SUMMARY
Inactivating mutations of the CREBBP and EP300 acetyltransferases are among the most common genetic alterations in diffuse large B cell lymphoma (DLBCL) and follicular lymphoma (FL). Here, we examined the relationship between these two enzymes in germinal center (GC) B cells, the normal counterpart of FL and DLBCL, and in lymphomagenesis by using conditional GC-directed deletion mouse models targeting Crebbp or Ep300. We found that CREBBP and EP300 modulate common as well as distinct transcriptional programs implicated in separate anatomic and functional GC compartments. Consistently, deletion of Ep300 but not Crebbp impaired the fitness of GC B cells in vivo. Combined loss of Crebbp and Ep300 completely abrogated GC formation, suggesting that these proteins partially compensate for each other through common transcriptional targets. This synthetic lethal interaction was retained in CREBBP-mutant DLBCL cells and could be pharmacologically targeted with selective small molecule inhibitors of CREBBP and EP300 function. These data provide proof-of-principle for the clinical development of EP300-specific inhibitors in FL and DLBCL.
In Brief
Loss-of-function mutations of CREBBP and EP300 are frequent and early events in the pathogenesis of FL and DLBCL, the two most common lymphoma subtypes. Meyer et al. uncover distinct as well as compensatory roles for these acetyltransferases in separate compartments of the germinal center and exploit this notion to document an EP300-dependency in CREBBP-deficient lymphoma cells that can be targeted therapeutically.
Graphical Abstract

INTRODUCTION
Diffuse large B cell lymphoma (DLBCL) and follicular lymphoma (FL) are the most common lymphoid malignancies, together accounting for ~60% of B cell lymphoma diagnoses (Swerdlow et al, 2016). Despite the significant progress made in the therapeutic management of these diseases, both remain partially unmet clinical needs. In particular, a substantial fraction of DLBCL patients do not achieve complete remission with current first-line chemo-immunotherapeutic approaches (Gisselbrecht et al., 2010) and FL, although indolent, is essentially incurable. Moreover, as many as 45% of FL cases transform into a high-grade malignancy, typically a DLBCL (also known as transformed FL or tFL), with dismal overall survival (Montoto et al., 2007). The development of treatments that can eradicate the reservoir of initiating cells responsible for resistance and transformation remains a high priority in the field.
Over the past decade, genomic analyses of FL and DLBCL have uncovered highly recurrent somatic mutations and deletions in the histone acetyl-transferase gene CREBBP (60% of FL and 25% of DLBCL); its paralogue EP300 is targeted at much lower frequencies (5% of FL and DLBCL) (Chapuy et al., 2018; Morin et al., 2011; Okosun et al., 2014; Pasqualucci et al., 2011a; Pasqualucci et al., 2014; Pasqualucci et al., 2011b; Schmitz et al., 2018). CREBBP and EP300 encode for ubiquitously expressed mammalian enzymes that act as global transcriptional co-activators by interacting with more than 400 transcription factors and by catalyzing the modification of lysines on both histone and non-histone proteins in a cell-context-dependent manner (Bannister and Kouzarides, 1996; Bedford et al., 2010; Dancy and Cole, 2015; Goodman and Smolik, 2000; Ogryzko et al., 1996).
In germinal center (GC) B cells, the normal counterpart of FL and DLBCL, two critical non-histone substrates of CREBBP- and EP300-mediated acetylation are the tumor suppressor p53, which requires acetylation for its transcriptional activity (Avantaggiati et al., 1997; Gu and Roeder, 1997; Lill et al., 1997), and the proto-oncogene BCL6, a potent transcriptional repressor that regulates the GC reaction and is functionally impaired by this modification (Bereshchenko et al., 2002). Additionally, by catalyzing H3K18 and H3K27 acetylation at promoter and enhancer regions, CREBBP modulates the expression of a selected number of genes that are implicated in GC exit including signaling pathways triggered by engagement of the B cell receptor (BCR) and CD40 receptor, the plasma cell regulator IRF4, and antigen processing and presentation through the major histocompatibility complex class II (MHC-II) complex (Green et al., 2015; Hashwah et al., 2017; Jiang et al., 2017; Zhang et al., 2017). Of note, the GC-specific CREBBP transcriptional network encompasses almost all BCL6 direct target genes, suggesting a critical role for this acetyltransferase in opposing the oncogenic activity of BCL6 while ensuring the rapid activation of programs that sustain terminal differentiation in the GC light zone (LZ) (Jiang et al., 2017; Zhang et al., 2017).
Mutations of CREBBP and EP300 inactivate the enzymatic function of these proteins by generating truncated forms that lack the histone acetyl-transferase (HAT) domain or by introducing amino acid changes, also within the HAT domain, which severely impair their affinity for AcetylCoA (Pasqualucci et al., 2011a). These mutations are acquired at an early stage of FL development by a common ancestral clone that subsequently progresses to FLor tFL throughdivergent evolution(Greenet al., 2015; Okosun et al., 2014; Pasqualucci et al., 2014). Accordingly, CREBBP-mutated B cells have been found in a pre-malignant condition known as FL in situ, often togetherwith the hallmark t(14;18) translocation deregulating BCL2 (Schmidt et al., 2018). Mutations in CREBBP are mono-allelic in 80% of DLBCL and over 50% of FL cases, leaving the residual wild-type (WT) allele expressed (García-Ramírez et al., 2017; Pasqualucci et al., 2011a). In mouse models, conditional GC-directed inactivation of Crebbp in both heterozygosis and homozygosis significantly increases the incidence of Bcl2-driven lymphomas (Jiang et al., 2017; Zhang et al., 2017). Reduced dosage of CREBBP (and EP300) is thus thought to facilitate malignant transformation by dysregulating signaling pathways that are important for terminal differentiation and by favoring the constitutive activity of the BCL6 oncogene at the expense of the p53 tumor suppressor.
Genetic alterations of CREBBP and EP300 are largely non-overlapping in FL and in DLBCL (Arthur et al., 2018; Chapuy et al., 2018; Green et al., 2015; Okosun et al., 2014; Pasqualucci et al., 2011a; Pasqualucci et al., 2014; Schmitz et al., 2018). This observation, together with the high structural and functional similarity, suggests a potential compensatory function of these two paralogues in GC B cells. This notion is corroborated by the contrast between the genome-wide binding pattern of CREBBP, which occupies virtually all predicted GC-specific super-enhancers, and the limited transcriptional changes observed after its deletion in GC B cells (Zhanget al., 2017). Accordingly, a functional screen for essential genes in lung adenocarcinoma, which also frequently harbors CREBBP mutations, has revealed a synthetic lethal role for EP300 in CREBBP-mutated cells, where its pharmacologic inhibition exerts anti-proliferative and anti-survival effects, although at very high concentrations of a tool compound with limited potency and selectivity (Ogiwara et al., 2016).
Here, we used mouse models where Ep300 and/or Crebbp were specifically deleted in GC B cells to explore the relationship between CREBBP and EP300 in GC physiology and lymphoma-genesis. We found that these two enzymes have common as well as distinct transcriptional targets in sub-compartments of the GC reaction, whereas their combined genetic deletion abrogated GC formation in vivo and impaired DLBCL cell line growth, suggesting that CREBBP-mutant DLBCL might depend on the residual EP300 activity. Treatment with small molecule inhibitors of CREBBP and EP300 was preferentially toxic to CREBBP-mutant DLBCL, establishing a paralogue lethality that could be explored as an actionable therapeutic target.
RESULTS
Crebbp and Ep300 Play Partially Distinct Roles in GC Development
The CREBBP and EP300 acetyltransferases share 60% amino acid identity and similar domain organization (Chan and La Thangue, 2001). To investigate whether EP300 is functionally equivalent to CREBBP in the GC, we compared the adaptive immune response in conditional, GC-specific Ep300-null vs Crebbp-null mice, obtained by crossing Ep300 (or Crebbp) floxed alleles (Kang-Decker et al., 2004; Kasper et al., 2006) with mice expressing the Cre recombinase under the control of the Cγ1 promoter (Cγ1Cre/+) (Casola et al., 2006).
As previously shown, loss of Crebbp led to increased GC formation when analyzed 10 days after intraperitoneal (i.p.) immunization with the T-cell-dependent antigen sheep red blood cells (SRBCs) (Figures 1A, top and 1B, left) (Hashwah et al., 2017; Zhang et al., 2017). In contrast, loss of Ep300 led to an approximately 50% decrease in both the percentage (Figures 1A, bottom and 1B, right) and absolute number (Figures S1A and S1B) of GC B cells, compared to littermate controls (on average 3.6% in Ep300+/+Cγ1Cre/+ mice vs 1.8%in Ep300fl/flCγ1Cre/+ mice; p < 0.01, Student’s t test). This difference was not due to variabilities in the deletion efficiencies of the two floxed alleles, as immunofluorescence analysis of Crebbp-deficient and Ep300-deficient spleen sections via antibodies specific for Crebbp or Ep300 and the GC marker peptide nucleic acid (PNA) documented loss of expression of the target protein in 100% (n = 36 out of 36) and 85% (n = 29 out of 34) of the GCs, respectively (Figure 1C). Consistent with the flow-cytometric data, immunohistochemistry staining of PNA (not shown) and the GC-marker BCL6 confirmed a significant dose-dependent reduction in both the number and size of GCs upon deletion of Ep300, with consequently reduced overall GC area (Figures 1D and 1E). Nonetheless, Ep300fl/flCγ1Cre/+ GCs were indistinguishable from those of WT animals in terms of relative BCL6 expression, measured by fluorescence activated cell sorting (FACS) analysis and RNA-sequencing (RNA-seq) analysis (Figures S1C and S1D). Interestingly, Crebbpfl/flCγ1Cre/+ mice displayed a statistically significant increase in the dark zone (DZ) to LZ ratio, recapitulating the previously reported requirement for this gene in LZ-specific signal transduction pathways (Figures S1E and S1F) (Zhang et al., 2017). An opposite trend was observed in Ep300-null GCs, which was not statistically significant (Figures S1E and S1F, right) and can be explained by the fact that DZ B cells feed the LZ (Victora and Nussenzweig, 2012); therefore, a decrease in DZ B cells is expected to cause a proportional decrease in LZ B cells. These data indicate that, although structurally and functionally similar, CREBBP and EP300 are not entirely interchangeable, and reveal distinct requirements for these two proteins in specific aspects of the GC reaction.
Figure 1. Crebbp and Ep300 Play Non-overlapping Roles in GC B Cells.

(A) Representative flow-cytometric analysis of splenic B220+ cells from Crebbp+/+, Crebbpfl/+, and Crebbpfl/fl (top) vs Ep300+/+, Ep300fl/+, and Ep300fl/fl (bottom) Cγ1Cre/+ mice, analyzed 10 days after SRBC immunization. GC B cells are identified as CD95+PNAhi cells, and numbers in each image indicate the percentage in the gate.
(B) Percentage of GC B cells in mice from the indicated genotypes, analyzed at 3 months of age, 10 days after SRBC immunization (n = 5–8 mice per genotype).
(C) Immunofluorescence staining of Crebbp (red) and Ep300 (purple) in representative spleen sections from SRBC-immunized Crebbpfl/flCγ1Cre/+ and Ep300fl/flCγ1Cre/+ mice. PNA (green) identifies the GC area (outlined). The total number of Crebbp or Ep300-null GCs, out of the total number of PNA+ GCs, is given on the right for the two mouse models (n = 3 animals per genotype). Scale bar, 30 μm.
(D) Immunohistochemical analysis of BCL6 in representative spleen sections from Ep300+/+, Ep300fl/+, and Ep300fl/fl Cγ1Cre/+ mice, analyzed 10 days after SRBC immunization. Scale bar, 500 μm.
(E) Mean GC number, GC size, and overall GC area (per spleen section) in mice of the indicated genotypes, measured in pixels using the ImageJ software on 3 sections per mouse (mean ± SD; n = 3–4 mice per genotype).
*p <0.05, **p <0.01; Student’s t-test. Only statistically significant p values are indicated.
