%, 12/14) and NTRK3 (75.0%, 6/8). With the available methods, we were confident that NTRK was the 3′ fusion partner in 40 out of 42 tumors. (Table 2) [24, 32].
Fig. 3.
NTRK fusions: (Top) Circos plot showing NTRK fusions from all 42 cases in the study cohort. There were a total of 29 unique fusion partners, with several recurrent fusions (BCAN-NTRK1, TPM3-NTRK1, ETV6-NTRK3, ARHGEF2-NTRK1, LMNA-NTRK1, BCR-NTRK2, and TRIM24-NTRK2). (Bottom): The frequencies of NTRK genes involved in the rearrangements varied by age group
Table 2.
NTRK rearrangements
| Case | Assay detecting NTRK fusion | NTRK fusion | 5’ fusion partner breakpoint or transcript | 3’ fusion partner breakpoint or transcript |
|---|---|---|---|---|
| 23 | DNA-based NGS | ARGLU1-NTRK1 | Exon 3 | Exon 11 |
| 29 | DNA-based NGS | ARHGEF11-NTRK1 | Exon 39 | Exon 10 |
| 10 | RNA-based fusion panel | ARHGEF2-NTRK1 | Exon 21 | Exon 10 |
| 40 | DNA-based NGS | ARHGEF2-NTRK1 | Exon 21 | Exon 9 |
| 27 | DNA-based NGS | BCAN-NTRK1 | Exon 13 | Exon 8 |
| 30 | DNA-based NGS | BCAN-NTRK1 | Exon 6 | Exon 8 |
| 31 | DNA-based NGS | BCAN-NTRK1 | Exon 14 | Exon 11 |
| 32 | DNA-based NGS | BCAN-NTRK1 | Exon 13 | Exon 9 |
| 8 | DNA-based NGS | CD247-NTRK1 | Intron 1 | Exon 15 |
| 42 | DNA-based NGS | CHTOP-NTRK1 | Exon 5 | Exon 8 |
| 21 | RNA-based fusion panel | KIF21B-NTRK1a | Unknown | Unknown |
| 9 | DNA-based NGS | LMNA-NTRK1 | Intron 5 | Intron 10 |
| 41 | DNA-based NGS | LMNA-NTRK1 | Exon 12 | Exon 11 |
| 26 | DNA-based NGS | NTRK1-NFASC | Exon 7 | Exon 3 |
| 24 | DNA-based NGS | NOS1AP-NTRK1 | Exon 10 | Exon 9 |
| 33 | DNA-based NGS | TPM3-NTRK1 | Exon 10 | Exon 12 |
| 36 | DNA-based NGS | TPM3-NTRK1 | Exon 7 | Exon 12 |
| 37 | DNA-based NGS | TPM3-NTRK1 | Exon 10 | Exon 9 |
| 38 | DNA-based NGS | TPM3-NTRK1 | Exon 7 | Exon 8 |
| 19 | RNA-based fusion panel | TPR and NTRK1a,b | Unknown | Unknown |
| 25 | DNA-based NGS | AFAP1-NTRK2 | Exon 14 | Exon 12 |
| 11 | DNA-based NGS | BCR-NTRK2 | Intron 1 | Intron 12 |
| 28 | DNA-based NGS | BCR-NTRK2 | Exon 1 | Exon 13 |
| 5 | RNA-based fusion panel | EML1-NTRK2 | Exon 2 | Exon 16 |
| 4 | RNA-based fusion panel | GKAP1-NTRK2 | Exon 9 | Exon 15 |
| 15 | RNA-based fusion panel | KANK1-NTRK2 | Exon 12 | Exon 3 |
| 16 | RNA-based fusion panel | KCTD16-NTRK2 | Exon 3 | Exon 16 |
| 1 | RNA-based fusion panel | NBPF20-NTRK2 | Exon 16 | Exon 15 |
| 39 | DNA-based NGS | PAIP1-NTRK2 | Exon 9 | Exon 13 |
| 18 | RNA-based fusion panel | QKI-NTRK2 | Exon 6 | Exon 16 |
| 2 | RNA-based fusion panel | SPECC1L-NTRK2 | Exon 11 | Exon 17 |
| 22 | RNA-based NGS | TNS3-NTRK2a | Exon 16 | Exon 12 |
| 3 | RNA-based fusion panel | TRIM24-NTRK2 | Exon 12 | Exon 15 |
| 17 | RNA-based fusion panel | TRIM24-NTRK2c | Exon 12 | Exon 15, exon 16 |
| 34 | DNA-based NGS | DLG1-NTRK3 | Exon 10 | Exon 11 |
| 6 | RNA-based fusion panel | ETV6-NTRK3 | Exon 4 | Exon 14 |
| 14 | RNA-based fusion panel | ETV6-NTRK3 | Exon 4 | Exon 14 |
| 20 | DNA-based NGS | ETV6-NTRK3 | Intron 4 | Intron 12 |
| 35 | DNA-based NGS | ETV6-NTRK3 | Exon 5 | Exon 15 |
| 12 | RNA-based fusion panel | FRY-NTRK3 | Exon 1 | Exon 14 |
| 13 | DNA-based NGS | KIAA1199-NTRK3 | Intron 1 | Intron 5 |
| 7 | DNA-based NGS | MYO5A-NTRK3 | Intron 33 | Exon 9 |
