Abstract
Dysregulated Th17 cell responses underlie multiple inflammatory and autoimmune diseases, including autoimmune uveitis and its animal model, EAU. However, clinical trials targeting IL-17A in uveitis were not successful. Here, we report that Th17 cells were regulated by their own signature cytokine, IL-17A. Loss of IL-17A in autopathogenic Th17 cells did not reduce their pathogenicity and instead elevated their expression of the Th17 cytokines GM-CSF and IL-17F. Mechanistic in vitro studies revealed a Th17 cell-intrinsic autocrine loop triggered by binding of IL-17A to its receptor, leading to activation of the transcription factor NF-κB and induction of IL-24, which repressed the Th17 cytokine program. In vivo, IL-24 treatment ameliorated Th17-induced EAU, whereas silencing of IL-24 in Th17 cells enhanced disease. This regulatory pathway also operated in human Th17 cells. Thus, IL-17A limits pathogenicity of Th17 cells by inducing IL-24. These findings may explain the disappointing therapeutic effect of targeting IL-17A in uveitis.
Keywords: IL-17, IL-24, GM-CSF, Th17, experimental autoimmune uveitis, encephalomyelitis, neuroinflammation, secukinumab
Graphical Abstract
Highlights
-
•
IL-17A deficiency does not reduce the pathogenicity of Th17 cells in uveitis
-
•
IL-17A binds to its own receptor on Th17 cells, activating NF-κB
-
•
NF-κB induces IL-24 production, repressing the Th17 cytokine program through SOCS1/3
-
•
Silencing or depleting IL-24 in Th17 cells exacerbates neuroinflammation
Loss of IL-17A, the signature cytokine of autoreactive Th17 cells, unexpectedly did not diminish the pathogenicity of Th17 cells in neuroinflammatory disease. This report demonstrates that IL-17A represses the Th17 cytokine program, primarily IL-17F and GM-CSF, by inducing autocrine production of IL-24. Thus, IL-17A has a dual role in Th17 cells: pathogenic as well as regulatory.
Introduction
Interleukin (IL)-17A is a critical cytokine for control of microbial pathogens but when dysregulated can also contribute to the pathogenesis of many autoimmune diseases, including multiple sclerosis, rheumatoid arthritis, Crohn’s disease, and autoimmune uveitis. This is achieved by its ability to induce the production of proinflammatory cytokines and chemokines from various cell types, including fibroblasts, endothelial cells, epithelial cells, and immune cells, and recruitment of neutrophils. IL-17A implicated in autoimmune disease is expressed by a subset of T cells, known as Th17 cells (Harrington et al., 2005; Park et al., 2005), which play a critical role in host defense.
Autoimmune uveitis represents a spectrum of ocular pathologies, of which some are restricted to the eye and others are part of a systemic syndrome (Caspi, 2010). Autoimmune uveitis and optic neuritis often precede or accompany multiple sclerosis and may be driven by some common mechanisms (Neurology Reviews, 2015). They are believed to involve an aberrant immune recognition of unique self-antigens found in the target tissue. In the case of the eye, these are antigens restricted to the retina and/or choroid, which are targets of a potentially blinding inflammation and destruction of the neural retina. Several types of uveitis, including Vogt-Koyanagi-Harada disease and Behçet’s disease, were reported to be associated with a Th17 response (Amadi-Obi et al., 2007; Chi et al., 2008; Wang et al., 2012). Notably, in the animal model representing these diseases, experimental autoimmune uveitis (EAU) induced in susceptible mice by immunization with retinal antigens, Th17 can be a dominant factor in disease development (Chong et al., 2015; Luger et al., 2008).
Our studies and those of others demonstrated that the auto-pathogenic Th17 response can be regulated at multiple levels, either in the priming or the effector phases (Chong et al., 2015; Horai and Caspi, 2011; Korn et al., 2009; Luger et al., 2008; Sandquist and Kolls, 2018; Stadhouders et al., 2018). Th17 cells produce a characteristic set of potentially proinflammatory effector cytokines, including IL-17A, IL-17F, IL-22, and GM-CSF, and each may contribute to inflammatory responses, depending on the context and the target tissue. This multifaceted and complex interaction between Th17-lineage cytokines and inflamed tissues may complicate treatment of autoimmune diseases, which is often based on targeting just a single cytokine. The most commonly targeted Th17-lineage cytokine is the signature cytokine, IL-17A. IL-17A neutralizing agents have been used in treating patients with autoimmune and inflammatory diseases. However, although anti-IL-17A treatment brings about improvement in psoriasis, it does not yield consistent results in other conditions, including multiple sclerosis, rheumatoid arthritis, Crohn’s disease, and autoimmune uveitis (Dick et al., 2013; Patel and Kuchroo, 2015).
In experiments that led up to this study, we observed that R161H mice on the B10.RIII background, which express a T cell receptor (TCR) specific to the retinal protein IRBP and develop spontaneous uveitis, had almost undiminished disease when crossed to an Il17a −/− background. This was unexpected in view of previous reports, including ours, that IL-17A was a major pathogenic cytokine in uveitis and in the EAU model induced by IRBP immunization (Amadi-Obi et al., 2007; Luger et al., 2008; Peng et al., 2007). Observations implicating other cytokines in the pathogenic effect of Th17 cells had also been reported in the model of experimental autoimmune encephalomyelitis (EAE) (Codarri et al., 2011; El-Behi et al., 2011; Haak et al., 2009; McGinley et al., 2020). In humans, clinical interventions targeting IL-17A in multiple sclerosis, rheumatoid arthritis, uveitis, and Crohn’s disease have yielded disappointing results (Dick et al., 2013; Patel and Kuchroo, 2015). In the aggregate, these observations suggested that there is a mechanism (or multiple mechanisms) by which Th17 cells compensate for the loss of the signature cytokine, IL-17A, to retain their pathogenicity. We therefore hypothesized that blocking of IL-17A in autoreactive Th17 cells may enhance other Th17-lineage cytokines, to compensate, reversing the benefit that might result from loss of IL-17A. In this study, we found that IL-17A deficiency in Th17 cells results in upregulation of other Th17-lineage cytokines, IL-17F, GM-CSF, and IL-22. Mechanistic studies revealed that IL-17A is responsible for reducing Th17 responses by autocrine feedback to Th17 cells for the induction of IL-24, which controls Th17 cytokines and thereby regulates the pathogenicity of this lineage.
Results
IL-17A Deficiency Does Not Limit the Pathogenicity of Autoreactive Th17 Cells
Current dogma, based on numerous studies in animal models and clinical conditions, implicates IL-17A as a central pathogenic cytokine in autoimmunity affecting the CNS and other tissues. Previous studies in immunization-induced models of autoimmune uveitis (Luger et al., 2008; Peng et al., 2007) and in clinical uveitis (Chi et al., 2008; Wang et al., 2012) were in line with this notion. We found that spontaneous uveitis in R161H mice, which express a retina-specific TCR transgene and characteristically develop 100% incidence of disease by 2 months of age, was essentially unaffected in terms of incidence and clinical scores by IL-17A deficiency (Figure 1 A). As all these mice were littermates that were bred in-house, this was unlikely to stem from environmental or microbiome differences. To examine the importance of IL-17A in autoreactive Th17 cells, we used a reductionist model in which IL-17A-sufficient or IL-17A-deficient R161H cells were polarized to Th17 phenotype and infused into wild-type (WT) recipients. Again, all mice, both donors and recipients, were from the same in-house colony and were reared in the same room, such that the only known difference between them was the ability of the donor cells to produce IL-17A. As shown in Figure 1B, there was no significant difference in the progression or severity of uveitis between the two groups, demonstrating that IL-17A deficiency did not limit the pathogenicity of auto-aggressive Th17 cells in this model. The notion that EAU in this genotype does not require IL-17A was also supported in WT B10.RIII mice: treatment with anti-IL-17A antibody (Ab) of actively immunized mice, or mice adoptively transferred with polarized IL-17A-sufficient R161H Th17 cells, did not reduce disease scores (data not shown).
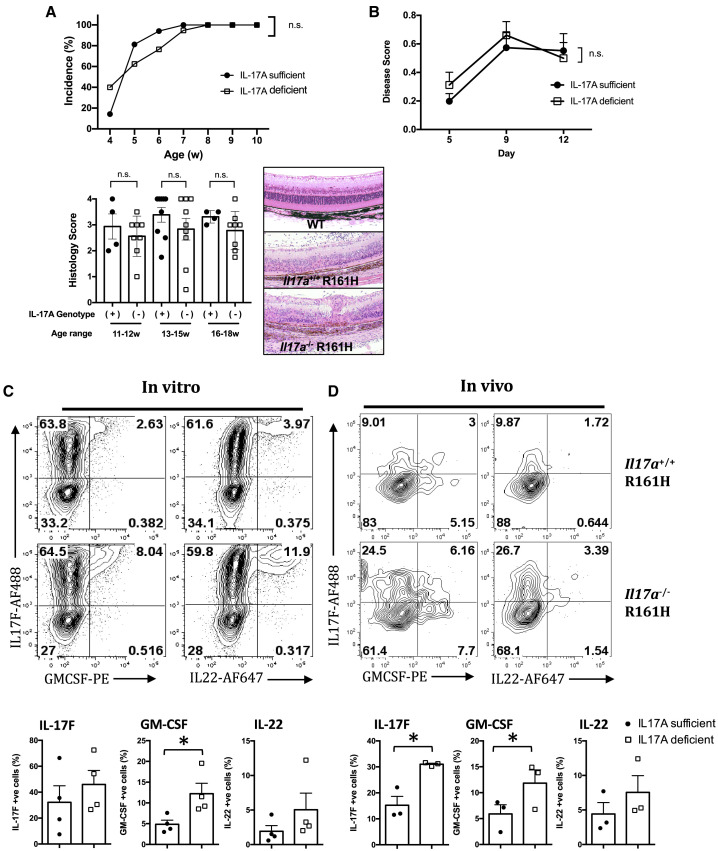
Figure 1.
Spontaneous Uveitis in IL-17A-Deficient Mice Develops in the Context of Increased Expression of Other Th17-Lineage Cytokines
(A) Incidence of spontaneous uveitis in Il17a−/− R161H mice and their Il17a+/+ or − R161H littermates was measured. Incidence was determined by fundus examination (upper panel; at least 5 mice per group at weaning at 4 weeks and at least 15 mice per group for all other time points), disease score by histology of eyes collected between 11 and 18 weeks of age (lower panel; at least 4 mice per group at each time point), and representative images of the H&E staining of the retina (100× original magnification).
(B) IL-17A-sufficient or IL-17A-deficient R161H cells were adoptively transferred into WT B10.RIII recipients. Data are combined from two experiments with at least 14 mice per point. Disease was scored at the indicated time points using fundoscopy.
(C and D) Cells were obtained from the eye-draining lymph nodes (LNs) of IL-17A-sufficient or IL-17A-deficient CD90.2 R161H mice and were polarized under Th17 conditions with IRBP161–180 peptide for 3 days and adoptively transferred into CD90.1 WT B10.RIII recipients. (C) Cytokine profiles of in vitro-polarized CD4+ T cells were analyzed using intracellular staining. Shown are representative fluorescence-activated cell sorting (FACS) plots (top) and compiled data (bottom) from four independent experiments. (D) Five days after adoptive transfer, eyes (6 or more) were pooled, and the cytokine profiles of eye-infiltrating donor (CD90.2) CD4+ T cells were determined using intracellular staining. Representative FCM plots (top) and compiled data (bottom) from three independent experiments.
∗p < 0.05, Student’s t test. Data are depicted as mean ± SEM.
IL-17A Limits the Production of Other Th17 Lineage Cytokines by Th17 Cells in a T Cell-Intrinsic Fashion
IL-17A induces inflammation and causes tissue damage by (1) activating nuclear factor kappa light-chain enhancer of activated B cells (NF-κB) signaling to enhance production of inflammatory molecules and (2) recruiting different types of immune cells into inflamed tissues (Gaffen, 2009). The finding that IL-17A deficiency in Th17 cells did not reduce their ability to induce EAU (Figure 1B) was therefore unexpected. One possible explanation is that the loss of IL-17A may be compensated for by other Th17-lineage cytokines. To test this hypothesis, we compared the cytokine profiles of Il17a +/+ and Il17a −/− Th17 cells. Cells from the lymph nodes of IL-17A−/− or IL-17A+/+ R161H mice were stimulated with IRBP161–180 peptide under Th17 polarization conditions. Il17a −/− CD4+ T cells produced substantially more IL-17F and GM-CSF and slightly more IL-22 compared with their Il17a +/+ counterparts (Figure 1C), with minimal IFN-γ expression (data not shown). Tracking these donor cells in vivo, in eyes of CD90.1 congenic recipients that developed undiminished disease, showed that 5 days after adoptive transfer, this cytokine profile of IL-17A-sufficient versus IL-17A-deficient Th17 cells became even more pronounced within the target tissue (Figure 1D).
