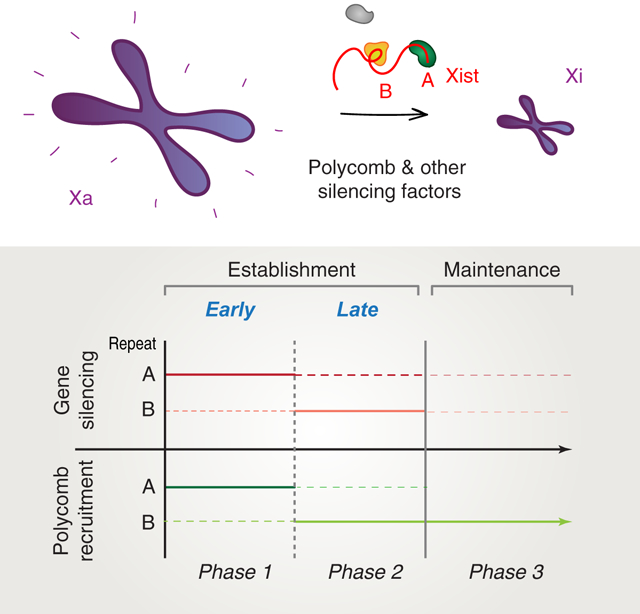
ETOC BLURB
Colognori, Sunwoo et al. show that X-inactivation establishment is a biphasic process with distinct genetic requirements. Repeat A initiates Polycomb recruitment and gene silencing, whereas Repeat B stabilizes them. Surprisingly, X-inactivation can initiate without Repeat B. Without Repeat A, however, differentiating female cells lose one X-chromosome to overcome loss of silencing.
Keywords: Epigenetics, lncRNA, X-inactivation, Xist, Repeat A, Repeat B, Polycomb, PRC1, PRC2
Graphical Abstract
SUMMARY
X-chromosome inactivation (XCI) is a global silencing mechanism by which XX and XY mammals equalize X-linked gene dosages. XCI begins with an establishment phase during which Xist RNA spreads and induces de novo heterochromatinization across a female X chromosome, and is followed by a maintenance phase when multiple epigenetic pathways lock down the inactive X (Xi) state. Involvement of Polycomb repressive complexes 1 and 2 in XCI has been intensively studied, but with conflicting conclusions regarding their recruitment and role in Xi silencing. Here we reveal that establishment of XCI has two phases and reconcile the roles that Xist Repeats A and B play in gene silencing and Polycomb recruitment. Repeat A initiates both processes, whereas Repeat B bolsters or stabilizes them thereafter. Once established, XCI no longer requires Repeat A during maintenance. These findings integrate disparate studies and present a unified view of Xist’s role in Polycomb-mediated silencing.
INTRODUCTION
XCI is initiated by a chromosome-counting mechanism that triggers dosage compensation only when there is more than one X-chromosome in the early mammalian embryo (Starmer and Magnuson, 2009; Disteche, 2016; Jegu et al., 2017). Once committed to XCI, one X-chromosome is selected to undergo a series of distinct epigenetic changes that are separable into establishment and maintenance phases. The long noncoding RNA Xist is instrumental in both phases. During establishment, Xist spreads across the future inactive X (Xi) and carries out several essential functions: (i) eviction of activating factors (Minajigi et al., 2015; Jegu et al., 2019), (ii) recruitment of silencing factors, including Polycomb repressive complexes 1 and 2 (PRC1, PRC2) (Wang et al., 2001; Plath et al., 2003; Silva et al., 2003; de Napoles et al., 2004; Kohlmaier et al., 2004; Schoeftner et al., 2006; Zhao et al., 2008; Pintacuda et al., 2017; Colognori et al., 2019), and (iii) global transformation of the 3D chromosomal structure (Rao et al., 2014; Minajigi et al., 2015; Giorgetti et al., 2016; Wang et al., 2018). The establishment phase involves building a chromosomal memory that persists through the ensuing maintenance phase and ensures stable retention of repressive heterochromatin (Kohlmaier et al., 2004; Simon et al., 2013). During this early window, the incipient Xi is easily perturbed and reactivated (Wutz and Jaenisch, 2000; Kohlmaier et al., 2004); but once established, the Xi is remarkably stable and becomes more difficult to reactivate (Brown and Willard, 1994; Csankovszki et al., 2001; Minajigi et al., 2015; Carrette et al., 2017; Adrianse et al., 2018). Even so, the Xi continues to require Xist to fully maintain its silent configuration. In somatic female cells, loss of Xist RNA results in reversal of some 3D chromosomal structures (Minajigi et al., 2015; Wang et al., 2019), loss of repressive Polycomb marks (Plath et al., 2003; Zhang et al., 2007; Nozawa et al., 2013; Pintacuda et al., 2017; Colognori et al., 2019)(Zhang et al., 2007; Nozawa et al., 2013), and partial reactivation of Xi genes (Zhang et al., 2007; Yildirim et al., 2013; Bhatnagar et al., 2014; Carrette et al., 2017; Adrianse et al., 2018). The establishment and maintenance phases are therefore biologically and functionally distinct.
There has been considerable interest in understanding the mechanistic differences between the earlier and more dynamic period of XCI (establishment) versus the later and more stable period of XCI (maintenance). Because Xist RNA and XCI are widely viewed as paradigms for understanding Polycomb-mediated epigenetic regulation (Starmer and Magnuson, 2009; Disteche, 2016; Jegu et al., 2017), similar concepts may extend to autosomal gene regulation as well. Moreover, a deeper understanding could inform growing interest in pharmacological Xi-reactivation as a method of treating X-linked neurodevelopmental disorders (Bhatnagar et al., 2014; Carrette et al., 2017; Sripathy et al., 2017; Adrianse et al., 2018; Carrette et al., 2018). Notably, for Rett Syndrome, reactivation of the wildtype MECP2 allele on the Xi could potentially restore expression of the missing protein for therapeutic impact. More comprehensive knowledge of how the Xi progresses through various stages would enormously benefit design of treatment approaches.