Crebbp and Ep300 Regulate Distinct Sets of Genes in the GC
In order to elucidate the mechanistic basis for the differential effects of Crebbp and Ep300 loss on the GC response, we performed transcriptomic analyses of purified GC B cell populations sorted from SRBC-immunized Crebbpfl/fl and Ep300fl/flCγ1Cre/+ mice (n = 3 animals per genotype and 5 WT littermates). Unsupervised hierarchical clustering separated the three genotypes on the basis of expression profiles (Figures S2A and S2B), indicating that individual loss of Crebbp and Ep300 imposes a sufficiently large number of unique changes on this population compared with normal cells. We then contrasted the genes differentially expressed in Crebbp-deficient cells with those modulated in Ep300-deficient cells, as obtained by independent comparison of their transcriptional profiles to that of WT cells and by direct supervised analysis of Crebbp-deficient vs Ep300-deficient transcriptional profiles (corrected false discovery rate [FDR] % 0.05, fold change [FC] R 1.2). Surprisingly, only 12% (n = 72 out of 591) of the genes showing significantly reduced expression (and 52 out of 387 genes with increased expression) were shared between the two genetic backgrounds (Figures 2A, 2B, S2C, and S2D; Table S1).
Figure 2. Crebbp and Ep300 Modulate Distinct Functional Programs Implicated in DZ to LZ Transition.
(A) Differentially expressed genes in Crebbpfl/fl and Ep300fl/flCγ1Cre/+ GC B cells, compared with WT (left and middle panel) or to each other (right). In the heatmaps, rows correspond to genes and columns correspond to different mice; the third category is shown as reference (blue, reduced expression; red, increased expression; FDR < 0.05, FC ≥ 1.2). Scale bar indicates the Z score. Representative transcripts are indicated, and the complete list is provided in Table S1. Only annotated genes are shown.
(B) Venn diagrams of genes differentially expressed in Crebbpfl/fl and Ep300fl/flCγ1Cre/+ GCB cells, compared with WT (only annotated genes considered). See also Figure S2.
(C) Top significantly enriched (p < 0.05 after correction for multiple hypothesis) biological pro-grams/signaling pathways identified among the list of genes showing reduced expression in Crebbp-deficient (left) and Ep300-deficient (right) GC B cells, compared with WT (see Method Details). The full list of differentially enriched categories is provided in Table S2.
(D) GSEA analysis of LZ-associated and DZ-associated genes along the T score rank of transcripts expressed in Crebbp+/+Ep300+/+ vs Crebbpfl/flCγ1Cre/+ (left) and Crebbp+/+Ep300+/+ vs Ep300fl/flCγ1Cre/+ (right) GC B cells. The reverse analysis showed no significant enrichment, indicating preferential modulation of LZ genes by Crebbp and of DZ genes by Ep300 (see Table S3).
Pathway analysis revealed that the lists of genes modulated by these two acetyltransferases were significantly and uniquely enriched in discrete biological programs. In particular, positive regulation of cell cycle (E2f1, Cdc25b, Anapc11, and Cdkn1a), DNA replication (Pcna, Cdt1, and Dna2), and DNA repair (Rad51 and Xrcc1) were under-represented in the Ep300fl/flCγ1Cre/+ transcriptional signature, whereas genes involved in antigen presentation/processing through the MHC-II complex (e.g., Ciita and H2-DM), receptor signaling, and terminal B cell differentiation (e.g. Irf4, SpiB, Nfkb2, and Cd40) were preferentially reduced in expression in Crebbp-defective GC B cells (Figure 2C; Table S2) (Jiang et al., 2017; Zhang et al., 2017). These differentially affected programs are reminiscent of functions that are normally compartmentalized between the DZ—namely proliferation, extended replicative potential, and the repair of various types of DNA damage associated with somatic hypermutation—and the LZ of the GC, where several signal transduction pathways become engaged downstream of the BCR, CD40, and Toll-like receptor (TLR) in association with recognition by T follicular-helper (Tfh) cells (Victora and Nussenzweig, 2012). We thus interrogated the transcriptional profiles of Crebbpfl/fl and Ep300fl/flCγ1Cre/+ GCs for the enrichmentin signatures previously identified as discriminators of DZ and LZ B cells (Victora et al., 2012). As illustrated inFigure 2D, gene set enrichment analysis (GSEA) uncovered opposite patterns in the two mouse models, with the expression of DZ genes being preferentially affected by Ep300 deletion and the expression of LZ genes being significantly decreased in the Crebbpfl/flCγ1Cre/+ mice (Table S3). The reduced expression of genes involved in cell cycle/DNA replication induced by loss of Ep300 but not Crebbp is also likely responsible for the lower percentage of GC B cells observed in this model.
The ability of CREBBP and EP300 to regulate distinct sets of genes was confirmed by chromatin immunoprecipitation sequencing (ChIP-seq) analysis of H3K27Ac in human isogenic cell lines engineered to lack CREBBP or EP300 (SUDHL4-CREBBP−/− and SUDHL4-EP300−/−; n = 2 clones per genotype, compared with WT control), which were readily separated upon unsupervised hierarchical clustering of the relative H3K27Ac density, indicating significant genome-wide redistribution of this activation mark (Pott and Lieb, 2015) (Figure S3A). In particular, of 21,091 H3K27Ac+ regions identified in WT cells, 932 were differentially enriched across the 3 genotypes (FDR ≤ 0.05;FC ≥ 2),681 showed significant depletion specifically upon CREBBP (n = 127) or EP300 (n = 554) loss (non-compensated targets), and 92 were significantly depleted in both CREBBP−/− and EP300−/− cells, suggesting co-regulatory roles or the requirement of both enzymes to achieve full gene activation (Figures S3B and S3C). This analysis also uncovered a number of regions where the H3K27Ac signal was higher after CREBBP or EP300 loss, possibly reflecting an indirect epigenetic re-organization (e.g., feedback-mechanism- and/or HAT-dependent reduced expression of transcriptional repressors). Functional annotation of the differentially acetylated regions showed that those lostin CREBBP−/− and/or EP300−/− cells were mainly represented by transcription start site (TSS)-distal domains predicted by the rank ordering of super enhancers (ROSE) algorithm as enhancers (Whyte et al., 2013) (Figures S3D and S3E) and located in proximity to genes critical for GC biology (e.g., BCL6, POU2F1, and AICDA) or implicated in pathways known to play key roles in normal and neoplastic GC B cells (e.g. BCR, MAPK, Gα13, and interleukin [IL] signaling) (Figure S3E).
Together, these data demonstrate that CREBBP and EP300 play common as well as distinct roles during the GC reaction by re-wiring diverse and shared enhancer networks.
Combined Loss of Crebbp and Ep300 Abrogates GC Formation
To examine the combined role of Crebbp and Ep300 in the GC, we generated compound Cγ1Cre/+ mouse models carrying variable combinations of partial or complete Crebbp and Ep300 loss and analyzed the GC response after immunization with SRBCs. We did not detect significant differences between mice with combined heterozygous deletion (Crebbpfl/+Ep300fl/+Cγ1Cre/+) and wild-type littermates (WT: Crebbp+/+Ep300+/+Cγ1Cre/+). In contrast, loss of three HAT alleles, as obtained in Crebbpfl/flEp300fl/+Cγ1Cre/+ and Crebbpfl/+Ep300fl/flCγ1Cre/+ mice, led to a severe reduction in the percentage of GC B cells, which in turn was significantly lower than that observed in single Crebbpfl/fl or Ep300fl/fl Cγ1Cre/+ mice (relative decrease compared with WT littermates: 60% in Crebbpfl/+ Ep300fl/flCγ1Cre/+ vs 40% in Ep300fl/flCγ1Cre/+; p < 0.05, Student’s t test) (Figures 3A and 3B). More importantly, GC B cells were completely absent in mice lacking both Crebbp and Ep300, where the residual GC structures were composed of cells that had escaped concurrent deletion of both genes (Figures 3A, 3B, S4A, and S4B). In line with the FACS data, the mean GC number, GC size, and total GC area were generally normal in Crebbpfl/+Ep300fl/+Cγ1Cre/+ mice, but significantly reduced in all compound strains (Figures 3C and 3D). We conclude that the fitness of GC B cells depends on the combined activity of CREBBP and EP300 and that, whereas these genes can partially compensate for each other, their complete loss is incompatible with GC B cell survival.
Figure 3. Crebbp-Deficient GC B Cells Are Dependent on the Residual Ep300 Protein.

(A) Representative flow-cytometric analysis of splenic B220+ cells from Crebbp+/+Ep300+/+, Crebbpfl/+Ep300fl/+, Crebbpfl/+Ep300fl/fl, Crebbpfl/flEp300fl/+, and Crebbpfl/flEp300fl/fl Cγ1Cre/+ mice, analyzed 10 days after SRBC immunization. GC B cells are identified as CD95+PNAhi cells, and numbers in each image indicate the percentage in the gate.
(B) Normalized percentage of splenic GC B cells in SRBC-immunized mice from the indicated genotypes, in relation to WT littermates (arbitrarily set as 1). Data correspond to 4 experiments, each performed with subsets of genotypes (vs WT) and 3 or 4 animals per genotype.
(C) Immunohistochemical staining of BCL6 in representative spleen sections from animals of the indicated genotypes, analyzed 10 days after SRBC immunization. Scale bar, 500 μm
(D) Mean GC number, GC size, and GC area (per spleen section) in the indicated mice, measured in pixels using the ImageJ software on 3 sections per mouse (Bars represent mean ± SD. The total number of animals analyzed is given inside the bars).
Only statistically significant p values are indicated. *p < 0.05, **p < 0.01; Student’s t test.
Crebbp-Defective Cells Require Ep300 for Proliferation
To understand the mechanism underlying the lack of GC formation in mice with combined loss of Crebbp and Ep300, we first measured the proliferative capacity of splenic B220+ cells isolated from the Cγ1Cre/+ compound mice and stimulated ex vivo by αCD40 and IL-4. In this system, engagement of the CD40 and IL-4 receptors mimics in part the signals delivered in vivo during a T-cell-dependent B cell response and is also needed to induce the expression of the Cre recombinase (Casola et al., 2006). We first confirmed efficient deletion of the floxed alleles at both the DNA (not shown) and protein level 2 days and 4 days after stimulation (see Figure 4A for day 4). Immunoblot analysis of chromatin extracts demonstrated a significant reduction in acetylated H3K18 and H3K27 (Weinert et al., 2018), when both genes were simultaneously ablated and, to a much lesser extent, upon single gene deletion (Figure 4A, bottom). As expected, 75% of the WT cell population had divided by day 4 after stimulation, and ~41% of them had undergone more than 3 cell divisions, as shown by flow-cytometric analysis after in vivo labeling with the CellTrace Violet dye; in contrast, only 9% of the Crebbp/Ep300 co-deleted cells were in this gate (p < 0.01, Student’s t test) (Figures 4B and 4C). Consistently, compared with WT, Crebbpfl/flEp300fl/flCγ1Cre/+ B lymphocytes showed significantly lower proliferation rates as measured by intracellular ATP-based enzymatic activity (Figure 4D). The other compound genotypes displayed variable effects in this assay, which were less evident than those observed in vivo in GC B cells. This difference could be explained in part by the short time-course involved in the ex vivo system (less than 4 days; note that at least one cell division is necessary for the conditional allele to be deleted) versus 10 days in the in vivo GC response. As a result, Crebbpfl/flEp300fl/flCγ1Cre/+ B cells were unable to undergo class switch recombination, a process that is known to require multiple cell divisions (Figures S5A and S5B) (Hodgkin et al., 1996). Thus, correct dosage of Crebbp and Ep300 might be critical to sustain the repeated rounds of cell division and proliferation that are required for proper GC formation. The percentage of dead cells, measured by size scatter and Annexin V staining, was increased in the Crebbpfl/flEp300fl/flCγ1Cre/+ culture at day 4 after stimulation (1.5-fold compared with WT), but not at earlier timepoints (Figures 4E, 4F, and S5C, and data not shown), suggesting that the complete loss of acetyltransferase activity primarily affects proliferation.
Figure 4. Combined Loss of Crebbp and Ep300 Blocks Cell Proliferation.
(A) WB analysis of Crebbp and Ep300 expression in Ficoll-separated splenic B cells of the indicated genotypes, cultured ex vivo in the presence of αCD40 and IL-4 for 4 days. Analysis of H3K27Ac and H3K18Ac monitors for the functional effects of Crebbp and/or Ep300 loss and is quantified on the bottom. Tubulin and total H3 serve as loading control for whole-cell and chromatin extracts, respectively.
(B) Representative histogram plots showing the number of cell divisions in cultured B cells from the indicated genotypes, measured on day 4 after labeling with the CellTraceViolet reagent (live cells gate).