We next addressed the question of whether other cell populations (e.g., B cells, macrophages, and/or dendritic cells) were necessary for upregulation of other Th17-lineage cytokines in Th17-polarized Il17a −/− cells. Toward that end, we polarized sorted naive (CD4+CD62L+) T cells (Figure S1A) from spleens of B10.RIII WT or Il17a −/− mice to Th17 conditions using anti-CD3 + anti-CD28 Ab activation. Il17a −/− Th17 cells, polarized in the absence of any other cells, also produced more IL-17F, IL-22, and GM-CSF (Figure 2 A). A similar phenomenon was observed in WT and Il17a −/− Th17 cells on the C57BL/6 background (Figure S1B).
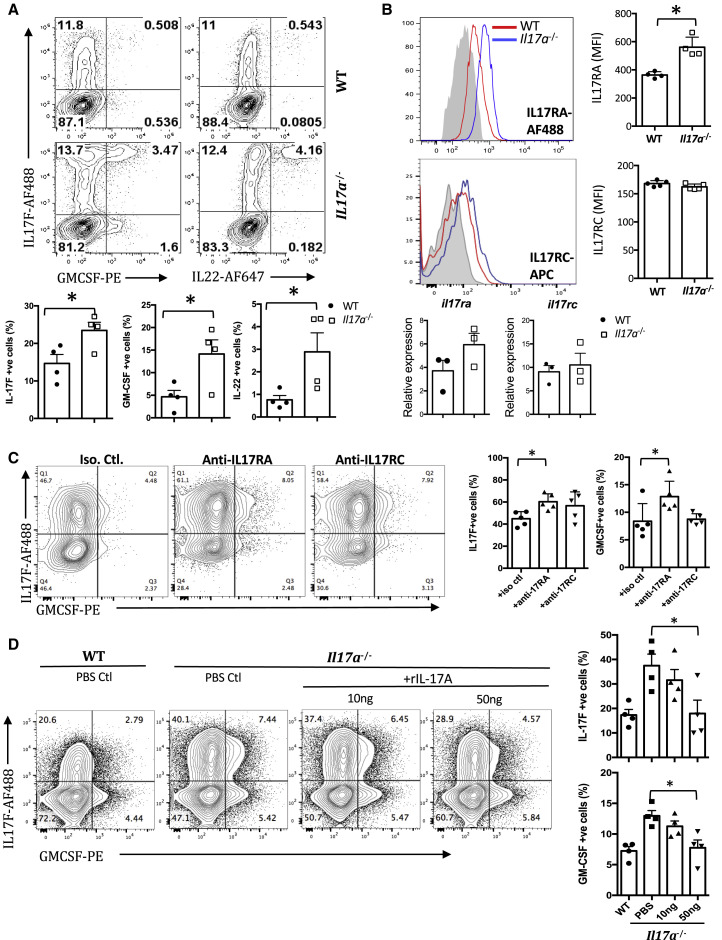
Figure 2.
IL-17A Inhibits Expression of Other Th17-Lineage Cytokines through the IL-17A Receptor
Cells were obtained from the spleen of IL-17A-sufficient or IL-17A-deficient B10RIII. CD4+CD62L+ T cells were isolated and polarized under Th17 conditions with anti-CD3/CD28 antibodies for 3 days.
(A) Cytokine profiles of polarized CD4+ T cells were analyzed using intracellular staining and shown as representative FCM plots (top) and compiled data (bottom) from three to five independent experiments.
(B) Th17-polarized CD4+ cells were stained with IL-17RA or IL-17RC. Gray filled histogram, isotype control; red histogram, WT Th17 cells; blue histogram, Il17a−/− Th17 cells. Shown are representative histograms (top left plots) and compiled mean fluorescence intensity (MFI) data (top right plots) from four or five independent experiments. Their gene expression was also determined using real-time PCR. Data were compiled from three independent experiments (bottom graphs) and were normalized to GAPDH and expressed as relative to WT Th0 cells.
(C) Th17-polarized WT CD4+ T cells with or without indicated isotype control, anti-IL-17RA, or anti-IL-17RC antibodies. Shown are representative FCM plots (left) and compiled data (right) from five independent experiments.
(D) Th17-polarized CD4+ T cells were cultured with or without recombinant IL-17A. Shown are representative FCM plots (left) and compiled data (right) from four independent experiments.
∗p < 0.05, Student’s t test (A and B) and one-way ANOVA (C and D). Data are depicted as mean ± SEM. See also Figures S1 and S2.
On the basis of this finding, we hypothesized that IL-17A can feed back onto Th17 cells in an autocrine manner. To address this, we first checked expression of the two subunits of the IL-17A receptor, IL-17RA and IL-17RC, in Th17 cells. Both subunits were expressed by Th17 cells, and IL-17RA was more highly expressed in Il17a −/− Th17 cells compared with Il17a +/+ Th17 cells (Figure 2B). Next, we blocked IL-17A signaling in WT cells by Abs against IL-17RA or IL-17RC during Th17 polarization. Similar to Il17a −/− Th17 cells, Th17 cells from WT mice expressed significantly more IL-17F and GM-CSF after IL-17 receptor blockade (IL-22 was not measured) (Figure 2C). Conversely, addition of recombinant IL-17A to Th17 polarization cultures of Il17a −/− CD4+CD62L+ cells inhibited production of these cytokines (Figure 2D, B10.RIII; Figure S1B, C57BL/6). These data suggest that IL-17A from Th17 cells regulates the production of other Th17-lineage cytokines by a negative feedback mechanism through the IL-17A receptor.
Because both IL-17A and IL-17F signal through the same IL-17RA-IL-17RC heterodimeric receptor (Gaffen, 2009), we investigated the effect of IL-17F on polarization of Th17 cells under similar conditions. IL-17F deficiency did not elicit upregulation of IL-17A and GM-CSF in Il17f −/− Th17 cells (Figure S1C), and addition of recombinant IL-17F did not suppress their production of IL-17A and GM-CSF (Figure S1C). These results indicate that although IL-17A and IL-17F share the same receptors, they display distinct biological effects, consistent with other functional differences reported in other studies (Ishigame et al., 2009; Tang et al., 2018; Yang et al., 2008).
Although IL-17F, GM-CSF, and possibly IL-22 all seem to be co-regulated by IL-24, it does not necessarily follow that they must all have a pathogenic role in the absence of IL-17A. To address which of the three Th17-associated cytokines might be implicated in driving EAU in Il17a −/− mice, we immunized Il17a −/− Il17f −/− mice for EAU or treated immunized Il17a −/− mice with anti-GM-CSF Ab. Anti-GM-CSF Ab treatment significantly reduced the EAU severity in Il17a −/− mice compared with those with isotype control treatment (Figure S2A). In contrast, Il17a −/− Il17f −/− mice developed similar disease to Il17a −/− mice (Figure S2B). These data support the notion that GM-CSF is the dominant cytokine that drives EAU in Il17a −/− mice. Because we recently found that IL-22 deficiency can be protective in EAU (Mattapallil et al., 2019), the contribution of IL-22 was not examined in this set of experiments.
IL-24 Is Induced in Th17 Cells by IL-17A and Negatively Regulates the Th17 Cytokine Program
The results thus far indicated that IL-17A can feed back to Th17 cells through IL-17RA-IL-17RC heterodimeric receptor and suppress Th17-lineage cytokine program. Because IL-17A typically exerts its effects by inducing secondary downstream mediators (Gaffen, 2009), we compared the transcriptomes of Il17a +/+ and Il17a −/− Th17 cells using RNA sequencing (RNA-seq) analysis to search for potential candidate genes involved in this negative feedback. Among the differentially expressed genes, IL-24 was selected for validation because of its comparatively higher expression in Il17a +/+ Th17 cells (Figure 3 A; Figure S3A) and because it is a member of the IL-10 family of cytokines and signals through the IL-20R, which has been implicated in T cell inhibitory effects (Wahl et al., 2009). We further confirmed the expression kinetics of IL-24 in WT Th17 cells (Figure 3B). No differences were detected in expression of other IL-20 receptor cytokines (i.e., IL-19 and IL-20) and their receptor subunits using RNA-seq (Figure S3B). As well, no difference was detected in gene expression of IL-10, and this was confirmed using intracellular staining (not shown).
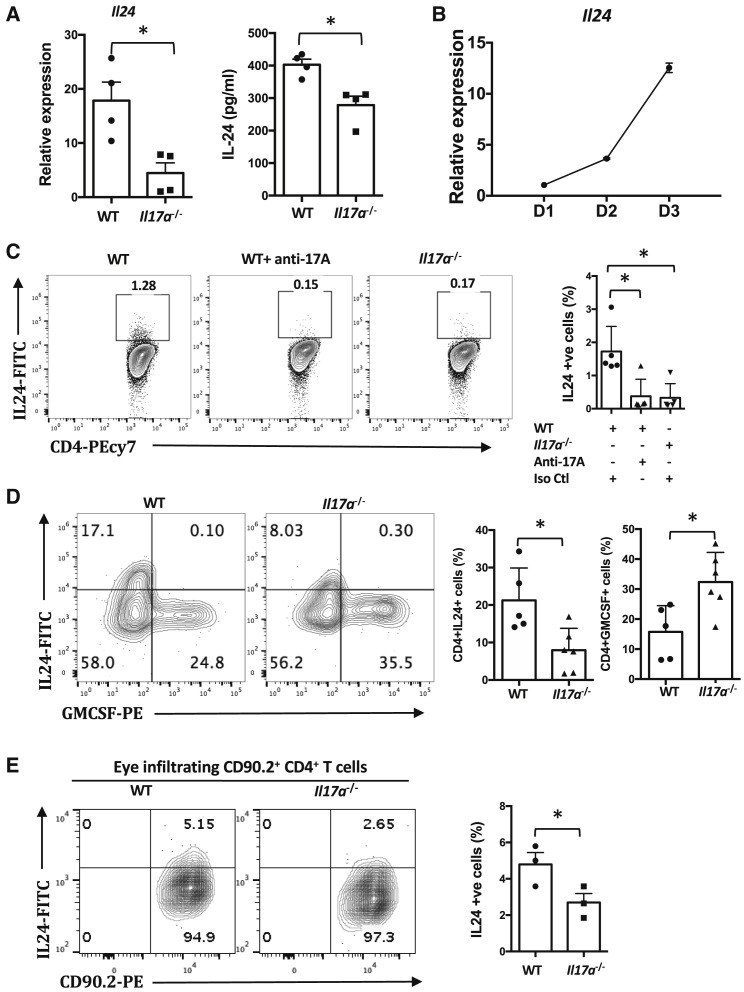
Figure 3.
IL-24 Is Expressed by Th17 Cells, and Its Expression Is Dependent on IL-17A
(A) IL-24 expression in WT and Il17a−/− Th17 cells as determined using q-PCR (left, n = 3 or 4, relative to resting CD4+ CD62L+ T cells) and ELISA (right, n = 4).
(B) Kinetics of IL-24 expression over 3 days of polarization in WT Th17 cells were studied (n = 3, relative to resting CD4+ CD62L+ T cells).
(C) CD4+CD62L+ T cells from WT and Il17a−/− mice were polarized under Th17 conditions with anti-CD3/CD28 antibodies for 3 days with or without an anti-IL-17A antibody. Shown are representative FCM plots (left) and compiled data (right) from five independent experiments.
(D) WT or Il17a−/− mice were immunized with IRBP161–180 peptide. Cytokine profiles of eye-infiltrating CD4+ T cells were determined using intracellular staining. Representative data of two independent experiments with at least five mice per group.
(E) Cells were obtained from the eye-draining LNs of IL-17A-sufficient or IL-17A-deficient CD90.2 R161H mice and were polarized under Th17 conditions with IRBP161–180 peptide for 3 days. Cells were adoptively transferred into CD90.1 WT B10.RIII recipients. Five days after adoptive transfer, eyeballs (six or more) were pooled, and the cytokine profiles of eye-infiltrating CD90.2+CD4+ T cells were determined using intracellular staining. Representative FCM plots (right) and compiled data (left) from three independent experiments.
∗p < 0.05, Student’s t test (A and E) or one-way ANOVA (C and D). Data are depicted as mean ± SEM. See also Figure S3.
