Abstract
The new virus of the of β-Coronaviruses genus, SARS-CoV-2, is the causative agent of coronavirus disease-2019 (COVID-19) and is winning a proverbial chess match against all players simultaneous, including physicians, clinicians, pathologists, doctors, scientists, economists, athletes and politicians. The COVID-19 outbreak has seriously threatened public health, killing the most vulnerable persons and causing general panic. To stop this disease, effective remedies (i.e., drugs, vaccines, personal protection elements, etc.) are urgently required. Unfortunately, no registered specific therapies (including antiviral therapies, immune-modulating agents and vaccines) are currently available to treat coronavirus infections, highlighting an urgent need for therapeutics targeting SARS-CoV-2. In this work, fourteen existing small molecule drugs or/and experimental drugs selected by experts and examined from the point of view of bioavailability via the Lipinski-Veber rules and assessment of their physicochemical descriptors. The aim of this study is to discover selected pattern similarities and peculiar characteristics that could be useful for antiviral drug optimization, drug combination or new antiviral agent design.
Keywords: SARS-CoV-2, Antiviral drugs, COVID-19, In silico ADMET properties, Lipinski’s parameters, Bioactivity scores
Graphical abstract
1. Introduction
The current pandemic coronavirus disease-2019 (COVID-19) is a new infectious pneumonia-like illness caused by a novel virus strain, so-called severe acute respiratory syndrome coronavirus 2 or SARS-CoV-2 [1,2]. This virus is a member of the β-Coronaviruses genus (family Coronaviridae) and is similar to severe acute respiratory syndrome coronavirus (SARS-CoV) [3] and Middle East respiratory syndrome coronavirus (MERS-CoV) [4]. The common symptoms of a person infected with SARS-CoV-2 include respiratory symptoms, fever, dry cough, shortness of breath, and central nervous system conditions leading to multiple organ failure. In more severe cases, infection causes bilateral pneumonia, acute respiratory distress syndrome, kidney failure, and ultimately, death [5,6]. Current data on COVID-19 indicate that the SARS-CoV-2 mortality rate is generally far lower (3–5%) than that of SARS-CoV (9–15%) and MERS-CoV (34–37%), but SARS-CoV-2 is much more transmissible/contagious than the SARS-CoV and MERS-CoV viruses and affects more people over the age of 60 or those with comorbidities that weaken the immune system [[7], [8], [9]]. At the height of the crisis, this virus is spreading at a rate and scale far worse than previous coronaviral epidemics.
To stop world circulation of SARS-CoV-2, we have only basic protective measures recommended by the World Health Organization (WHO) [10], as in the case of SARS-CoV. The phase “Using barrier precautions, the transmission of SARS-CoV can be prevented” was written in 2004 [8], and moreover, experts warned that sooner or later a new outbreak might occur [11,12]. This event occurred in 2019, but no one expected that it would cover the entire world, i.e., growing to pandemic size, and the pharmaceutical industry was taken by surprise. To date (June 04, 2020), more than 11million worldwide cases of infection and 525.491 deaths have been attributed to the novel coronavirus, SARS-CoV-2, since its emergence in December 2019. To cure hundreds of thousands of infected people, it is necessary to develop effective specific and selective antiviral drugs. The truth is that to date, there are no safe and specific antiviral agents against SARS-CoV-2, and the current methods for treatment of COVID-19 are still controversial. Fortunately, after analyzing the complete genome of SARS-CoV-2 from Wuhan, China [13,14], important structural and biochemical details are now available on this coronavirus and its clinical features:
-
1.
The viral genome consists of more than 29,000 bases and encodes nonstructural 29 proteins, including RNA-dependent RNA polymerase (RdRp), coronavirus main protease (3CLpro), and papain-like protease (PLpro) [[15], [16], [17]];
-
2.
The SARS-CoV-2 virus, similar to its cousin SARS-CoV, uses a glycosylated spike protein S1 that binds the virion to a receptor protein known as angiotensin-converting enzyme 2 (ACE2) located on the surface membrane of the host [[18], [19], [20], [21]], whereas the MERS-CoV binds to the human dipeptidyl peptidase 4 (DPP4) receptor through its spike glycoprotein [22]. Viral spike glycoproteins (SARS-CoV-2 virus) can also bind to the serine protease TMPRSS2, another host cell surface receptor that uses the SARS-CoV virus [21];
-
3.
Although both SARS-CoV-2 and SARS-CoV are close relatives (79.5–82% identity) and are highly comparable at the amino acid level, they are not identical, especially with respect to the spike proteins that might be the reason for the higher binding affinity of the SARS-CoV-2 spike protein to the human ACE2 receptor, which is abundantly present in the lungs. This feature apparently gives the virus its high ability to cause pneumonia [20,23];
-
4.
Upon entering the host cells, in the cytoplasm, the viral genome is released as a positive sense single-stranded RNA, and its antigens are detected by the antigen-presenting cells (APCs), which are the major cells of the immune system (dendritic cells or macrophages, especially alveolar macrophages) involved in protection against invading antigens. This detection triggers a downstream cascade of many T-cell stimulators and inflammatory molecules, the so-called cytokine storm, leading to activation of transcription factor nuclear factor-κB (NF-κB) and interferon regulatory factor 3 (IRF3), with the subsequent production of type I interferons (IFN-α/β) and a series of pro-inflammatory cytokines including interleukin (IL)-1β and IL-6 [[24], [25], [26], [27]]. The cytokine storm produces a violent attack on the body by the immune system, initiating acute respiratory distress syndrome and multiple organ failure.
Based on this information and their experiences, many scientific researchers have begun to contribute to the design and development of molecules against coronavirus. The largest obstacle is that there is simply no time to create a new drug, and the need exists to use old drugs or experimental drug candidates, i.e., to find such compounds for which preclinical trials have been conducted and safety has been proven, and it remains “only” to check activity against SARS-CoV-2, thus realizing a drug repurposing strategy [28]. As such, rapid screening of safe and effective drugs available in drug libraries is the first important step in the anti-SARS-Co-V2 drug search.
Most of the drugs examined for treatment of this disease fall into the following drug classifications: antiviral with broad spectrum, anti-HIV (antiretroviral), antimalarial, antibiotic, antiparasitic, anticancer, anti-inflammatory and immunosuppressive drugs (including monoclonal antibodies such as tocilizumab, adalimumab, etc.) [29,30].
This work addresses potential anti-SARS-CoV-2 drugs based on small heterocyclic molecules. In this work, fourteen promising approved drugs or/and experimental drugs selected by experts in antiviral research and clinical parasitic infections were analyzed from the point of view of bioavailability by applying the Lipinski-Veber rules and determining their physicochemical parameters to find similar patterns and peculiar characteristics that could be valuable in antiviral drug optimization and drug combination or new antiviral agent design.
2. The question is, how do we find a suitable cure against COVID-19?
Using the main potential targets and their roles in viral infection, many researchers have attempted to collect lists of possible synthetic and natural compounds via molecular dynamics simulations that can supply a roadmap for future studies [[31], [32], [33], [34], [35], [36], [37]]. However, due to the critical situation of coronavirus disease 2019, the WHO launched a megatrial to test selected repurposed drugs and experimental drug candidates [38]. The selection is based on i) previous biochemical, pharmacological and medical information on a drug and ii) new information on in silico molecular modeling screening and in vitro and in vivo anti-SARS-Co-V2 activities of a drug [[37], [38], [39], [40], [41], [42], [43], [44]]. Using these criteria, the race to identify treatments has dramatically accelerated over the past three months. Nevertheless, even if a drug candidate is discovered quickly, which is far from certain, preclinical and clinical tests are expected to take years.
However, numerous published articles are currently available on a wide variety of aspects of studying and combatting SARS-CoV-2 via open access websites or as reprints in scientific journals such as chemRxiv, bioRxiv, medRxiv, etc. Certainly, these papers have not been reviewed and should not be considered as final conclusions or recommendations for treatment or prevention and should not be covered in the media as proven information. Moreover, promising results obtained from molecular docking of selected drugs with potential targets do not guarantee the effectiveness of a drug in treatment under clinical conditions. However, these works provide a general idea of where research should go.