Two motifs within Xist RNA have been linked to the processes of gene silencing and Polycomb recruitment: Repeats A and B. Regarding gene silencing, the importance of Repeat A is universally accepted (Wutz et al., 2002; Minks et al., 2013), whereas reports on Repeat B have argued for varying degrees of silencing defects upon its deletion―though there is agreement that the defects are not as severe as those associated with loss of Repeat A (Pintacuda et al., 2017; Bousard et al., 2019; Colognori et al., 2019; Nesterova et al., 2019). Regarding Polycomb recruitment, the relative roles of Repeats A and B have yet to be resolved, with some observations supporting Repeat A as an important factor (Kohlmaier et al., 2004; Zhao et al., 2008; Davidovich et al., 2013; Kaneko et al., 2013; Cifuentes-Rojas et al., 2014; da Rocha et al., 2014; Davidovich et al., 2015; Lee et al., 2019) and others arguing that the Repeat B region alone is responsible (da Rocha et al., 2014; Pintacuda et al., 2017; Nesterova et al., 2019). Finally, the functional relationship between Polycomb recruitment and gene silencing remains unclear. Some reports show that loss of Polycomb recruitment has only a minor effect on Xi silencing (Kalantry and Magnuson, 2006; Leeb and Wutz, 2007; Bousard et al., 2019), while others show a significant effect (Wang et al., 2001; Kalantry and Magnuson, 2006; Almeida et al., 2017; Pintacuda et al., 2017; Colognori et al., 2019; Nesterova et al., 2019).
These disparate findings have been difficult to reconcile and urge further investigation to elucidate the functional relationships between Repeats A and B in Xi gene silencing and Polycomb recruitment. Here, we reconcile disparate models by demonstrating the existence of two discrete phases during XCI establishment. We ascribe Repeats A and B functions in the two establishment phases, with Phase 1 being primarily Repeat A-dependent and Phase 2 being primarily Repeat B-dependent. We provide evidence that XCI can thus best be characterized as a 3-part process with discrete genetic requirements and epigenetic outcomes.
RESULTS
Xi gene silencing is initiated but not maintained in female cells lacking Xist Repeat B
Recent work has shown that Xist Repeat B plays a major role in recruiting PRC1 and PRC2 to the Xi for proper establishment of silencing (Pintacuda et al., 2017; Colognori et al., 2019; Nesterova et al., 2019). In these studies, deleting Repeat B significantly impaired Xist-mediated gene silencing and essentially abolished deposition of PRC1/PRC2 histone modifications (H2AK119ub/H3K27me3) across the Xi. A separate study, however, reported that Xist transcripts lacking Repeat B do not show significantly impaired silencing and exhibit some residual H2AK119ub/H3K27me3 at Xi regions (Bousard et al., 2019). These seemingly contradictory results warrant closer examination. Indeed, because many of these studies were conducted using an autosomal transgene or inducible Xist system in male embryonic stem cells (ESCs), or on the already-established Xi in fibroblasts, the effect of deleting Repeat B has not been explored fully in a physiological context during ESC differentiation when XCI is established de novo.
Here, we examined female ESCs carrying WT Xist or Xist lacking the Repeat B region (ΔRepB, Fig. S1A (Colognori et al., 2019)) as they underwent differentiation. Importantly, our parental cell line is a Mus musculus/Mus castaneus hybrid, enabling us to distinguish the two X chromosomes by genetic variants between strains. Moreover, Xmus carries a Tsix mutation forcing it to become the future Xi in ~95% of cells, rather than a random choice between Xmus and Xcas (Ogawa et al., 2008). All cell lines were extensively validated by several means to ensure: (i) the identity of the deletion, (ii) that it occurs on Xmus, (iii) that Xist RNA levels and splicing are not altered, (iv) that it does not affect preferential Xist expression from (and thus inactivation of) Xmus over Xcas, and (v) that it does not interfere with ESC differentiation. First, Sanger sequencing across the deleted region, while using SNPs to determine allelic identity, ensured deletion of the expected sequence on Xmus (Table S1). Second, two-color RNA fluorescence in situ hybridization (FISH) using one probe specifically targeting the deleted sequence and another targeting a control exon 7 sequence ensured Xist transcripts lacked the deleted sequence (Fig. S1B). Third, RT-qPCR for different regions across Xist again confirmed loss of the deleted region without affecting adjacent ones or overall Xist levels (Fig. S1C). Fourth, RT-qPCR for Xist using allele-specific (SNP-targeting) primer sets verified ~95% expression from Xmus (Fig. S1D). Lastly, characteristic expression patterns of common differentiation markers (Sox2, Oct4, Gata4) were indistinguishable between WT Xist and deletion cells, suggesting normal progression of differentiation across 14 days (Fig. S1E).