We first wished to confirm that Th17 cells express IL-24 at the protein level. Using intracellular Ab staining, IL-24 was detectable in in vitro-polarized IL-17A-sufficient Th17 cells (Figure 3C), and its expression was reduced by IL-17A neutralization or its genetic deficiency. Furthermore, IL-24 was present in ocular-infiltrating effector T cells from mice immunized for EAU (Figure 3D) and was similarly reduced by IL-17A deficiency (Figure 3D). Finally, adoptively transferred R161H Th17 cells from IL-17A-sufficient donors expressed more IL-24 than parallel cells from Il17a −/− donors when retrieved from recipient eyes 5 days after transfer, confirming that this phenotype was maintained in vivo under conditions of disease (Figure 3E).
To directly address the question whether IL-24 suppresses production of Th17 cytokines, we polarized naive (CD4+CD62L+) Il17a −/− T cells to the Th17 phenotype in the presence or absence of recombinant IL-24 and measured expression of IL-17F and GM-CSF. Addition of IL-24 to Il17a −/− Th17 cultures normalized the elevated gene expression of IL-17F and GM-CSF to levels comparable with those of Il17a +/+ Th17 cells (Figures 4A and 4B). Conversely, neutralization of IL-24 or deletion of the IL-24 gene in Il17a +/+ Th17 cultures increased production of IL-17F and GM-CSF (Figure 4C).
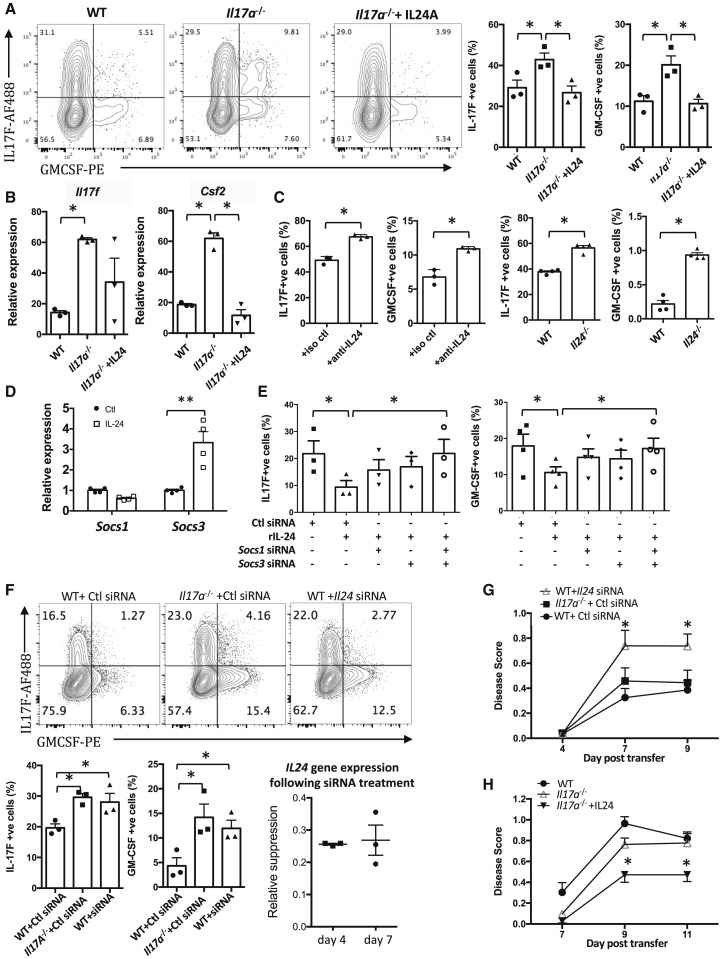
Figure 4.
IL-24 Inhibits the Effector Functions of Th17 Cells
(A–E) CD4+CD62L+ T cells from WT, Il17a−/− mice were isolated and polarized under Th17 conditions with anti-CD3/CD28 antibodies for 3 days with or without (A and B) recombinant IL-24 or (C) anti-IL-24 antibody or isotype control in WT. CD4+CD62L+ T cells were also isolated from Il24−/− mice for Th17 polarization. (D) Il17a−/− Th17 cells were re-stimulated with anti-CD3/CD28 antibodies for 24 h with or without IL-24. The mRNA expression of SOCS1 and SOCS3 was determined using real-time PCR. (E) Il17a−/− Th17 cells were polarized in presence of different combinations of IL-24 and siRNAs; expression of IL-17F and GM-CSF was determined using flow cytometry.
(F–H) Cells were obtained from the draining LNs of IL-17A-sufficient or IL-17A-deficient R161H mice and were polarized under Th17 conditions with IRBP161–180 peptide for 3 days with Il24 siRNA or scrambled control. (F) The expression of IL-17F and GM-CSF was determined by intracellular cytokine staining. Stability of the knockdown was confirmed using RT-PCR (normalized to control siRNA). (G) Cells were polarized in the presence of control siRNA or Il24 siRNA and were transferred to naive WT recipients. (H) After the adoptive transfer, the recipient mice received recombinant IL-24 (intraperitoneal [i.p.] injection) every other day.
In (A)–(F), data are combined from at least three independent experiments. In (G) and (H), data are combined from two independent experiments with at least seven mice per group. ∗p < 0.05; ∗∗p < 0.01, Student’s t test (A and C), one-way ANOVA (B, E, and F), and two-way ANOVA with Dunnett’s correction for multi-group comparison (G and H). Data are depicted as mean ± SEM. See also Figures S4–S7.
We next sought to address the mechanism whereby IL-24 suppresses Th17-lineage cytokine production. It has been reported that IL-24 regulates production of other cytokines by inducing suppressor of cytokine signaling (SOCS) proteins (Andoh et al., 2009). We therefore examined the effects of the IL-17A-IL-24 circuit on expression of SOCS genes and measured the effect of manipulating SOCS on expression of IL-17F and GM-CSF (IL-22 was not examined). Th17-polarized Il17a −/− cells had lower expression of SOCS genes compared with Il17a +/+ Th17 cells, with reduction of Socs1 and Socs3 being the most prominent (Figure S4). Conversely, if polarized Il17a −/− Th17 cells were re-stimulated by anti-CD3 + anti-CD28 Abs in the presence of IL-24, the expression of Socs3 (though not Socs1) was restored (Figure 4D). Finally, in line with a role for SOCS in mediating the effect of IL-24 in control of IL-17F and GM-CSF, the ability of IL-24 to normalize their elevated expression in Il17a −/− cells was reversed by small interfering RNA (siRNA) knockdown of Socs1 and Socs3 (Figure 4E). These in vitro data support the interpretation that IL-24 negatively regulates production of IL-17F and GM-CSF through SOCS1 and SOCS3. However, IL-17A itself may not be subject to regulation through IL-24, because neutralization of IL-24 in Th17 polarization cultures of Il17a +/+ cells did not increase the expression of IL-17A (Figure S5).
To examine the in vivo consequences of the IL-17A-IL-24 regulatory pathway on autoimmunity, we investigated the effect of IL-24 on pathogenicity of retina-specific Th17 cells in the adoptive transfer system. Because Il24 −/− R161H Th17 cells could not be generated (the appropriate Cre-lox combinations do not exist on the B10.RIII background to cross with R161H TCR Tg mice), we used siRNA knockdown in donor cells or recombinant IL-24 treatment of recipients as two complementary approaches. In the first scenario, knockdown of Il24 in Il17a +/+ cells, which stably reduced IL-24 gene expression to about one-quarter of control for at least 1 week, enhanced production of IL-17F and GM-CSF compared with control siRNA (Figure 4F). When adoptively transferred into naive WT recipients, these cells elicited enhanced disease compared with the control siRNA-treated cells (Figure 4G). Conversely, treatment of recipients with recombinant IL-24 ameliorated disease induced by Il17a −/− Th17 cells (Figure 4H). To address the general nature of this pathway in regulation of Th17 driven autoimmunity, we examined the effect of IL-24 deficiency on disease in the classical immunization-induced models of EAU and EAE. Toward that end, we immunized Il24 −/− or WT C57BL/6 mice with IRBP651–670 or with MOG35–55 in complete Freund’s adjuvant (CFA). IL-24 deficiency resulted in more severe disease in both EAU and EAE models (Figures S6A and S7A, respectively) with increased expression of Th17-lineage cytokines in CD4+ T cells infiltrating the target tissues (Figures S6B, S7B, and S7C). Furthermore, we blocked IL-17A during the course of EAU in Il24 −/− mice by treating them with an anti-IL-17A neutralizing Ab. These mice developed EAU disease scores only slightly lower compared with isotype control (Figure S6C). This suggests that the pathogenic role of IL-17A in Il24 −/− mice is not dominant, likely because other Th17-related cytokines (i.e., IL-17F and GM-CSF) were elevated and could have compensatory roles. In the aggregate, these in vivo data support the generality of IL-24 as a regulator of autoreactive Th17 cells by demonstrating its role in two different models of uveitis (passive transfer and active immunization), in different experimental autoimmune diseases (EAU and EAE), and in two different strains of mice (B10.RIII and C57BL/6).
Binding of IL-17A to Its Receptor Elicits NF-κB Signaling in Th17 Cells and Drives Transcription from the IL-24 Promoter
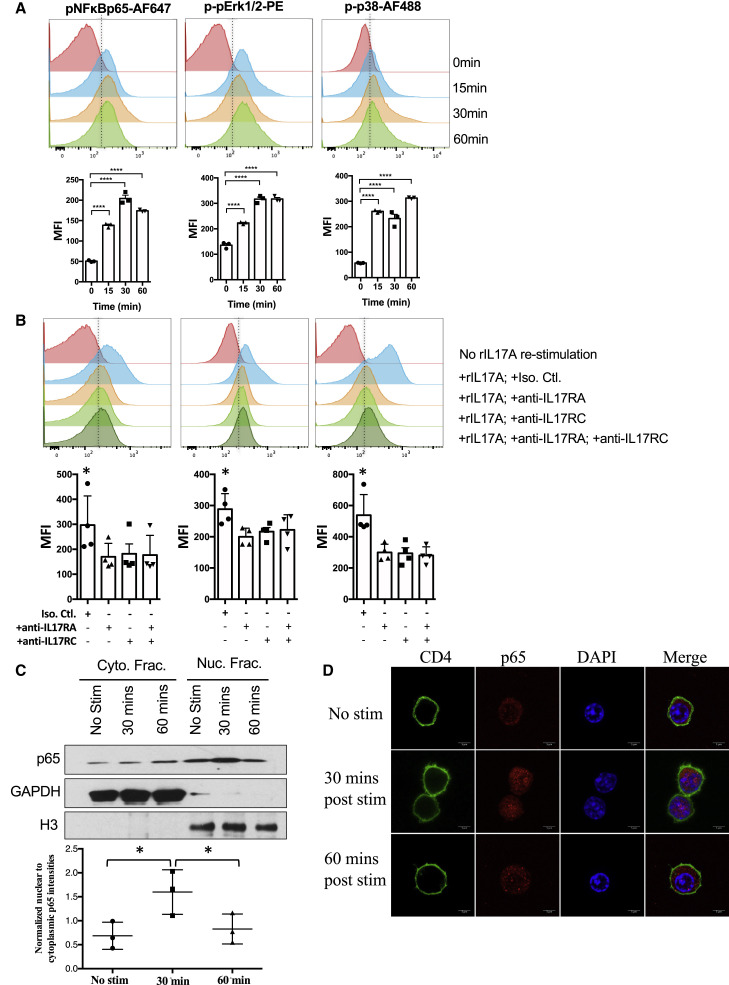
We demonstrated that IL-17A could bind to IL-17R on Th17 cells and regulate their lineage-specific cytokine program (Figures 2A–2C). To dissect the signaling events involved, we examined activation of IL-17A downstream signaling molecules, NF-κB, ERK, and MAPK (Gaffen, 2009). A pulse with recombinant IL-17A resulted in a significant increase in the phosphorylation of the NF-κBp65 subunit, Erk1/2 and p38 MAPK in Th17 cells (Figure 5 A). This was dependent on the IL-17R, as blocking Abs to IL-17RA or IL-17RC reduced phosphorylation of these molecules (Figure 5B). Western blotting (Figure 5C) and immunohistochemistry (Figure 5D) demonstrated presence of NF-κBp65 in the nucleus of Th17 cells as early as 15 min after IL-17A pulse, with peak responses at 30 min (Figures 5C and 5D). These data not only confirmed that IL-17A signaling occurs within Th17 cells but also indicated that it uses the NF-κB signaling pathway, similarly to other cell types.
Figure 5.