aNTRK rearrangement confirmed by fluorescence in situ hybridization (FISH). b Unknown if NTRK is 5′ or 3′ fusion partner. cTwo NTRK2 fusions detected, with the major form fused to exon 16 of NTRK2 and the minor form fused to exon 15 of NTRK2. NGS Next generation sequencing
Histology of NTRK-fused gliomas
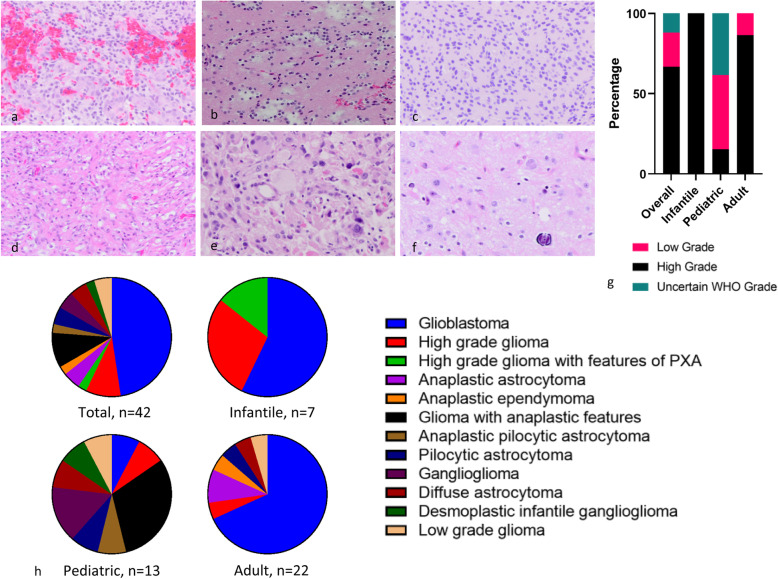
The histologic grade and diagnosis of NTRK-fused gliomas were heterogeneous. All infantile cases were histologically high-grade (100%, 7/7), including one with features of pleomorphic xanthoastrocytoma (PXA). Most adult cases were also histologically high-grade (86.4%, 19/22), with the majority being diagnosed as GBM (68.2%, 15/22). Interestingly, the adult cohort included a tumor with morphology and immunohistochemical profile indistinguishable from anaplastic ependymoma, but the molecular test results were more in keeping with a glioblastoma. The pediatric cohort was enriched in cases with low (46.2%, 6/13) or uncertain WHO grade (38.5%, 5/13), with fewer cases demonstrating high-grade histology (15.4%, 2/13). This was the group with the most histologic diversity, and the diagnoses included ganglioglioma (GG), diffuse astrocytoma (DA), glioblastoma (GBM), anaplastic pilocytic astrocytoma (APA), and desmoplastic infantile ganglioglioma (DIGG). The distribution of histologic grade and diagnosis by age group, as well as representative photos illustrating various histologic diagnoses are included in Fig. 4a-f. The distribution of NTRK-fused gliomas by histologic grade (low, high, and uncertain WHO grade) is summarized in Fig. 4g and the distribution of NTRK-fused gliomas by histologic diagnosis is summarized in Fig. 4h. The histologic diagnosis of each case is also listed in Table 3.
Fig. 4.
(Top-left) The histologic spectrum of NTRK-fused gliomas can include (a) glioblastoma (GBM), (b) infiltrative low-grade glioma, (c) glioma with anaplastic features and uncertain WHO grade, (d) pilocytic astrocytoma, (e) pleomorphic xanthoastrocytoma, and (f) ganglioglioma. (Top right) Histologic grades of NTRK-fused gliomas: most NTRK-fused gliomas demonstrate high-grade histology, particularly in tumors diagnosed in infant and adult patients. In contrast, the vast majority of NTRK-fused gliomas diagnosed in pediatric patients are of low-grade histology or of uncertain WHO grade. (Bottom) Histologic diagnoses of NTRK-fused gliomas: there were 12 unique histologic diagnoses assigned to NTRK-fused glioma in the study. Slightly less than half of all cases were diagnosed as GBM. Within the infantile and adult age cohorts, the majority of cases were diagnosed as GBM. A more diverse spectrum of tumors was diagnosed in the pediatric age cohort for which there was no single predominant histologic diagnosis
Table 3.
Histologic diagnosis and methylation class (most with low confidence score)
| Case | Age (years) | Histologic diagnosis | NTRK fusion | Match to known methylation family? | Closest methylation family match | Methylation family score |
|---|---|---|---|---|---|---|
| 1 | 24 | Low grade glioma | NBPF20-NTRK2 | No | LGG, DNT | 0.6959 |
| 2 | 16 | Glioma with anaplastic features | SPECC1L-NTRK2 | No | DMG, K27 | 0.4858 |
| 3 | 0.42 | High grade glioma with features of pleomorphic xanthoastrocytoma | TRIM24-NTRK2 | Yes | PXA | 0.989 |
| 4 | 6 | Low grade glioma | GKAP1-NTRK2 | No | MTGF_PA | 0.3584 |