(C) Quantification of the data shown in (B) (mean ± SD; n = 3 mice per genotype).
(D) Cell growth in the same cells, measured by enzymatic activity and expressed as fold changes relative to day 0 (mean ± SD; n = 3 mice per genotype);
(E and F) Analysis of cell viability, assessed on the basis of the percentage of dead cells in the forward scatter versus side scatter (FSC/SSC) (E) and the percentage of AnnexinV+ cells (F). Data are from one experiment where all genotypes were simultaneously analyzed and are representative of at least two independent experiments performed with subsets of genotypes (n = 3 each) that gave analogous results and were combined for statistical analyses. Note that the ex vivo assay is associated with an intrinsic elevated cell death. Bars represent mean ± SD.
Only statistically significant p values are indicated. *p < 0.05, **p < 0.01; Student’s t test.
To corroborate this finding in an in vivo context, we then analyzed the kinetics of GC formation in the same cohorts by measuring the percentage of B220+CD95+PNAhi cells at days 7 and 4 after immunization, i.e., when early GCs can first be histologically identified within secondary follicles (De Silva and Klein, 2015) and Cγ1Cre-mediated recombination of the floxed allele is complete in at least 50% of GC B cells (Casola et al., 2006). As shown in Figures S5D and S5E, Crebbpfl/flEp300fl/flCγ1Cre/+ mice exhibited a measurable drop in the GC population already 4 days after antigenic stimulation, supporting a role for these two enzymes in the initial proliferative phases of the GC reaction.
CREBBP-Mutant DLBCL Cell Lines Are Addicted to EP300 Activity
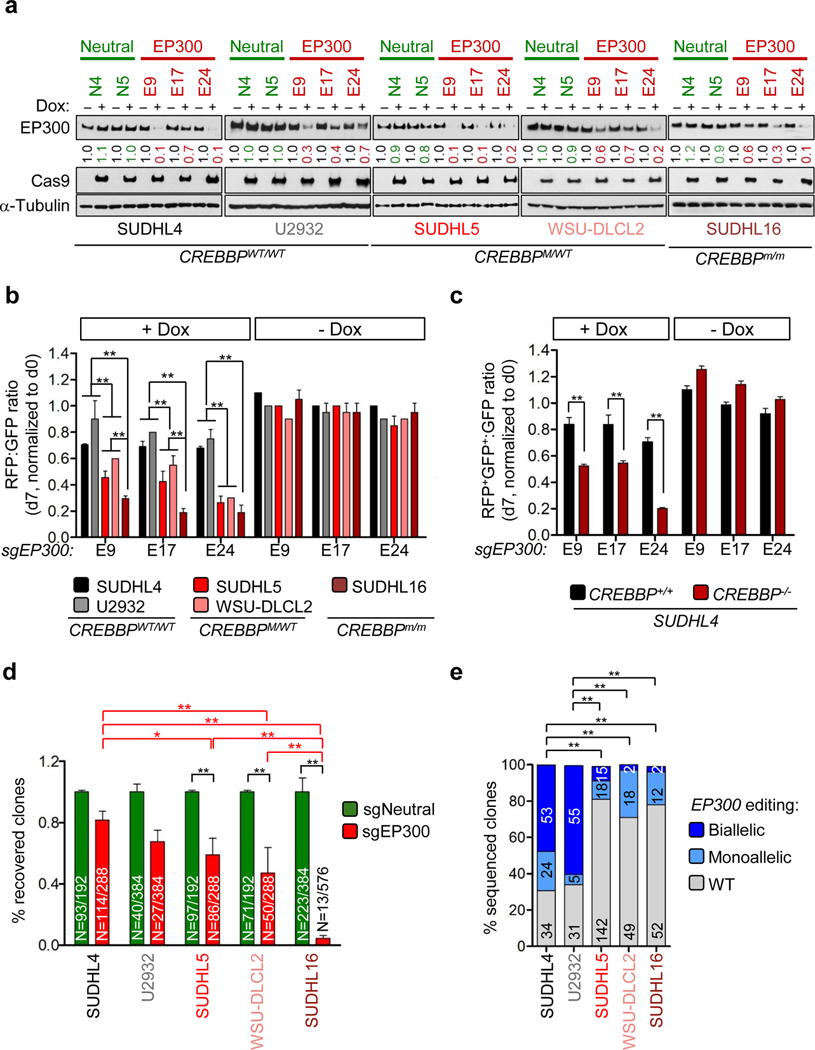
We next sought to determine whether, analogous to normal GC B cells, CREBBP-mutant DLBCL cells remain addicted to the residual EP300-mediated acetyltransferase activity. We selected five DLBCL cell lines representative of different CREBBP mutational states in the context of an intact EP300 locus, including CREBBPWT (SUDHL4 and U2932), heterozygous mutant (SUDHL5, carrying a heterozygous deletion spanning CREBBP; and WSU-DLCL2, harboring a splice-site mutation leading to a frameshift) (CREBBPM/+), and one of the rare biallelically mutated cases (SUDHL16, carrying one inactivating missense mutation and one in-frame deletion in the HAT domain that render the protein enzymatically inactive) (CREBBPm/m) (Pasqualucci et al., 2011a). Cells were engineered to express a doxycycline (Dox)-inducible Cas9 protein (iCas9) together with lentiviral vectors carrying single guide RNAs (sgRNAs) targeting three different EP300 exons (E9, E17, and E24) linked to a constitutive red fluorescent protein (RFP) protein, or, as control, neutral sgRNAs (N4 and N5) designed against an intron of the PPP1R12C gene, linked to green fluorescent protein (GFP) (n = 2 iCas9 derivatives/each).
To test whether deletion of EP300 had a differential effect on CREBBP WT vs mutant backgrounds, we first performed competition assays in which we mixed equal numbers of sorted RFP+ and GFP+ cells and monitored the changes in their ratio over time, after treatment with Dox or control vehicle. Efficient induction of Cas9 protein expression and deletion of EP300 were verified by western blot (WB) analysis of bulk populations 3 days after addition of Dox (Figure 5A). As expected, the 1:1 RFP:GFP ratio was maintained in vehicle-treated cells regardless of the sgRNA used, ruling out the presence of leakage (Figure 5B) (−DOX). Moreover, analogous to what had been observed in vivo, EP300 deletion led to a modest impairment in cell growth kinetics even in a CREBBPWT background, reflected by the ~30% reduction in RFP cells observed at day 7 after Dox-induction (Figure 5B, black and gray bars). However, compared with WT cells, lines carrying monoallelic (light red shade) and, to a larger extent, biallelic (dark red shade) CREBBP inactivation were significantly counter-selected, suggesting dose-dependent susceptibility to EP300 withdrawal (relative reduction: 60% and 80%, respectively; p < 0.01, two-way ANOVA with Bonferroni post-test) (Figure 5B) (+DOX). The same competitive fitness experiment performed in an isogenic setting (SUDHL4-CREBBP+/+ and SUDHL4-CREBBP−/−), where the presence or absence of an intact CREBBP locus represented the single variable, confirmed the prominent detrimental effect of EP300 loss in CREBBP-mutated compared with WT cells (Figure 5C and not shown).
Figure 5. CREBBP Mutant DLBCL Cells Are Significantly Counter-Selectedupon EP300 Deletion.
(A) Immunoblot analysis of EP300 and Cas9 expression in five DLBCL cell lines carrying wild-type (wt/wt) or mutant (M indicates truncating mutation; m indicates missense mutation or in frame deletion) CREBBP alleles, treated with Dox for 3 days to induce Cas9-mediated disruption of the EP300 gene or a control intronic region. Values indicate normalized EP300 protein levels in relation to uninduced, set as 1; α-Tubulin, loading control.
(B) Relative fraction of RFP+ (sgEP300-transduced) to GFP+ (sgNeutral-transduced) cells in the same lines, measured on day 7 after Dox-induction (mean ± SD; n = 2 assays performed by using different sg-neutral transduced clones). Significance was calculated by using two-way ANOVA with Bonferroni post-test; 1 representative experiment out of 2 that gave similar results. Only statistically significant p values are indicated; **p < 0.01.
(C) Relative fraction of RFP+GFP+ (sgEP300-transduced) to GFP+ (sgNeutral-transduced) cells in isogenic SUDHL4 clones engineered to carry WT (+/+) or disrupted (−/−) CREBBP alleles, measured on day 7 after Dox induction (mean ± SD; n = 3). **p < 0.01, two-way ANOVA with Bonferroni post-test.
(D) Percentage of recovered clones in the indicated cell lines after Dox-induced deletion of EP300 (red) vs a control region (green) arbitrarily set as 1. Bars represent the average ± SD of independent transductions using 3 different EP300-sgRNAs and 2 neutral-sgRNAs, except for the U2932 cell line, where only 2 EP300-sgRNAs and 1 neutral-sgRNA gave informative results because of its general poor growth as single clones. The absolute number of clones recovered over the total plated is provided inside the bars. Only statistically significant p values are indicated; *p < 0.05, **p < 0.001; Fisher’s exact test. In U2932, no significant differences were found between the number of clones recovered in EP300- and neutral-sgRNAs; comparisons with other cell lines (red parentheses) are not reported because they were not informative given the distinct growth characteristics of this cell line.
(E) Pattern of EP300 editing in the recovered clones, as determined by PCR amplification and Sanger sequencing. Color codes denote biallelically edited, monoallelically edited, and unedited (WT) clones. Data are expressed as percentage of total sequenced clones, and the absolute number is shown inside the bars. Note that the two biallelically edited SUDHL16 clones harbored in-frame deletions that did not disrupt EP300 protein expression. Only statistically significant p values are indicated. **p < 0.001; Fisher’s exact test.
To corroborate these results further, we performed “drop-out” assays in which we plated single cells from the five DLBCL line models (at least 96 clones per sgRNA per cell line, comprising the three EP300-sgRNAs and both Neutral-sgRNAs) and monitored the number and targeted allele configuration of the recovered clones up to 21 days after Dox induction. In the CREBBPWT SUDHL4 and U2932 cell lines, clones grew at similar frequencies independently of whether the cells had been transduced with EP300-directed or control guides (Figure 5D). In contrast, CREBBP heterozygous mutant and, more evidently, CREBBP biallelic mutant cells showed significantly inferior recovery rates upon disruption of EP300 (p < 0.001; Fisher’s exact test). Moreover, Sanger sequencing of the surviving clones revealed a markedly smaller number of bi-allelically edited clones in all CREBBP-mutant cells than in SUDHL4 and U2932; in particular, none of the SUDHL16 derivatives carried frameshift events in both EP300 alleles and/or lacked EP300 protein expression (Figure 5E and data not shown). We conclude that, analogous to normal GC B cells, transformed CREBBP-mutant lymphoma cells remain sensitive to EP300 dosage.
Pharmacologic Targeting of CREBBP-Mutant DLBCL Cells
The observation that tumors harboring CREBBP genetic defects depend on EP300, that is, require a threshold level of acetyltransferase activity, prompted us to explorethis dependency for therapeutic purposes. Although no EP300-specific inhibitors are available, we took advantage of two recently developed small molecule inhibitors that are selective for CREBBP/EP300: CCS1477, a clinical candidate bromodomain (BRD) inhibitor (Pegg et al., 2017), and CU329, a pre-clinical HAT domain inhibitor (Lasko et al., 2017). We chose compounds with different mechanisms of action because a large number of CREBBP mutant alleles harbor missense rather than truncating mutations and, although we have previously demonstrated that the encoded proteins are enzymatically inactive (Pasqualucci et al., 2011a), they could still potentially bind to their substrates; thus, the HAT inhibitor would have no effect on such non-enzymatic functions, whereas BRD inhibitors could offer the advantage of evicting the mutant protein from its substrates. RNA-seq and WB analysis of SUDHL4 cells treated with CCS1477 or CU329 (vs DMSO) confirmed their on-target activity by showing reduced CREBBP/EP300 self-acetylation, H3K18Ac and H3K27Ac, as well as a significant negative enrichment in both CREBBP- and EP300-modulated programs, including the expression of HLA-DR (Figures S6A–S6D). Nonetheless, the two compounds displayed both quantitative differences in the commonly induced responses and qualitative differences in a number of transcripts that were uniquely affected, in line with their different modes of action (Table S4; Figure S6E; and not shown).