3. Promising drug candidates in the fight against COVID-19
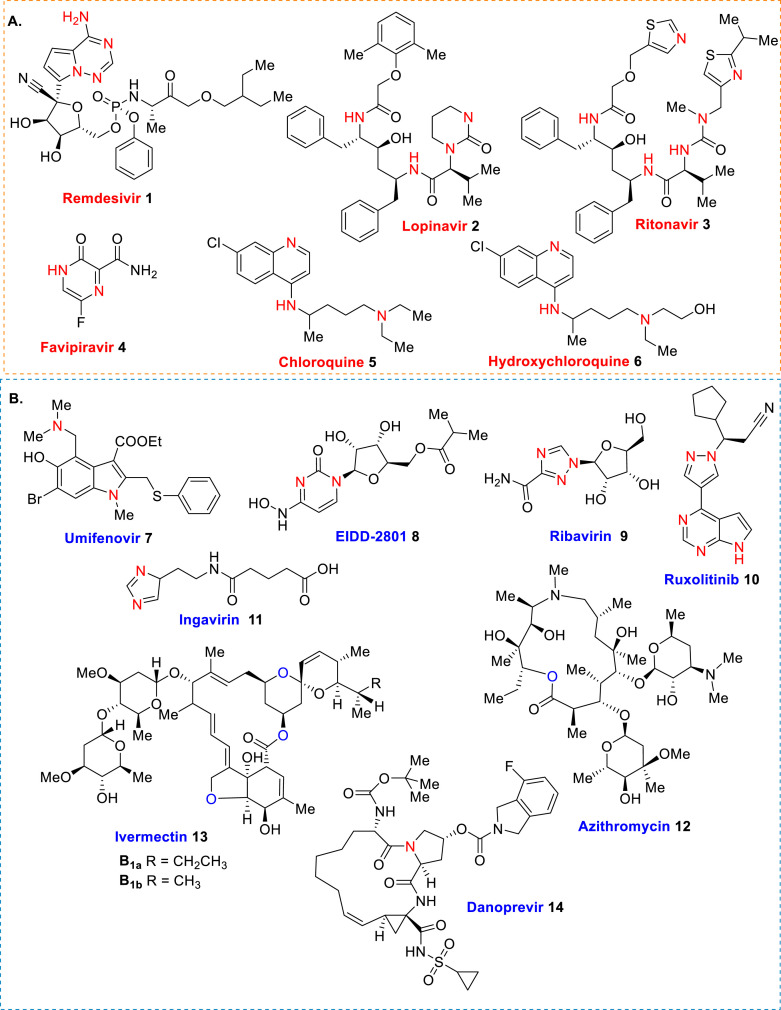
Certainly, the risk from the use of an approved drug should be significantly lower than from the disease itself. Therefore, the medicine must be safe. This fact greatly complicates the search: it is likely that at the stage of large clinical trials, it will be discovered that too much risk exists from the drug, risk that is incomparable with the danger of the disease. However, remdesivir (1), lopinavir (2) and ritonavir (3), favipiravir (4), chloroquine (5) and hydroxychloroquine (6) (Fig. 1 A) and their combinations [[45], [46], [47]] are currently the most promising small molecule drugs for use in COVID-19 treatment. Clinical efforts to find appropriate, efficient and safe therapeutics are still ongoing [30,48].
Fig. 1.
Structures of promising drugs and experimental drug candidates 1–14 in COVID-19 treatment.
Remdesivir (GS-5734) is an antivirus unapproved drug candidate that was developed by Gilead Sciences (Foster City, CA, USA) to combat the West African Ebola virus disease epidemic in the mid-2010s. This nucleotide prodrug has also shown activity against the hemorrhagic fever Marburg virus (MARV), the SARS-coV and MERS-coV). However, it is administrated only intravenously [[49], [50], [51]]. Anti-HIV drug lopinavir is developed from ritonavir. Both are created against HIV-1 protease. Lopinavir/ritonavir combination as a single medication is known as Kaletra®, effective drug, approved by FDA in 2001 for the treatment of HIV infection in adults [52,53]. Favipiravir (T-705, Avigan®) is a pyrazinecarboxamide molecule, capable of converting into the cell into a nucleoside analog, ribonucleotide T-705-4-ribofuranosyl-5′-monophosphate, which inhibits the activity of viral RNA polymerase without affecting cellular synthesis of RNA or DNA. This antiviral drug was developed in Japan and approved in 2014 for treating viral strains unresponsive to current antivirals [54,55]. Chloroquine (Resochin®, Aralen®) and hydroxychloroquine (Dolquine®, Plaquenil®, Axemal®) are antimalarial drugs. The latter drug is also used in rheumatoid arthritis and lupus treatments. Both drugs possess a high level of toxicity, especially, cardiotoxicity [[56], [57], [58], [59]].
However, because the epidemiological situation in the world is changing rapidly, literally daily, the following chemical entities of umifenovir 7, agent EIDD-2801 8, ribavirin 9, ruxolitinib 10, ingavirin 11, azithromycin 12, ivermectin 13 and danoprevir 14 (Fig. 1B) must be considered in ongoing trials.
Umifenovir is name of arbidol, which is an original Russian antiviral drug widely used for etiotropic therapy of influenza and flu. In 2011, the WHO rendered an international nonproprietary name Umifenovir to an active ingredient of arbidol but the original name given to this drug by its creators is still much more common and familiar both to patients and researchers. Up to now, its primary therapeutic action has not yet been known for certain: whether it is a direct antiviral activity, stimulation of immune system or anti-inflammatory action [60,61]. Agent EIDD-2801 works similarly to remdesivir, acting as nucleoside analogs that metabolize into an active form that blocks RNA polymerase, an essential component of viral replication. It is being studied in five Phase III trials against COVID-19. This drug candidate can be taken in pill form [62,63]. Ribavirin (Rebetol®, Virazole®, etc.) is a synthetic nucleoside antiviral drug, which is approved by FDA for treatment of respiratory syncytial virus and hepatitis C virus infection. This drug, discovered in 1972, can be administered to humans by aerosol, oral and intravenous routes with a range of dosing regimens in different clinical settings. Ribavirin was found clinically effective against influenza, herpesvirus infections, hemorrhagic fever and Lassa fever but ineffective against Ebola and Marburg viruses [[64], [65], [66], [67]].
Ruxolitinib (Akavi®) is approved in 2011 to treat certain rare bone marrow/blood cancers (e.g., myeloproliferative neoplasm). The drug works by blocking enzymes JAK1/JAK2 in the JAK-STAT pathway, which is overactive in these blood cancers. At the same time, the JAK-STAT pathway is fundamental to many biological processes related to immunity and inflammation, including marshaling a cytokine response [68,69]. Ingavirin® is an original Russian antiviral drug for the treatment and prevention of acute respiratory viral infections by influenza and non-influenza with a unique mechanism of action. This drug provides early recognition of infection and the formation of the antiviral state of cells that stops the virus from reproducing and spreading in the body. It is effective against type A and B viruses, adenoviruses, parainfluenza viruses, respiratory syncytial viruses, coronaviruses, metapneumoviruses, enteroviruses, including Coxsackie virus and rhinovirus [[70], [71], [72]]. Azithromycin (Zithromax®) is an inexpensive semisynthetic antibiotic drug used for the treatment of several bacterial infection. It was discovered 1980 and approved for medical use in 1988. In 2019, this drug was found active against pandemic influenza 2009 (A(H1N1)pdm09) virus acting by interfering with virus internalization process [73,74]. Ivermectin (Stromectol®), an old antiparasitic drug, is widely used in veterinary medicine as an anthelminthic medication since 1981. It is used as a mixture of two macrocyclic lactone molecules, which were isolated from the bacterium Streptomyces avermitilis in 1975. This drug works by paralyzing and killing parasites [[75], [76], [77], [78]]. Danoprevir (ITMN-191/R7227, Ganovo®) is one the antivirals against hepatitis C virus belonging to the second-generation group so-called, direct-acting antiviral agents that act as NS3/4A protease inhibitors. It was developed in 2008; and in 2018, danoprevir was approved as a drug in combination with ritonavir, peginterferon alfa and ribavirin for the treatment of treatment-naive patients with non-cirrhotic genotype 1b chronic hepatitis C. It is a potent and selective noncovalent reversible NS3 inhibitor, a chymotrypsin-like serine protease that plays an essential role in the HCV viral replication process [[79], [80], [81], [82]].