| 5 | 0.23 | High grade glioma | EML1-NTRK2 | No | MTGF_PLEX_T | 0.4474 |
| 6 | 4 | Glioma with anaplastic features | ETV6-NTRK3 | No | MTGF_PLEX_T | 0.5473 |
| 7 | 42 | Anaplastic ependymoma, WHO grade 3 | MYO5A-NTRK3 | No | MTGF_PLEX_T | 0.19 |
| 8 | 42 | Glioblastoma, WHO grade 4 | CD247-NTRK1 | N/A | N/A | N/A |
| 9 | 43 | Glioblastoma, WHO grade 4 | LMNA-NTRK1 | No | MTGF_GBM | 0.6923 |
| 10 | 31 | High grade glioma | ARHGEF2-NTRK1 | No | DLGNT | 0.2434 |
| 11 | 27 | Glioblastoma, WHO grade 4 | BCR-NTRK2 | N/A | N/A | N/A |
| 12 | 38 | Anaplastic astrocytoma, WHO grade 3 | FRY-NTRK3 | No | MTGF_PLEX_T | 0.4062 |
| 13 | 54 | Glioblastoma, WHO grade 4 | KIAA1199-NTRK3 | N/A | N/A | N/A |
| 14 | 0.25 | Glioblastoma, WHO grade 4 | ETV6-NTRK3 | Yes | IHG | 0.9836 |
| 15 | 2.7 | Anaplastic pilocytic astrocytoma | KANK1-NTRK2 | No | PXA | 0.7403 |
| 16 | 7 | Ganglioglioma, WHO grade 1 | KCTD16-NTRK2 | No | MTGF_PA | 0.2699 |
| 17 | 9 | Ganglioglioma, WHO grade 1 | TRIM24-NTRK2 | No | MTGF_PA | 0.6814 |
| 18 | 16 | Diffuse astrocytoma, WHO grade 2 | QKI-NTRK2 | N/A | N/A | N/A |
| 19 | 2 | Glioma with anaplastic features | TPR and NTRK1 | No | MTGF_PA | 0.3765 |
| 20 | <1 | Glioblastoma, WHO grade 4 | ETV6-NTRK3 | No | MTGF_PLEX_T | 0.1241 |
| 21 | 1.83 | High grade glioma | KIF21B-NTRK1 | No | MTGF_GBM | 0.773 |
| 22 | 7 | Glioma with anaplastic features | TNS3-NTRK2 | No | DLGNT | 0.6739 |
| 23 | 55 | Anaplastic astrocytoma, WHO grade 3 | ARGLU1-NTRK1 | N/A | N/A | N/A |
| 24 | <1 | Glioblastoma, WHO grade 4 | NOS1AP-NTRK1 | N/A | N/A | N/A |
| 25 | 16 | Pilocytic astrocytoma, WHO grade 1 | AFAP1-NTRK2 | N/A | N/A | N/A |
| 26 | 26 | Diffuse astrocytoma, WHO grade 2 | NTRK1-NFASC | N/A | N/A | N/A |
| 27 | 48 | Glioblastoma, WHO grade 4 | BCAN-NTRK1 | N/A | N/A | N/A |
| 28 | 8 | Glioblastoma, WHO grade 4 | BCR-NTRK2 | N/A | N/A | N/A |
| 29 | 79 | Glioblastoma, WHO grade 4 | ARHGEF11-NTRK1 | N/A | N/A | N/A |
| 30 | 59 | Glioblastoma, WHO grade 4 | BCAN-NTRK1 | N/A | N/A | N/A |
| 31 | 54 | Glioblastoma, WHO grade 4 | BCAN-NTRK1 | N/A | N/A | N/A |
| 32 | 24 | Pilocytic astrocytoma, WHO grade 1 | BCAN-NTRK1 | N/A | N/A | N/A |
| 33 | 3 | Desmoplastic infantile ganglioglioma, WHO grade 1 | TPM3-NTRK1 | N/A | N/A | N/A |
| 34 | 52 | Glioblastoma, WHO grade 4 | DLG1-NTRK3 | N/A | N/A | N/A |
| 35 | 1 | High grade glioma | ETV6-NTRK3 | N/A | N/A | N/A |
| 36 | <1 | Glioblastoma, WHO grade 4 | TPM3-NTRK1 | N/A | N/A | N/A |
| 37 | 63 | Glioblastoma, WHO grade 4 | TPM3-NTRK1 | N/A | N/A | N/A |
| 38 | 78 | Glioblastoma, WHO grade 4 | TPM3-NTRK1 | N/A | N/A | N/A |
| 39 | 45 | Glioblastoma, WHO grade 4 | PAIP1-NTRK2 | N/A | N/A | N/A |
| 40 | 31 | Glioblastoma, WHO grade 4 | ARHGEF2-NTRK1 | N/A | N/A | N/A |
| 41 | 81 | Glioblastoma, WHO grade 4 | LMNA-NTRK1 | N/A | N/A | N/A |
| 42 | 44 | Glioblastoma, WHO grade 4 | CHTOP-NTRK1 | N/A | N/A | N/A |
N/A Not applicable, LGG, DNT Low-grade glioma, dysembryoplastic neuroepithelial tumor, DMG, K27 Diffuse midline glioma H3 K27M mutant, PXA Pleomorphic xanthoastrocytoma, MTGF_PA Methylation group family_pilocytic astrocytoma, MTGF_PLEX_T Methylation group family_plexus tumor, MTGF_GBM Methylation group family_glioblastoma IDH wildtype, DLGNT Diffuse leptomeningeal glioneuronal tumor, IHG Infantile hemispheric glioma.