To investigate whether pharmacologic inhibition of CREBBP/EP300 has preferential toxicity toward CREBBP-mutated lymphomas, we utilized the same CrispR/Cas9 platform to generate isogenic stable SUDHL4 clones carrying CREBBP WT or disrupted alleles (n = 4 clones each, obtained by 2 independent sgRNAs per locus) (Figure S7A). Immunoblot analysis confirmed the lack of full-length CREBBP protein expression with comparable EP300 levels across all lines, documenting the specificity of the CREBBP sgRNAs (Figure S7B; see also C–F for extensive epigenetic and phenotypic characterization of the CREBBP-deficient clones).
Importantly, both inhibitors were significantly more toxic to CREBBP−/− than to CREBBP+/+ SUDHL4 cells in 48 h dose response assays over a broad range of doses (10 μM to 0.002 μM) (Figures S7E–S7H) and, to a greater extent, upon a 6-day treatment using sub-micromolar concentrations (100 nM for CCS1477 and 50 nM for the CU329; p value < 0.01, Student’s t test) (Figure 6A). This difference in sensitivity was explained in part by the more profound effect of both compounds on cell cycle arrest, indicated by the accumulation of cells in the G0/G1 phase, with consequent reduction in the percentage of cells in G1/S and G2/M (CREBBP+/+ vs CREBBP−/−: 18% vs 8% in CCS1477-treated cells, and 17% vs 8% upon treatment with CU329; p < 0.05, Student’s t-test) (Figure 6B). Indeed, although more prominently associated with the BRD inhibitor, cell cycle positive regulators (e.g., PCNA, E2F1, and E2F2) as well as the MYC gene emerged as major transcriptional components of the response to both drugs (Figures 6C and 6D). Compared with CREBBP+/+ cells, CREBBP-null cells also exhibited more robust suppression of global H3K27Ac, H3K18Ac, and CREBBP/EP300 self-acetylation upon WB analyses (Figure 6E). Altogether, these results demonstrate that CREBBP-mutant cells are preferentially susceptible to pharmacologic inhibition of the residual acetyltransferase activity over WT cells and provide a proof-of-principle for the use of drugs exploiting this dependency in CREBBP-mutated B cell lymphomas.
Figure 6. CREBBP Mutant Cells Are Preferentially Vulnerable to CREBBP/EP300 Inhibition.
(A) Cell proliferation of CREBBP+/+ and CREBBP−/− SUDHL4 clones grown in the presence of 100 nM CCS1477 or 50 nM CU329 over the course of 6 days (mean ± SD; n = 4); **p < 0.01; Student’s t-test.
(B) Quantification of cell cycle analysis in the same cells, assessed after 72 h of treatment with CCS1477, CU329, or control DMSO (mean ± SD; n = 4); only statistically significant p values indicated. *p < 0.05; Student’s t-test.
(C) Differentially expressed transcripts involved in cell cycle/DNA replication, as identified by DESeq2 in SUDHL4 cells treated with DMSO versus CCS1477 or CU329 for 48 h. In the heatmap, rows correspond to genes and columns represent 3 independent clones cultured in the presence or absence of the inhibitor as indicated (FDR ≤ 0.05%, FC ≥ 2 in at least one of the compounds and ≥ 1.2 in both compounds, except for MYC that showed a 1.5-fold reduction upon treatment with CU329 and 1.3-fold reduction upon treatment with CCS1477). Scale bar indicates the Z score, with blue representing decreased expression and red representing increased expression. Only representative transcripts are highlighted.
(D) GSEA of cell cycle and DNA replication genes in the rank of transcripts differentially expressed between DMSO-treated and BRDi- or HATi-treated SUDHL4 cells. See also Table S4.
(E) Western blot analysis of CREBBP and EP300 expression in isogenic SUDHL4 cell lines carrying intact (grey) or disrupted (red) CREBBP alleles and treated with DMSO, CCS1477, or CU329. Analysis of global H3K18 and H3K27 acetylation documents the stronger effect of the two inhibitors in CREBBP-deficient clones, as quantified at the bottom after normalization for total H3.
DISCUSSION
The studies presented here revealed distinct functions for the CREBBP and EP300 acetyltransferases in GC B cells and provided direct genetic evidence for the dependence of CREBBP-deficient cells on its paralogue EP300 in vivo and ex vivo. This addiction was maintained in malignant DLBCL cells and could be pharmacologically exploited, thereby offering a proof-of-principle for the specific targeting of EP300 as a promising therapeutic strategy in CREBBP-mutant lymphomas.
The first finding of our study was the ability of CREBBP and EP300 to influence distinct transcriptional programs in the GC through extensive chromatin rewiring at enhancers/super-enhancers, which might explain the opposite phenotype observed in the conditional knockout mouse models. Although their high structural similarity has often led to the assumption of an inter-changeable role between these two proteins, it is becoming increasingly clear that CREBBP and EP300 serve several non-redundant functions in development and cancer. For instance, Crebbp heterozygous mice display reduced self-renewal capacity of hematopoietic stem cells, whereas Ep300 heterozygous mice do not (Rebel et al., 2002). Additionally, mice with point mutations in the Ep300 KIX domain have severe hematopoietic abnormalities, whereas mice with identical mutations in the Crebbp KIX domain are largely normal (Kasper et al., 2002). The results herein conclusively demonstrated that CREBBP and EP300 have divergent effects on GC physiology and defined a set of transcripts that are differentially influenced by these two proteins, suggesting their preferential recruitment of specific transcription factors. These data provide important information on the distinct programs that regulate DZ versus LZ development and can be further explored in the context of GC biology and the DLBCL cell of origin.
The unique roles uncovered for these two enzymes during the physiological GC reaction suggest that CREBBP and EP300 might have evolved to allow the tight yet dynamic regulation of separate functional programs operating in the DZ and LZ compartments, which need to rapidly switch between transcription factor networks sustaining proliferation and somatic hypermutation in the DZ and antigen-driven selection/differentiation in the LZ (De Silva and Klein, 2015; Mesin et al., 2016). This model also suggests that the disrupted balance between the activities of CREBBP and EP300 caused by inactivating mutations of either enzyme could be a major contributor to malignant transformation by perturbing GC homeostasis. Consistent with this notion, recent single-cell analysis of FL cells, which harbor CREBBP/EP300 mutations in as many as 60% of patients, has revealed a de-synchronization of the normal GC transcriptional program characterized by the loss of this cyclic continuum of DZ-LZ transitional states (Milpied et al., 2018). The functional separation of CREBBP and EP300 is also of evolutionary interest when considering that both CREBBP and EP300 are present in mammalian cells, whereas only CREBBP is present in Drosophila (Akimaru et al., 1997; Dancy and Cole, 2015). Given that a major difference between invertebrates and mammals is the lack of a lymphocyte-based adaptive immune system in the former, which rely on innate immunity for protection against pathogens, a divergence between the roles of CREBBP and EP300 might reflect the selective pressure imposed with the appearance of adaptive immunity, including the GC reaction.
In addition to controlling distinct transcriptional programs, our in vivo data indicated that CREBBP and EP300 must have a common program for which they can partially substitute for each other, as combined deletion of both genes was incompatible with GC formation from its very early stages. These findings are in line with a number of previous studies indicating strong counter selection of Crebbp/Ep300 double-deficient cells in other cellular contexts, such as mouse T lymphocytes and immature B lymphocytes (Kasper et al., 2006; Xu et al., 2006). Our approach failed to unequivocally identify critical shared targets that could explain the synthetic lethal effect. This result might not be surprising, as the expression and chromatin pattern of these targets would not be expected to change in the presence of compensatory mechanisms. Furthermore, CREBBP and EP300 acetylate and modulate the activity of numerous non-histone protein substrates, including transcription factors, that might be affected by their reduced dosage and were not investigated in our study. Nonetheless, a number of likely candidates can be suggested among the loci that are bound by CREBBP specifically in GC B cells (Zhang et al., 2017), but did not show significant transcriptional changes upon Crebbp deletion in mice when Ep300 expression was retained. These genes include the GC master regulator BCL6 (Basso and Dalla-Favera, 2012), the transcription factors E2A, MEF2C, and MEF2B; the cell cycle regulator CCND3; and the B cell co-activator POU2AF1/OCAB (Brescia et al., 2018; De Silva and Klein, 2015; Khiem et al., 2008; Kwon et al., 2008; Peled et al., 2010; Schubart et al., 1996; Wilker et al., 2008; Zhang et al., 2017), all of which are required for GC formation. In this context, the redundant functions of CREBBP and EP300 might serve as a safety mechanism to guarantee the expression of proteins that are critical for the GC reaction. This compensatory mechanism appeared to be co-opted by CREBBP-mutant lymphoma cells for their own survival advantage.
Finally, a potentially clinically relevant finding of our studies was the demonstration that EP300 paralogue dependency is maintained in CREBBP-mutant malignant B cells and could be pharmacologically targeted with the use of specific inhibitors directed against this class of acetyltransferases. The differential response observed in mutant vs WT cells suggests the existence of a therapeutic window for the clinical application of these approaches. Importantly, CCS1477 is currently being tested in a phase I clinical trial for the treatment of solid tumors (NCT03568656), and a clinical trial for patients with hematologic malignancies is scheduled to start mid-2019. Therapeutic targeting of these acetyltransferases would be particularly valuable in the context of FL and DLBCL, where CREBBP mutations represent “truncal” events that are already present in the putative common precursor clone. Thus, elimination of CREBBP-mutant cells is expected to lead to the eradication of not only the dominant tumor clone but also the reservoir of initiating cells that are responsible for transformation and relapse.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Laura Pasqualucci (lp171@cumc.columbia.edu). The applicant’s laboratory and institution adhere to the NIH Grants Policy on Sharing of Unique Research Resources.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse models and strains
The conditional Ep300 floxed and Crebbp floxed mouse models have been reported (Kang-Decker et al., 2004; Kasper et al., 2006). Deletion of Ep300 and/or Crebbp was directed to GC B cells by breeding Crebbpfl/+ and Ep300fl/+ mice with the Cγ1Cre/+ deletor strain (Casola et al., 2006) (all backcrossed into C57BL/6 background for at least 6 generations), followed by offspring intercrossing to generate compound mice. Immunological responses were evaluated in immune-competent mice at 3–4 months of age. Both females and males were included in the experiments. Mice were housed in a dedicated pathogen-free environment, and all animal work was performed according to protocols revised and approved by the National Cancer Institute and Columbia University Institutional Animal Care and Use Committee. Genotyping was performed by PCR analysis, and the protocol is available upon request.
Cell lines
The human DLBCL cell lines SUDHL4, U2932, SUDHL5, WSU-DLCL2, and SUDHL16 and their derivatives were grown in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin and 100 μg/ml streptomycin. HEK293T cells (American Type Culture Collection) were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% FCS, 100 U/ml penicillin and 100 mg/ml streptomycin. Cells were maintained at 37°C in humidified incubators under 5% CO2. All cell lines tested negative for Mycoplasma contamination and were verified for identity by STR profiling and/or by analysis of somatic single nucleotide variants as obtained by whole genome sequencing.
METHOD DETAILS
Small molecule CREBBP/EP300 inhibitors
CCS1477, a selective and orally active small molecule inhibitor of the single bromodomain of CREBBP and EP300, was obtained from CellCentric (Pegg et al., 2017). The specific CREBBP/EP300 HAT inhibitor was independently synthesized from example #715 in patent WO 2016/044770 PCT/US2015/051028 (Lasko et al., 2017). Both inhibitors were tested for their on-target activity by immunoblotting analysis of CREBBP/EP300 self-acetylation, H3K18Ac and H3K27Ac, as well as by FACS analysis of HLA-DR and RNA-seq analysis.
Expression vectors and sgRNA design
The inducible Cas9 expression construct pCW-Cas9 was acquired from Addgene (Addgene: 50661) (Wang et al., 2014). The pLKO5-sgRNA-EFS-GFP (Addgene: 57822) and pLKO5-sgRNA-EFS-tRFP (Addgene: 57823) vectors (Heckl et al., 2014) were used to clone sgRNAs targeting early exons and/or active domains of the human CREBBP and EP300 genes, and a neutral control region in the PPP1R12C intron 1, as described (Sanjana et al., 2014; Shalem et al., 2014). SgRNAs (n = at least two each) were designed using the Benchling web tool (http://benchling.com) to have high efficiency and an off-target score above 47 for CREBBP and above 62 for EP300, to minimize off-target effects (Note that due to the homology of CREBBP andEP300, higher off target scores could not be achieved for all sgRNAs). All sgRNA sequences used in this study are provided in Table S5.