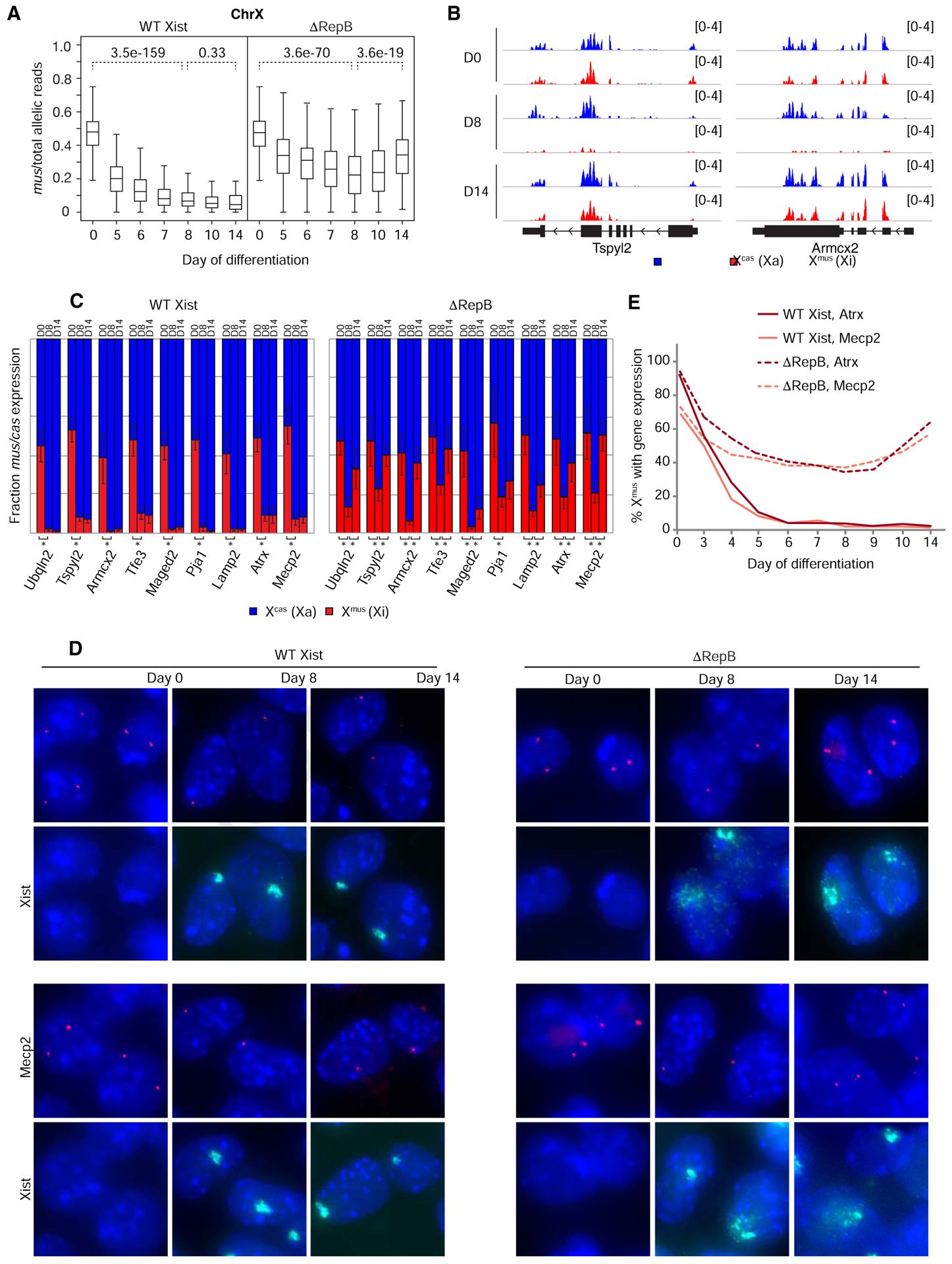
We began by performing a transcriptomic timecourse for Xmus (Xi) expression. In WT Xist cells, XCI occurred as expected between days 0–14 of cell differentiation, evidenced by the progressive decrease in activity from Xmus (Fig. 1A). By contrast, ΔRepB cells showed a biphasic profile: Between days 0–8, the ΔRepB chromosome could initiate noticeable silencing, as demonstrated by the progressive decrease in Xmus transcripts to <20% X-linked gene expression by day 8 (Fig. 1A). Beyond day 8, this initial silencing eroded such that, by day 14, Xmus activity reverted back to ~40% of X-linked allelic reads. Autosomes, as represented by Chromosome 13, did not display any sign of skewed gene expression in ΔRepB cells (Fig. S2A). Examination of specific X-linked genes (e.g., Tspyl2 and Armcx2) confirmed the overall trends (Fig. 1B). These findings suggest a hidden biphasic dynamic during XCI establishment.
Figure 1. Xi gene silencing can be initiated, but not maintained, without Xist Repeat B.
See also Figures S1 and S2.
(A) Boxplots of allele-specific RNA-seq timecourse showing abortive Xi gene silencing in ΔRepB versus WT Xist differentiating female ESCs. Two-tailed student t-test, p values for pairwise comparison as shown.
(B) Zoom-in of allele-specific RNA-seq tracks showing reads from individual X-linked genes.
(C) Allele-specific RT-qPCR showing relative expression from each allele for several X-linked genes in WT Xist and ΔRepB differentiating female ESCs. Error bars show standard deviation between 3 biological replicates. Two-tailed student t-test, asterisks indicate p < 0.05.
(D) Atrx and Mecp2 nascent RNA FISH combined with Xist RNA FISH in WT Xist and ΔRepB differentiating female ESCs. Diffuse Xist cloud morphology caused by disruption of Repeat B/Polycomb was previously described (Colognori et al., 2019). Arrowheads mark positions of Xist cloud.
(E) Quantification of (D). Note that no Xist clouds are present at day 0 to indicate Xmus, but expression is inferred by the presence of two pinpoint signals. n > 100 per time point.
We validated these findings using two orthogonal approaches. First, we performed timecourse allele-specific RT-qPCR to quantify Xmus:Xcas expression for nine genes located at various positions along the X and that are normally subject to XCI (Fig. 1C). Consistent with our transcriptome-wide analysis, all nine genes showed a statistically significant drop from biallelic expression at day 0 to monoallelic expression from Xmus at days 8 and 14 in WT Xist cells. By contrast, ΔRepB cells showed a similar drop from day 0 to 8, but a statistically significant reversion back toward biallelic expression at day 14 for eight out of nine genes assayed. Second, we performed nascent RNA FISH for two of the above genes, Atrx and Mecp2, in combination with Xist RNA FISH to indicate the Xmus (Fig. 1D,E). In WT cells, there was a progressive loss of nascent transcription overlapping the Xist cloud between days 0–14, as expected. On the other hand, ΔRepB cells demonstrated an initial silencing of Atrx and Mecp2 between days 0–8, but regained expression between days 8–14. Notably, Xist clouds appeared dispersed on the ΔRepB chromosome, as previously reported (Colognori et al., 2019). Together these data show that XCI does initiate―though transiently and not to a full extent―despite the absence of Repeat B, thereby implicating additional Xist motifs in initiating silencing. However, the data also demonstrate that the establishment process cannot be completed without Repeat B.