In analyzing the main structural details of selected drugs/drug candidates, it was noted that of the 14 compounds, two are oxygenated macrocyclic compounds obtained directly or via sequential transformations from Actinomycete bacteria (comp. 12 and comp. 13 as a B1a/B1b mixture, 80/20), and twelve are nitrogen-containing heterocycles are synthetic molecules. Among these, three candidates are nucleoside-based heterocycles (pyrrolo[2,1-f] [1,2,4]triazine 1, 2-oxopyrimidine 8 and 1,2,4-triazole 9), which resemble the respective RNA bases of adenosine and cytidine, two are so-called peptide-heterocyclic chimera derivatives (comp. 2 and 3), and others are functionalized derivatives of classic heteroaromatic rings, i.e., pyrazine (comp. 4), quinoline (comp. 5 and 6), indole (comp. 7), pyrrolo[2,3-d]pyrimidin-pyrazol hybrid (comp. 10), imidazole (comp. 11) and macrocyclic lactam (comp. 14).
Our list contains only fourteen compounds, but the molecular and skeletal diversity is so great that it guarantees their specific interactions with the different above-mentioned targets of SARS-Co-V2 virus. Certainly, each structure of these drugs has been optimized, but for their specific profiles, we followed the approach based on monotherapy (“one-target, one-disease” approach). We are facing a novel virus that can mutate rapidly [83,84]. Similar to HIV/AIDS treatment, which consists of the so-called “AIDS cocktail” therapy (combination antiretroviral therapy) [85], future COVID-19 treatment could be based on combination drugs that impact multiple targets simultaneously (i.e., “double drug cocktail” or “triple drug cocktail”, polytherapy). This approach could be generally more effective than the monotherapy method, and it is currently implemented in practice in this case [86,87], but unsuccessful individual experiments [45,88] cannot rule out the significance of this approach. Moreover, multicomponent drugs that are co-formulated in a single tablet, which alters the ability of the other components to reach its target, or better, single drugs that are based on two different frameworks responsible for pharmacological effects (pharmacophores) and able to modulate multiple targets simultaneously [89,90], could be highly advantageous in the future. However, from old magic bullets to modern “binary or trinary weapons”, a treatment against this coronavirus is still far away.
4. Bioavailability via the Lipinski-Veber rules and assessment of physicochemical descriptors
It is well known that the balance between solubility and polar/hydrophobic properties is critical for a drug that must be absorbed, penetrate numerous biological barriers and reach the desired site of action without affecting the biological balance of the entire organism. Based on a literature analysis, almost all of the abovementioned small molecule drugs could bind four highly different targets of SARS-Co-V2 virus, i.e., RNA-dependent RNA polymerase (RdRp), coronavirus main chymotrypsin-like protease (3CLpro), papain-like protease (PLpro) and spike protein S1. Additionally, human angiotensin-converting enzyme 2 (ACE2) receptor [21,91], JAK 1 and JAK 2 tyrosine kinases [92] or cytokines including interleukin (IL)-1β and IL-6 [93] are also crucial targets in COVID-19 treatment. The latter targets might be the most important because they are principal players in the immune system, which is overactive under viral infection [94,95].
In this context, it is of interest to quickly compare molecular descriptors, i.e., physicochemical properties and bioactivity scores (drug-likeness), of these molecules to identify selected similar patterns and peculiar characteristics. This simple and rapid approach could be helpful in antiviral drug optimization and drug combination and possible identification of molecular hybrid entities, the so-called chimeric molecules. These physicochemical properties, which form the well-known Lipinski “rule of five” (RO5) [96] and Veber’s rule [97], can be easily established using the basic tools of open access, e.g., Molinspiration [98]. The importance of RO5 (MW < 500, cLogP < 5, number of hydrogen-bond acceptors HBA < 10 and number of hydrogen-bond donors HBD < 5) is associated primarily with the solubility of the drug and the permeation of in vivo barriers. It is known that a) calculated descriptors are only simple filters that could aid selection of drug-like compounds; b) RO5 compliance is not a guarantee that a compound will be drug-like; and c) certain of the pharmacological targets in current drug discovery do not have high affinity and selectivity ligands that comply with RO5, and several studies do focus on series of compounds beyond the RO5 chemical space with the expected consequences on the properties of ADMET (absorption, distribution, metabolism, excretion and toxicity) of the identified ligands. Furthermore, according to Veber’s rule, good bioavailability is more likely for compounds with ≤10 rotatable bonds (ROTB), total HBA + HBD < 12 and topological polar surface area (TPSA) ≤ 140 Å2. These two parameters reflect that a) as the number of rotatable bonds increases, the molecule becomes more flexible and more adaptable for efficient interaction with a particular binding pocket of the target, and b) the TPSA descriptor correlates well with passive molecular transport through membranes and can predict the transport properties of drugs in the intestines and permeation of the blood-brain barrier. TPSA values less than 140 Å2 are associated with good cell membrane permeability.
Considering these facts, we calculated the principal molecular descriptors of these small molecule drugs and examined them (Table 1 ). The calculated data indicate that among 14 drugs, seven compounds with MW < 500 Da fulfill the RO5 for oral absorption, and the other half of the compounds with MW > 600 Da do not fulfill the RO5, failing two or three criteria. The case of chloroquine (5) is a border zone; although it has small MW, it fails only one criterion. This observation is not surprising because ca. 16% of drugs, especially antiparasitic, anticancer and antiviral drugs that have oral bioavailability violate at least one of the criteria, and 6% fail in two or more criteria.
Table 1.
Molecular descriptors calculated for drugs and drug candidates 1–14.
| Comp. | Key target | MWa | pKab | LogSc | % Abs.d | cLogPe | HBAf | HBDg | ROTBh | TPSAi | MVj | n violationsk |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RdRp | 616.61 | 15.98 | −4.96 | 38.77 | 2.12 | 14 | 5 | 15 | 203.57 | 539.85 | 2 |
| 2 | 3CLpro/PLpro | 628.81 | 13.59 | −7.14 | 67.60 | 5.69 | 9 | 4 | 15 | 119.99 | 607.96 | 2 |
| 3 | 3CLpro/PLpro | 706.93 | 13.63 | −6.49 | 58.71 | 7.14 | 11 | 4 | 17 | 145.78 | 646.30 | 3 |
| 4 | RdRp | 157.10 | N/A | −1.84 | 78.26 | −0.52 | 5 | 3 | 1 | 89.11 | 118.95 | 0 |
| 5 | ACE2 | 319.88 | 10.06 | −4.53 | 99.28 | 5.00 | 3 | 1 | 8 | 28.16 | 313.12 | 1 |
| 6 | ACE2 | 335.87 | 9.22 | −3.77 | 92.31 | 4.00 | 4 | 2 | 9 | 48.38 | 321.38 | 0 |
| 7 | S protein/ACE2 | 477.42 | 5.24 | −6.04 | 90.13 | 4.86 | 5 | 1 | 8 | 54.70 | 386.24 | 0 |
| 8 | RdRp | 329.31 | 12.15 | −1.28 | 59.61 | −0.26 | 10 | 4 | 6 | 143.15 | 280.87 | 0 |
| 9 | RdRp | 244.21 | 14.81 | 0.60 | 59.41 | −2.77 | 9 | 5 | 3 | 143.73 | 197.68 | 0 |
| 10 | JAK1/JAK2l | 306.37 | N/A | −4.14 | 80.30 | 1.83 | 6 | 1 | 4 | 83.19 | 281.01 | 0 |
| 11 | RdRp | 225.25 | 4.55 | −0.11 | 76.20 | −0.26 | 6 | 3 | 7 | 95.08 | 207.27 | 0 |
| 12 | UNm | 749.00 | 8.63 | −4.08 | 46.87 | 3.10 | 14 | 5 | 7 | 180.09 | 736.45 | 2 |
| 13a | 3CLpro/PLpro | 873.09 | 15.44 | −5.47 | 50.32 | 2.61 | 14 | 3 | 8 | 170.09 | 825.84 | 2 |
| 13b | 3CLpro/PLpro | 859.06 | 15.24 | −5.06 | 50.32 | 2.11 | 14 | 2 | 7 | 170.09 | 809.04 | 2 |
| 14 | 3CLpro/PLpro | 731.84 | N/A | −7.67 | 46.72 | 4.65 | 14 | 3 | 8 | 180.52 | 637.73 | 2 |
Bold items indicate violations of the Lipinski’s rule.
Molecular weight (g/mol).
pKa calculated using commercially available ChemDraw 15.0 program.
Aqueous solubility.
Percentage of absorption calculated by % Absorption = 109-(0.345 × TPSA).
Logarithm of the partition coefficient between n-octanol and water.
Number of hydrogen-bond acceptors.
Number of hydrogen-bond donors.
Number of rotatable bonds.
Total molecular polar surface area (Å2).
Molecular volume.
Violations of the Lipinski’s rule and did not include the violations of Verb’s rule.
JAK, Janus kinases play a central role in signal transduction in hematopoietic cells as well as in cells of the immune system.