IL-17A Induces NF-κB Signaling in Th17 Cells
(A and B) CD4+CD62L+ T cells from WT mice were isolated and polarized under Th17 conditions with anti-CD3/CD28 antibodies for 3 days. Cells were pulsed with IL-17A. (A and B) Phosphorylation of NF-κBp65, Erk1/2, and p38 was determined using flow cytometry. Data are shown as representative histogram and as compiled data from three (A) and four (B) independent experiments.
(C and D) NF-κBp65 translocated into the nucleus after IL-17A re-stimulation. Representative of three (C) and two (D) independent experiments.
∗p < 0.05 and ∗∗∗∗p < 0.0001, one-way ANOVA (A–C). Data are depicted as mean ± SEM.
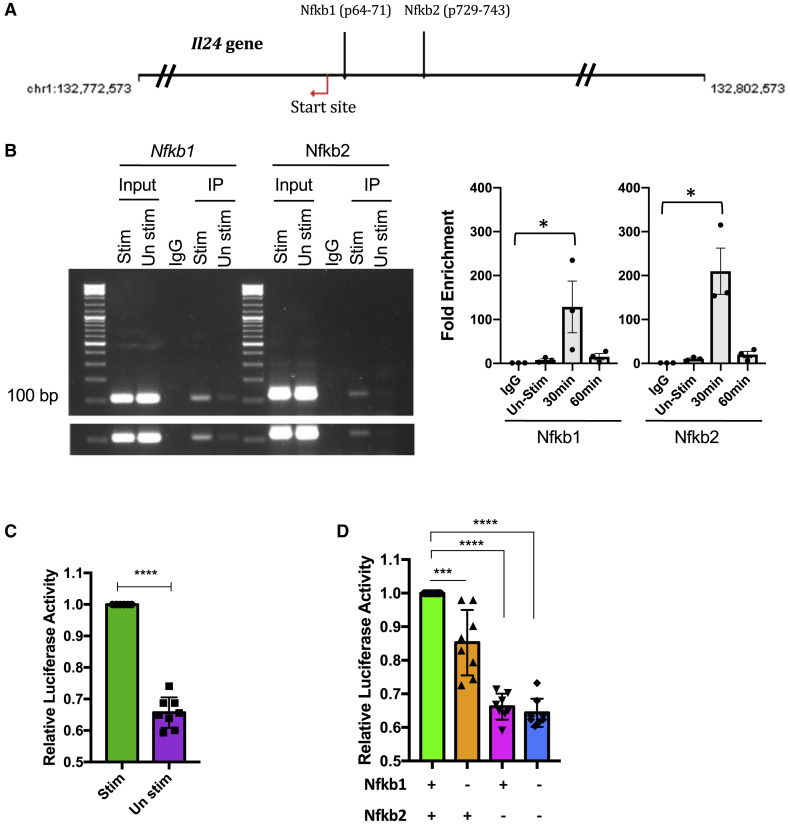
NF-κB regulates the transcription of a large array of cytokines and chemokines. We therefore hypothesized that IL-17A utilized NF-κB to induce IL-24. A bioinformatic analysis of the Il24 promoter region for canonical NF-κB binding sequences identified two potential sites that could be targeted by NF-κB: Nfkb1 (position −64 to −71) and Nfkb2 (position −729 to −743) (Figure 6 A). Chromatin immunoprecipitation (ChIP) assay confirmed binding of NF-κB with the Nfkb1 and Nfkb2 sites (Figure 6B). To examine the functional consequences of this binding on the Il24 promoter, we performed a transcriptional reporter assay. Polarized Th17 cells were transfected with a reporter construct in which a NF-κB-responsive Il24 promoter controls luciferase expression. Expression of luciferase following an IL-17A pulse confirmed promoter activity (Figure 6C), and a single mutation introduced into the Nfkb1 or Nfkb2 sequences in the construct reduced luciferase expression, confirming specificity (Figure 6D). These data indicate that IL-17A signals through its receptor to cause NF-κB translocation into the nucleus where it binds to, and activates, the IL 24 promoter in Th17 cells.
Figure 6.
IL-17A Induces the Il24 Gene Promoter Activity through NF-κB
(A) The Il24 gene has two potential binding sites for NF-κB (predicted by Genomatix).
(B) PCR and real-time PCR showing the result of the two Il24 promoter sequences after the DNA was pulled down by anti-p65 antibody. Left panel shows representative data after 30 min; right panel shows average data compiled from three independent experiments.
(C and D) CD4+CD62L+ T cells from WT mice were isolated and polarized under Th17 conditions with anti-CD3/CD28 antibodies for 3 days. Cells were pulsed with IL-17A. Cells were transfected with the indicated reporter constructs (C) Il24 promoter and (D) Il24 promoter with mutation at Nfkb1 and/or Nfkb2. Fold change in Firefly/Renilla (FL/RL) ratio is plotted with respect to control (n = 8).
∗p < 0.05, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, one-way ANOVA (B and D) and Student’s t test (C). Data are depicted as mean ± SEM.
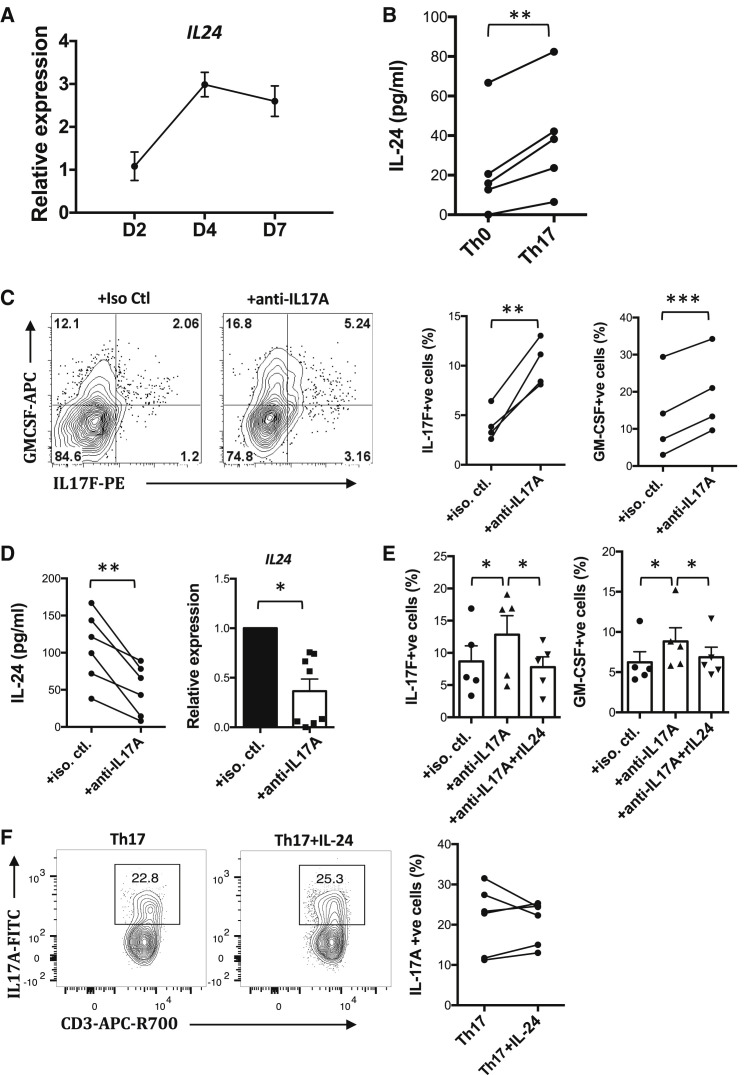
The IL-17-IL-24 Pathway Controls Lineage-Specific Cytokines in Human Th17 Cells
To examine potential clinical relevance of this regulatory pathway within Th17 cells, we investigated the effect of IL-17A on cytokine production by human Th17 cells. Naive CD4+ T cells were isolated from peripheral blood of healthy human volunteers and polarized to Th17 using standard protocol. Expression of IL-24 by human Th17 cells peaked on day 4 and was still high on day 7 (Figure 7 A). Th17 cells expressed significantly more IL-24 than control non-polarized Th0 cells from the same donors, indicating that IL-24 induction was not due simply to T cell activation but was connected to Th17 polarization (Figure 7B). Neutralization of IL-17A using monoclonal Abs during Th17 polarization abolished the expression of IL-24 at the mRNA and protein levels and at the same time increased the production of IL-17F and GM-CSF (Figures 7C and 7D). Conversely, addition of recombinant IL-24 normalized the elevated production of these cytokines (Figure 7E). IL-24 supplementation did not inhibit the expression of IL-17A by human Th17 cells (Figure 7F). These data suggest that, similarly to mouse, human IL-17A elicits negative feedback regulation to inhibit the Th17 cytokine program through autocrine induction of IL-24.
Figure 7.
IL-17A Induces Human Th17 Cells to Produce IL-24, Which Suppresses Expression of Th17-Lineage Cytokines
(A–F) Naive human CD4+ T cells were isolated using naive CD4+ T cell isolation kit (Miltenyi) and were polarized under Th17 conditions for 14 days. (A) Kinetics of IL-24 expression during Th17 polarization were studied. Cells were collected at the indicated time points and were analyzed using real-time PCR (n = 4). (B) Expression of IL-24 by human Th0 and Th17 cells was determined from supernatant collected by ELISA on day 5 (n = 5). (C, E, and F) Cells were harvested on day 14 and subjected to intracellular staining for the indicated cytokines (C, n = 4; E, n = 5; F, n = 6). (D) Supernatant and cells were harvested on day 5 for ELISA (n = 6) and real-time PCR (n = 8), respectively, to detect IL-24 expression.
∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, paired t test (A, C, and D) and one-way ANOVA (E). (F) Not significant. Data are depicted as mean ± SEM.
Discussion
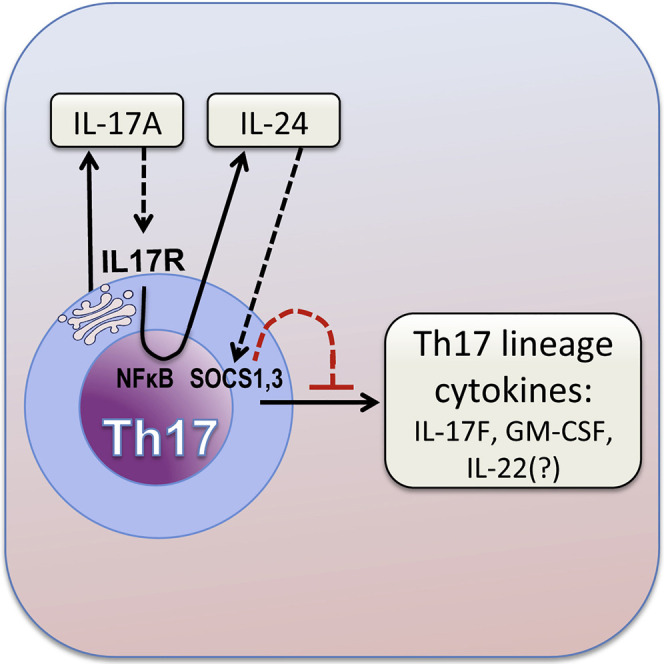
In the present study, we uncover a circuit that regulates the Th17 response, which is driven by the lineage signature cytokine IL-17A. We show that differentiated Th17 cells express both IL-17RA and IL-17RC for the full heterodimeric IL-17A receptor to bind IL-17A, leading to activation of NF-κB and induction of IL-24, which feeds back through SOCS1 and SOCS3 to repress the Th17-lineage cytokines GM-CSF and IL-17F. Notably, this suppression did not seem to operate on IL-17A itself, which would seem untypical of the classical notion of feedback inhibition that we expect to affect the eliciting cytokine itself. Our study provides compelling evidence that IL-17A deficiency leads to the increased production of other Th17-lineage cytokines from Th17 cells and identifies a regulatory axis whereby IL-17A regulates the Th17 cytokine program in an autocrine manner. This occurs at least in part via IL-17A-driven IL-24 production from Th17 cells and leads to dampening of the expression of other Th17-lineage cytokines through a negative feedback mechanism.