Polycomb recruitment to Xi is initiated but cannot be maintained without Xist Repeat B
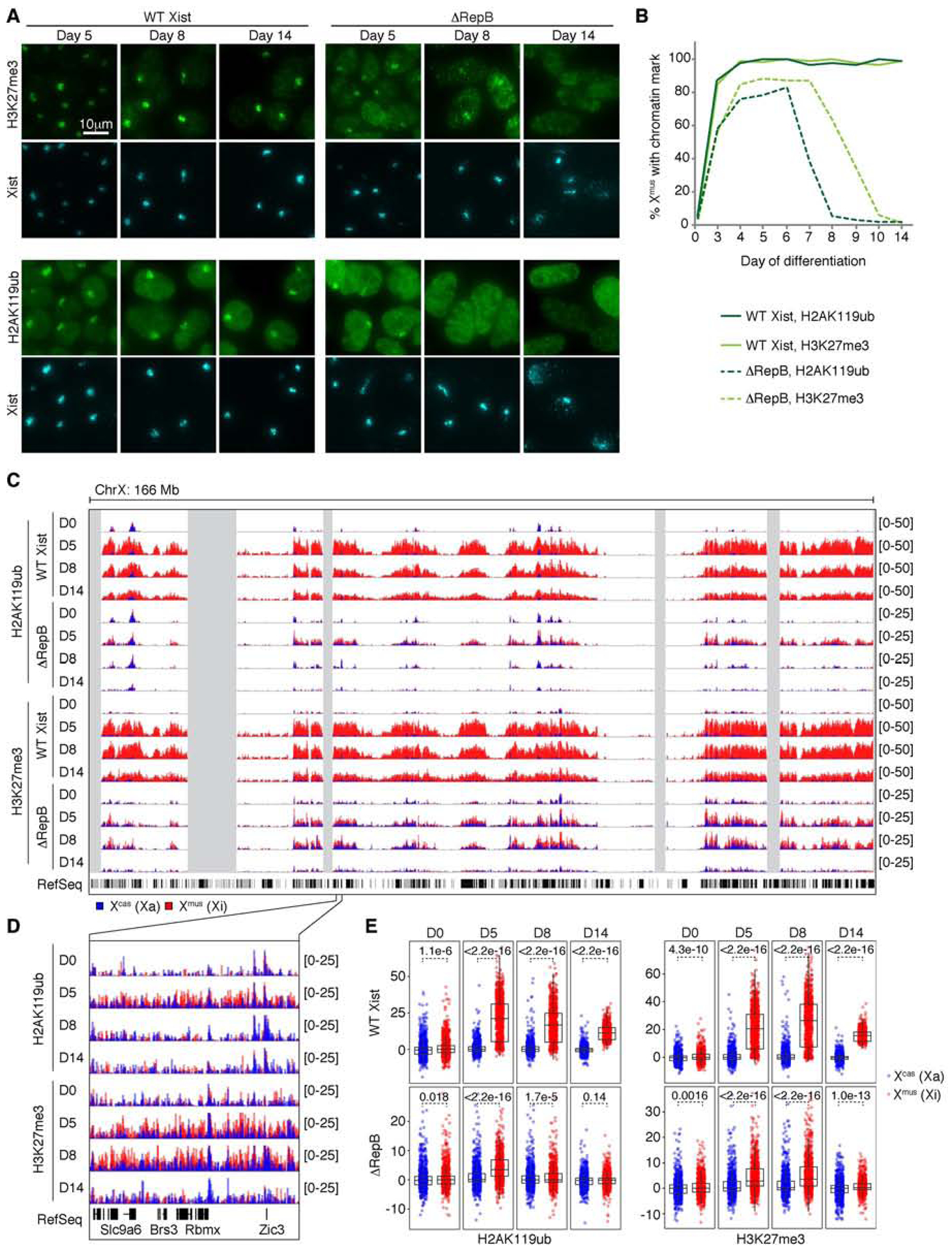
As Repeat B has now been associated with Polycomb binding to the Xi (Pintacuda et al., 2017; Bousard et al., 2019; Colognori et al., 2019; Nesterova et al., 2019), we performed immunofluorescence (IF) for the H2AK119ub and H3K27me3 histone marks (deposited by PRC1 and PRC2, respectively) across a timecourse beginning with the earliest appearance of Xist clouds at day 3. Among nearly all WT Xist-positive cells, strong foci of H2AK119ub and H3K27me3 staining were observed on the Xmus (Fig. 2A,B), as expected. Unexpectedly, ΔRepB cells also displayed noticeable enrichment of both repressive marks in Xist-positive cells during early timepoints (Fig. 2A,B). These data suggest that, contrary to recent reports, deleting Repeat B does not fully abrogate the initiation of Polycomb recruitment.
Figure 2. Xi Polycomb recruitment can be initiated, but not maintained, without Xist Repeat B.
See also Figure S2.
(A) H2AK119ub and H3K27me3 IF combined with Xist RNA FISH in WT Xist and ΔRepB differentiating female ESCs.
(B) Quantification of (A). Note that no Xist clouds are present at day 0 to indicate Xmus, but cells accordingly show no focal enrichment of H2AK119ub or H3K27me3. n > 100 per time point.
(C) Allele-specific H2AK119ub/H3K27me3 ChIP-seq timecourse in WT Xist and ΔRepB differentiating female ESCs. Note the 2-fold difference in y-axis scaling between WT Xist and ΔRepB tracks.
(D) Zoom-in of allele-specific ChIP-seq tracks in (C).
(E) Boxplots quantifying allele-specific ChIP-seq coverage over X-linked genes. Wilcoxon rank-sum test, p values as shown.