Mouse Immunizations
For the analysis of T cell dependent immune responses, age-matched 10 to 16-week-old mice were immunized by intraperitoneal injection of sheep red blood cells (SRBC) (Cocalico Biologicals) (n=500 million/mouse in PBS) and analyzed 10 days post-immunization. To achieve a higher yield of GC B cells (e.g. for sorting and RNA-seq studies), mice were immunized with two sequential injections of SRBC (day 0, 1×108 cells; day 5, 13109 cells) and sacrificed at day 12 for B cell isolation.
Flow cytometric analysis of mouse B cell subsets
Multi-color flow cytometric analysis of the B cell lymphoid compartment was performed at 3 months of age as previously reported (Zhang et al., 2015), using 3–4 mice/genotype/experiment. Briefly, single cell suspensions prepared from lymphoid organs were stained for 20 minutes on ice using different combinations of fluorescent-labeled antibodies (see Key Resources Table for the complete list). Data were acquired on either a FACSCanto™ II or a FACSCalibur™ (BD Biosciences) and analyzed using the FlowJo software (TreeStar). For detection of intracellular proteins, cells were fixed and permeabilized using the BD Cytofix/Cytoperm buffer (BD Biosciences) following the manufacturer’s instructions, and subsequently stained for 60 minutes at room temperature with the appropriate antibodies (Key Resources Table). B cell subpopulations were identified according to established gating strategies (Victora et al., 2010; Zhang et al., 2015; Zhang et al., 2017). To calculate the absolute numbers of cells within splenic B cell subsets, spleen fragments were weighed, and erythrocyte-depleted cell suspensions were counted by Trypan blue exclusion using the Countess Automated Cell Counter (Thermo Fisher Scientific). The total number of counted splenic B cells was then multiplied by the fraction of each subpopulation, as identified by the cytofluorimetric analyses.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-CD45R/B220 (clone RA3-6B2), PercP/Cy5.5-conjugated | BD Biosciences | Cat#103236, RRID: AB_893354 |
| Rat monoclonal anti-CD45R/B220 (clone RA3-6B2), PE-conjugated | BD Biosciences | Cat#561878, RRID: AB_10893353 |
| Rat monoclonal anti-CD45R/B220 (clone RA3-6B2), Biotin-conjugated | BD Biosciences | Cat#553086; RRID: AB_394616 |
| Mouse monoclonal anti-BCL6, Alexa Fluo 647-conjugated | BD Biosciences | Cat#561525, RRID: AB_10898007 |
| Rat monoclonal anti-IgG1 (clone X56), APC-conjugated | BD Biosciences | Cat#550874, RRID: AB_398470 |
| Rat monoclonal anti-IgM (clone 11/41), APC-conjugated | BD Biosciences | Cat#550676, RRID: AB_398464 |
| Rat monoclonal anti-IgD, VioGreen-conjugated | Miltenyi Biotech | Cat#130-103-005, RRID: AB_2659783 |
| Rat monoclonal anti CD93 (Clone AA4.1), PE-conjugated | BioLegend | Cat#136503, RRID: AB_1967094 |
| Rat monoclonal anti CD23 (Clone B3B4), PE/Cy7-conjugated | BioLegend | Cat#101613, RRID: AB_2103037 |
| Rat monoclonal anti I-A/I-E antibody (Clone M5/114.15.2), APC/Cy7-conjugated | BioLegend | Cat#107628, RRID: AB_2069377 |
| Hamster monoclonal anti-CD95 (clone Jo2), PE/Cy7-conjugated | BD Biosciences | Cat#557653,RRID: AB_396768 |
| Mouse monoclonal anti-CD95 (clone Jo2), PE-conjugated | BD Biosciences | Cat#554258, RRID: AB_395330 |
| Anti-PNA, Biotin-conjugated | Vector Laboratories | Cat#B-1075,RRID: AB_2313597 |
| Anti-PNA, FITC-conjugated | Vector Laboratories | Cat#FL-1071,RRID: AB_2315097 |
| Rat monoclonal anti-CD86 (clone B7-2), APC-conjugated | Thermo Fisher Scientific | Cat#17-0862-82,RRID: AB_469419 |
| Rat monoclonal anti-CD184 (CXCR4), PerCP eFluor 710-conjugated | Thermo Fisher Scientific | Cat#46-9991-80,RRID: AB_10670192 |
| Rat monoclonal anti-CD19 (clone 1D3), FITC-conjugated | BD Biosciences | Cat#553785, RRID: AB_395049 |
| Rat monoclonal anti-CD138 (Syndecan 1) (clone 281-2), PE-conjugated | BioLegend | Cat#142504, RRID: AB_10916119 |
| Annexin V Antibody, FITC-conjugated | BD Biosciences | Cat#556419, RRID: AB_2665412 |
| Mouse monoclonal anti-HLA-DR (Clone L243), Alexa-Fluor 700-conjugated | BioLegend | Cat#307626, RRID: AB_493771 |
| Rabbit polyclonal anti-BCL6 (N3 clone) | Santa Cruz Biotechnology | Cat#sc-858,RRID: AB_2063450 |
| Rabbit monoclonal anti-CD3 (clone SP7) | Lab Vision | Cat#RM-9107-S1, RRID: AB_149924 |
| Rabbit polyclonal anti-H3K27Ac | Diagenode | Cat#C15410196, RRID: AB_2637079 |
| Rabbit polyclonal anti-Histone H3 (acetyl K27ac) antibody | Abcam | Cat#ab4729, RRID: AB_2118291 |
| Rabbit polyclonal Histone H3 (acetyl K18) antibody | Abcam | Cat#ab1191, RRID: AB_298692 |
| Rabbit monoclonal anti-Histone H3 (D1H2) | Abcam | Cat#4499, RRID: AB_10544537 |
| Rabbit polyclonal anti-CREBBP (Clone C-20) | Santa Cruz Biotechnology | Cat#sc-583, RRID: AB_2245237 |
| Rabbit polyclonal anti-CREBBP (Clone A-22) | Santa Cruz Biotechnology | Cat#sc-369, RRID: AB_631006 |
| Rabbit polyclonal anti-Ep300 (Clone C-20) | Santa Cruz Biotechnology | Cat#sc-585, RRID: AB_2231120 |
| Rabbit polyclonal anti-Ep300 (Clone N15) | Santa Cruz Biotechnology | Cat#sc-584, RRID: AB_2293429 |
| Rabbit monoclonal anti-EP300 (D2X6N) | Cell Signaling Technology | Cat#54062, RRID: AB_2799450 |
| Rabbit polyclonal Acetylated-Lysine Antibody, unconjugated | Cell Signaling Technology | Cat#9441, RRID: AB_331805 |
| Mouse monoclonal CRISPR/Cas9 Antibody (Clone 7A9), unconjugated | EpiGentek | Cat#A-9000-010,N/A |
| Rat monoclonal anti-Mouse Ig, HRP-conjugated (TrueBlot) | Rockland | Cat#18-8817-33; RRID: AB_2610851 |
| Donkey anti-Rabbit IgG, HRP-conjugated | GE Healthcare | Cat#NA934;RRID: AB_772206 |
| Goat polyclonal anti-Rabbit IgG (H+L), HRP-conjugated | Thermo Fisher Scientific | Cat#31460, RRID: AB_228341 |
| Mouse monoclonal anti-Rabbit IgG, HRP-conjugated (TrueBlot) | Rockland | Cat#18-8816-33, RRID: AB_2610848 |
| Goat anti-Rabbit IgG, EnVision HRP-conjugated | Agilent | Cat#K4003, RRID: AB_2630375 |
| Rat absorbed, made in horse anti-mouse IgG (H+L), biotinylated | Vector Laboratories | Cat#BA-2001, RRID: AB_2336180 |
| TSA Cyanine 3 System - antibody amplification kit | PerkinElmer | Cat#NEL704A001KT, RRID: AB_2572409 |
| Armenian Hamster Anti-CD40 Monoclonal Antibody, Unconjugated, Clone HM40-3 | BD Biosciences | Cat#553721, RRID: AB_395006 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Doxycyclin | Sigma-Aldrich | D9891-25G |
| CCS1477 | Pegg et al., 2017 | N/A |
| CU329 | WO 2016/044770 PCT/US2015/ | N/A |
| Sheep Red Blood Cells in Alsevere’s | Cocalico Biologicals | Cat#20-1334A |
| NP-KLH (Keyhole Limpet Hemocyanin) | Biosearch Technologies | Cat#N-5060-5 |
| Recombinant mouse IL-4 | R&D Systems | Cat#404-ML-010 |
| TRIzol Reagent | Life Technologies | Cat#15596-018 |
| Dynabeads Protein A | Novex | Cat#10002D |
| Lipopolysaccharides | Sigma-Aldrich | Cat#L2630 |
| Streptavidin, Cy3-conjugated | Molecular Probes | Cat#43-4315 |
| Streptavidin, Alkaline Phosphatase (AP)-conjugated | Vector Laboratories | Cat#SA-5100 |
| 3-Amino-9-ethylcarbazole (AEC), tablets | Sigma-Aldrich | Cat#A6926 |
| NBT/BCIP Stock Solution | Roche | Cat#11681451001 |
| Prolong Gold Anti-Fade Mountant with DAPI | Molecular Probes | Cat#P36935 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dilactate) | BioLegend | Cat#422801 |
| Proteinase Inhibitor Cocktail | Sigma-Aldrich | Cat#P8340 |
| Quant-iT PicoGreen dsDNA Reagent | Life Technologies | Cat#P11496 |
| Critical Commercial Assays | ||
| Cytofix/Cytoperm buffer | BD Biosciences | Cat#554714 |
| CellTrace Violet cell proliferation kit | Thermo Fisher Scientific | Cat#C34557 |
| Mouse B Cell Isolation kit | Miltenyi Biotec | Cat#130-090-862 |
| CellTiter-Glo Luminescent cell viability assay | Promega | Cat#PR-G7572 |
| Pierce ECL Western Blotting Substrate | Thermo Fisher Scientific | Cat#PI32106 |
| RNeasy Mini Kit | Qiagen | Cat#74104 |
| Nucleospin RNA XS | Macherey-Nagel | Cat#740902.10 |
| Superscript First Strand Synthesis System for RT-PCR | Life Technologies | Cat#11904-018 |
| TruSeq RNA Library Preparation Kit v2 (Illumina) | Illumina | Cat#RS-122-2001 |
| TruChIP High Cell Chromatin Shearing Kit | Covaris | Cat#PN520154 |
| Agencourt AMPure XP beads (Protein A magnetic beads) | Beckman Coulter | Cat#A63881 |
| MiniElute Reaction Cleanup Kit | Qiagen | Cat#28204 |
| KAPA SYBR FAST Universal qPCR Kit | KAPA Biosystems | Cat#KK5503KK4824 |
| Deposited Data | ||
| Raw and analyzed RNA-seq data | This paper | GEO: GSE110669 |
| Raw and analyzed ChIP-seq data | This paper | GEO: GSE132365 |
| Experimental Models: Cell Lines | ||
| Human: SU-DHL-4 | ATCC | Cat#CRL-2957, RRID: CVCL_0539 |
| Human: SU-DHL-5 | DSMZ | Cat#ACC-633,RRID: CVCL_1896 |
| Human: SU-DHL-16 | DSMZ | Cat#ACC-539,RRID: CVCL_1168 |
| Human: U2932 | DSMZ | Cat#ACC-633, RRID: CVCL_1896 |
| Human: WSU-DLCL2 | DSMZ | Cat#ACC-575,RRID: CVCL 1902 |
| Human: HEK293T | ATCC | Cat#CRL-3216,RRID: CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Crebbpfl/+ | Kang-Decker et al, 2004 | N/A |
| Mouse: Ep300fl/+ | Kasper et al, 2006 | N/A |
| Mouse: Cγ1cre/+ | Casola et al., 2006 | N/A |
| Oligonucleotides | ||
| mEp300_Exon8_F: AGTGAAAATGCTGGTGTGGC | This paper | N/A |
| mEp300_Exon11_R: TAGACGGGTCAGGTACAGGA | This paper | N/A |
| sgEP300_E9:CCGGCGTAGGAAATATGGCT | This paper | N/A |
| sgEP300_E17:GGGTCCACAGGTTGACGAAA | This paper | N/A |
| sgEP300_E24:TCATGCTTCTGACAAAACCG | This paper | N/A |
| sgCREBBP_E13:TGTGCACCCATCATGTTCGG | This paper | N/A |
| sgCREBBP_E20:CAGACGTAAGTACCGTCCTG | This paper | N/A |
| sgPPP1R12C_4 (also called Neutral-4):CCAGCGAG TGAAGACGGCAT | This paper | N/A |
| sgPPP1R12C_5 (also called Neutral-5):AGGGAGAC ATCCGTCGGAGA | This paper | N/A |
| Recombinant DNA | ||
| pCW-Cas9 | Wang et al., 2014 | addgene: 50661; RRID: Addgene_50661 |
| pLKO5-sgRNA-EFS-GFP | Heckl et al., 2014 | addgene: 57822; RRID: Addgene_57822 |
| pLKO5-sgRNA-EFS-tRFP | Heckl et al., 2014 | addgene: 57823; RRID: Addgene_57823 |
| pCMV-VSVg | Stuart et al., 2003 | addgene: 8454; RRID: Addgene_8454 |
| pCMV-Δ8.91 | Dr. J. Luban (University of Massachusetts Medical School) |
N/A |
| pLKO5-sgEP300-E9-EFS-GFP | This paper | N/A |
| pLKO5-sgEP300-E9-EFS-tRFP | This paper | N/A |
| pLKO5-sgEP300-E17-EFS-GFP | This paper | N/A |
| pLKO5-sgEP300-E17-EFS-tRFP | This paper | N/A |
| pLKO5-sgEP300-E24-EFS-GFP | This paper | N/A |
| pLKO5-sgEP300-E24-EFS-tRFP | This paper | N/A |