Molecular features of NTRK-fused gliomas
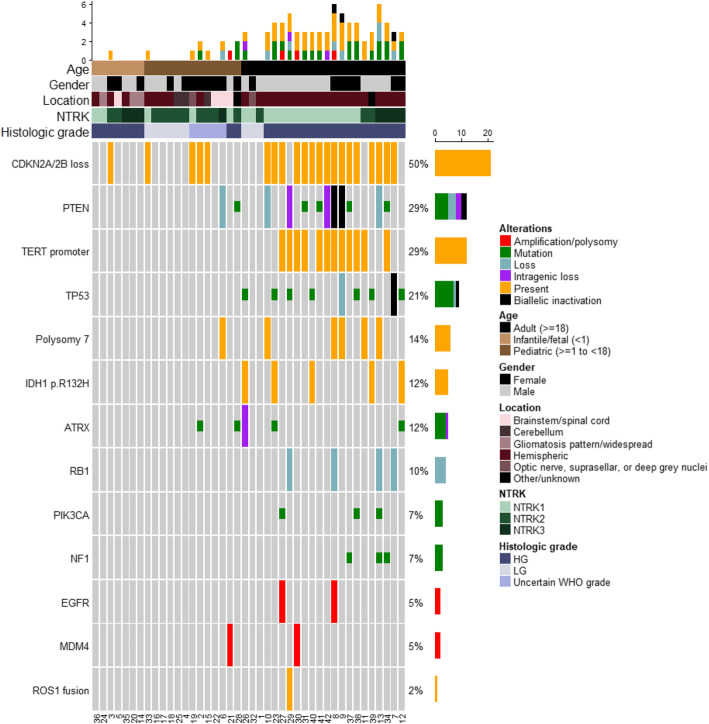
NTRK-fused gliomas in our cohort demonstrated concurrent aberrations involving CDKN2A/2B, TERT promoter, TP53, PTEN, EGFR, ATRX, RB1, IDH1, polysomy 7, ROS1, PIK3CA, NF1, and MDM4. The frequency of these genetic aberrations in NTRK-fused gliomas increased with patient age cohort and seemed to correlate with histological grade in the pediatric and adult cohorts. Within the infantile cohort, there were no significant mutations and copy number changes beyond the NTRK fusion; the exception was case 3, a high-grade glioma with histology most consistent with anaplastic PXA that showed CDKN2A/2B loss. Within the pediatric cohort, detected aberrations included CDKN2A/2B loss (30.8%, 4/13), ATRX mutation (15.4%, 2/13), PTEN loss/mutation (15.4%, 2/13), polysomy 7 (7.7%, 1/13), and MDM4 amplification (7.7%, 1/13); these were restricted to cases with high-grade histology or concerning histology of uncertain WHO grade; the only exception was case 33, diagnosed as DIGG, that had CDKN2A/B homozygous deletion. Within the adult cohort, detected aberrations included CDKN2A/2B loss (72.7%, 16/22), TERT promoter mutation (54.5%, 12/22), PTEN mutation/biallelic inactivation/(intragenic) loss (45.5%, 10/22), TP53 mutation/ biallelic inactivation/loss (40.9%, 9/22), IDH1 p.R132H mutation (22.7%, 5/22), polysomy 7 (22.7%, 5/22), RB1 loss (18.2%, 4/22), ATRX mutation/intragenic loss (13.6%, 3/22), PIK3CA mutation (13.6%, 3/22), EGFR amplification (9.1%, 2/22), and MDM4 amplification (4.5%, 1/22). The IDH-mutated gliomas were all negative for 1p/19q co-deletion, in keeping with diffuse astrocytomas. An oncoprint containing the major co-occurring genetic alterations along with clinicopathologic characteristics and tumor histology is provided in Fig. 5. Supplemental Table 2 provides all the genes with molecular alterations and chromosomal copy number changes for each case.
Fig. 5.
Oncoprint detailing common molecular alterations in NTRK-fused gliomas and patient clinical characteristics
Methylation profiling of NTRK-fused gliomas
Methylation profiling with clustering analysis was performed on 18 cases with available material. Two tumors matched to known methylation class families with high confidence (Fig. 6, Table 3): case 3, an infantile HGG with features of PXA, matched to methylation class family PXA (calibrated score = 0.989) and case 14, a 3-month-old with a histologic diagnosis of GBM, matched to infantile hemispheric glioma (IHG, calibrated score = 0.9836). In both instances, the histology was consistent with the matched methylation class family. All other cases matched with low confidence or not at all to known methylation class families (i.e. scores were lower than the recommended threshold value of ≥0.9 [7] or the less conservative threshold of ≥0.84 [8]. Seven cases had methylation classifier scores between 0.5 and 0.84 [8], with 2 (cases 9 and 21) having histology consistent with the closest methylation class family. Overall, a disproportionately high number of case either classified with calibrated score < 0.9 or did not classify with any reference cohort compared to previously published data [7] suggesting perhaps that NTRK fusions alter the DNA methylation pattern from non-NTRK driven cases of similar histology (Table 3; also please see Supplemental Table 3 for link to all methylation reports and t-sne plots).
Fig. 6.
Methylation clustering analysis t-distributed stochastic neighbor embedding (tSNE) plots: of the 18 NTRK-fused gliomas with methylation profiling data, only two matched with high confidence to known methylation class families. (a) Case 3 (histologic diagnosis: high-grade glioma with features of pleomorphic xanthoastrocytoma (PXA)) matched to methylation class PXA, and (b) case 14 (histologic diagnosis: glioblastoma) matched to methylation class infantile hemispheric glioma (IHG). LGG, GG, low grade glioma, ganglioglioma; LGG, PA/GG ST, low grade glioma, pilocytic astrocytoma ganglioglioma spectrum in supratentorial compartment
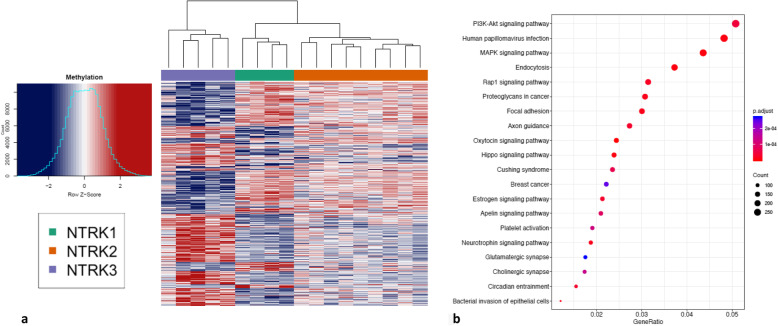
While some newly described CNS tumor entities driven by gene fusions form unique entities [45], unsupervised PCA of the methylation profiles of NTRK-fused cohort showed no obvious grouping by NTRK gene involved, histologic grade, or age (Supplemental Fig. 1). KEGG pathway analysis of the top 10,000 most variably methylated genes/probes in the cohort demonstrated enrichment in pathways involving PI3K-AKT signaling (and related human papillomavirus infection signaling) and MAPK signaling, among others (Fig. 7).
Fig. 7.