The PRC2 mark, H3K27me3, was observed overlapping with Xist clouds in ~85% of Xist-positive ΔRepB cells and remained stable between days 4–7, after which time the signal began to fade away to undetectable levels by day 10, suggesting that Repeat B is required for maintaining PRC2. Yet at the same time, PRC2 must come independently of Repeat B during the initiation phase. In parallel, the PRC1 mark, H2AK119ub, remained stable between days 4–6 in ~80% of Xist-positive cells, after which point it also faded away and became undetectable by day 8, suggesting Repeat B’s role in stabilizing PRC1 as well. The differential kinetics for H3K27me3 with respect to H2AK119ub are similar to the previous observation that depleting HNRNPK (Repeat B’s direct binding partner) has a more immediate effect on PRC1 than PRC2 (Pintacuda et al., 2017; Colognori et al., 2019; Zylicz et al., 2019). This delay may reflect order of recruitment, difference in turnover rate for each mark, or differential requirements for PRC1/2 recruitment. Importantly, the rise and fall of repressive histone marks coincided with the initiation and erosion of gene silencing (inflection around day 8) (Fig. 1), and support the idea of a biphasic dynamic during XCI establishment.
At higher resolution, allele-specific chromatin immunoprecipitation (ChIP)-seq for H2AK119ub and H3K27me3 agreed with the IF data (Fig. 2C–E). WT Xist cells showed characteristic enrichment of H2AK119ub and H3K27me3 across Xmus between days 5–14 (Fig. 2C). Consistent with IF data, ΔRepB cells also accumulated the two marks on Xmus (red track) compared to Xcas (blue track), although overall levels were lower than on the WT Xist Xmus (Fig. 2D). Quantification of coverages over genes revealed significant enrichment of H2AK119ub at day 5, but less so at days 8–14 (Fig. 2E)―consistent with IF data. The H3K27me3 mark was enriched on the ΔRepB Xmus on day 5 and persisted longer to day 8, but less so by day 14 (Fig. 2E). When examined individually, there were no obvious differences in coverage dynamics for genic versus intergenic regions, or genes silenced by XCI versus those non-expressed to begin with (at least at this temporal resolution), though genes silenced by XCI exhibited a stronger gain in H2AK119ub and H3K27me3 during the inactivation process (Fig. S2B,C).
Taken together, these data demonstrate several crucial points: (i) Polycomb complexes can be recruited and XCI can initiate without Repeat B; (ii) Without Repeat B, however, Polycomb recruitment and gene silencing cannot be fully established or stabilized; (iii) The establishment of gene silencing and Polycomb recruitment can therefore be characterized as “biphasic”. The first phase is primarily Repeat B-independent, while the second is Repeat B-dependent. This discovery left open the question of what additional Xist motifs are essential for the first phase of establishing silencing.
Failed Xi gene silencing and selective pressure towards X-aneuploidy in cells lacking Xist Repeat A
The Repeat A motif has been reported to play a role in both Xi silencing and Polycomb recruitment (Wutz et al., 2002; Zhao et al., 2008; Minks et al., 2013; Cifuentes-Rojas et al., 2014; Bousard et al., 2019; Lee et al., 2019; Nesterova et al., 2019). However, various deletions of Repeat A have produced different findings, resulting in a lack of consensus regarding its in vivo role. This is due partly to the fact that existing deletions cover different sequences around Repeat A and have been analyzed in different contexts, such as on an autosomal transgene and/or in male cells. Previous observations also suggested that sequences within or around Repeat A can influence Xist expression and/or splicing (Wutz et al., 2002; Zhao et al., 2008; Hoki et al., 2009; Royce-Tolland et al., 2010; Colognori et al., 2019).
In vivo investigation of Repeat A’s role in XCI necessitates a discrete deletion in the endogenous context in female cells. Here, we created a distinct ΔRepA clone in female ESCs that removes a minimal region containing the motif without affecting Xist splicing or expression levels (Fig. S1A–C). As earlier, we carried out extensive validation of our cell line to ensure ΔRepA Xist was expressed selectively from Xmus during differentiation (Table S1, Fig. S1B–E)―though ΔRepA Xist clouds often appeared smaller than WT Xist clouds, as noted previously (Ha et al., 2018). Interestingly, roughly half of cells lacked any Xist cloud over the course of differentiation (Fig. S1C,S3A). Follow-up analysis showed that this was due to loss of an X chromosome in most cells, occurring specifically during cell differentiation. While 95% of ΔRepA cells carried two X chromosomes on day 0, only 9% retained them both by day 14 (Fig. S3B). This contrasted sharply with differentiation of WT Xist or ΔRepB cells, which consistently retained their XX status. We suspect that a crucial function for Repeat A during differentiation precludes survival (or competitive advantage) of XX cells lacking it. Indeed, because the ΔRepA mutation renders Xist unable to properly silence the Xi in cis (see below), there may be strong selective pressure to lose one X during differentiation in order to achieve proper 1X gene dosage (Fig. 4A). Interestingly, Xmus (selectively expressing ΔRepA Xist) and Xcas (carrying a WT but non-expressed copy of Xist) were lost with roughly equal probability, as determined by allele-specific PCR of genomic DNA from the differentiated cell population (Fig. S3B). This lack of discrimination could explain why roughly half of cells were Xist-positive (XmusO) and half were Xist-negative (XcasO), and further supports the idea that XiΔA and Xa are functionally interchangeable in supplying cells with the necessary X-linked gene products (Fig. 4A). We also point out that, at least in the context of our Tsix mutant background, once an early decision was made to express (mutant) Xist from and silence Xmus (even if unsuccessful), this decision appeared irreversible: Xist expression did not “switch” alleles despite a capable WT Xist copy on Xcas, and continued to be expressed from Xmus even after subsequent loss of Xcas. Notably, the Xist allele expressed from Xmus is mutated (ΔRepA) and cannot initiate silencing on Xmus. Thus, in these XmusO cells, an overall “count” of one Xa and dosage compensation were preserved (Fig. S1B–D). The acute tendency to become XO is further testament to the importance of RepA for female cells undergoing dosage compensation.
Figure 4. Repeat B, but not A, continues to play a role in maintaining Polycomb across Xi.