UN, unknown detailed anti-influenza virus mechanism; azithromycin does not affect attachment of viruses onto the cell surface but blocks internalization into host cells during the early phase of infection (in mice).
One of the most parameters of this rule is lipophilicity, which is expressed by the cLogP feature. Almost all selected compounds have an appropriate cLogP (less than 5) for Lipinski’s rule, excluding two drugs, i.e., lopinavir (2) and ritonavir (3), which are highly lipophilic molecules with respective cLogP values of 5.69 and 7.14. In contrast, the drugs favipiravir (4), EIDD-2801 (8), ribavirin (9), ruxolitinib (10) and ingavirin (11) are highly hydrophilic molecules with cLogP values ranging from −2.27 to 1.83. These are notably small molecules that are highly soluble in an aqueous medium and have a poor capacity for permeability through membranes (weak lipophilicity), but they are probably capable of transport by certain membrane proteins. Interestingly, three macrocyclic compounds displaying an appropriate lipophilicity, i.e., azithromycin (12), ivermectin (13) and danoprevir (14), are less lipophilic than lopinavir (2) and ritonavir (3), which possess an alicyclic peptide backbone. All of these molecules are the “heaviest” (MW > 628–873 Da) and bulkiest (MV > 607–825 Da). Comparing both antimalarial drugs 5 and 6, although they have almost identical molecular volumes, hydroxychloroquine is more polar, less lipophilic, and thus encounters more difficulty diffusing across cell the membrane, as confirmed by kinetic and thermodynamic experiments [99].
It is important to note that remdesivir (1), the most probable drug according to new information from clinical studies [49,50,100], has a similar MW (616 Da), the highest TPSA value (203.57 Å2) and exhibits an ideal partition coefficient between polar and lipid layers (cLogP = 2.12), but its absorption (38%) is the worst among the selected drugs. More precisely, this nucleotide analog prodrug, which inhibits viral RNA polymerase, is only administered intravenously, and compassionate use in treatment of patients hospitalized for severe Covid-19 showed clinical improvement [101].
The best absorption (% ABS) according to our calculations (92–99%) was found for antimalarial 4-aminoquinoline drugs 5 and 6. Other small nitrogen-containing molecule drugs such as favipiravir (4), umifenovir (7), ruxolitinib (10) and ingavirin (11) also demonstrated good absorption (76–90%), which is an indication of good bioavailability by the oral route. This feature is consequence of their good TPSA values, ranking from 28.16 to 95.08 Å2, which also indicates notably good cell membrane permeability. It is worth mention that the presence of a 3,4-dihydroxy-5-(hydroxymethyl)-tetrahydrofuran-2-yl fragment in the heterocyclic ring increases the TPSA parameter of nucleoside-based drugs EIDD-2801 (8), ribavirin (9) and remdesivir (1) (TPSA = 143.15–203.57 Å2). Increased TPSA values are also observed for macrocyclic molecule drugs azithromycin (12), ivermectin (13) and danoprevir (14). Among these three drugs, ivermectin is the most hydrophilic molecule, is well resorbed after oral administration, and its distribution to the brain is discouraged by the blood-brain-barrier (TPSA > 140 Å2) because of its molecular size, which is not conducive to passive diffusion, but it can be transferred in the presence of efflux pumps, for which ivermectin is a substrate [102]. In addition, these drugs have the appropriate molecular flexibility (ROTB = 7–8) according to the Veber’s rule. Curiously, chloroquine (5) and hydroxychloroquine (6) show the same capacity, and remdesivir (1), lopinavir (2) and ritonavir (3) are much more flexible molecules (ROTB = 15–17).
In addressing the pKa values of the selected drug molecules, it can be noted that among the 14 studied drugs, seven drugs do not present essentially acidic properties in water (pKa > 12), and are strong conjugated bases (comp. 1–3, 8, 9, 13 and 14). Three drugs (comp. 5, 6 and 12) show notably weak acid strength (pKa = 8–12), and the two drugs umifenovir (7) and ingavirin (11) are weak acids with 5.24 and 4.55 pKa values, respectively, but they are stronger than the other evaluated drugs. In this context, it is well known that the pH environment to which an orally administered drug molecule is subjected is highly changeable (stomach pH ∼ 3.5, intestinal tract pH ∼ 8.5 and plasma pH = 7.4), and the absorption and transport capacities of these molecules also depend on their pKa, which can have a pronounced effect on the pharmacokinetics. From the data of Table 1, we can easily observe a good correlation between pKa and % ABS parameters: drugs with notably low acidity (pKa = 12–15) have generally poor to moderate absorption in the alkaline intestine (% ABS < 67.60) (comp. 1–3, 8, 9, 13 and 14), except for the azithromycin molecule, which exhibits a 8.63 value of pKa, and its intestinal absorption is expected to be quite poor (% ABS = 46.87), comparable to that of remdesivir.
It is noteworthy that as ACE2 inhibitors, two small drugs of chloroquine (5) and hydroxychloroquine (6), with respective pKa values of 10.06 and 9.22, show the best intestinal absorption. Unfortunately, pKa calculations from the ChemDraw program do not supply a second ionization constant for chloroquine and hydroxychloroquine. However, in practice, it was found that chloroquine has two basic groups corresponding to the quinoline-ring nitrogen and the diethylamino side-chain nitrogen with ionization constants of 8.1 and 10.2, respectively. Moreover, at a physiologic pH of 7.4 (plasma), 18% of chloroquine is monoprotonated but still soluble in lipid and able to traverse cell membranes, and when it enters a lysosome (pH ∼ 4–5), it is biprotonated [103,104]. In practice, chloroquine is administered as a phosphate salt, whereas hydroxychloroquine is processed as a sulfate, and both drugs are usually absorbed in the upper intestinal tract [105]. Interestingly, these drugs possess molecular polar surface area values below 60 Å2 that allow them to cross the hematoencephalic barrier. Umifenovir (7) is also a possible ACE2 inhibitor, but it is more acidic (pKa = 5.24) and less soluble in water (LogS = −6.04) than chloroquine and hydroxychloroquine drugs (LogS = −4.53 and −3.77, respectively) (Table 1).
5. Bioactivity scores
The Molinspiration tool also permits (to a certain degree) prediction of the bioactivity of the compounds as a ligand for the G-protein coupled receptor (GPCR) and the nuclear receptor, as an ion channel modulator, and as an enzyme inhibitor. With an indicator of the activity of a molecule, one can assess its inhibitory ability with respect to the vital proteins and enzymes of the human body. The data obtained can be used to estimate how harmful these active antiviral molecules are to the human body. Hence, the predicted relative affinity values to vital proteins of compounds 1–14 are presented in Table 2 .
Table 2.
Molinspiration bioactivity scoresa for compounds 1–14.
| Comp. | GPCR ligand | Ion channel modulator | Kinase inhibitor | Nuclear receptor ligand | Protease inhibitor | Enzyme inhibitor |
|---|---|---|---|---|---|---|
| 1 | 0.17 | −0.55 | 0.05 | −0.57 | 0.54 | 0.22 |
| 2 | 0.04 | −0.78 | −0.55 | −0.66 | 0.42 | −0.37 |
| 3 | 0.04 | −1.24 | −0.82 | −1.25 | 0.42 | −0.51 |
| 4 | −0.43 | 0.42 | −0.35 | −1.14 | −0.58 | −0.18 |
| 5 | 0.32 | 0.32 | 0.38 | −0.19 | 0.05 | 0.11 |
| 6 | 0.35 | 0.30 | 0.44 | −0.12 | 0.12 | 0.15 |
| 7 | −0.19 | −0.44 | −0.39 | −0.34 | −0.46 | −0.07 |
| 8 | 0.64 | −0.06 | 0.19 | −0.76 | 0.28 | 0.82 |
| 9 | 0.31 | 0.21 | −0.21 | −1.46 | −0.20 | 0.71 |
| 10 | 0.49 | 0.10 | 0.91 | −0.67 | −0.13 | 0.24 |
| 11 | 0.46 | 0.31 | −0.20 | −1.05 | 0.33 | 0.59 |
| 12 | −0.68 | −1.59 | −1.45 | −1.41 | −0.27 | −0.73 |
| 13a | −2.50 | −2.89 | −3.21 | −2.94 | −1.91 | −2.53 |
| 13b | −2.31 | −2.79 | −3.08 | −2.80 | −1.72 | −2.41 |
| 14 | −0.05 | −1.35 | −1.06 | −1.15 | 0.72 | −0.49 |
Items marked in bold parameters indicate strong drug-target interaction.