IL-24 is a relatively little studied cytokine that was originally described as a Th2 cytokine (Schaefer et al., 2001). It is a member of the IL-20 subfamily cytokines, along with IL-19 and IL-20, and has been implicated in the regulation of T cells (Anuradha et al., 2016; Wahl et al., 2009). It can inhibit Th1 and cytotoxic CD8+ T cell responses (Anuradha et al., 2016; Golan et al., 2018). However, its role in the context of the Th17 response had not been studied previously. The groups of Regev and Kuchroo identified expression of the IL-24 gene during the late phase of Th17 polarization (Yosef et al., 2013) but did not pursue this observation further. IL-20 subfamily cytokines bind the same heterodimeric type I IL-20 receptor (IL-20R1 and IL-20R2), which is expressed by Th17 cells, while IL-22R1, which together with IL-20R2 forms an alternative IL-20 and IL-24 receptor (Dumoutier et al., 2001), was not detected (Figure S3). Our RNA-seq analysis indicated that IL-19 (but not IL-20) is expressed by Th17 cells, but there was no significant difference in its expression between WT and Il17a −/− Th17 cells, suggesting that it is not regulated by IL-17A, therefore, it was not pursued further.
Although it is well accepted that the IL-17RA/IL-17RC heterodimeric receptor is expressed on non-hematopoietic cells (Gaffen, 2009), it is not common knowledge that it can also be expressed by T cells. The naive T cells and hematopoietic compartments have been reported to express barely detectable levels of IL-17RA and IL-17RC (Haudenschild et al., 2002; Ishigame et al., 2009; O’Connor et al., 2009). In contrast, our data demonstrate expression of both IL-17RA and RC on in vitro differentiated Th17 cells at the level of RNA, protein, and function. As another example of negative feedback of IL-17A on Th17 cells, O’Connor et al. (2009) demonstrated that Il17a −/− or Il17ra −/− effector precursor (CD45RBhi CD4+) T cells elicit exacerbated colitis in Rag1 −/− recipient mice with an increased Th1 response. Several other studies reported effects of IL-17A on lymphocytes, including T cells (von Vietinghoff and Ley, 2009), B cells (Milovanovic et al., 2010), and natural killer (NK) cells (Al Omar et al., 2013), but the signaling pathway and functional downstream consequences were not characterized, although it was suggested that IL-17A-induced genes in lymphocytes are distinct from those in non-hematopoietic cells (Hsu et al., 2008; Ishigame et al., 2009). Our data thus demonstrate a new role for hematopoietic IL-17R expression, namely, controlling the Th17-lineage cytokine program via cell-intrinsic induction of IL-24.
A number of additional questions remain. First, we are unsure whether IL-17A, and for that matter, IL-24, actually must be secreted from the Th17 cell to bind to receptors on the cell surface in order to elicit this regulatory loop, or whether the circuit can operate, at least in part, intracellularly. Given that addition of exogenous IL-17A or IL-24 affects the outcome of the responses, the extracellular route seems relevant. This raises the possibility that in vivo, the autocrine nature of this loop might potentially be supplemented by IL-17A and IL-24 produced by other cells that can produce these cytokines (Chen et al., 2018). However, the relative contribution of a putative paracrine versus autocrine pathway is difficult to evaluate and would depend on the concentration of these cytokines in the immediate microenvironment of the Th17 cells being regulated (likely being highest in an autocrine situation).
Although it appears that IL-17A, via IL-24, co-regulates IL-17F, GM-CSF, and possibly IL-22, dissecting their individual contributions is not straightforward. It is known that in the IL-17A-sufficient setting, single deficiency of IL-17F does not affect neuropathology (Haak et al., 2009). In contrast, multiple reports in the literature support that GM-CSF plays a pathogenic role even in presence of the other cytokines (Codarri et al., 2011; El-Behi et al., 2011). Our data, showing that depletion of GM-CSF in Il17a −/− mice inhibits disease, whereas deficiency of IL-17F does not (Figure S2), support the notion that GM-CSF is a dominant pathogenic cytokine in the Il17a −/− model. IL-17F appears non-dominant, as its elimination when GM-CSF is still present is insufficient. That said, in mice deficient in both IFN-γ and IL-17A, elimination of IL-17F does inhibit disease, indicating that under some conditions, IL-17F can contribute to pathogenicity (Bing et al., 2020). Finally, of the three Th17-associated cytokines, IL-22 appeared to be the least effectively and least consistently controlled by the IL-17-IL-24 circuit. In our hands, IL-22 actually appears to be neuroprotective (Mattapallil et al., 2019). Therefore, even though an increase in IL-22 may occur as a result of reduced IL-17/IL-24 regulation, it is uncertain what would be the net contribution to pathology.
Another question is whether IL-17A, via IL-24, only regulates other Th17-associated cytokines or also its own expression. There is an apparent discrepancy in IL-17A expression in the absence of IL-24 in vitro versus in vivo. It is conceivable that in vivo, there are additional factors that could come into play as a result of lack of IL-24 in cells other than Th17 cells, which could have indirect effects on IL-17A production that would not be seen in the minimalistic setting of T cell culture in vitro. We also noted that the overall frequency of GM-CSF-producing CD4+ T cells was not elevated in the EAU-immunized Il24 −/− mice (although it was elevated in the IL-17A+ population and across the board in Il24 −/− EAE mice). Possibly, this is dynamic and could be affected by the time point at which the tissue was collected.
These complexities notwithstanding, our findings help reconcile and provide a mechanistic explanation for sporadic observations in the literature, including our own, that did not fit the well-documented proinflammatory role of IL-17A in tissue specific autoimmunity. Thus, recombinant IL-17A treatment was paradoxically reported to inhibit EAU (Ke et al., 2009), whereas IL-17A deficiency upregulated IL-17F in Th17 cells that could be reversed by addition of recombinant IL-17A (Smith et al., 2008; von Vietinghoff and Ley, 2009). This is not to say, as some have suggested, that IL-17A is not a proinflammatory cytokine. A study just published (McGinley et al., 2020) showed that IL-17A triggers recruitment of IL-1β-producing myeloid cells and exacerbates inflammation. Our interpretation, based on the data reported here, is that IL-17A has both pathogenic and regulatory roles and that these opposing effects balance each other. Future studies will be required to dissect out which role may prevail under a given set of conditions, which may also include genetic and environmental influences.
Importantly, the regulatory pathway described here appears to operate not only in mice but also in human Th17 cells, suggesting that it may be relevant to clinical targeting of IL-17A in a variety of inflammatory and autoimmune situations. With the notable exception of psoriasis (Wasilewska et al., 2016), in which the responding cells are keratinocytes rather than lymphoid cells, clinical trials using IL-17A antagonists in autoimmune and inflammatory diseases have been less effective than expected. Neutralization of IL-17A has had marginal, if any, effects in multiple sclerosis, rheumatoid arthritis, and autoimmune uveitis (Dick et al., 2013; Patel and Kuchroo, 2015). Although Th17 responses were associated with uveitis (Chi et al., 2008; Wang et al., 2012), three multicenter double-masked placebo controlled clinical trials of anti-IL-17A treatment in uveitis failed to meet their primary efficacy endpoints and were terminated early (Dick et al., 2013). Our data would predict that these patients would have reduced IL-24 in their serum and elevated IL-17F and GM-CSF. Finally, anti-IL-17A agents unexpectedly aggravated Crohn’s disease and failed to affect Behçet’s disease, in which IL-17A was thought, by association, to be particularly pathogenic (Dincses et al., 2019; Hueber et al., 2012; Mozaffari et al., 2015; Shiga et al., 2017). Although we now know that IL-17A is needed to control gut commensals, our current findings, combined with a recent report that IL-17F, and not IL-17A, is responsible for microbiota-mediated colitis in mice (Tang et al., 2018), suggest an additional or alternative explanation. Namely, on the basis of the present findings, aggravation of Crohn’s pathology by IL-17A neutralization (Hueber et al., 2012) could be due not so much to reduction of IL-17A as to upregulation of IL-17F. Appropriate clinical studies will be needed to confirm this hypothesis.
In conclusion, our present data identify a regulatory circuit that controls Th17 cells and is driven by IL-17A, which feeds back to control the Th17 response by inducing the inhibitory cytokine IL-24. Our findings offer a possible explanation why results of anti-IL-17A agents such as secukinumab, in treating autoimmune diseases have been disappointing, and suggest that this paradigm could be re-evaluated and potentially improved by modulating IL-24 in addition to IL-17A.
Limitations of Study
This study has a number of limitations. First, conditional-knockout Il17ra fl/flCD4cre mice should ideally be used to support the effect of IL-17A on Th17 cells. However, these mice are currently available only in C57BL/6 background, whereas the R161H spontaneous uveitis model is on the B10.RIII background. Second, although co-housed controls and WT littermates indicate that microbiome was not a factor in the EAU experiments with Il17a −/− R161H mice and Il24 −/− mice, bacterial 16S analysis would provide further support to this conclusion. Unfortunately, because of the coronavirus disease 2019 (COVID-19) pandemic, this analysis could not be completed. Third, although depletion of IL-17A does not alter the development of EAU in B10.RIII mice, we previously reported that this treatment does ameliorate EAU in mice on the C57BL/6 background (Luger et al., 2008). The reason for this strain-specific difference is unknown. Fourth, IL-22 was not included in most of the experiments because of the limited cell numbers and the low quality of the anti-IL-22 Ab available for intracellular cytokine staining. This limits our ability to conclude regarding the effect of IL-24 in regulating IL-22 expression. Finally, our data predict that uveitis patients treated with secukinumab to target IL-17A would have less IL-24 and more IL-17F and GM-CSF. Unfortunately, we were unable to obtain patient sera from these clinical trials to support the contention that loss of the IL-17-IL-24 regulatory pathway contributed to the unsatisfactory therapeutic outcome.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Hamsters anti-CD3e (clone 500A2) | BD Biosciences | Cat#: 560771; RRID:AB_1937314 |
| Rat anti-CD4 (clone RM4-5) | Biolegend | Cat#: 100528; RRID:AB_312729 |
| Mouse anti-CD90.1 (clone OX-7) | Biolegend | Cat#: 202506 RRID:AB_492882 |
| Rat anti-CD90.2 (clone 30-H12) | Biolegend | Cat#: 105308; RRID:AB_313179 |
| Rat anti-IL-17A (clone TC11-18H10.1) | Biolegend | Cat#: 506926; RRID:AB_2632611 |
| Mouse anti-IL-17F (clone 9D3.1C8) | Biolegend | Cat#: 517006; RRID:AB_10661903 |
| Rat anti-GM-CSF (clone MP1-22E9) | Biolegend | Cat#: 505406; RRID:AB_315382 |
| Anti-IL-22 (clone 3F11.3) | Genentech | N/A |
| Goat anti-IL-24 (polyclonal) | R&D systems | Cat#: AF2786; RRID:AB_2124803 |
| Rat anti-IL-17RA/IL-17R (clone 657603) | R&D systems | Cat#: FAB4482G; RRID:AB_10891099 |
| Goat anti-IL-17RC (polyclonal) | R&D systems | Cat#: FAB2270A; RRID:AB_2125672 |
| Rat anti-IFN-γ (clone XMG1.2) | eBioscience | Cat#: 25-7311-82; RRID:AB_469680 |
| Mouse anti-CD3 (clone SK7) | BD Biosciences | Cat#: 563219; RRID:AB_2738076 |
| Mouse anti-CD4 (clone RPA-T4) | BD Biosciences | Cat#: 562424; RRID:AB_11154417 |
| Mouse anti-IL-17A (clone BL168) | Biolegend | Cat#: 512304; RRID:AB_961390 |
| Mouse anti-IL-17F (clone 033-782) | BD Biosciences | Cat#: 561197; RRID:AB_10611873 |
| Rat anti-GM-CSF (clone BVD2-21C11) | BD Biosciences | Cat#: 562257; RRID:AB_11152076 |
| Rabbit anti-phospho-NF-κB p65 (clone 93H1) | Cell Signaling Technology | Cat#: 4887; RRID:AB_561198 |