| pLKO5-sgPPP1R12C_4-EFS-GFP | This paper | N/A |
| pLKO5-sgPPP1R12C_4-EFS-tRFP | This paper | N/A |
| pLKO5-sgPPP1R12C_5-EFS-GFP | This paper | N/A |
| pLKO5-sgPPP1R12C_5-EFS-tRFP | This paper | N/A |
| pLKO5-sgCREBBP_E13-EFS-tRFP | This paper | N/A |
| pLKO5-sgCREBBP_E20-EFS-tRFP | This paper | N/A |
| Softwares and Algorithms | ||
| FlowJo (v.10.4.0) | TreeStar | https://www.flowjo.com |
| IMGT/HighV-Quest | Lefranc et al., 2011 | http://www.imgt.org |
| GraphPad Prism (v.6.0) | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | Schindelin et al., 2015 | http://imagej.nih.gov/ij/ |
| NIS Elements software | Nikon | https://www.nikoninstruments.com/Products/Software |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| ChIPseeqer (v2.0) | Giannopoulou and Elemento, 2011 | http://physiology.med.cornell.edu/faculty/elemento/lab/chipseq.shtml |
| ROSE | Loven et al, 2013 | https://bitbucket.org/young_computation/rose |
| SAMtools (v0.1.19) | Li and Durbin, 2009 | http://samtools.sourceforge.net/ |
| FeatureCounts | Liao et al., 2014 | http://bioinf.wehi.edu.au/featureCounts/ |
| DEseq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Hisat2 | Pertea et al. 2016 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| TIDE | Brinkman et al., 2014 | https://tide.deskgen.com/ |
| CRISP-ID | Dehairs et al., 2016 | http://crispid.gbiomed.kuleuven.be/ |
| Hisat2 | Pertea et al. 2016 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| GSEA (v2.2.0) | Subramanian et al., 2005 | https://software.broadinstitute.org/gsea/index.jsp |
| NCBI Homologene database (2016) | NCBI | ftp://ftp.ncbi.nih.gov/pub/HomoloGene/ |
| NCBI Gene database (2016) | NCBI | ftp://ftp.ncbi.nih.gov/gene/ |
| DAVID (v6.7) | DAVID | https://david.ncifcrf.gov/ |
| Adobe Photoshop (v10.0) | Adobe | Adobe Photoshop, RRID: SCR_014199 |
| VENNY 2.1 | Oliveros, JC (2007-2015) | http://bioinfogp.cnb.csic.es/tools/venny/index.html |
| MSigDB v6.2 | MSigDB | http://software.broadinstitute.org/gsea/msigdb/annotate.jsp |
| Extended GSEA | Lim et al., 2009 | https://github.com/antonybholmes/libgsea |
Immunofluorescence analysis
Double-immunofluorescence analysis of Crebbp or Ep300 and PNA was performed on formalin-fixed paraffin-embedded (FFPE) material from mouse spleens. Serial sections (3mm-thick) were stained using a combination of two anti-CREBBP antibody (each at 1:400 dilution) (A22 and C20, rabbit polyclonal, Santa Cruz Biotechnologies) or an anti-EP300 antibody (1:400) (N15, rabbit polyclonal, Santa Cruz Biotechnologies) and biotinylated PNA (1:300 dilution) (Vector Laboratories, cat#B-1075). Detection of CREBBP and EP300 was obtained using the EnVision System–HRP-Rabbit antibody (Dako) followed by Tyramide Signal Amplification system (PerkinElmer); NeutrAvidin®, FITC conjugated (cat#A2662, Invitrogen) was used at 1:300 dilution to detect PNA. Images were captured using a Nikon Eclipse microscope and the NIS Elements software (Nikon). All images were colored, resized, and merged using Adobe Photoshop (version 10.0).
Histological and immunohistochemical analysis of mouse tissues
Histological analysis of mouse lymphoid organs was performed on 3-μm-thick FFPE tissue sections, stained with Hematoxylin & Eosin (Thermo Scientific) following standard procedures. The following primary antibodies were used for immunohistochemical analysis: anti-Bcl6 (1:300) (N3, rabbit polyclonal, Santa Cruz Biotechnology); biotin-conjugated anti-PNA (1:200) (Vector Laboratories); biotin-conjugated anti-B220 (1:400) (RA3–6B2, rat monoclonal, Pharmingen 553086) and anti-CD3 (1:800) (SP7, rabbit monoclonal, NeoMarkers RM9107). GC numbers, size, and overall area were calculated using the ImageJ software on scanned images obtained with a Leica SCN400 slide scanner (Schindelin et al., 2015).
Ex vivo stimulation of splenic B lymphocytes with anti-CD40 and IL4
Splenic murine B-cells were isolated from age-matched 10- to 16-week old mice using the mouse B-cell isolation kit (Miltenyi Biotec), according to the manufacturer’s instructions, plated at 2 × 106 cells/ml, and cultured for 4 days in RPMI 1640 medium supplemented with 15% FBS, 55 μM β-mercaptoethanol, 50ng/ml recombinant mouse IL4 (R&D systems) and 1μg/ml Hamster monoclonal anti-CD40 antibody (clone HM40–3, BD Pharmingen). Cells were harvested at days 2, 3, and 4 and processed for flow cytometric analysis and RNA or protein isolation.
Cell viability and proliferation assays
Cell proliferation was analyzed with the CellTrace™ Violet Cell Proliferation Kit (Life Technologies), which monitors distinct generations of proliferating cells by a fluorescent dye dilution. Data were acquired at day 2–4 from stimulation on a FACSCanto™ II (BD Biosciences) flow cytometer with 405 nm excitation and an emission filter in the 450nm range. Experiments were performed at least twice. To assess cell viability and proliferation, the CellTiter-Glo® Luminescent Cell Viability Assay (Promega) was used according to the manufacturer’s instructions. Specifically, cells were plated in triplicates in 96 well plates at 7,500 cells/well and CellTiter-Glo® readings were acquired after 2h, 48h, 72h and 96h. The raw data were normalized to the reading at 2 h and fold changes were plotted using Prism v.5.0 (GraphPad Software, San Diego, CA). The viability of ex vivo cultured mouse B cells was also determined based on analysis of forward vs side scatter plots.
Assessment of apoptosis
To assess apoptosis, cells were stained with AnnexinV-FITC (BD Pharmingen, Cat51–658741) and 7AAD (BD Pharmingen, Cat 51–2359KC) at day 3 and 4 after stimulation with CD40 and IL4, according to standard protocols. Data were acquired on a FACSCanto™ II or a FACSCalibur™ (BD Biosciences) and analyzed in FlowJo (TreeStar).
RNA extraction, cDNA synthesis and real-time PCR
Total RNA was extracted from sorted murine GC B cells using the NucleoSpin XS kit (Marchery-Nagel), and from SUDHL4 cells exposed for 48hrs to DMSO (0.1%), CCS1477 (1 μM), or CU329 (0.1 μM) using the TRIzol reagent (Invitrogen), as per manufacturer’s instructions. cDNA synthesis was performed using the SuperScript® First-Strand Synthesis System (Life Technologies). Oligonucleotides annealing to exon 8 (AGTGAAAATGCTGGTGTGGC) and exon 11 (TAGACGGGTCAGGTACAGGA) of the murine Ep300 locus were used to determine the relative abundance of the Ep300 mRNA before and after Cre-mediated recombination (see Figure S1A).
RNA-seq analysis
The transcriptional profiles of Crebbp-null and Ep300-null mouse GC B cells were obtained using total RNA extracted from sorted B220+CD95+PNAhi splenocytes as described above (n = 3 animals/genotype, and 5 wild-type littermate controls) and verified for integrity on a BioAnalyzer 2100 (Agilent). Samples (100–200 ng each) with RNA integrity numbers (RIN) >9 were processed to generate RNA-seq libraries using the TruSeq RNA Library Preparation Kit v2 (Illumina). Sequencing was performed on an Illumina NOVASeq 6000 or HiSeq 4000 instrument using a paired-end 150 bp protocol. RNA-seq reads were mapped to the Mus musculus (mm10/GRCm38) or the Homo sapiens (GRCh38) genome assembly using the hisat2 prebuilt genome index (Pertea et al., 2016). Genome-mapped reads were aligned to exons on the mm10 (or GRCh38) transcriptome reference (Harrow et al., 2006) based on the information in the genomic BAM files, using featureCounts (Liao et al., 2014) to produce abundance tables. We sanitized the transcriptome references (exon only) by removing read-through genes, anti-sense elements, miRNA, and rRNA. The count tables were subsequently normalized to produce transcript per million (TPM) tables. Differentially expressed genes were determined using the DESeq2 software with the following filters: FDR < 0.05 (after Benjamini-Hochberg correction) and absolute fold change (FC) ≥ 1.2 (for data generated from the heterogeneous mouse GC B cell population) and ≥ 2 (for data generated in the SUDHL4 cell line).
Gene set enrichment analysis and extended GSEA
Transcriptomic data generated from Crebbp-null and Ep300-null murine GC B cells and from treated SUDHL4 cells were analyzed for enrichment in pre-defined sets of genes using the GSEA software tool 2–2.2.0 (Subramanian et al., 2005) on log2 (TPM+1) transformed data, with 1000 gene set permutations and the Canonical Pathway (C2) Molecular Signature Database v6.2 gene sets collection (http://software.broadinstitute.org/gsea/msigdb/), as well as the gene sets available in the SignatureDB (https://lymphochip.nih.gov/signaturedb/). To interrogate the enrichment of LZ- and DZ-upregulated genes in the rank of genes differentially expressed between CrebbpKO and CrebbpWT or Ep300KO and Ep300WT GC B cells, we used the list of genes differentially expressed in DZ vs LZ subpopulations, identified as described in (Victora et al., 2012; Zhang et al., 2015). The false discovery rate cutoff was set at <0.05. Cross-comparison of the signatures obtained in the two genetic backgrounds (Figure S2E) was performed by an extended GSEA approach as described (Lim et al., 2009). To verify the on-target activity of the HAT and BRD inhibitors and the phenotype of the isogenic CREBBP−/− SUDHL4 clones, we used the list of genes identified as downregulated in Crebbpflfl and Ep300fl/flCγ1Cre/+ GC B cells and reported in Table S1.
Pathway enrichment analysis
To determine whether genes down-regulated in Crebbp- or Ep300-null GC B cells were enriched in annotated functional categories, we used a hypergeometric test assessing the significance of the overlap between the list of significantly down-regulated genes, as obtained by supervised analysis of transcriptomic data (FDR<0.05, and FC ≥ 1.2), and pre-defined gene collection lists (C2, C6, C7) provided in the MSigDB GSEA page (http://software.broadinstitute.org/gsea/msigdb/annotate.jsp) or in the SignatureDB webpage (https://lymphochip.nih.gov/signaturedb/). Only pathways with a significant P value (<0.05 after Benjamini-Hochberg correction) were retained. Pathway enrichment analysis was also performed using the DAVID 6.8 tool and KEGG, Biocarta, and Reactome databases (Huang et al., 2009), giving analogous results. The same approach was used to identify biological signatures and signaling pathways preferentially enriched in SUDHL4 cells treated with CCS1477 or CU329.