The results in Table 2 show certain interesting data. First, there are only three drugs with highly different chemical natures and modes of action, i.e., umifenovir (7), azithromycin (12) and ivermectin (13), that do not virtually alter any vital proteins, and five drugs that interact with one human target, i.e., remdesivir (1), lopinavir (2), ritonavir (3) and danoprevir (14), which bind to protease enzymes, whereas favipiravir (4) can act as an ion channel modulator similar to many other drugs (comp. 5, 6, 8–11). However, these latter candidates could switch to 2-3 additional human targets. Second, as viral anti-3CLpro/PLpro activities, drugs 2, 3 and 14 could also alter human protease enzymes, and it is logical but remdesivir and favipiravir, as viral RdRp inhibitors involved in viral replication inhibition, can act on human protease enzymes and ion channels, respectively, according to the obtained data. Both drugs are currently in Phase III clinical trials as a possible treatment for COVID-19 in the USA (remdesivir) and Japan (favipiravir). Ruxolitinib (10), an anticancer drug tested for its potential to dampen the cytokine storm, could act as a modulator of signaling proteins, such as GPCR and enzyme inhibitors, in addition to its kinase inhibition activity.
According to recent work on control of the SARS-CoV-2 infection in vitro, hydroxychloroquine was less toxic and more efficient as a viral inhibitor than chloroquine [106], which confirms the current in silico calculation, but both drugs display retinal toxicity, which is an ophthalmologic concern because it is not treatable [107]. However, the short-term administration of these drugs rarely causes severe side effects, and longer exposure has been associated with certain serious although uncommon adverse events, including cardiomyopathy, bone marrow suppression and hypoglycemia [108]. In addition, it was found early on that these drugs are neither embryotoxic nor fetotoxic when used at the usual dosage for malaria prophylaxis [109]. The recent work of Wolfram et al. which reports new details on viral cell entry inhibition of (hydroxy) chloroquine, offers a cautiously optimistic report that (hydroxy) chloroquine might have a prophylactic and/or therapeutic effect against COVID-19 [110], though there is sufficient preclinical rationale and evidence on their effectiveness for treatment of COVID-19 to validate their use in the clinic [111].
6. Concluding remarks
Reprofiling a drug or experimental agent for a concrete disease is a complex task that requires time and fundamental dedication, and the repurposed pipeline is finite. Despite immense efforts, no good clinical evidence currently exists for any specific therapies (including antiviral and immune modulating agents). The repurposed drugs that are under investigation for COVID-19 should be used only in randomized and controlled trials. The revised physicochemical properties and bioactivity scores of potential small molecule drugs studied in this work indicate that no molecule is perfect: if certain small molecule drugs (comp. 4, 6, 8–11) meet Lipinski’s rule, they do not pass the standards for bioactivity scores, and if notably few drugs show a good drug-likeness criterion (comp. 12 and 13), they violate two criteria of Lipinski’s rule.
However, it should be noted that umifenovir (arbidol) (7) seems to be a privileged molecule because this polyfunctionalized indole derivative does not infringe on either the Lipinski-Veber rules or the bioactivity score criteria. It is a pity that information on this drug in COVID-19 treatment is minimally available, and its use against coronavirus is still controversial. Although selected Chinese scientists have asked the WHO to include arbidol in the list of drugs recommended for combating coronavirus, experts have doubts related to the usefulness of this drug, which has not proved its effectiveness in several tests, and tests are still ongoing [112]. Until now, arbidol has shown efficacy against influenza viruses by targeting the hemagglutinin fusion machinery that suggests stimulation of the immune system or anti-inflammatory action. However, the structural basis of the mechanism underlying fusion inhibition by arbidol has remained obscure [113].
Special attention has been focused on use of (hydroxy) chloroquine drugs for a COVID-19 cure. Both drugs, which show remarkable absorption, cell membrane permeability and transport properties, are important drugs in the treatment of malaria as well as severe rheumatoid arthritis and systemic lupus erythematosus diseases in which they bind to a heme moiety and interfere with normal hemoglobin metabolism. These drugs also interfere with lysosomal activity, autophagy, and membrane stability and alter signaling pathways and transcriptional activity, which could result in reduction of the cytokine storm by modulating T-cell stimulators and inflammatory molecules [114]. It is noteworthy that hydroxychloroquine has been found to be a more potent SARS-CoV-2 inhibitor than chloroquine, with considerable contribution to suppression of the cytokine storm [115,116].
Another promising drug candidate for coronavirus treatment is ivermectin, which is in phase III clinical trials in dengue patients, quickly avoids replication of SARS-CoV-2 in vitro [78]. This antiparasitic drug is safe for human use at relatively high doses, is widely available and is a relatively low-cost medicine that prompts further evaluation for possible benefits in COVID-19 patients [117,118].
It is almost certain that novel and repurposed drugs will be needed in the fight against COVID-19 [119,120]. In this context, cocktail drugs with optimized relationships seem to be more realistic [[121], [122], [123]]. The combination of anti-HIV Kaletra® (Lopinavir/Ritonavir) drug (3CLpro/PLpro inhibition) and α-interferon (immunomodulatory properties) has been used, but the therapeutic effect remains highly limited, and toxic side effects can occur [124]. On the other hand, arbidol monotherapy has been shown superior to lopinavir/ritonavir in treating COVID-19 [125] and contrariwise, combination therapy was associated with a significantly improved the chest CT scans in 7-day [126].
Returning to the idea of single drugs with multitargeting properties, drug combination based on a hybridization approach could be useful and efficient in creation of an antiviral molecule hybrid drug that possesses two or even three pharmacophore groups that act selectively on different viral replication targets (RdRp/3CLpro and RdRp/PLpro), viral cell entry (S protein/ACE2) and regulation of the human immune response to viral infection (JAK 1/JAK 2/interleukin (IL)-1β and IL-6). Understanding the mechanisms by which these drugs affect SARS-CoV-2 is expected to be critical to optimization and development of preventative and therapeutic strategies [127,128].
6.1. Post datum
Potential drug candidates for COVID-19 therapy are continuously substituted and changed, similar to basketball players in the last minutes of the final game to save the championship. Remdesivir, the most awaited and most promising candidate, has been fervently supported by doctors and politicians as a potential cure for COVID-19 [129] but appears to be inefficient according to the first randomized trial (158 patients) in China on patients with severe COVID-19 symptoms [130]. In addition, it was found that a high dosage of remdesivir may induce testicular toxicity and result in deterioration of sperm parameters in mice [131].
Thus, more investigation on the reproductive toxicity of this drug is required. Nevertheless, ClinicalTrials.gov indicated that five randomized trials involving remdesivir are recruiting globally, with one in severe COVID-19 from Gilead (NCT04292899), the drug manufacturer, with a target of 6000 participants; naively, this trial should be adequately powered [132], and on 02 May, the Food and Drug Administration issued an emergency use authorization for remdesivir for treating COVID-19 [133]. Thus, it is the first drug shown to help fight COVID-19. However, other drugs, such as favipiravir (Avigan®), a viral RNA polymerase inhibitor, or tilarone (Amixin®), an inducer of interferon, are also gaining attention [134,135]. During the last two months, the activity related to the drug design and development for suitable anti-SARS-Co-V2 agents has been constantly ongoing, but without new significant achievements [136,137], while the biological-structural and medical studies of the virus, disease and its symptoms have progressed [[138], [139], [140], [141], [142]]. Remdesivir production is increasing, however this drug (approved only in USA) and treatment is very expensive, perhaps due to its complicated and ineffective synthesis [143]. Its combination with tocilizumab (monoclonal antibodies drug, Actemra/RoActemra®) is being studying for hospitalized patients with severe COVID-19 pneumonia (phase 3). Favipiravir, an inexpensive antiviral drug, was approved for a COVID-19 treatment in several countries. Recently, it was found that dexamethasone (glucocorticoid) reduced deaths in hospitalized patients with severe COVID-19 disease [144], but once again, more investigation on this drug is required [145].
Therefore, the current methods for treatment of COVID-19 and the criterions of drug selection are still debatable. The story with hydroxychloroquine tests is very meaningful [[146], [147], [148], [149]], it’s believed it will continue until now when all the questions of how this Corona virus works inside our body will be clear. Unfortunately, a Roman historian and politician, Cornelius Tacitus was right saying that “Tardiora sunt remedia quam mala” (Remedies are slower in their operation than diseases). It is still very actual, especially in the case of COVID-19.
Declaration of competing interest
The authors declare no competing interest.