| Rabbit anti-phospho-Erk1/2 (clone 197G2) | Cell Signaling Technology | Cat#: 14095; RRID:AB_2728834 |
| Mouse anti-phospho-p38 MAPK (clone 28B10) | Cell Signaling Technology | Cat#: 4551; RRID:AB_331302 |
| Mouse anti-NF-κB p65 (clone F-6) | Santa Cruz Biotechnology | Cat#: sc-8008; RRID:AB_628017 |
| Rat anti-CD4 (clone RM4-4) | eBioscience | Cat#: 11-0043-82; RRID:AB_464900 |
| Rabbit anti-Histone H3 (polyclonal) | EMD Millipore Corporation | Cat#: 06-755; RRID:AB_2118461 |
| Rabbit anti-GAPDH (clone 14C10) | Cell Signaling Technology | Cat#: 2118S; RRID:AB_561053 |
| Mouse anti-IL-17A (clone 17F3) | BioXcell | Cat#: BE0173; RRID:AB_10950102 |
| Mouse anti-IgG1 (clone MOPC1-21) | BioXcell | Cat#: BE0083; RRID:AB_1107784 |
| Rat anti-IL-4 (clone 30340) | R&D systems | Cat#: MAB404; RRID:AB_2128960 |
| Hamster anti-IFN-γ (clone H22) | R&D systems | Cat#: MAB4851; RRID:AB_2123046 |
| Mouse anti-IL-4 (clone 34019) | R&D systems | Cat#: MAB204; RRID:AB_2126745 |
| Mouse anti-IFN-g (clone K3.53) | R&D systems | Cat#: MAB2852; RRID:AB_2123304 |
| Bacterial and Virus Strains | ||
| M. tuberculosis H37 Ra | BD | 231141 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Complete Freund’s Adjuvant | Sigma | F5881 |
| Bordetella pertussis toxin | Sigma | P7208-50UG |
| Recombinant mouse TGF-beta 1 | R&D systems | Cat#: 7666-MB |
| Recombinant mouse IL-6 | R&D systems | Cat#: 406-ML |
| Recombinant mouse IL-24 | R&D systems | Cat#: 7807-ML |
| Recombinant mouse IL-23 | R&D systems | Cat#: 1887-ML |
| Recombinant human TGF-beta 1 | R&D systems | Cat#: 240-B |
| Recombinant human IL-1beta | R&D systems | Cat#: 201-LB |
| Recombinant human IL-6 | R&D systems | Cat#: 206-1L |
| Recombinant human IL-24 | R&D systems | Cat#: 1965-1L |
| Recombinant human IL-23 | R&D systems | Cat#: 1290-1L |
| Recombinant human TGF-beta 1 | Peprotech | Cat#: 100-21 |
| Recombinant human IL-1beta | Peprotech | Cat#: 200-01B |
| Recombinant human IL-6 | Peprotech | Cat#: 200-06 |
| Recombinant human IL-23 | Peprotech | Cat#: 200-23 |
| MOG35-55 | Bio Basic, Inc | Custom synthesis |
| IRBP161-180 | Bio Basic, Inc | Custom synthesis |
| IRBP1-20 | Bio Basic, Inc | Custom synthesis |
| Critical Commercial Assays | ||
| TruSeq RNA Sample Prep Kit | Illumina | FC-122-1001 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE152534 |
| Experimental Models: Organisms/Strains | ||
| Mouse: R161H. B10.RIII | Horai et al., 2013 | N/A |
| Mouse: Il17A−/−, B6 | Nakae et al., 2002 | N/A |
| Mouse: Il17F−/−, B6 | Ishigame et al., 2009 | N/A |
| Mouse: Il24−/−, B6 | Chan et al., 2006 | N/A |
| Oligonucleotides | ||
| Primer for IL-24 promoter Fwd 5′TAGCTAGCTAGCTTTAGCTTCTGGGCT 3′ | This paper | N/A |
| Primer for IL-24 promoter Rev 5′ ATAAGCTTTCAAGGCAGCAGCCTAACA 3′ |
This paper | N/A |
| Accell siRNA for IL-24 | Dharmacon | N/A |
| Accell siRNA for Socs1 | Dharmacon | N/A |
| Accell siRNA for Socs3 | Dharmacon | N/A |
| Software and Algorithms | ||
| Bowtie2 v2.1.0 | Langmead et al., 2009 | https://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.1.0/ |
| TopHat2 v2.0.9 | Trapnell et al., 2009 | https://ccb.jhu.edu/software/tophat/index.shtml |
| Integrative Genomics Viewer | Robinson et al., 2011 | http://software.broadinstitute.org/software/igv/ |
| SAMtools v0.1.19 | Robinson et al., 2011 | https://sourceforge.net/projects/samtools/files/samtools/0.1.19/ |
| FastQC v0.10.1 | Andrews S: FastQC: A quality control tool for high throughput sequence data | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| eXpress v1.3.1 | Roberts et.al., 2013 | https://pachterlab.github.io/eXpress/ |
| EdgeR | Robinson et al., 2010 | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Genome Analyzer IIx (SCS v2.10 software) | Illumina | https://www.illumina.com/content/dam/illumina-marketing/documents/products/specifications/specification_genome_analyzer.pdf |
| Flowjo v10 | FlowJo | N/A |
| Prism 7 | Graphpad | N/A |
| Genomatix software | GGA suite | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Rachel R Caspi (caspir@nei.nih.gov)
Materials Availability
This study did not generate new unique reagents.
Data and Code availability
The RNaseq datasets generated during this study are available at GEO: GSE152534 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152534.
Experimental Model and Subject Details
Mice
In the study we used both male and female mice, 6-8 weeks old, unless otherwise specified. C57BL/6J and B10.RIII mice were purchased from The Jackson Laboratory (Bar Harbor, ME). R161H mice were generated as described previously (Horai et al., 2013). Il17a −/− R161H mice were generated by backcrossing Il17a −/− mice on the C57BL/6 background (a kind gift from Dr. Yoichiro Iwakura, Tokyo University of Science, Japan(Nakae et al., 2002)) to R161H mice on the B10.RIII background for at least 8 generations. Il17f −/− and Il17a −/− Il17f −/− mice on the C57BL/6 background were from Dr. Iwakura (Ishigame et al., 2009). Il24 −/− mice were obtained from Merck Sharp & Dohme Corp (New Jersey) (Chan et al., 2006). All mice were kept in a specific pathogen-free facility and fed standard laboratory chow ad libitum. Animal care and use were in compliance with institutional and ARVO guidelines. The animal study protocol was approved by the Animal Care and Use Committee of the National Eye Institute.
Human samples
Peripheral blood was collected from 14 healthy blood donors (7 males and 7 females, ages ranging from 20 to 36 years) by venipuncture upon approval by the Institutional Review Board of the Zhongshan Ophthalmic Center (2014MEKY045). Peripheral blood mononuclear cells (PBMC) were isolated as described ahead. The study was performed at the Zongshan Ophthalmic Center and adhered to the tenets of the Declaration of Helsinki.
Method Details
Induction of EAU and disease scoring
Induction of EAU by active immunization was described previously (Mattapallil et al., 2015; Silver et al., 1995). In brief, C57BL/6J and Il24 −/− mice were immunized subcutaneously with 200-300 μg IRBP651-670 peptide emulsified in an equal volume of Complete Freund’s Adjuvant (CFA) containing 2.5 mg/ml M. tuberculosis. In addition, these mice also received 1.0 μg of Bordetella pertussis toxin (Sigma-Aldrich, St. Louis, MO) intraperitoneally on the day of immunization as reported previously (Mattapallil et al., 2015). B10.RIII mice were immunized with 7-8 μg IRBP161-180 peptide emulsified in an equal volume of Complete Freund’s Adjuvant (CFA) containing 2.5 mg/ml M. tuberculosis subcutaneously without pertussis toxin. In some experiments, an anti-IL-17A antibody (clone 17F3, BioXcell) or an isotype IgG1 control (MOPC-21, BioXcell) was injected i.p. (250 μg) every other day from day −1 of immunization.
For induction of EAU by adoptive transfer, lymph nodes from naive Il17a +/+ R161H or Il17a −/− R161H mice (B10.RIII background) (Horai et al., 2015; Horai et al., 2013) were dispersed into single-cell suspensions and cultured in 12-well plates at 2 × 106 cells/ml (5 × 106 cells/well) or 6-well plates at 2-3 × 106 cells/ml (10x 106 cells/well). Cells were activated with 2 μg/ml of IRBP161-180 peptide in the presence of 25 ng/ml IL-6, 2.5 ng/ml of TGF-β, 10 μg/ml of anti–IFN-γ and 10 μg/ml of anti–IL-4 Abs (R&D systems, Minneapolis, MN) for the Th17 polarization condition. After 48 hr., 10 ng/ml of IL-23 was added. After 72 hr., activated live cells were purified by centrifugation over Lympholyte M (Cedarlane, Burlington, NC) and washed with 1X PBS. Four to 5 × 106 cells were injected i.p. into naive congenic B10.RIII mice. In some experiments, the recipient mice received recombinant IL-24 (R&D systems) at the rate of 100 μg per mouse by i.p. injection every other day.
Clinical EAU was evaluated by fundus examination on a scale of 0–4 based on the severity of inflammation (Agarwal and Caspi, 2004). Eyes, harvested at the end of experiments, were processed for histopathology and stained with standard hematoxylin and eosin stain. Disease severity by histology was scored by a masked observer on a scale of 0–4, based on the number, type, and extent of lesions, as described previously (Agarwal and Caspi, 2004).
Induction of EAE and disease scoring
Mice on the C57BL/6 background were immunized with 200 μg of Myelin Oligodendrocyte Glycoprotein peptide (MOG35-55) emulsified in equal volume of CFA containing 4.0 mg/ml M. tuberculosis and two doses of 0.3 μg of Bordetella pertussis toxin given intraperitoneally at the time of and 48 hr. after immunization. Mice were observed for clinical signs of EAE starting from day 8 after immunization and disease was scored on a scale of 0-5 using a standard scoring system; 0 = no disease, 1 = limp tail, 2 = hind limb weakness, 3 = hind limb paralysis, 4 = hind limb and fore limb paralysis, and 5 = moribund or death.
Isolation and analysis of tissue-infiltrating cells
Eyes were collected from mice with EAU 19-21 days after immunization or 4–8 days after adoptive transfer, as specified. After removal of external tissues, the eyes were carefully dissected along the limbus for lens removal. The remaining tissue was minced with scissors in cold RPMI medium. After centrifugation, the resultant cell pellet was resuspended in RPMI medium containing 1 mg/ml collagenase D and incubated for 45 min at 37°C. Samples were then dispersed by pipetting several times, washed, filtered, and suspended in RPMI medium with 10% FCS. Brains and spinal cords from mice with EAE were removed, minced and incubated in RPMI medium containing Collagenase D and incubated at 37°C for 45 min. Tissue pieces were then crushed, filtered through 70 μm cell strainer and washed. Resuspended cells were then mixed with Percoll and centrifuged at 15,000 x g at 4°C for 5 min to separate leukocytes from erythrocytes and myelin. Tissue infiltrating cells from eyes or brains or spinal cords were then pulsed with phorbol myristate acetate (PMA) (50 ng/ml) and ionomycin (500 ng/ml) in the presence of brefeldin A (GolgiPlug; BD Biosciences, San Diego, CA) for 4 hr., stained for cell surface markers, fixed with 4% paraformaldehyde and permeabilized with PBS containing 0.1% BSA and 0.05% Triton X-100 for intracellular cytokine staining. Anti–IFN-γ, anti–IL-17A, anti-IL-17F, anti-GM-CSF antibodies were purchased from eBioscience (San Diego, CA) and/or BioLegend (San Diego, CA). Primary anti–IL-24 goat antibody was purchased from R&D systems and secondary anti-goat antibody was purchased from Thermofisher Scientific. Mouse anti-IL-22 antibody (clone 3F11.3 from Genentech) was conjugated with AF647 dye (Molecular Probes, Eugene, OR) according to manufacturer’s instructions. IL-22 was not examined in some of the experiments due to the limited number of cells available from ocular tissue.
T cell differentiation
Mouse
CD4+CD62L+ T cells were purified from spleen by using CD4+CD62L+ T Cell Isolation Kit II (Miltenyi Biotech, Cambridge, MA). Cells were stimulated by plate bound anti-CD3 (2 μg/ml) and soluble anti-CD28 (1 μg/ml) Abs. For Th17 polarization, culture media were supplemented with 10 ng/ml IL-6, 10 ng/ml IL-23, 1 ng/ml TGF-β, 10 μg/ml anti-IFN-γ and 10 μg/ml anti-IL-4 antibodies. Where specified, 50 ng/ml of IL-17A, IL-17F or IL-24 (R&D systems) was added to the culture. On day 3, cells were pulsed with PMA and ionomycin in the presence of brefeldin A for 4 hr., surface stained, fixed and permeabilized for intracellular cytokine staining as described above.
Human
PBMCs were isolated from peripheral blood using Ficoll (Sigma-Aldrich). Naive CD45RA+CD4+ T cells were isolated from the PBMCs using naive CD4+ T cell isolation kit II (Miltenyi Biotech). Cells were stimulated with anti-human CD3/CD28 coated beads (Invitrogen) in a bead-to-cell ratio of 1:10. For Th17 polarization, cultures were supplemented with 10 ng/ml each of IL-1β, IL-6, IL-23, 1 ng/ml TGF-β, 10 μg/ml anti-IFN-γ and 10 μg/ml anti-IL-4. Where specified, 10 μg/ml of anti-human IL-17A antibody or isotype control (R&D systems) was added to the cultures. Half of the culture medium was replaced with fresh cytokine-containing medium every 4-5 days. On day 14, cells were pulsed with PMA/Ionomycin and stained for intracellular cytokine analysis as described above Supernatant was collected for IL-24 ELISA (R&D systems).