Chromatin Immuno precipitation and sequencing (ChIP-Seq)
Chromatin Immunoprecipitation (ChIP) was performed on 15 million cells/sample as previously described (Zhang et al., 2017). Briefly, cells were cross-linked with 1% formaldehyde for 10 min at RT, quenched by the addition of glycine to a final concentration of 0.125 M, and frozen. The TruChIP High Cell Chromatin Shearing Kit with SDS (Covaris) was used for cell lysis and nuclei isolation, followed by sonication in an S220 ultrasonicator (Covaris, Woburn, MA) to a chromatin fragment size distribution of 200–500 bp. Sheared chromatin was incubated overnight with 4μg of anti-H3K27Ac antibody (Diagenode, cat#A1723–0041D). The immune-complexes were collected with protein A magnetic beads over a 4 hr incubation and washed sequentially at increasing stringency before reverse cross-linking. Following RNAse and proteinase K treatment, DNA fragments were purified using the MiniElute Reaction Clean Up Kit (Qiagen) and quantified by Quant-iT PicoGreen dsDNA Reagent (Life Technologies). The specificity of the antibodies against H3K27 acetylation has been extensively documented (not shown). Barcoded ChIP-seq libraries were constructed starting from 3 ng of immunoprecipitated or input DNA as reported (Zhang et al., 2015), quantified using the KAPA SYBR FAST Universal qPCR Kit (KAPA Biosystems), normalized to 15nM, and pooled for sequencing on an Illumina HiSeq 4000 instrument as paired-end 150 bp reads,obtainingonaverage25 × 106reads/sample. All experiments were performed in two biological replicates (two independent clones each for WT, CREBBP−/−, and EP300−/− SUDHL4 cells).
ChIP-seq analysis
Sequencing data were processed according to the default Illumina pipeline using Casava V1.8. Raw reads were mapped to the human genome GRC37 assembly using the Bowtie2 aligner v2.1.0 (Langmead and Salzberg, 2012), allowing up to two mismatches. Duplicate reads (i.e., reads of identical length mapping to exactly the same genomic locations) were removed with SAM tools v0.1.19 using the rmdup option (Li et al., 2009), and the remaining reads were normalized to total reads aligned and displayed as read counts per million mapped reads. Peaks were identified using ChIPseeqer v2.0 (Giannopoulou and Elemento, 2011), enforcing a minimum fold change of 2 between ChIP and input reads, a minimum peak width of 100 bp, and a minimum distance of 100 bp between peaks. The p value threshold for statistical significance of peaks was set at 10−15, and peaks overlapping with Encode Blacklist or an internal manually curated signal artifact blacklist were discarded. Only peaks (regions) detected in both biological replicates (i.e. overlapping peaks) were considered in downstream analyses. H3K27Ac peaks located within ±12.5 kb were subject to stitching unless mapping around a TSS (±2 kb). Unsupervised hierarchical clustering of H3K27Ac regions was performed using the union of E/SEs identified by ROSE in the three genetic backgrounds. To identify differentially acetylated regions in WT, CREBBPKO and EP300KO cells, we applied the DESeq2 algorithm (Love et al., 2014) to data obtained from the three isogenic cell lines (n=2 clones/each) using the list of H3K27Ac regions detected in SUDHL4-WT cells. Differentially acetylated regions were identified as those with absolute [log2 transformed H3K27ac counts] fold-change ≥ 2 and Benjamini-Hochberg adjusted P-value < 0.05. ChIP-Seq data have been deposited in the GEO database under accession number GSE132365.
Functional annotation of H3K27Ac-marked regions
Significantly enriched H3K27Ac peaks were annotated as promoters if located within 2 kb from the transcription start site (TSS) of an annotated gene, and intragenic or intergenic if distal to a TSS, using the GRCh37 assembly. Active enhancers and super-enhancers were defined by ROSE as published (Whyte et al., 2013). In brief, ROSE identifies enhancers as all H3K27Ac peaks that do not overlap with known gene promoters (±2 kb from TSS), after concatenating those located within ±12.5 kb from each other, and then ranks them by their input-subtracted H3K27Ac signal. The cut-point between enhancers and super-enhancers was defined on the enrichment profile as the inflection point of H3K27Ac signal intensity versus concatenated enhancer rank. H3K27Ac peaks located within ±2 kb from a TSS were assigned to promoters.
Assignment of active enhancers/super-enhancers to genes
H3K27Ac-marked enhancers and super-enhancers, identified as described above, were assigned to the nearest transcriptionally active gene (distance from enhancer center to TSS) as the most likely candidate target (Zhang et al., 2015).
Virus production and lentiviral transduction of DLBCL cell lines
The SUDHL4, U2932 SUDHL5, WSU-DLCL2 and SUDHL16 cell lines were engineered to express an inducible Cas9 (iCas9) by lentiviral transduction. Briefly, lentiviral particles were generated by co-transfecting HEK293T cells with pCW-Cas9, pVSVg and psΔ8.9 plasmids in a 4:1:3 ratio using a standard Calcium Phosphate transfection protocol (Zhang et al., 2017). Viral supernatants, collected between 36h and 72h post-transfection, were filtered through 0.45mM-pore-size nitrocellulose membranes and used to transduce DLBCL cell lines following a spinoculation procedure. Successfully transduced cells were then selected in complete IMDM medium containing puromycin (1mg/ml) to obtain single cell–derived clones, which were screened for high levels of Cas9 expression at 48h, 72h, and 96h from doxycycline induction by immunoblot analysis with an anti-CRISPR/Cas9 antibody (mouse monoclonal 7A9, EpiGentek, A-9000). Two independent clones displaying similar Cas9 induction efficiency were selected for delivery of lentiviral vectors carrying the sgRNA of interest, which was obtained as described above. GFP and/or RFP-positive populations were isolated in a SH800 cell sorter (Sony Biotechnology) and used for cell competition assays or single cell plating (n = at least 96/sgRNA) in order to determine the percentage of recovered clones and to isolate individual EP300-null (or CREBBP-null) clones. Induction of Cas9 expression and disruption of the target gene were verified in the bulk population at day 3 post-induction by immunoblotting and PCR amplification/sequencing of the edited site, followed by TIDE analysis, a specifically developed decomposition algorithm that uses sequence traces to identify the major induced mutations in the predicted editing site and accurately determine their frequency in a cell population (Brinkman et al., 2014). Single clones were individually analyzed for CREBBP or EP300 editing by PCR amplification and direct sequencing, followed by inspection of the chromatograms both manually and using the Crisp-ID tool (http://crispid.gbiomed.kuleuven.be/) (Dehairs et al., 2016). Clones carrying bi-allelic frameshift mutations were expanded and deletion of CREBBP or EP300 was confirmed by immunoblotting.
Protein Extraction and Immunoblot analysis
Whole cell extracts were obtained from purified mouse B cells or human cell lines in log phase of growth using NP-40 lysis buffer according to a previously described protocol (Bereshchenko et al., 2002). Histones were extracted using an acid extraction method; briefly, chromatin pellets were resuspended in 0.2N HCl, incubated overnight at 4°C, and cleared by centrifugation at 12,000 r.p.m. for 10 min. Protein extracts were resolved on NuPAGE Tris-acetate 3%–8% gels (for CREBBP and EP300) or Tris-glycine 4%–20% gels (for histone H3) (Life Technologies) and transferred to nitrocellulose membranes (GE Healthcare) according to the manufacturer’s instructions. Antibodies used were: rabbit monoclonal anti-CREBBP (D6C5, Cell Signaling), rabbit monoclonal anti-EP300 (D2X6N, Cell signaling), rabbit polyclonal anti-acetylated Lysine (9441, Cell signaling), mouse monoclonal anti-CRISPR/Cas9 antibody (7A9, EpiGentek, A-9000), mouse monoclonal anti-ß-Actin (A5441, Sigma-Aldrich), mouse monoclonal anti-α-tubulin (clone B512, Sigma-Aldrich), rabbit polyclonal antiH3K18Ac (Abcam, cat#ab1191) and anti-H3K27Ac (Abcam, cat#ab4729), and rabbit monoclonal anti-Histone H3 (clone D1H2, Cell Signaling Technology). Quantification of signal intensity was obtained in the ImageJ software, and values are expressed as fold differences relative to the wild-type protein sample, set at 1, after normalization for the loading control.
Growth competition assays
For competition assays, sorted RFP+ (sgEp300-transduced; n = 3 independent pools, obtained using 3 different sgRNAs) and GFP+ (sgNeutral-transduced, n = 2 independent pools, obtained using 2 different sgRNAs) populations from iCas9 isogenic DLBCL cell lines (SUDHL4, U2932, SUDHL5, WSU-DLCL2 and SUDHL16) were allowed to recover for 48h, mixed in equal numbers, and seeded at 0.5 3 106 cells/ml in a 48 well plate, followed by treatment with Doxycycline (1 μg/ml at d0 and d2) to induce Cas9 expression, or vehicle as control. The ratio of RFP+ vs GFP+ cells was monitored at d3 and then every 48h on a FACSCanto™ II (BD Biosciences), and data were analyzed using the FlowJo software (TreeStar) to calculate depletion of the RFP+ cells relativeto d0(dN %RFP+/d0%RFP+). All experiments were performed at least twice. Competition assays were also performed on isogenic CREBBPWT and CREBBPKO SUDHL4 clones, generated as described in the previous section and subsequently transduced with pLKO5-sgEP300-EFS-tRFP (or -tGFP) (n = 3 independent pools using 3 different sgRNAs), and pLKO5-sgNeutral-EFS-tRFP (or -tGFP) as controls (n = 2 independent pools using 2 different sgRNAs). Sorted cells were allowed to recover for 48–96h, and experiments were performed according to the protocol described for the native DLBCL cell lines, using the ratio of double positive vs single positive cells as a readout for fitness of the EP300-deleted cells.
Cell viability and drug dose response assays
The response of isogenic CREBBPWT and CREBBPKO SUDHL4 cells (n = 4 clones each) to increasing drug concentrations (range: 10 μM-0.002μM) was determined after 48h incubation, using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega), according to the manufacturer’s instructions. Specifically, cells were plated at 500 cells/well in 50ml complete IMDM in a 384-well plate (Greiner, USA scientific, FL) using a Matrix WellMate microplate dispenser (Thermo Fisher Scientific). After 24 h, the CCS1477 and HATi329 compounds were added in 4 replicates using a HP D300 digital dispenser (HP), with 12 wells of DMSO-treated control cells. CellTiter-Glo® readings were acquired after 48 h using a Tecan Infinite 200 plate reader. The raw data for all compounds were normalized to the average of the DMSO-treated control wells. Dose response curves were generated by plotting the resulting values against the log of the concentration of the inhibitor, using the Prism v5.0 function log(Inhibitor) vs Response with top plateau set to 1. The Area under the curve (AUC) was calculated in Prism v5.0. For the longer time-courses, the same clones were plated at 0.5×106 cells/ml in complete IMDM medium (300 μl/well of a 48-well plate), and fixed concentrations of the CREBBP/EP300 small molecule inhibitors, selected based on the dose-response curve (CCS1477, 0.1μM; HATi329, 0.05mM), were added in triplicate after 6 h, with DMSO as control. Cell number and viability were assessed every 48 h for 8 days using Countess® counting chambers and Trypan blue to distinguish live from dead cells. DMSO treated cells were split every second day at 1:3 ratio, whereas CCS1477 or CU329 treated cells were split according to the CREBBPWT cell growth, specifically 1:3 at d2 and 1:2 at d4.
Cell cycle analysis
Cell pellets were fixed using 70% ethanol at 4°C for at least 1h and stained with DAPI (1 μg/ml) for 30min at 4°C in the presence of RNase A and 0.05% Triton X-100. Cell cycle distribution was measured using a FACSCanto™ II flow cytometer (BD Biosciences), and data were analyzed using the FlowJo (Treestar) software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
To assess statistically significant differences between groups, p values were calculated with the Student’s t test, Fisher’s exact test, or two-way ANOVA with Bonferroni post-test in the Graphpad Prism v5.0 software, unless described otherwise. Mutual exclusivity of CREBBP and EP300 mutations in human FL and DLBCL was computed using CoMEt (Leiserson et al., 2015). Results were considered statistically significant at P<0.05.