Acknowledgements
Constant financial support from MinCiencias (Colciencias, project No. RC-007-2017, Cod. 110274558597) is gratefully acknowledged.
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R., Drosten C., Gulyaeva A.A., et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–554. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R., Drosten C., Gulyaeva A.A., et al. BioRxiv; 2020. Severe acute respiratory syndrome-related coronavirus-The species and its viruses, a statement of the Coronavirus Study Group. [DOI] [Google Scholar]
- 3.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Chhikara B.S., Rathi B., Singh J., Poonam F.N.U. Corona virus SARS-CoV-2 disease COVID-19: infection, prevention and clinical advances of the prospective chemical drug therapeutics. Chem. Biol. Lett. 2020;7(1):63–72. [Google Scholar]
- 6.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang B., Bragazzi N.L., Li Q., Tang S., Xiao Y., Wu J. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov) Infect. Dis. Model. 2020;5:248–255. doi: 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J.T., Chang S.C. Severe acute respiratory syndrome. Curr. Opin. Infect. Dis. 2004;17:143–148. doi: 10.1097/00001432-200404000-00013. [DOI] [PubMed] [Google Scholar]
- 9.Middle East respiratory syndrome coronavirus (MERS-CoV) https://www.who.int/emergencies/mers-cov/en/ Available at:
- 10.Coronavirus disease (COVID-19) advice for the public. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/advice-for-public Available at:
- 11.Menachery V., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21(12):1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuhan Seafood Market Pneumonia Virus Isolate Wuhan-Hu-1, Complete Genome. Nucleotide, National Center for Biotechnology Information (NCBI) National Library of Medicine (US), National Center for Biotechnology Information, Bethesda; 2009. https://www.ncbi.nlm.nih.gov/nuccore/1798174254 Available at: [Google Scholar]
- 15.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81(4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 16.Baez-Santos Y.M., John S.E.S., Mesecar A.D. The SARScoronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Y. Gao, L. Yan, Y. Huang, F. Liu, Y. Zhao, L. Cao, Structure of the RNA-dependent RNA polymerase from COVID-19 virus, Science 368 (6492) 779-782. [DOI] [PMC free article] [PubMed]
- 18.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen T.S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song W., Wang Y., Wang N., Wang D., Guo J., Fu L. Identification of residues on human receptor DPP4 critical for MERS-CoV binding and entry. Virology. 2014;471:49–53. doi: 10.1016/j.virol.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baig A.M., Khaleeq A., Ali U., Syeda H. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host–virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 2020;11(7):995–998. doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- 24.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat. Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 25.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020;10(2):102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conti P., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Frydas I., et al. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by COVID-19: anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34(2) doi: 10.23812/conti-e. [DOI] [PubMed] [Google Scholar]
- 28.Khan S., Siddique R., Shereen M.A., Ali A., Liu J., Bai Q., et al. The emergence of a novel coronavirus (SARS-CoV-2), their biology and therapeutic options. J. Clin. Microbiol. 2020;58(5) doi: 10.1128/JCM.00187-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vellingiri B., Jayaramayya K., Iyer M., Narayanasamy A., Govindasamy V., Giridharan B. COVID-19: a promising cure for the global panic. Sci. Total Environ. 2020;725(10) doi: 10.1016/j.scitotenv.2020.138277. July. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aronson J.K., Ferner R.E., DeVito N., Heneghan C. 2020. COVID-19 Trials Registered up to 8 March 2020-an Analysis of 382 Studies.https://www.cebm.net/oxford-covid-19/covid-19-registered-trials-and-analysis/ Retrieved from: [Google Scholar]
- 31.J. Wang, Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study, J. Chem. Inf. Model. 60 (6) 3277–3286. [DOI] [PMC free article] [PubMed]
- 32.Smith M., Smith J.C. ChemRxiv; 2020. Repurposing Therapeutics for COVID-19: Supercomputer-Based Docking to the SARS-CoV-2 Viral Spike Protein and Viral Spike Protein-Human ACE2 Interface. [DOI] [Google Scholar]
- 33.ul Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020:26. doi: 10.1016/j.jpha.2020.03.009. March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhavoronkov A., Aladinskiy V., Zhebrak A., Zagribelnyy B., Terentiev V., Bezrukov D.S., et al. ChemRxiv; 2020. Potential COVID-2019 3C-like Protease Inhibitors Designed Using Generative Deep Learning Approaches. [DOI] [Google Scholar]
- 35.Durdagi S., Aksoydan B., Dogan B., Sahin K., Shahraki A. ChemRxiv; 2020. Screening of Clinically Approved and Investigation Drugs as Potential Inhibitors of COVID-19 Main Protease: A Virtual Drug Repurposing Study. [DOI] [Google Scholar]
- 36.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248(1) doi: 10.1016/j.lfs.2020.117477. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farag A., Wang P., Ahmed M., Sadek H. Identification of FDA approved drugs targeting COVID-19 virus by structure-based drug repositioning. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12003930. [DOI] [Google Scholar]
- 38.Kupferschmidt K., Cohen J. Race to find COVID-19 treatments accelerates. Sciences. 2020;367(6485):1412–1413. doi: 10.1126/science.367.6485.1412. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., et al. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63(9):4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., et al. Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication. BioRxiv. 2020 doi: 10.1101/2020.02.10.936898. [DOI] [PubMed] [Google Scholar]
- 41.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez M.A. Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrob. Agents Chemother. 2020;64(5) doi: 10.1128/AAC.00399-20. http://aac.asm.org/content/64/5/e00399-20.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ekins S., Mottin M., Ramos P.R., Sousa B.K., Neves B.J., Foil D.H., et al. Déjà vu: stimulating open drug discovery for SARS-CoV-2, Drug Discov. Today Off. 2020;25(5):928–941. doi: 10.1016/j.drudis.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gautret P., Lagier J.C., Parola P., Meddeb L., Mailhe M., Doudier B., et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;20 doi: 10.1016/j.ijantimicag.2020.105949. March. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Molina J.M., Delaugerre C., Goff J.L., Mela-Lima B., Ponscarme D., Goldwirt L., et al. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Maladies Infect. 2020;50(4):384. doi: 10.1016/j.medmal.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lythgoe M.P., Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol. Sci. 2020;41(6):363–382. doi: 10.1016/j.tips.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ko W.C., Rolain J.M., Lee N.Y., Chen P.L., Huang C.T., Lee P.I., et al. Arguments in favor of remdesivir for treating SARS-CoV-2 infections. Int. J. Antimicrob. Agents. 2020;55(4) doi: 10.1016/j.ijantimicag.2020.105933105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Tawfiq J.A., Al-Homoud A.H., Memish Z.A. Remdesivir as a possible therapeutic option for the COVID-19, Travel Med. Inf. Disp. 2020;34 doi: 10.1016/j.tmaid.2020.101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9(396) doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chandwani A., Shuter J. Lopinavir/ritonavir in the treatment of HIV-1 infection: a review. Therapeut. Clin. Risk Manag. 2008;4(5):1023–1033. doi: 10.2147/tcrm.s3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Croxtall J.D., M Perry C. Lopinavir/ritonavir, Drugs. 2010;70(14):1885–1915. doi: 10.2165/11204950-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 54.Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013;100(2):446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad-spectrum inhibitor of viral RNA polymerase. Proc. Japan Acad., Series B. 2017;93(7):449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piscianz E., Cuzzoni E., Sharma R., Tesser A., Sapra P., Tommasini A. Reappraisal of antimalarials in interferonopathies: new perspectives for old drugs. Curr. Med. Chem. 2018;25(24):2797–2810. doi: 10.2174/0929867324666170911162331. [DOI] [PubMed] [Google Scholar]
- 57.Plantone D., Koudriavtseva T. Current and future use of chloroquine and hydroxychloroquine in infectious, immune, neoplastic, and neurological diseases: a mini-review. Clin. Drug Invest. 2018;38(8):653–671. doi: 10.1007/s40261-018-0656-y. [DOI] [PubMed] [Google Scholar]