Knockdown of Il24 by siRNA
Purified mouse CD4+CD62L+ T cells or splenocytes were polarized under Th17 conditions with anti-CD3/CD28 Abs, or IRBP161-180, in the presence of Accell siRNA oligos (Dharmacon, Lafayette, CO) or random control siRNA in medium containing 3 % FCS as described previously (Chong et al., 2014). Cells were harvested on day 3 for follow-up experiments, or on days 4 and 7 to examine the efficiency and duration of IL-24 gene expression knockdown.
RNaseq analysis
RNA samples were purified, fragmented, and the library was constructed as described (Brooks et al., 2012). Sequencing was performed on Illumina GAIIx instrument with SCS v2.10 software. QC was confirmed with FastQC v0.10.1 and was within expected levels for RNA-seq libraries (< 1% of reads). For visualization of aligned sequences, Bowtie2 v2.1.0 (Langmead et al., 2009) was used to align the reads first to transcriptome and then to genome, and TopHat2 v2.0.9 (Trapnell et al., 2009) was used to align to the mouse genome assembly GRCm38.p2. For annotation of mouse RNA, Ensemble v73 was used. SAMtools v0.1.19 was used to create BAM files for visualization in Integrative Genomics Viewer (Robinson et al., 2011). For quantitation of aligned reads to transcriptome, Bowtie2 v2.1.0 was used to align the reads to all possible transcripts and eXpress v1.3.1 was used to assign the reads to transcript isoforms and quantitation (Roberts and Pachter, 2013).
Principal Component Analysis (for reproducibility of replicates), expression plots and clustering were done using the statistical environment ‘R’ (version 3.3.1) and Differential Expression (DE) analysis using EdgeR (Robinson et al., 2010). Generalized Linear Model was used for multiple factor testing and batch correction before DE analysis.
Nuclear Cytoplasmic fractionation and western blotting
Naive T cells were isolated from B10.RIII mice using CD4+CD62L+ T Cell Isolation Kit II. Cells were polarized under the Th17 condition with anti-CD3 and anti-CD28 stimulation for 3 days. Cells were harvested and washed with PBS, suspended in serum-free medium and rested on ice for 1.5 hours. Cells were then treated with IL-17A for 30 min or 60 min and collected into 1.5 mL microcentrifuge tubes. Nuclear and cytoplasmic fractions were prepared according to the manufacture’s protocol using the NE-PER kit (Promega, Madison, WI). Protein lysates were separated by SDS-PAGE. Anti-Histone H3 (EMD Millipore, 06-755 at a dilution of 1:1000) and Anti-GAPDH antibodies (Cell Signaling, 2118S at a dilution of 1:1000) were used as markers for nuclear and cytoplasmic fractions. Anti-p65 (Santa Cruz, SC-109 at a dilution of 1:500) was used to detect p65 subunit of NFkB. HRP-conjugated secondary antibodies and SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) were used for detection.
Immunofluorescence
Naive T cells from B10.RIII mice were isolated and polarized to Th17, harvested, rested on ice for 1.5 hr., and treated with IL-17A as described above. T cells were then fixed using 4% paraformaldehyde at room temperature for 10 minutes. After 2 washes with PBS, cells were permeabilized using 0.3% Triton X-100 in PBS at room temperature for 10 minutes. Cells were incubated with indicated primary antibodies overnight at 4°C in PBS containing 3% BSA and 0.1% Tween 20. After 3 alternate washes with PBS and PBS containing 0.1% Tween 20, cells were incubated with Donkey anti-rabbit Alexa Fluor 568 secondary antibody (ThermoFisher Scientifc) in 1X PBS with 3% BSA and 0.1% Tween 20. After 3 alternate washes with PBS and PBS containing 0.1% Tween 20, cells were mounted using Vectashield on a glass slide with #1.5 coverslips (Leica Biosystems). NFkB p65 Antibody (Santa Cruz, SC-109) was used at dilution of 1:20, FITC-conjugated anti-CD4 Ab (eBioscience, RM4-4) was used at a dilution of 1:100, donkey anti-rabbit Alexa Fluor 568 secondary antibody was used at a dilution of 1:500 and DAPI (ThermoFisher Scientific) at 0.5 ng/ml in PBS was used to stain the nuclei. The samples were imaged using a Zeiss LSM 880 Confocal Microscope with a 63 × 1.4 N.A. Plan-Apochromat objective, 4X zoom and at 1 airy unit using excitation with 405 nm, 488 nm and 568 nm laser line. Images were processed and analyzed using Fiji.
Plasmids and constructs
Genomatix software (GGA suite) was used for predicting the binding sites of NFkB to the Il24 promoter (NC_000067.5). Two putative p65 binding sites were identified as NFKB1 = gtcagaaactcccta and NFKB2 = gaaggagcttcccac. The 2074 bp long Il24 promoter was amplified starting at −216 bp from the transcription start site using total genomic DNA isolated from the tail snip of B10.RIII mice by PCR primers Fwd 5′TAGCTAGCTAGCTTTAGCTTCTGGGCT 3′ and Rev 5′ ATAAGCTTTCAAGGCAGCAGCCTAACA 3′. The WT and the 3 deletion mutants of Il24 promoter were PCR amplified and cloned in pGL4 vector (Promega) using NheI and HindIII sites.
CHIP assays
Naive T cells from B10.RIII mice were isolated and polarized toward Th17 by stimulating with anti-CD3 and anti-CD28 Abs. After 3 days of polarization and 1.5 hr. resting on ice, cells were pulsed with IL-17A for 30 min or 60 min. For CHIP assays, at least 10 million cells were used for each immunoprecipitation conditions. Cells were processed as per manufacturer’s protocols of ChIP-IT Express Enzymatic kit (Active Motif, 103295) and Chromatin IP DNA purification kit (Active Motif, 103500). Anti-p65 (Santa Cruz, SC-109) was used to immunoprecipitate p65 subunit of NFkB. Genomatix software (GGA suite) was used for predicting the binding sites of NFkB to the IL-24 promoter. Real-time PCR was performed using SYBR green (ThermoFisher Scientific) for detection of putative p65 binding sites in the IL-24 promoter in the CHIP DNA. The following primer sequences were used for PCR, promoter region 1: Fwd 5′ TGAGACATTAGCCCAGAGGC 3′, Rev 5′ CTGGAGACTGTGGGTGGTTG 3′; promoter region 2: Fwd 5′ AAAAGTTGGGCAGGCTCAGT 3′, Rev 5′ CAGTTCTCCCAGCCTTGCTT 3′.
Luciferase promoter assay
Naive cells from B10.RIII mice were polarized under the Th17 condition with anti-CD3 and anti-CD28 Abs. Three days later, cells were harvested and transfected with the indicated constructs using Nucleofector (LONZA) according to manufacturer’s instructions. IL-17A was added 6 hr. post transfection. After overnight incubation, cells were harvested, and cell lysates were prepared using passive lysis buffer and Luciferase assays were performed using Dual Luciferase Reporter system from Promega.
Quantification and Statistical Analysis
Graphpad Prism 7 was used for graphing and statistical analyses. Mann-Whitney U test, unpaired t test or paired t test was used for two-group comparison of parametric data. Linear Regression was used for two-group comparison of non-parametric data (EAE and EAU scores). One-way or Two-way ANOVA with Dunnett’s or Tukey’s correction was used for multi-group non-parametric analyses. A p value of ≤ 0.05 was considered statistically significant. Data are depicted as mean ± SEM.
Acknowledgments
We thank Dr. Yoichiro Iwakura for Il17a −/−, Il17f −/−, and Il17a −/− Il17f −/− mice and Dr. Renee de Waal Malefyt of Merck (formerly DNAX) for his gift of Il24 −/− mice. We are grateful to Matthew Brooks, BS, of N-NRL, NEI, for performing RNA-seq. We thank the staff of the Zhongshan Ophthalmic Center Flow Cytometry Core and the NEI Flow Cytometry Core for flow cytometry support. This study was funded by NIH/National Eye Institute (NEI) intramural funding (project number EY000184); the Fundamental Research Funds of the State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center; and the Natural Science Foundation of Guangdong Province (grants 201604020082, 2017A030313836, 2018A030313877 and 2019A1515010956).
Author Contributions
W.P.C. designed, performed, and analyzed experiments and wrote the draft of the manuscript. K.R., M.J.M., S.J.B., W.W.W., Z.C., S.W., Y.Z., Y.J., and R.H. performed and analyzed experiments. P.B.S., and Y.J. evaluated and interpreted fundoscopy data. J.C. analyzed data, provided resources, and secured funding. C.-C.C. and R.H. evaluated and interpreted histopathology data. All authors contributed to writing the manuscript. R.R.C. conceptualized the study, interpreted the data, provided resources, secured funding, and edited and finalized the manuscript.
Declaration of Interests
U.S. Patent No. 10,512,671, issued December 24, 2019 (R.R.C., W.P.C., R.H., and M.J.M.).
Published: July 15, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2020.06.022.