(a) Heatmap of the top 10,000 differentially methylated genes/probes by NTRK gene involved (blue indicates hypomethylation, and red indicates hypermethylation). (b) KEGG pathway analysis reveals several pathways enriched in the top differentially methylated gene/probes. The dot plots represent the ratio of genes (x-axis) involved in each signaling pathway (y-axis). The size of the dots shows the gene counts, and the color denotes the significance level
Discussion
With the recent FDA approval of larotrectinib, a selective pan-TRK inhibitor, and entrectinib, a selective pan-TRK, ROS1, and ALK inhibitor [42], there has been much interest in characterizing and diagnosing tumors with NTRK fusions. Both therapies have demonstrated significant treatment responses in NTRK-fused tumors [4, 13, 15–17, 23, 30], including CNS metastases [13, 16, 17, 23] and primary CNS tumors [1, 9, 16, 50, 57], and are generally well tolerated [4, 13, 15–17, 23, 30]. Our study addresses gaps in the knowledge of the clinical and molecular features of NTRK-fused gliomas. In addition, the study serves to corroborate features of NTRK-fused gliomas that have been previously described.
One of the findings of this multi-institutional study is the considerable clinicopathologic and molecular heterogeneity of NTRK-fused gliomas. NTRK fusions do not appear to define ipso facto a single glial entity but rather are a genetic feature occurring in multiple tumor types.
Clinically, we found that NTRK-fused gliomas can involve all CNS compartments but are primarily hemispheric in adults (90.9%) and infants (85.7%); the anatomic distribution of pediatric NTRK-fused gliomas is less predictable. During a median follow-up period of 23 months after diagnosis, 28.6% of patients died and 58.0% of patients showed evidence of recurrence/progression, events that were mostly associated with tumors with high-grade histology. A prior study showed a 5 year overall survival of 42.9% in young patients with hemispheric NTRK-fused gliomas [22]. However, NTRK-fused gliomas with low-grade histology may still exhibit an aggressive clinical course [26]. Ultimately, the prognosis of NTRK-fused gliomas may rapidly change with the more widespread use of targeted TRK inhibitors.
The diverse histology of NTRK-fused gliomas overlaps with entities such as DIGG, GG, PXA, PA (including anaplastic), DA grades 2 and 3, and GBM in our study. In keeping with the wide spectrum of NTRK-fused CNS tumor histology, NTRK rearrangements have been previously reported in GBM [9, 18, 19, 21, 28, 34, 38, 39, 41, 43, 44, 52, 53, 56], gliosarcoma [21], AA [9, 18, 21, 52], diffuse midline glioma / DIPG [9, 52], HGG [9, 22, 34, 57], glioneuronal tumor (including high grade) [1, 16, 29], pilocytic astrocytoma (PA) (including anaplastic) [9, 18, 21, 25, 35], low grade astrocytroma with features of PA [26], PXA [55], GG [1, 9, 36, 37], DIGG [6, 9], LGG [18, 34, 44, 50, 53], glioma, not otherwise specified [9, 18], neuroepithelial neoplasm [43], CNS fibroblastic tumor [47], primitive neuroectodermal tumor (PNET) [12], CNS embryonal tumor [14], and tumors with oligodendroglial or oligoastrocytic-like histology [12, 21, 29, 37, 55].
The present study adds to the literature by demonstrating that the histology and histologic grade of NTRK-fused gliomas vary by patient age. NTRK-fused gliomas in all infantile and most adult patients were histologically high-grade, with the majority diagnosed as GBM. In contrast, pediatric NTRK-fused gliomas were more likely to be of low-grade (46.2%) or uncertain WHO grade (38.5%), and there was no single predominant histologic diagnosis in this cohort. These features of the pediatric NTRK-fused gliomas make their diagnosis and clinical management difficult.
Gliomas with NTRK fusions have been previously reported to possess co-occurring genetic alterations such as IDH [18, 21, 39, 56], H3.3 K27M [52], H3F3A [9], EGFR amplification [21, 28, 56], EGFRvIII [21], PTEN [9, 21, 28], CDKN2A/2B deletion [9, 22, 28, 39, 52, 55, 57], CDKN2C deletion [28], TP53 mutations/inactivation [21, 39, 52, 57], and ATRX [39], among others [21]. Our study matches many of these molecular findings and further demonstrates that the frequency of pathologically significant mutations in NTRK-fused gliomas appears to increase with patient age. In addition, TERT promoter mutations are observed only in histologically high-grade adult tumors, PTEN alterations are almost exclusively seen in histologically high-grade tumors, and CDKN2A/2B loss is rare in histologically low-grade tumors.
Notably, 22.7% of adult NTRK-fused gliomas in our cohort are IDH1 p.R132H mutated, raising questions about the oncogenic driver event in these specific tumors and whether they display oncogenic dependence on the NTRK fusion. In one case report of secondary IDH-mutant GBM [39], an NTRK fusion was detected in only a subclonal tumor population and was absent in the original AA, suggesting that the NTRK fusion was a secondary alteration. In contrast, in NTRK-fused gliomas without co-occurring pathologically significant mutations [9, 21, 52], typically arising in younger patients, NTRK fusions are almost certainly the oncogenic driver event. This is supported by multiple in vivo models that have demonstrated the capability of NTRK fusions to drive gliomagenesis/tumorgenesis [11, 28, 33, 51, 52].
In our series, gliomas with rearrangements involving the same NTRK gene and fusion partner do not necessarily have the same histology or methylation class, which suggests that other factors such as age, co-occurring genetic events, cell of origin and microenvironment potentially play an important role in tumor biology. We have also observed that the NTRK gene involved in the rearrangement differs in frequency by age, with pediatric gliomas having a high percentage of rearrangements involving NTRK2 (69.2%), and adult gliomas having a high percentage of rearrangements involving NTRK1 (68.2%).
Most NTRK-fused gliomas in our cohort were not a perfect match with known methylation entities. Only two cases matched with high confidence to methylation class families. Furthermore, cases that matched with low confidence generally had histology that was not characteristic of the methylation class family, mirroring the experience described in a prior case study of NTRK-fused glioneuronal tumor [29]. Another study reported a proportion of NTRK-fused gliomas matching to methylation classes including IHG, diffuse midline glioma H3 K27M mutant, PXA, and GBM, IDH wildtype, subclass midline [9]. Overall, the findings from our study and prior studies with methylation data [9, 29] suggest that better methylation profile classifier guidelines are needed to account for NTRK-fused. Our unsupervised PCA demonstrating no obvious grouping by NTRK gene involved, age, or histology highlights the heterogeneity within the NTRK-fused glioma methylome.