- 58.Touret F., de Lamballerie X. Of chloroquine and COVID-19. Antivir. Res. 2020;177 doi: 10.1016/j.antiviral.2020.104762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yazdany J., Kim A.H.J. Use of hydroxychloroquine and chloroquine during the COVID-19 pandemic: what every clinician should know. Ann. Intern. Med. 2020 doi: 10.7326/M20-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Balakin K.V., Filosa R., Lavrenov S.N., Mkrtchyan A.S., Nawrozkij M.B., Novakov I.A. Arbidol: a quarter-century after. Past, present and future of the original Russian antiviral. Russ. Chem. Rev. 2018;87(6):509–552. [Google Scholar]
- 61.Blaising J., Polyak S.J., Pécheur E.I. Arbidol as a broad-spectrum antiviral: an update. Antivir. Res. 2014;107:84–94. doi: 10.1016/j.antiviral.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toots M., Yoon J., Hart M., Natchus M.G., Painter G.R., Plemper R.K. Quantitative efficacy paradigms of the influenza clinical drug candidate EIDD-2801 in the ferret model. Transl. Res. 2020;218:16–28. doi: 10.1016/j.trsl.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emory-discovered antiviral is poised for COVID-19 clinical trials. 2020. https://cen.acs.org/biological-chemistry/infectious-disease/Emory-discovered-antiviral-poised-COVID/98/i12 Available at:
- 64.Graci J.D., Cameron C.E. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2006;16(1):37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loustaud-Ratti V., Rousseau A., Marquet P., Denis F., Alain S. Ribavirin in chronic hepatitis C: past and future. Expert Rev. Anti-infect. Ther. 2009;7(3):249–253. doi: 10.1586/eri.09.5. [DOI] [PubMed] [Google Scholar]
- 66.Xia Y., Qu F., Peng L. Triazole nucleoside derivatives bearing aryl functionalities on the nucleobases show antiviral and anticancer activity. Mini Rev. Med. Chem. 2010;10(9):806–821. doi: 10.2174/138955710791608316. [DOI] [PubMed] [Google Scholar]
- 67.Khalili J.S., Zhu H., Mak A., Yan Y., Zhu Y. Novel coronavirus treatment with ribavirin: groundwork for evaluation concerning COVID-19. J. Med. Virol. 2020:30. doi: 10.1002/jmv.25798. March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plosker G.L. Ruxolitinib: a review of its use in patients with myelofibrosis. Drugs. 2015;75(3):297–308. doi: 10.1007/s40265-015-0351-8. [DOI] [PubMed] [Google Scholar]
- 69.Lussana F., Cattaneo M., Rambaldi A., Squizzato A. Ruxolitinib-associated infections: a systematic review and meta-analysis. Am. J. Hematol. 2018;93(3):339–347. doi: 10.1002/ajh.24976. [DOI] [PubMed] [Google Scholar]
- 70.Kolobukhina L.V., Merkulova L.N., Shchelkanov M., Burtseva E.I., Isaeva E.I., Malyshev N.A. Efficacy of ingavirin in adults with influenza. Ter. Arkh. 2009;81(3):51–54. [Article in Russian] [PubMed] [Google Scholar]
- 71.Loginova S., Borisevich S.V., Maksimov V.A., Bondarev V.P., Nebol’sin V.E. Therapeutic efficacy of Ingavirin, a new domestic formulation against influenza A virus (H3N2) Antib. Khimioterap. 2008;53(7–8):27–30. [Article in Russian] [PubMed] [Google Scholar]
- 72.Ingavirin New - well forgotten old? Evaluation of real effectiveness. https://ostit.ru/en/coronary-artery-disease/ingavirin-novoe-horosho-zabytoe-staroe-ocenka-realnoi-effektivnosti/ Available at:
- 73.Parnham M.J., Haber V.E., Giamarellos-Bourboulis E.J., Perletti G., Verleden G.M., Vos R. Azithromycin: mechanisms of action and their relevance for clinical applications. Pharmacol. Therapeut. 2014;143(2):225–245. doi: 10.1016/j.pharmthera.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 74.Tran D.H., Sugamata R., Hirose T., Suzuki S., Noguchi Y., Sugawara A. Azithromycin, a 15-membered macrolide antibiotic, inhibits influenza A (H1N1) pdm09 virus infection by interfering with virus internalization process. J. Antibiot. 2019;72(10):759–768. doi: 10.1038/s41429-019-0204-x. [DOI] [PubMed] [Google Scholar]
- 75.Ohe M., Shida H., Jodo S., Kusunoki Y., Seki M., Furuya K., Goudarzi H. Macrolide treatment for COVID-19: will this be the way forward? BioScience Trends. 2020;14(2):159–160. doi: 10.5582/bst.2020.03058. [DOI] [PubMed] [Google Scholar]
- 76.Geary T.G. Ivermectin 20 years on: maturation of a wonder drug. Trends Parasitol. 2005;21(11):530–532. doi: 10.1016/j.pt.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 77.Omura S. Ivermectin: 25 years and still going strong. Int. J. Antimicrob. Agents. 2008;31(2):91–98. doi: 10.1016/j.ijantimicag.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 78.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seiwert S.D., Andrews S.W., Jiang Y., Serebryany V., Tan H., Kossen K. Preclinical characteristics of the hepatitis C virus NS3/4A protease inhibitor ITMN-191 (R7227) Antimicrob. Agents Chemother. 2008;52(12):4432–4441. doi: 10.1128/AAC.00699-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang Y., Andrews S.W., Condroski K.R., Buckman B., Serebryany V., Wenglowsky S. Discovery of danoprevir (ITMN-191/R7227), a highly selective and potent inhibitor of hepatitis C virus (HCV) NS3/4A proteas. J. Med. Chem. 2014;57(5):1753–1769. doi: 10.1021/jm400164c. [DOI] [PubMed] [Google Scholar]
- 81.Markham A., Keam S.J. Danoprevir: first global approval. Drugs. 2018;78(5):1271–1276. doi: 10.1007/s40265-018-0960-0. [DOI] [PubMed] [Google Scholar]
- 82.Chen H., Zhang Z., Wang L., Huang Z., Gong F., Li X., Chen Y. First clinical study using HCV protease inhibitor danoprevir to treat naive and experienced COVID-19 patients. MedRxiv. 2020 doi: 10.1101/2020.03.22.20034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grubaugh N.D., Petrone M.E., C Holmes E. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020;5(4):529–530. doi: 10.1038/s41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Antiretroviral Therapy Cohort Collaboration Life expectancy of individuals on combination antiretroviral therapy in high-income countries: a collaborative analysis of 14 cohort studies. Lancet. 2008;372(9635):293–299. doi: 10.1016/S0140-6736(08)61113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 2020;20(4):400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020;38(4):379–391. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 88.Rajeshkumar N.V., Yabuuchi S., Pai S.G., Maitra A., Hidalgo M., Dang C.V. Fatal toxicity of chloroquine or hydroxychloroquine with metformin in mice. BioRxiv. 2020 doi: 10.1101/2020.03.31.018556. [DOI] [Google Scholar]
- 89.Foucquier J., Guedj M. Analysis of drug combinations: current methodological landscape. Pharm. Res. Perspect. 2015;3(3) doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zimmermann G.R., Lehar J., Keith C.T. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov. Today. 2007;12(1–2):34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 91.Wang P.H., Cheng Y. Increasing host cellular receptor-angiotensin-converting enzyme 2 (ACE2) expression by coronavirus may facilitate 2019-nCoV infection. BioRxiv. 2020 doi: 10.1101/2020.02.24.963348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Malavolta M., Giacconi R., Brunetti D., Provinciali M., Maggi F. Exploring the relevance of senotherapeutics for the current SARS-CoV-2 emergency and similar future global health threats. Cells. 2020;9(4):909. doi: 10.3390/cells9040909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ledford H. How does covid-19 kill? uncertainty hampers doctors’ ability to choose treatments. Science. 2020;580(7803):311–312. doi: 10.1038/d41586-020-01056-7. [DOI] [PubMed] [Google Scholar]
- 95.Fu Y., Cheng Y., Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol. Sin. 2020:3. doi: 10.1007/s12250-020-00207-4. March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23(1–3):3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 97.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 98.Cheminformatics M. Bratislava (Slovak Republic), Calculation of molecular properties and bioactivity score. 2011. http://www.Molinspiration.com Available at:
- 99.Ferrari V., Cutler D.J. Kinetics and thermodynamics of chloroquine and hydroxychloroquine transport across the human erythrocyte membrane. Biochem. Pharmacol. 1991;41(1):23–30. doi: 10.1016/0006-2952(91)90006-q. [DOI] [PubMed] [Google Scholar]
- 100.Cao Y.C., Deng Q.X., Dai S.X. Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19: an evaluation of the evidence. Trav. Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101647. 2 Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., et al. Compassionate use of remdesivir for patients with severe covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chaccour C., Hammann F., Rabinovich N.R. Ivermectin to reduce malaria transmission I. Pharmacokinetic and pharmacodynamic considerations regarding efficacy and safety. Malar. J. 2017;16(1):161. doi: 10.1186/s12936-017-1801-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Browning D.J. In: Hydroxychloroquine and Chloroquine Retinopathy in: Hydroxychloroquine and Chloroquine Retinopathy. Browning D.J., editor. Springer Science+Business Media New York; 2014. pp. 35–63. [Google Scholar]