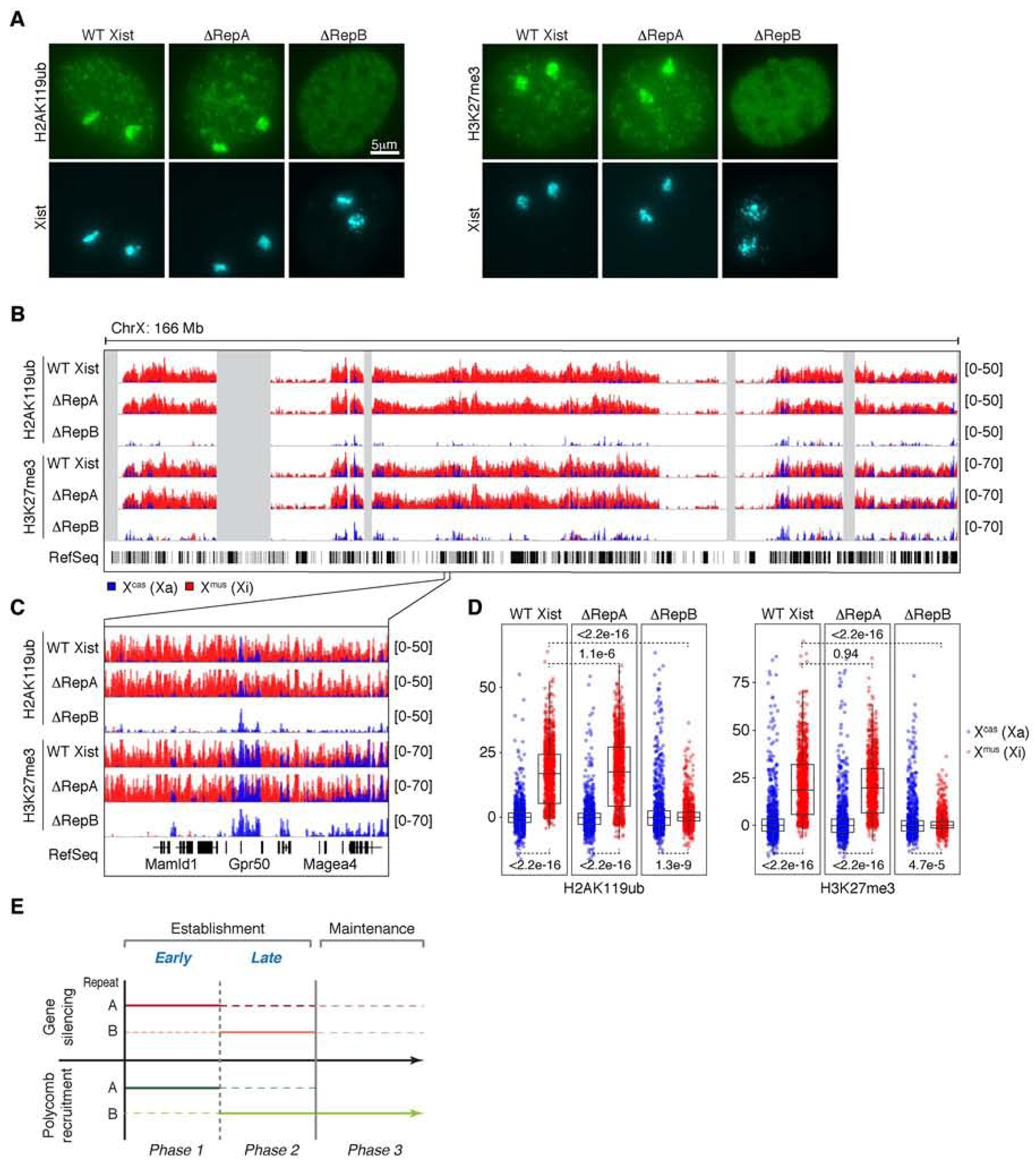
See also Figures S1 and S4.
(A) H2AK119ub and H3K27me3 IF combined with Xist RNA FISH in WT Xist, ΔRepA, and ΔRepB female MEFs. n > 100 per deletion cell line, with nearly all cells showing the indicated pattern of enrichment for each mark.
(B) Allele-specific H2AK119ub/H3K27me3 ChIP-seq in WT Xist, ΔRepA, and ΔRepB MEFs.
(C) Zoom-in of allele-specific ChIP-seq tracks in (B).
(D) Boxplots quantifying allele-specific ChIP-seq coverage over X-linked genes. Wilcoxon rank-sum test, p values as shown.
(E) Diagram summarizing roles of Repeats A and B in gene silencing and Polycomb recruitment throughout XCI. Repeat A is required for initial gene silencing and an early wave of Polycomb recruitment; Repeat B is required for sustained gene silencing and Polycomb recruitment. Dashed line indicates uncertain or minimal contribution.
Due to the stochastic loss of Xcas or Xmus and mixture of XO/XX cells during ΔRepA ESC differentiation, we could not pursue genomic analyses such as RNA-seq and ChIP-seq. However, in spite of large variation between replicates, we were clearly able to see expression from both alleles for nine X-linked genes using allele-specific RT-qPCR (Fig. 4B), indicative of failed Xi silencing. To rule out that apparent biallelic expression could be an artifact of cellular mosaicism, we examined nascent transcription at the single-cell level using RNA FISH. Within the fraction of Xist-positive cells (most being XmusO, some being XmusXcas), Atrx and Mecp2 demonstrated clear failure to be silenced throughout the entire 14-day differentiation time-course, despite an overlying Xist cloud (Fig, 4C,D). These results reaffirm the consensus in the field that Repeat A is critical for gene silencing (Wutz et al., 2002).
An early wave of Xi Polycomb recruitment requires Xist Repeat A
To address whether Repeat A is required for Polycomb targeting, we inspected H2AK119ub and H3K27me3 modifications in IF experiments. In Xist-positive ΔRepA differentiating ESCs, a focus of H2AK119ub and H3K27me3 could still be observed over the Xist domain throughout the time-course (Fig. 4E,F). We conclude that, at the cytological level, loss of Repeat A alone is insufficient to abolish bulk enrichment of these marks, consistent with previous transgenic studies on autosomes (Plath et al., 2003; Kohlmaier et al., 2004; da Rocha et al., 2014). Without ChIP-seq analysis, however, we could not rule out finer defects that might be present at the molecular level despite clear foci at the cytological level, as demonstrated previously (Wang et al., 2018; Wang et al., 2019; Zylicz et al., 2019). Indeed, recent data have suggested that Repeat A is necessary for spreading both H2AK119ub and H3K27me3 into active genic regions (Bousard et al., 2019; Zylicz et al., 2019).