In summary, NTRK-fused gliomas are clinically, histologically, and molecularly diverse, with notable differences by age group and associated genetic alterations. Additional studies are needed to develop clinical guidelines for the diagnostic workup of potential NTRK-fused CNS tumors and to improve methylation classifier guidelines for NTRK-fused gliomas. Further mechanistic work is required to determine the role of NTRK fusions in driving gliomagenesis in the setting of concurrent oncogenic drivers such as IDH mutations and to demonstrate how downstream TRK signaling pathways may be mediated by different NTRK gene involved, location of NTRK fusion breakpoint, fusion partner, and cell of origin.
Supplementary information
Additional file 1: Supplemental figure 1. Unsupervised principal component analysis (PCA) of methylation profiles of NTRK-fused gliomas demonstrates that no homogenous groups form when correlated with (a) NTRK gene involved, (b) patient age, or (c) histologic grade.
Additional file 2: Supplemental tables. (1) methods for each case; (2) single nucleotide variants and copy number variants encountered in each case; (3) legend for the methylation reports and t-sne plots corresponding to each case.
Acknowledgements
We would like to acknowledge the staff members of the pathology, clinical cytogenetics, and molecular laboratories at Brigham and Women’s Hospital, Boston Children’s Hospital, Dana-Farber Cancer Institute, Children’s Hospital of Philadelphia, NYU Langone Health, and Foundation Medicine for their contributions and support. MT currently receives salary support from an NIH Institutional Training Grant (T32 HL007627). NYU DNA methylation profiling was in part supported by a grant from the Friedberg Charitable Foundation (to MSn).
Authors’ contributions
MT co-wrote the manuscript, designed figures, and contributed to the interpretation of results and design of the project. VV, JS, MD, and MSn performed the methylation analysis and interpreted the methylation data. VV provided figures and methods related to this aspect of the manuscript. DMM, FD, and AHL provided expertise in interpretation of select molecular or cytogenetic results. SM provided the oncoprint and methods for producing it. AFB, MP, MSa, MPN, CH, SD, SHR, and KLL contributed to the cohort. SA, MT, SHR, MSa, and CH evaluated pathology slides. MGF provided critical review and edits on the manuscript. SA designed the project, oversaw all aspects of it, and co-wrote the manuscript. All authors critically reviewed the manuscript and approved it for submission.
Competing interests
The other authors have no interests to declare.
Footnotes
SHR is employed by Foundation Medicine Inc. and has ownership interests in the company. The other authors have no interests to declare.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mariella G Filbin, Matija Snuderl and Sanda Alexandrescu are Co-senior authors
Contributor Information
Matthew Torre, Email: mgtorre@partners.org.
Varshini Vasudevaraja, Email: varshini.vasudevaraja@nyulangone.org.
Jonathan Serrano, Email: Jonathan.Serrano@nyulangone.org.
Michael DeLorenzo, Email: Michael.Delorenzo@nyulangone.org.
Seth Malinowski, Email: SethW_Malinowski@dfci.harvard.edu.
Anne-Florence Blandin, Email: Anne-Florence_Blandin@dfci.harvard.edu.
Melanie Pages, Email: m.pages@ghu-paris.fr.
Azra H. Ligon, Email: aligon@bwh.harvard.edu
Fei Dong, Email: fdong1@bwh.harvard.edu.
David M. Meredith, Email: dmmeredith@bwh.harvard.edu
MacLean P. Nasrallah, Email: maclean.nasrallah@uphs.upenn.edu
Craig Horbinski, Email: craig.horbinski@northwestern.edu.
Sonika Dahiya, Email: sdahiya@wustl.edu.
Keith L. Ligon, Email: Keith_Ligon@dfci.harvard.edu
Mariarita Santi, SANTIM@email.chop.edu.
Shakti H. Ramkissoon, Email: sramkissoon@foundationmedicine.com
Mariella G. Filbin, Email: Mariella_GruberFilbin@dfci.harvard.edu
Matija Snuderl, Email: Matija.Snuderl@nyulangone.org.
Sanda Alexandrescu, Email: Sanda.Alexandrescu@childrens.harvard.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40478-020-00980-z.