Supplemental Information
References
- Agarwal R.K., Caspi R.R. Rodent models of experimental autoimmune uveitis. Methods Mol. Med. 2004;102:395–419. doi: 10.1385/1-59259-805-6:395. [DOI] [PubMed] [Google Scholar]
- Al Omar S., Flanagan B.F., Almehmadi M., Christmas S.E. The effects of IL-17 upon human natural killer cells. Cytokine. 2013;62:123–130. doi: 10.1016/j.cyto.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Amadi-Obi A., Yu C.R., Liu X., Mahdi R.M., Clarke G.L., Nussenblatt R.B., Gery I., Lee Y.S., Egwuagu C.E. TH17 cells contribute to uveitis and scleritis and are expanded by IL-2 and inhibited by IL-27/STAT1. Nat. Med. 2007;13:711–718. doi: 10.1038/nm1585. [DOI] [PubMed] [Google Scholar]
- Andoh A., Shioya M., Nishida A., Bamba S., Tsujikawa T., Kim-Mitsuyama S., Fujiyama Y. Expression of IL-24, an activator of the JAK1/STAT3/SOCS3 cascade, is enhanced in inflammatory bowel disease. J. Immunol. 2009;183:687–695. doi: 10.4049/jimmunol.0804169. [DOI] [PubMed] [Google Scholar]
- Anuradha R., Munisankar S., Dolla C., Kumaran P., Nutman T.B., Babu S. Modulation of CD4+ and CD8+ T-cell function by interleukin 19 and interleukin 24 during filarial infections. J. Infect. Dis. 2016;213:811–815. doi: 10.1093/infdis/jiv497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing S.J., Silver P.B., Jittayasothorn Y., Mattapalli M.J., Chan C.-C., Horai R., Caspi R.R. Autoimmunity to neuroretina in the concurrent absence of IFN-γ and IL-17A is mediated by a GM-CSF-driven eosinophilic inflammation. J. Autoimmun. 2020 doi: 10.1016/j.jaut.2020.102507. Published online June 24, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks M.J., Rajasimha H.K., Swaroop A. Retinal transcriptome profiling by directional next-generation sequencing using 100 ng of total RNA. Methods Mol. Biol. 2012;884:319–334. doi: 10.1007/978-1-61779-848-1_23. [DOI] [PubMed] [Google Scholar]
- Caspi R.R. A look at autoimmunity and inflammation in the eye. J. Clin. Invest. 2010;120:3073–3083. doi: 10.1172/JCI42440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.R., Blumenschein W., Murphy E., Diveu C., Wiekowski M., Abbondanzo S., Lucian L., Geissler R., Brodie S., Kimball A.B. IL-23 stimulates epidermal hyperplasia via TNF and IL-20R2-dependent mechanisms with implications for psoriasis pathogenesis. J. Exp. Med. 2006;203:2577–2587. doi: 10.1084/jem.20060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Caspi R.R., Chong W.P. IL-20 receptor cytokines in autoimmune diseases. J. Leukoc. Biol. 2018;104:953–959. doi: 10.1002/JLB.MR1117-471R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W., Zhu X., Yang P., Liu X., Lin X., Zhou H., Huang X., Kijlstra A. Upregulated IL-23 and IL-17 in Behçet patients with active uveitis. Invest. Ophthalmol. Vis. Sci. 2008;49:3058–3064. doi: 10.1167/iovs.07-1390. [DOI] [PubMed] [Google Scholar]
- Chong W.P., Horai R., Mattapallil M.J., Silver P.B., Chen J., Zhou R., Sergeev Y., Villasmil R., Chan C.C., Caspi R.R. IL-27p28 inhibits central nervous system autoimmunity by concurrently antagonizing Th1 and Th17 responses. J. Autoimmun. 2014;50:12–22. doi: 10.1016/j.jaut.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong W.P., van Panhuys N., Chen J., Silver P.B., Jittayasothorn Y., Mattapallil M.J., Germain R.N., Caspi R.R. NK-DC crosstalk controls the autopathogenic Th17 response through an innate IFN-γ-IL-27 axis. J. Exp. Med. 2015;212:1739–1752. doi: 10.1084/jem.20141678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codarri L., Gyülvészi G., Tosevski V., Hesske L., Fontana A., Magnenat L., Suter T., Becher B. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 2011;12:560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- Dick A.D., Tugal-Tutkun I., Foster S., Zierhut M., Melissa Liew S.H., Bezlyak V., Androudi S. Secukinumab in the treatment of noninfectious uveitis: results of three randomized, controlled clinical trials. Ophthalmology. 2013;120:777–787. doi: 10.1016/j.ophtha.2012.09.040. [DOI] [PubMed] [Google Scholar]
- Dincses E., Yurttas B., Esatoglu S.N., Melikoglu M., Hamuryudan V., Seyahi E. Secukinumab induced Behçet’s syndrome: a report of two cases. Oxf. Med. Case Rep. 2019;2019:omz041. doi: 10.1093/omcr/omz041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoutier L., Leemans C., Lejeune D., Kotenko S.V., Renauld J.C. Cutting edge: STAT activation by IL-19, IL-20 and mda-7 through IL-20 receptor complexes of two types. J. Immunol. 2001;167:3545–3549. doi: 10.4049/jimmunol.167.7.3545. [DOI] [PubMed] [Google Scholar]
- El-Behi M., Ciric B., Dai H., Yan Y., Cullimore M., Safavi F., Zhang G.X., Dittel B.N., Rostami A. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat. Immunol. 2011;12:568–575. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffen S.L. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 2009;9:556–567. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan H., Shukrun R., Caspi R., Vax E., Pode-Shakked N., Goldberg S., Pleniceanu O., Bar-Lev D.D., Mark-Danieli M., Pri-Chen S. In vivo expansion of cancer stemness affords novel cancer stem cell targets: malignant rhabdoid tumor as an example. Stem Cell Reports. 2018;11:795–810. doi: 10.1016/j.stemcr.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak S., Croxford A.L., Kreymborg K., Heppner F.L., Pouly S., Becher B., Waisman A. IL-17A and IL-17F do not contribute vitally to autoimmune neuro-inflammation in mice. J. Clin. Invest. 2009;119:61–69. doi: 10.1172/JCI35997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L.E., Hatton R.D., Mangan P.R., Turner H., Murphy T.L., Murphy K.M., Weaver C.T. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- Haudenschild D., Moseley T., Rose L., Reddi A.H. Soluble and transmembrane isoforms of novel interleukin-17 receptor-like protein by RNA splicing and expression in prostate cancer. J. Biol. Chem. 2002;277:4309–4316. doi: 10.1074/jbc.M109372200. [DOI] [PubMed] [Google Scholar]
- Horai R., Caspi R.R. Cytokines in autoimmune uveitis. J. Interferon Cytokine Res. 2011;31:733–744. doi: 10.1089/jir.2011.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai R., Silver P.B., Chen J., Agarwal R.K., Chong W.P., Jittayasothorn Y., Mattapallil M.J., Nguyen S., Natarajan K., Villasmil R. Breakdown of immune privilege and spontaneous autoimmunity in mice expressing a transgenic T cell receptor specific for a retinal autoantigen. J. Autoimmun. 2013;44:21–33. doi: 10.1016/j.jaut.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai R., Chong W.P., Zhou R., Chen J., Silver P.B., Agarwal R.K., Caspi R.R. Spontaneous ocular autoimmunity in mice expressing a transgenic T cell receptor specific to retina: a tool to dissect mechanisms of uveitis. Curr. Mol. Med. 2015;15:511–516. doi: 10.2174/1566524015666150731095201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.C., Yang P., Wang J., Wu Q., Myers R., Chen J., Yi J., Guentert T., Tousson A., Stanus A.L. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat. Immunol. 2008;9:166–175. doi: 10.1038/ni1552. [DOI] [PubMed] [Google Scholar]
- Hueber W., Sands B.E., Lewitzky S., Vandemeulebroecke M., Reinisch W., Higgins P.D., Wehkamp J., Feagan B.G., Yao M.D., Karczewski M., Secukinumab in Crohn’s Disease Study Group Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61:1693–1700. doi: 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigame H., Kakuta S., Nagai T., Kadoki M., Nambu A., Komiyama Y., Fujikado N., Tanahashi Y., Akitsu A., Kotaki H. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Ke Y., Liu K., Huang G.Q., Cui Y., Kaplan H.J., Shao H., Sun D. Anti-inflammatory role of IL-17 in experimental autoimmune uveitis. J. Immunol. 2009;182:3183–3190. doi: 10.4049/jimmunol.0802487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M., Kuchroo V.K. IL-17 and Th17 cells. Annu. Rev. Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger D., Silver P.B., Tang J., Cua D., Chen Z., Iwakura Y., Bowman E.P., Sgambellone N.M., Chan C.C., Caspi R.R. Either a Th17 or a Th1 effector response can drive autoimmunity: conditions of disease induction affect dominant effector category. J. Exp. Med. 2008;205:799–810. doi: 10.1084/jem.20071258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil M.J., Silver P.B., Cortes L.M., St Leger A.J., Jittayasothorn Y., Kielczewski J.L., Moon J.J., Chan C.C., Caspi R.R. Characterization of a new epitope of IRBP that induces moderate to severe uveoretinitis in mice with H-2b haplotype. Invest. Ophthalmol. Vis. Sci. 2015;56:5439–5449. doi: 10.1167/iovs.15-17280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil M.J., Kielczewski J.L., Zárate-Bladés C.R., St Leger A.J., Raychaudhuri K., Silver P.B., Jittayasothorn Y., Chan C.C., Caspi R.R. Interleukin 22 ameliorates neuropathology and protects from central nervous system autoimmunity. J. Autoimmun. 2019;102:65–76. doi: 10.1016/j.jaut.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinley A.M., Sutton C.E., Edwards S.C., Leane C.M., DeCourcey J., Teijeiro A., Hamilton J.A., Boon L., Djouder N., Mills K.H.G. Interleukin-17A serves a priming role in autoimmunity by recruiting IL-1beta-producing myeloid cells that promote pathogenic T cells. Immunity. 2020;52:342–356.e6. doi: 10.1016/j.immuni.2020.01.002. [DOI] [PubMed] [Google Scholar]
- Milovanovic M., Drozdenko G., Weise C., Babina M., Worm M. Interleukin-17A promotes IgE production in human B cells. J. Invest. Dermatol. 2010;130:2621–2628. doi: 10.1038/jid.2010.175. [DOI] [PubMed] [Google Scholar]
- Mozaffari S., Nikfar S., Abdollahi M. Inflammatory bowel disease therapies discontinued between 2009 and 2014. Expert Opin. Investig. Drugs. 2015;24:949–956. doi: 10.1517/13543784.2015.1035432. [DOI] [PubMed] [Google Scholar]
- Nakae S., Komiyama Y., Nambu A., Sudo K., Iwase M., Homma I., Sekikawa K., Asano M., Iwakura Y. Antigen-specific T cell sensitization is impaired in IL-17-deficient mice, causing suppression of allergic cellular and humoral responses. Immunity. 2002;17:375–387. doi: 10.1016/s1074-7613(02)00391-6. [DOI] [PubMed] [Google Scholar]
- Neurology Reviews What is the relationship between MS and uveitis? Neurol. Rev. 2015;23:9. [Google Scholar]
- O’Connor W., Jr., Kamanaka M., Booth C.J., Town T., Nakae S., Iwakura Y., Kolls J.K., Flavell R.A. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat. Immunol. 2009;10:603–609. doi: 10.1038/ni.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Li Z., Yang X.O., Chang S.H., Nurieva R., Wang Y.H., Wang Y., Hood L., Zhu Z., Tian Q., Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D.D., Kuchroo V.K. Th17 cell pathway in human immunity: lessons from genetics and therapeutic interventions. Immunity. 2015;43:1040–1051. doi: 10.1016/j.immuni.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Peng Y., Han G., Shao H., Wang Y., Kaplan H.J., Sun D. Characterization of IL-17+ interphotoreceptor retinoid-binding protein-specific T cells in experimental autoimmune uveitis. Invest. Ophthalmol. Vis. Sci. 2007;48:4153–4161. doi: 10.1167/iovs.07-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandquist I., Kolls J. Update on regulation and effector functions of Th17 cells. F1000Res. 2018;7:205. doi: 10.12688/f1000research.13020.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer G., Venkataraman C., Schindler U. Cutting edge: FISP (IL-4-induced secreted protein), a novel cytokine-like molecule secreted by Th2 cells. J. Immunol. 2001;166:5859–5863. doi: 10.4049/jimmunol.166.10.5859. [DOI] [PubMed] [Google Scholar]
- Shiga H., Fukuda S., Iijima K. Interleukin-17A inhibitor-induced Crohn’s disease/Behçet’s disease-like lesions. Inflamm. Bowel Dis. 2017;23:E38–E39. doi: 10.1097/MIB.0000000000001142. [DOI] [PubMed] [Google Scholar]
- Silver P.B., Rizzo L.V., Chan C.C., Donoso L.A., Wiggert B., Caspi R.R. Identification of a major pathogenic epitope in the human IRBP molecule recognized by mice of the H-2r haplotype. Invest. Ophthalmol. Vis. Sci. 1995;36:946–954. [PubMed] [Google Scholar]
- Smith E., Stark M.A., Zarbock A., Burcin T.L., Bruce A.C., Vaswani D., Foley P., Ley K. IL-17A inhibits the expansion of IL-17A-producing T cells in mice through “short-loop” inhibition via IL-17 receptor. J. Immunol. 2008;181:1357–1364. doi: 10.4049/jimmunol.181.2.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R., Lubberts E., Hendriks R.W. A cellular and molecular view of T helper 17 cell plasticity in autoimmunity. J. Autoimmun. 2018;87:1–15. doi: 10.1016/j.jaut.2017.12.007. [DOI] [PubMed] [Google Scholar]
- Tang C., Kakuta S., Shimizu K., Kadoki M., Kamiya T., Shimazu T., Kubo S., Saijo S., Ishigame H., Nakae S., Iwakura Y. Suppression of IL-17F, but not of IL-17A, provides protection against colitis by inducing Treg cells through modification of the intestinal microbiota. Nat. Immunol. 2018;19:755–765. doi: 10.1038/s41590-018-0134-y. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Vietinghoff S., Ley K. IL-17A controls IL-17F production and maintains blood neutrophil counts in mice. J. Immunol. 2009;183:865–873. doi: 10.4049/jimmunol.0804080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl C., Müller W., Leithäuser F., Adler G., Oswald F., Reimann J., Schirmbeck R., Seier A., Weiss J.M., Prochnow B., Wegenka U.M. IL-20 receptor 2 signaling down-regulates antigen-specific T cell responses. J. Immunol. 2009;182:802–810. doi: 10.4049/jimmunol.182.2.802. [DOI] [PubMed] [Google Scholar]
- Wang C., Tian Y., Lei B., Xiao X., Ye Z., Li F., Kijlstra A., Yang P. Decreased IL-27 expression in association with an increased Th17 response in Vogt-Koyanagi-Harada disease. Invest. Ophthalmol. Vis. Sci. 2012;53:4668–4675. doi: 10.1167/iovs.12-9863. [DOI] [PubMed] [Google Scholar]
- Wasilewska A., Winiarska M., Olszewska M., Rudnicka L. Interleukin-17 inhibitors. A new era in treatment of psoriasis and other skin diseases. Postepy Dermatol. Alergol. 2016;33:247–252. doi: 10.5114/ada.2016.61599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.O., Chang S.H., Park H., Nurieva R., Shah B., Acero L., Wang Y.H., Schluns K.S., Broaddus R.R., Zhu Z., Dong C. Regulation of inflammatory responses by IL-17F. J. Exp. Med. 2008;205:1063–1075. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N., Shalek A.K., Gaublomme J.T., Jin H., Lee Y., Awasthi A., Wu C., Karwacz K., Xiao S., Jorgolli M. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNaseq datasets generated during this study are available at GEO: GSE152534 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152534.