DATA AND CODE AVAILABILITY
Data Availability
The raw RNA-seq data from the Crebbpfl/fl and Ep300fl/flCγ1Cre/+ murine GC B cells were deposited in the GEO database under accession number GSE124192. H3K27Ac ChIP-Seq data are available in GEO as series GEO: GSE132365.
Supplementary Material
Highlights.
CREBBP and EP300 control distinct as well as shared transcriptional targets in the GC
Deletion of Crebbp and Ep300 in B cells abrogates GC formation, revealing paralog lethality
CREBBP-mutant DLBCL cells are preferentially sensitive to EP300 deletion
EP300-dependency can be pharmacologically targeted by CREBBP and EP300 inhibitors
ACKNOWLEDGEMENTS
We would like to thank T. Mo for assistance with the mouse colony maintenance; H. Tang for technical support; R. Kumar for bioinformatics support with the genomic validation of DLBCL cell lines; F. Bosch for critically reading the manuscript; N. Compagno for the generation of the neutral sgRNA control; Izabela Krupska at the JP Sulzberger Genome Center of Columbia University and Genewiz, Inc. for help with high-throughput sequencing; and Charles Karan at the High-Throughput Screening Facility of the Herbert Irving Comprehensive Cancer Center for help with the drug treatment assays. We also thank K. Rajewsky (Max Delbrück Center for Molecular Medicine, Berlin, Germany) for the Cγ1Cre/+ mice and Paul Brindle (Department of Biochemistry, St. Jude Children’s Research Hospital, 332 N. Lauderdale, Memphis, TN 38105, USA) for the Crebbp and Ep300 floxed mice. This study was supported by grants from the National Institutes of Health (NIH) RO1-CA172492 to L.P. and P50-CA192937 to R.D.F., a Sanofi Innovator Award to L.P., and a Leukemia & Lymphoma Society Translational Research Project award to L.P. This research was also funded in part through the NIH/NCI Cancer Center Support Grant P30CA013696 and used the Flow Cytometry and the Genomics and High-Throughput Screening Shared Resources. S. Meyer is a fellow of the Swiss National Science Foundation.
Footnotes
DECLARATION OF INTEREST
L. Pasqualucci was a recipient of a Sanofi innovation award. N. Brooks is a full-time employee and stockholder at CellCentric.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2019.08.006.
REFERENCES
- Akimaru H, Chen Y, Dai P, Hou DX, Nonaka M, Smolik SM, Armstrong S, Goodman RH, and Ishii S. (1997). Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature 386, 735–738. [DOI] [PubMed] [Google Scholar]
- Arthur SE, Jiang A, Grande BM, Alcaide M, Cojocaru R, Rushton CK, Mottok A, Hilton LK, Lat PK, Zhao EY, et al. (2018). Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat. Commun 9, 4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, and Kelly K. (1997). Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89, 1175–1184. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, and Kouzarides T. (1996). The CBP co-activator is a histone acetyltransferase. Nature 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Bedford DC, Kasper LH, Fukuyama T, and Brindle PK (2010). Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics 5, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereshchenko OR, Gu W, and Dalla-Favera R. (2002). Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet 32, 606–613. [DOI] [PubMed] [Google Scholar]
- Brescia P, Schneider C, Holmes AB, Shen Q, Hussein S, Pasqualucci L, Basso K, and Dalla-Favera R. (2018). MEF2B instructs germinal center development and acts as an oncogene in B cell lymphomagenesis. Cancer Cell 34, 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, and van Steensel B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Hao Z, Waisman A, Egert A, Ghitza D, and Rajewsky K. (2006). Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. USA 103, 7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HM, and La Thangue NB (2001). p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci 114, 2363–2373. [DOI] [PubMed] [Google Scholar]
- Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, Lawrence MS, Roemer MGM, Li AJ, Ziepert M, et al. (2018). Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med 24, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancy BM, and Cole PA (2015). Protein lysine acetylation by p300/CBP. Chem. Rev 115, 2419–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva NS, and Klein U. (2015). Dynamics of B cells in germinal centres. Nat. Rev. Immunol 15, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehairs J, Talebi A, Cherifi Y, and Swinnen JV (2016). CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Sci. Rep 6, 28973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ramírez I, Tadros S, González-Herrero I, Martín-Lorenzo A, Rodríguez-Hernández G, Moore D, Ruiz-Roca L, Blanco O,Alonso-López D, Rivas JL, et al. (2017). Crebbp loss cooperates with Bcl2 overexpression to promote lymphoma in mice. Blood 129, 2645–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannopoulou EG, and Elemento O. (2011). An integrated ChIP-seq analysis platform with customizable workflows. BMC Bioinformatics 12, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselbrecht C, Glass B, Mounier N, Singh Gill D, Linch DC, Trneny M, Bosly A, Ketterer N, Shpilberg O, Hagberg H, et al. (2010). Salvage regimens with autologous transplantation for relapsed large B-cell lymphoma in the rituximab era. J. Clin. Oncol 28, 4184–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RH, and Smolik S. (2000). CBP/p300 in cell growth, transformation, and development. Genes Dev. 14, 1553–1577. [PubMed] [Google Scholar]
- Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, Irish J, Stehr H, Vicente-Dueñas C, Romero-Camarero, et al. (2015). Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc. Natl. Acad. Sci. USA 112, E1116–E1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, and Roeder RG (1997). Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606. [DOI] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. (2006). GENCODE: producing a reference annotation for ENCODE. Genome biology 7 Suppl. 1, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashwah H, Schmid CA, Kasser S, Bertram K, Stelling A, Manz MG, and Müller A. (2017). Inactivation of CREBBP expands the germinal center B cell compartment, down-regulates MHCII expression and promotes DLBCL growth. Proc. Natl. Acad. Sci. USA 114, 9701–9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, and Ebert BL (2014). Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol 32, 941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin PD, Lee JH, and Lyons AB (1996). B cell differentiation and iso-type switching is related to division cycle number. J. Exp. Med 184, 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Ortega-Molina A, Geng H, Ying HY, Hatzi K, Parsa S, McNally D, Wang L, Doane AS, Agirre X, et al. (2017). CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov.7, 38–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang-Decker N, Tong C, Boussouar F, Baker DJ, Xu W, Leontovich AA, Taylor WR, Brindle PK, and van Deursen JM (2004). Loss of CBP causes T cell lymphomagenesis in synergy with p27Kip1 insufficiency. Cancer Cell 5, 177–189. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Boussouar F, Ney PA, Jackson CW, Rehg J, van Deursen JM, and Brindle PK (2002). A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature 419, 738–743. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Fukuyama T, Biesen MA, Boussouar F, Tong C, de Pauw A, Murray PJ, van Deursen JM, and Brindle PK (2006). Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol. Cell. Biol 26, 789–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khiem D, Cyster JG, Schwarz JJ, and Black BL (2008). A p38 MAPK-MEF2C pathway regulates B-cell proliferation. Proc. Natl. Acad. Sci. USA 105, 17067–17072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, and Busslinger M(2008). Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28, 751–762. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V, et al. (2017). Discovery of a selective catalytic p300/CBP inhibitor that targets line-age-specific tumours. Nature 550, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Wu HT, Vandin F, and Raphael BJ (2015). CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol. 16, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lill NL, Grossman SR, Ginsberg D, DeCaprio J, and Livingston DM (1997). Binding and modulation of p53 by p300/CBP coactivators. Nature 387, 823–827. [DOI] [PubMed] [Google Scholar]
- Lim WK, Lyashenko E, and Califano A. (2009). Master regulators used as breast cancer metastasis classifier. Pac. Symp. Biocomput 504–515. [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesin L, Ersching J, and Victora GD (2016). Germinal center B cell dynamics. Immunity 45, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milpied P, Cervera-Marzal I, Mollichella ML, Tesson B, Brisou G, Traverse-Glehen A, Salles G, Spinelli L, and Nadel B. (2018). Human germinal center transcriptional programs are de-synchronized in B cell lymphoma. Nat. Immunol 19, 1013–1024. [DOI] [PubMed] [Google Scholar]
- Montoto S, Davies AJ, Matthews J, Calaminici M, Norton AJ, Amess J, Vinnicombe S, Waters R, Rohatiner AZ, and Lister TA (2007). Risk and clinical implications of transformation of follicular lymphoma to diffuse large B-cell lymphoma. J. Clin. Oncol 25, 2426–2433. [DOI] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. (2011). Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, Tominaga Y, and Kohno T. (2016). Targeting p300 Addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov. 6, 430–445. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, and Nakatani Y. (1996). The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Okosun J, Bödör C, Wang J, Araf S, Yang CY, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, et al. (2014). Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet 46, 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, et al. (2011a). Inactivating mutations of acetyltransferase genes in B-cell lymphoma.Nature 471, 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O, et al. (2011b). Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet 43, 830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, Ouillette P, Trifonov V, Rossi D, Tabbò F, et al. (2014). Genetics of follicular lymphoma transformation. Cell Rep. 6, 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg N, Brooks N, Worthington J, Young B, Prosser A, Lane J, Taddei D, Brown R, Harbottle G, Shannon J, et al. (2017). Characterisation of CCS1477: A novel small molecule inhibitor of p300/CBP for the treatment of castration resistant prostate cancer. J. Clin. Oncol 35, ---–-−−. [Google Scholar]
- Peled JU, Yu JJ, Venkatesh J, Bi E, Ding BB, Krupski-Downs M, Shaknovich R, Sicinski P, Diamond B, Scharff MD, and Ye BH(2010). Requirement for cyclin D3 in germinal center formation and function. Cell Res. 20, 631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, and Salzberg SL (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc 11, 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott S, and Lieb JD (2015). What are super-enhancers? Nat. Genet 47, 8–12. [DOI] [PubMed] [Google Scholar]
- Rebel VI, Kung AL, Tanner EA, Yang H, Bronson RT, and Livingston DM (2002). Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl. Acad. Sci. USA 99, 14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Rueden CT, Hiner MC, and Eliceiri KW (2015). The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev 82, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J,Ramis-Zaldivar JE,Bonzheim I,Steinhilber J,Müller I, Haake A, Yu SC, Raffeld M, Fend F, Salaverria I, et al. (2018). CREBBP gene mutations are frequently detected in in situ follicular neoplasia. Blood 132, 2687–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, Roulland S, Kasbekar M, Young RM, Shaffer AL, et al. (2018). Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med 378, 1396–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, and Matthias P. (1996). B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature 383, 538–542. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, and Zhang F. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow SHCE, Harris NL, Jaffe ES, Pileri SA, Stein H, and Thiele J. (2016). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, Vol 2 Lyon: International Agency for Research on Cancer (IARC). [Google Scholar]
- Victora GD, and Nussenzweig MC (2012). Germinal centers. Annu. Rev. Immunol 30, 429–457. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, and Nussenzweig MC (2010). Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, and Nussenzweig MC (2012). Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood 120, 2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, and Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Narita T, Satpathy S, Srinivasan B, Hansen BK, Scholz C, Hamilton WB, Zucconi BE, Wang WW, Liu WR, et al. (2018). Timeresolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell 174, 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker PR, Kohyama M, Sandau MM, Albring JC, Nakagawa O, Schwarz JJ, and Murphy KM (2008). Transcription factor Mef2c is required for B cell proliferation and survival after antigen receptor stimulation. Nat. Immunol 9, 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Fukuyama T, Ney PA, Wang D, Rehg J, Boyd K, van Deursen JM, and Brindle PK (2006). Global transcriptional coactivators CREB-binding protein and p300 are highly essential collectively but not individually in peripheral B cells. Blood 107, 4407–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, Vlasevska S, Mo T, Tang H, Basso K, et al. (2015). Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat. Med 21, 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, Meyer SN, Mo T, Basso K, Brindle PK, et al. (2017). The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 7, 322–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data Availability
The raw RNA-seq data from the Crebbpfl/fl and Ep300fl/flCγ1Cre/+ murine GC B cells were deposited in the GEO database under accession number GSE124192. H3K27Ac ChIP-Seq data are available in GEO as series GEO: GSE132365.
The raw RNA-seq data from the Crebbpfl/fl and Ep300fl/flCγ1Cre/+ murine GC B cells were deposited in the GEO database under accession number GSE124192. H3K27Ac ChIP-Seq data are available in GEO as series GEO: GSE132365.