References
- 1.Alvarez-Breckenridge C, Miller JJ, Nayyar N, Gill CM, Kaneb A, D'Andrea M, et al. Clinical and radiographic response following targeting of BCAN-NTRK1 fusion in glioneuronal tumor. NPJ Precis Oncol. 2017;1(1):5. doi: 10.1038/s41698-017-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amatu A, Sartore-Bianchi A, Bencardino K, Pizzutilo EG, Tosi F, Siena S. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann Oncol. 2019;30 Suppl 8:viii5–viii15. doi: 10.1093/annonc/mdz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amatu A, Sartore-Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1(2):e000023. doi: 10.1136/esmoopen-2015-000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardini E, Menichincheri M, Banfi P, Bosotti R, De Ponti C, Pulci R, et al. Entrectinib, a pan-TRK, ROS1, and ALK inhibitor with activity in multiple molecularly defined cancer indications. Mol Cancer Ther. 2016;15(4):628–639. doi: 10.1158/1535-7163.MCT-15-0758. [DOI] [PubMed] [Google Scholar]
- 5.Arevalo JC, Conde B, Hempstead BL, Chao MV, Martin-Zanca D, Perez P. TrkA immunoglobulin-like ligand binding domains inhibit spontaneous activation of the receptor. Mol Cell Biol. 2000;20(16):5908–5916. doi: 10.1128/mcb.20.16.5908-5916.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blessing MM, Blackburn PR, Krishnan C, Harrod VL, Barr Fritcher EG, Zysk CD, et al. Desmoplastic infantile Ganglioglioma: a MAPK pathway-driven and microglia/macrophage-rich Neuroepithelial tumor. J Neuropathol Exp Neurol. 2019;78(11):1011–1021. doi: 10.1093/jnen/nlz086. [DOI] [PubMed] [Google Scholar]
- 7.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capper D, Stichel D, Sahm F, Jones DTW, Schrimpf D, Sill M, et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol. 2018;136(2):181–210. doi: 10.1007/s00401-018-1879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke M, Mackay A, Ismer B, Pickles JC, Tatevossian RG, Newman S et al (2020) Infant high grade gliomas comprise multiple subgroups characterized by novel targetable gene fusions and favorable outcomes. Cancer Discov; 2020. 10.1158/2159-8290.CD-19-1030. Online ahead of print [DOI] [PMC free article] [PubMed]
- 10.Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15(12):731–747. doi: 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook PJ, Thomas R, Kannan R, de Leon ES, Drilon A, Rosenblum MK, et al. Somatic chromosomal engineering identifies BCAN-NTRK1 as a potent glioma driver and therapeutic target. Nat Commun. 2017;8:15987. doi: 10.1038/ncomms15987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng MY, Sill M, Chiang J, Schittenhelm J, Ebinger M, Schuhmann MU, et al. Molecularly defined diffuse leptomeningeal glioneuronal tumor (DLGNT) comprises two subgroups with distinct clinical and genetic features. Acta Neuropathol. 2018;136(2):239–253. doi: 10.1007/s00401-018-1865-4. [DOI] [PubMed] [Google Scholar]
- 13.Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF, et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol. 2020;21(2):271–282. doi: 10.1016/S1470-2045(19)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drilon A. TRK inhibitors in TRK fusion-positive cancers. Ann Oncol. 2019;30 Suppl 8:viii23–viii30. doi: 10.1093/annonc/mdz282. [DOI] [PubMed] [Google Scholar]
- 15.Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of Larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378(8):731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor Entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7(4):400–409. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farago AF, Le LP, Zheng Z, Muzikansky A, Drilon A, Patel M, et al. Durable clinical response to Entrectinib in NTRK1-rearranged non-small cell lung cancer. J Thorac Oncol. 2015;10(12):1670–1674. doi: 10.1097/01.JTO.0000473485.38553.f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson SD, Zhou S, Huse JT, de Groot JF, Xiu J, Subramaniam DS, et al. Targetable gene fusions associate with the IDH wild-type astrocytic lineage in adult gliomas. J Neuropathol Exp Neurol. 2018;77(6):437–442. doi: 10.1093/jnen/nly022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45(10):1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia EP, Minkovsky A, Jia Y, Ducar MD, Shivdasani P, Gong X, et al. Validation of OncoPanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med. 2017;141(6):751–758. doi: 10.5858/arpa.2016-0527-OA. [DOI] [PubMed] [Google Scholar]
- 21.Gatalica Z, Xiu J, Swensen J, Vranic S. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol. 2019;32(1):147–153. doi: 10.1038/s41379-018-0118-3. [DOI] [PubMed] [Google Scholar]
- 22.Guerreiro Stucklin AS, Ryall S, Fukuoka K, Zapotocky M, Lassaletta A, Li C, et al. Alterations in ALK/ROS1/NTRK/MET drive a group of infantile hemispheric gliomas. Nat Commun. 2019;10(1):4343. doi: 10.1038/s41467-019-12187-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong DS, Bauer TM, Lee JJ, Dowlati A, Brose MS, Farago AF, et al. Larotrectinib in adult patients with solid tumours: a multi-Centre, open-label, phase I dose-escalation study. Ann Oncol. 2019;30(2):325–331. doi: 10.1093/annonc/mdy539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiao SJ, Zehir A, Sireci AN, Aisner DL. Detection of tumor NTRK gene fusions to identify patients who may benefit from tyrosine kinase (TRK) inhibitor therapy. J Mol Diagn. 2019;21(4):553–571. doi: 10.1016/j.jmoldx.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones DT, Hutter B, Jager N, Korshunov A, Kool M, Warnatz HJ, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones KA, Bossler AD, Bellizzi AM, Snow AN (2019) BCR-NTRK2 fusion in a low-grade glioma with distinctive morphology and unexpected aggressive behavior. Cold Spring Harb Mol Case Stud. 2019;5(2):a003855. 10.1101/mcs.a003855. Print 2019 Apr [DOI] [PMC free article] [PubMed]
- 27.Khotskaya YB, Holla VR, Farago AF, Mills Shaw KR, Meric-Bernstam F, Hong DS. Targeting TRK family proteins in cancer. Pharmacol Ther. 2017;173:58–66. doi: 10.1016/j.pharmthera.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Lee Y, Cho HJ, Lee YE, An J, Cho GH, et al. NTRK1 fusion in glioblastoma multiforme. PLoS One. 2014;9(3):e91940. doi: 10.1371/journal.pone.0091940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurozumi K, Nakano Y, Ishida J, Tanaka T, Doi M, Hirato J, et al. High-grade glioneuronal tumor with an ARHGEF2-NTRK1 fusion gene. Brain Tumor Pathol. 2019;36(3):121–128. doi: 10.1007/s10014-019-00345-y. [DOI] [PubMed] [Google Scholar]