Additional support for Repeat A function came from female ESCs carrying deletions of both Repeats A and B on Xmus (Table S1, Fig. S1A–E). Similar to ΔRepA, ΔRepAB differentiating ESCs exhibited a cluster of Xist RNA in roughly half of cells (Fig. S3A) and had heightened tendency to become XO during differentiation, with only 8% of cells retaining both X chromosomes by day 14 (Fig. 4A,S3B). Furthermore, ΔRepAB cells failed to undergo gene silencing, as shown by persistence of biallelic Mecp2 and Atrx expression in allele-specific RT-qPCR and nascent RNA FISH experiments (Fig. 4B–D). Notably, Xist cloud dispersal seen in ΔRepB cells (Fig 1D,S1B) (Colognori et al., 2019) became exacerbated by the simultaneous deletion of Repeat A, suggesting Repeat A may also play a role in localizing and/or spreading Xist on the Xi. However, in contrast to both the ΔRepA and ΔRepB single deletions, the ΔRepAB double deletion completely abolished H2AK119ub and H3K27me3 signals on the Xi, as determined by IF (Fig. 4E,F). This was true throughout the entire differentiation timecourse. Thus, Repeats A and B both contribute to establishing Polycomb recruitment and gene silencing during the early window of XCI.
Continued Polycomb recruitment during XCI maintenance requires Xist Repeat B but not A
Given the unexpected relationship between Repeats A and B during XCI establishment, we next investigated their roles in XCI maintenance by examining similar ΔRepA and ΔRepB deletions in female MEFs. Our parental MEF cell line is again a Mus musculus/Mus castaneus hybrid, but one that became tetraploid post-XCI and thus carries 2 Xi’s (Xmus) and 2 Xa’s (Xcas)(Yildirim et al., 2011). IF for H2AK119ub/H3K27me3 revealed that loss of Repeat B alone was sufficient to abolish both marks from Xi (Fig. 4A), as previously observed (Colognori et al., 2019). On the other hand, deletion of Repeat A alone had no apparent cytological effect (Fig. 4A). To obtain higher resolution information, we performed allele-specific ChIP-seq for H2AK119ub and H3K27me3. In agreement with IF data, there was chromosome-wide depletion of both marks from Xmus (becoming indistinguishable from Xcas) in ΔRepB cells (Colognori et al., 2019), but no similar effect in ΔRepA cells (Fig. 4B-D). Furthermore, we detected no obvious differences in coverage over genic versus intergenic regions, or non-expressed genes versus those subject to XCI, besides again higher coverage over genes subject to XCI (except in ΔRepB cells where the marks are both absent) (Fig. S4A,B). Together, these data indicate that, while Repeat A plays a role in establishing Polycomb recruitment during the first phase of XCI, it appears dispensable once recruitment has been established. On the other hand, Repeat B is required for Polycomb recruitment during establishment and remains relevant throughout the XCI maintenance phase.
DISCUSSION
Here our work has addressed the longstanding confusion over the roles of Xist Repeats A and B for Polycomb recruitment and Xi gene silencing. In doing so, we found that XCI can best be characterized as having three distinct phases (Fig. 4E): (i) an early establishment phase [days 0–8 of ESC differentiation] during which Repeat A is required to initiate gene silencing and an early wave of Polycomb recruitment; (ii) a late establishment phase [days 8–14] during which Repeat B is essential for stabilizing Polycomb proteins and gene silencing on the Xi; and (iii) a maintenance phase [in somatic cells] in which Repeat A is no longer required but Repeat B continues to play a role in Polycomb maintenance. During the maintenance phase, gene silencing is stabilized but may still depend on continued expression of Xist in a context-dependent manner, as post-XCI deletions of Xist (in part or in whole) can cause either major physiological perturbations (Yildirim et al., 2013) or minimal reactivation unless combined with other pharmacological agents (Csankovszki et al., 2001; Minajigi et al., 2015; Carrette et al., 2017; Adrianse et al., 2018; 2015; Colognori et al., 2019). Of note, our data do not rule out that Repeat A may also function during late establishment (dotted lines, Fig. 4E), since we did not conditionally remove it during this time frame. Similarly, Repeat B may also contribute somewhat to early establishment (dotted lines, Fig. 4E), since initial gene silencing and Polycomb recruitment were less robust upon its removal, and deletion of both Repeats A and B was necessary to abolish early Polycomb enrichment. Thus, during the early critical stages, Repeats A and B may work together and both be required to establish the typical Polycomb binding patterns, associated enrichment of H3K27me3 and H2AK119ub, and full genic silencing on the Xi.
Our data show that deleting Repeat B alone does not preclude initiation of Polycomb recruitment and gene silencing during early XCI between days 0–8 (Fig. 1,2). Thus, in contrast with recent proposals (Pintacuda et al., 2017; Nesterova et al., 2019), Repeat B is not the only motif involved in these activities. However, without Repeat B, gene silencing and Polycomb recruitment cannot proceed to completion and are also unstable, exhibiting a reversion to biallelic expression and an inability to retain the Polycomb marks beyond day ~8. Thus, Repeat B functions as a parallel pathway to stabilize and/or bolster gene silencing and Polycomb on Xi. In this regard, our study reconciles disparate conclusions of prior studies in which deletions of Repeat B have been shown to both significantly affect Xi silencing (Pintacuda et al., 2017; Colognori et al., 2019; Nesterova et al., 2019) or oppositely, to have little effect (Bousard et al., 2019). Our work explains this disparity in that Repeat B’s impact on Xi silencing changes over the timecourse of XCI, being more pronounced at later versus earlier timepoints. It also explains the residual amounts of H2AK119ub and H3K27me3 detected in one study after deleting Repeat B (Bousard et al., 2019).