- 104.Titus E.O. Recent developments in the understanding of the pharmacokinetics and mechanism of action of chloroquine, Ther. Drug Monit. 1989;11(4):369–379. [PubMed] [Google Scholar]
- 105.Tett S.E., Cutler D.J., Day R.O., Brown K.F. Bioavailability of hydroxychloroquine tablets in healthy volunteers, Brit. J. Clin. Pharm. 1989;27(6):771–779. doi: 10.1111/j.1365-2125.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6(1):16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marmor M.F., Kellner U., Lai T.Y., Melles R.B., Mieler W.F. Recommendations on screening for chloroquine and hydroxychloroquine retinopathy (2016 revision) Ophthalmol. Times. 2016;123(6):1386–1394. doi: 10.1016/j.ophtha.2016.01.058. [DOI] [PubMed] [Google Scholar]
- 108.Verbaanderd C., Maes H., Schaaf M.B., Sukhatme V.P., Pantziarka P., Sukhatme V. Repurposing drugs in oncology (ReDO)-chloroquine and hydroxychloroquine as anti-cancer agents. Ecancermedicalscience. 2017;11:781. doi: 10.3332/ecancer.2017.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McGready R., Thwai K.L., Cho T., Looareesuwan S., White N.J., Nosten F. The effects of quinine and chloroquine antimalarial treatments in the first trimester of pregnancy. Trans. R. Soc. Trop. Med. Hyg. 2002;96(2):180–184. doi: 10.1016/s0035-9203(02)90297-x. [DOI] [PubMed] [Google Scholar]
- 110.Hu T.Y., Frieman M., Wolfram J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat. Nanotechnol. 2020;15(4):247–249. doi: 10.1038/s41565-020-0674-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care. 2020 doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lian N., Xie H., Lin S., Huang J., Zhao J., Lin Q. Umifenovir treatment is not associated with improved outcomes in patients with coronavirus disease 2019: a retrospective study. Clin. Microbiol. Infect. 2020;26(7):917–921. doi: 10.1016/j.cmi.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kadam R.U., Wilson I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. U.S.A. 2017;114(2):206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schrezenmeier E., Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 115.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020:12. doi: 10.1016/j.ijantimicag.2020.105938. March 105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa237. ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chaccour C., Hammann F., Ramón-García S., Rabinovich N.R. Ivermectin and novel coronavirus disease (COVID-19): keeping rigor in times of urgency. Am. J. Trop. Med. Hyg. 2020;102(6):1156–1157. doi: 10.4269/ajtmh.20-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Momekov G., Momekova D. Ivermectin as a potential COVID-19 treatment from the pharmacokinetic point of view. MedRxiv. 2020 doi: 10.1101/2020.04.11.20061804. [DOI] [Google Scholar]
- 119.Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses, Drug Discov. Today Off. 2020;25(4):668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ghosh A.K., Brindisi M., Shahabi D., Chapman M.E., Mesecar A.D. Drug development and medicinal chemistry efforts toward SARS-coronavirus and covid-19 therapeutics. ChemMedChem. 2020 doi: 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Duan Y., Zhu H.L., Zhou C. Advance of promising targets and agents against 2019-nCoV in China, Drug Discov. Today Off. 2020;25(5):810–812. doi: 10.1016/j.drudis.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Patrì A., Fabbrocini G. Hydroxychloroquine and ivermectin: a synergistic combination for COVID-19 chemoprophylaxis and/or treatment? J. Am. Acad. Dermatol. 2020 doi: 10.1016/j.jaad.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yu J., Wang M., Zhou K., Zhang S., Xiang B. Clinical characteristics of Coronavirus Disease 2019 in Hangzhou, China: the combination of lopinavir/ritonavir, interferon, and arbidol may be a well choice for antiviral therapy in common cases. Res. Square. 2020 doi: 10.21203/rs.3.rs-19184/v1. [DOI] [Google Scholar]
- 124.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020 doi: 10.7326/M20-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu Z., Lu Z., Xu T., Chen C., Yang G., Zha T., Xue Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020;81(1):e21–e23. doi: 10.1016/j.jinf.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Deng L., Li C., Zeng Q., Liu X., Li X., Zhang H. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: a retrospective cohort study. J. Infect. 2020;81(1):e1–e5. doi: 10.1016/j.jinf.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Scavone C., Brusco S., Bertini M., Sportiello L., Rafaniello C., Zoccoli A. Current pharmacological treatments for COVID-19: what’s next? Br. J. Pharmacol. 2020 doi: 10.1111/bph.15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McKee D.L., Sternberg A., Stange U., Laufer S., Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020 doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: a review of its discovery and development leading to human clinical trials for treatment of COVID-19. Preprints. 2020 doi: 10.20944/preprints202004.0299.v1. 2020040299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fan J., Zhao D., Li X., Deng T., Sun Y., Luo J., Weng Y. A Preliminary Study on the Reproductive Toxicity of GS-5734 on Male Mice. bioRxiv. 2020 doi: 10.1101/2020.04.21.050104. [DOI] [Google Scholar]
- 132.Norrie J.D. Remdesivir for COVID-19: challenges of underpowered studies. Lancet. 2020;395(10236):1525–1527. doi: 10.1016/S0140-6736(20)31023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.FDA Allows For ’Emergency Use’ of Remdesivir Experimental coronavirus drug. https://time.com/5831062/fda-allows-emergency-use-remdesivir/ Available at:
- 134.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020 doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ekins S., Lane T.R., Madrid P.B. Tilorone: a broad-spectrum antiviral invented in the USA and commercialized in Russia and beyond. Pharm. Res. (N. Y.) 2020;37:71. doi: 10.1007/s11095-020-02799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Saxena A. Drug targets for COVID-19 therapeutics: ongoing global efforts. J. Bio. Sci. 2020;45:87. doi: 10.1007/s12038-020-00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M.A., et al. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 138.Zhang Y., Kutateladze T.G. Molecular structure analyses suggest strategies to therapeutically target SARS-CoV-2. Nat. Commun. 2020;11(1):1–4. doi: 10.1038/s41467-020-16779-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically-sensitive activation loop. J. Mol. Biol. 2020;432(10):3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang W., Stephen P., Thériault J.F., Wang R., Lin S.X. Novel coronavirus polymerase and nucleotidyl-transferase structures: potential to target new outbreaks. J. Phys. Chem. Lett. 2020;11(11):4430–4435. doi: 10.1021/acs.jpclett.0c00571. [DOI] [PubMed] [Google Scholar]
- 141.Osorio-Mogollon C., Olivos-Ramirez G.E., Otazu K., Chenet-Zuta M.E., Ropon-Palacios G., Camps I., et al. Attacking the SARS-CoV-2 replication machinery with the pathogen box’s molecules. ChemRxiv. Preprint. 2020 doi: 10.26434/chemrxiv.12501791.v1. [DOI] [Google Scholar]
- 142.Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Marrero M.C., et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020 doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., et al. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo [2, 1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J. Med. Chem. 2017;60(5):1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 144.Oxford University News Release https://www.recoverytrial.net/files/recovery_dexamethasone_statement_160620_v2final.pdf
- 145.Theoharides T.C., Conti P. Dexamethasone for COVID-19? Not so fast. J. Biol. Regul. Homeost. Agents. 2020;34(3) doi: 10.23812/20-EDITORIAL_1-5. [DOI] [PubMed] [Google Scholar]
- 146.Mehra M.R., Desai S.S., Ruschitzka F., Patel A.N. Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: a multinational registry analysis. Lancet. 2020 doi: 10.1016/S0140-6736(20)31180-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 147.Funck-Brentano C., Salem J.E. Chloroquine or hydroxychloroquine for COVID-19: why might they be hazardous? Lancet. 2020 doi: 10.1016/S0140-6736(20)31174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mahase E. Covid-19: 146 researchers raise concerns over chloroquine study that halted WHO trial. BMJ. 2020;369 doi: 10.1136/bmj.m2197. [DOI] [PubMed] [Google Scholar]
- 149.Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., et al. Observational study of hydroxychloroquine in hospitalized patients with Covid-19. N. Engl. J. Med. 2020;382:2411–2418. doi: 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]