- 30.Laetsch TW, DuBois SG, Mascarenhas L, Turpin B, Federman N, Albert CM, et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol. 2018;19(5):705–714. doi: 10.1016/S1470-2045(18)30119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez GY, Perry A, Harding B, Li M, Santi M. CDKN2A/B loss is associated with anaplastic transformation in a case of NTRK2 fusion-positive Pilocytic astrocytoma. Neuropathol Appl Neurobiol. 2019;45(2):174–178. doi: 10.1111/nan.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luberg K, Wong J, Weickert CS, Timmusk T. Human TrkB gene: novel alternative transcripts, protein isoforms and expression pattern in the prefrontal cerebral cortex during postnatal development. J Neurochem. 2010;113(4):952–964. doi: 10.1111/j.1471-4159.2010.06662.x. [DOI] [PubMed] [Google Scholar]
- 33.Ni J, Xie S, Ramkissoon SH, Luu V, Sun Y, Bandopadhayay P, et al. Tyrosine receptor kinase B is a drug target in astrocytomas. Neuro-Oncology. 2017;19(1):22–30. doi: 10.1093/neuonc/now139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamura R, Boichard A, Kato S, Sicklick JK, Bazhenova L, Kurzrock R (2018) Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: implications for NTRK-targeted therapeutics. JCO Precis Oncol; 2018;2018:10.1200/PO.18.00183. 10.1200/PO.18.00183. Epub 2018 Nov 15. [DOI] [PMC free article] [PubMed]
- 35.Pattwell SS, Konnick EQ, Liu YJ, Yoda RA, Sekhar LN, Cimino PJ. Neurotrophic receptor tyrosine kinase 2 (NTRK2) alterations in low-grade gliomas: report of a novel gene fusion partner in a Pilocytic astrocytoma and review of the literature. Case Rep Pathol. 2020;2020:5903863. doi: 10.1155/2020/5903863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prabhakaran N, Guzman MA, Navalkele P, Chow-Maneval E, Batanian JR (2018) Novel TLE4-NTRK2 fusion in a ganglioglioma identified by array-CGH and confirmed by NGS: potential for a gene targeted therapy. Neuropathology. Neuropathology. 2018. 10.1111/neup.12458. Online ahead of print. [DOI] [PubMed]
- 37.Qaddoumi I, Orisme W, Wen J, Santiago T, Gupta K, Dalton JD, et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016;131(6):833–845. doi: 10.1007/s00401-016-1539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosen EY, Goldman DA, Hechtman JF, Benayed R, Schram AM, Cocco E, et al. TRK fusions are enriched in cancers with uncommon Histologies and the absence of canonical driver mutations. Clin Cancer Res. 2020;26(7):1624–1632. doi: 10.1158/1078-0432.CCR-19-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schram AM, Jonsson P, Drilon A, Bale TA, Hechtman JF, Benayed R et al (2018) Genomic heterogeneity underlies mixed response to tropomyosin receptor kinase inhibition in recurrent glioma. JCO Precis Oncol. 2018;2:10.1200/PO.18.00089. 10.1200/PO.18.00089. Epub 2018 Aug 8 [DOI] [PMC free article] [PubMed]
- 40.Serrano J, Snuderl M. Whole genome DNA methylation analysis of human glioblastoma using Illumina BeadArrays. Methods Mol Biol. 2018;1741:31–51. doi: 10.1007/978-1-4939-7659-1_2. [DOI] [PubMed] [Google Scholar]
- 41.Shah N, Lankerovich M, Lee H, Yoon JG, Schroeder B, Foltz G. Exploration of the gene fusion landscape of glioblastoma using transcriptome sequencing and copy number data. BMC Genomics. 2013;14:818. doi: 10.1186/1471-2164-14-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solomon JP, Hechtman JF. Detection of NTRK fusions: merits and limitations of current diagnostic platforms. Cancer Res. 2019;79(13):3163–3168. doi: 10.1158/0008-5472.CAN-19-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solomon JP, Linkov I, Rosado A, Mullaney K, Rosen EY, Frosina D, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol. 2020;33(1):38–46. doi: 10.1038/s41379-019-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164(5):1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torre M, Alexandrescu S, Dubuc AM, Ligon AH, Hornick JL, Meredith DM. Characterization of molecular signatures of supratentorial ependymomas. Mod Pathol. 2020;33(1):47–56. doi: 10.1038/s41379-019-0329-2. [DOI] [PubMed] [Google Scholar]
- 47.Torre M, Jessop N, Hornick JL, Alexandrescu S. Expanding the spectrum of pediatric NTRK-rearranged fibroblastic tumors to the central nervous system: a case report with RBPMS-NTRK3 fusion. Neuropathology. 2018;38(6):624–630. doi: 10.1111/neup.12513. [DOI] [PubMed] [Google Scholar]
- 48.Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5(1):25–34. doi: 10.1158/2159-8290.CD-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viaene AN, Santi M, Rosenbaum J, Li MM, Surrey LF, Nasrallah MP. SETD2 mutations in primary central nervous system tumors. Acta Neuropathol Commun. 2018;6(1):123. doi: 10.1186/s40478-018-0623-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter AW, Kandula VVR, Shah N. Larotrectinib imaging response in low-grade glioma. Pediatr Blood Cancer. 2020;67(1):e28002. doi: 10.1002/pbc.28002. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Diaz AK, Shaw TI, Li Y, Niu M, Cho JH, et al. Deep multiomics profiling of brain tumors identifies signaling networks downstream of cancer driver genes. Nat Commun. 2019;10(1):3718. doi: 10.1038/s41467-019-11661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34(37):4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45(6):602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20(12):1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 57.Ziegler DS, Wong M, Mayoh C, Kumar A, Tsoli M, Mould E, et al. Brief report: potent clinical and radiological response to larotrectinib in TRK fusion-driven high-grade glioma. Br J Cancer. 2018;119(6):693–696. doi: 10.1038/s41416-018-0251-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental figure 1. Unsupervised principal component analysis (PCA) of methylation profiles of NTRK-fused gliomas demonstrates that no homogenous groups form when correlated with (a) NTRK gene involved, (b) patient age, or (c) histologic grade.
Additional file 2: Supplemental tables. (1) methods for each case; (2) single nucleotide variants and copy number variants encountered in each case; (3) legend for the methylation reports and t-sne plots corresponding to each case.