Moreover, while all reports agree that Repeat A is required for Xi silencing (Wutz et al., 2002; Bousard et al., 2019; Nesterova et al., 2019), there has been a lack of consensus regarding its role in targeting Polycomb complexes (Plath et al., 2003; Kohlmaier et al., 2004; Zhao et al., 2008; Hoki et al., 2009; Davidovich et al., 2013; Kaneko et al., 2013; Cifuentes-Rojas et al., 2014; da Rocha et al., 2014; Davidovich et al., 2015). Although we were unable to pursue a more detailed epigenomic analysis of the ΔRepA mutant due to its instability and propensity to become XO during differentiation, our cytological comparison of ΔRepB versus ΔRepAB cells promote the idea that Repeat A contributes to early targeting of Polycomb complexes. Deleting either Repeat A or B alone does not fully abolish initiation of Polycomb recruitment, but simultaneous deletion of both does. Therefore, by inference, Repeat A must collaborate with Repeat B during XCI establishment for full recruitment of Polycomb complexes, with Repeat A being more critical for the early phase and Repeat B being more critical for subsequent phases. This was likely missed in the past because the requirement is revealed only in conjunction with a Repeat B deletion (Fig. 4).
Although on its own, the ΔRepA mutant shows Xi enrichment of PRC1 and PRC2 marks in most Xist-positive cells, there could be underlying local defects that cannot be discerned by cytological assays. Indeed, SMCHD1-depleted cells also demonstrate an apparently normal Xist cloud and enrichment of H2AK119ub/H3K27me3 by IF, but regional defects become clear in higher-resolution molecular assays (Wang et al., 2018; Wang et al., 2019). Together, our data affirm a role for Repeat A in the initiation of Polycomb recruitment, consistent with previous studies (Kohlmaier et al., 2004; Zhao et al., 2008; Cifuentes-Rojas et al., 2014; Davidovich et al., 2015), and are also conceptually consistent with work attributing to Repeat A the recruitment of Polycomb to initially active genes in ESCs (Simon et al., 2013; Bousard et al., 2019; Zylicz et al., 2019). Mechanistically, how this recruitment occurs is still under debate. One possibility is through direct RNA-mediated recruitment of Polycomb complexes (Zhao et al., 2008; Cifuentes-Rojas et al., 2014). An alternative is through indirect recruitment as a consequence of de novo gene silencing by other Repeat A-interacting proteins such as SPEN (Nesterova et al., 2019). It is also possible that both types of mechanisms are at play. Our findings also have implications for the role of Polycomb complexes during Xi gene silencing. Previous literature suggests Polycomb may be dispensable for initial silencing (Kalantry and Magnuson, 2006; Leeb and Wutz, 2007), but is required for its stabilization (Kalantry and Magnuson, 2006; Wang et al., 2001) until additional mechanisms such as DNA methylation solidify the silenced state in maintenance phase (Csankovszki et al., 2001). Our observation that deleting Repeat B leads to reversal of gene silencing coincident with loss of Polycomb marks at differentiation day 8 supports the latter half of this hypothesis (Figs. 1,2). As for a role in initial silencing, Polycomb mark enrichment was observed on Xi in a Repeat A-dependent manner coincident with initial transient silencing in ΔRepB cells (Fig. 2). However, the marks were also enriched on Xi in ΔRepA cells despite failure to initiate silencing (Fig. 4). Thus, it is less clear whether Polycomb recruitment in this case is a cause or consequence of initial silencing.
The functional importance of Repeat A is further underscored by our inability to derive stable XX female differentiated ESCs lacking it. The instability occurred only during differentiation and not in undifferentiated cells, suggesting that the propensity towards aneuploidy is caused by selective pressure to make up for failed dosage compensation. Intriguingly, loss of Repeat B does not similarly lead to aneuploidy, potentially because of the less drastic effect on Xi gene silencing (still some silencing at day 14 [Fig. 1A]) or compensation by partial downregulation of Xa (Colognori et al., 2019). It is also tempting to speculate that proper X dosage may be more critical during early differentiation (when Repeat A is critical) than late (when Repeat B is critical).
In a broader context, our study reveals establishment of gene silencing and Polycomb domains to be more complex than previously thought, entailing distinct molecular requirements compared to their maintenance. Xi gene silencing and Polycomb domains occurring in the presence of Xist Repeat A eventually disappear without Repeat B. This is consistent with burgeoning evidence that pre-existing Polycomb marks alone are insufficient to recruit the complexes and maintain Polycomb domains on autosomes; de novo recruitment occurs via different means (Kahn et al., 2016; Hojfeldt et al., 2018; Oksuz et al., 2018). Notably, the division of labor between Repeats A and B in recruiting Polycomb during XCI establishment does not persist into the maintenance phase, when Repeat B but not A continues to be required (Fig. 4). Perhaps Repeat A is required during de novo XCI establishment to silence initially active genes, but becomes dispensable in the maintenance phase when Xi genes are already silenced. Whether similar multi-phasic and/or context-dependent mechanisms are required to establish and maintain epigenetic silencing over autosomal Polycomb targets would be of high interest to future investigations.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeannie T. Lee (lee@molbio.mgh.harvard.edu).
Materials Availability
Cell lines generated in this study will be available upon request following completion of an MTA.
Data and Code Availability
Original unprocessed microscope images in this manuscript have been deposited at Mendeley Data and are available at: https://doi.org/10.17632/77fjk9p346.1
Raw high-throughput sequencing data and processed files for RNA-seq and ChIP-seq reported in this paper have been deposited at GEO under accession number: GSE135389
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Xist deletion cell lines
Xist deletions were generated by CRISPR/Cas9 using a pair of gRNAs flanking the target region. gRNA sequences (Table S1) were designed using tools available online (http://crispr.mit.edu) and cloned into pSpCas9(BB)-2A-GFP or pSpCas9(BB)-2A-Puro vectors (Ran et al., 2013). gRNA/Cas9 plasmid was delivered into ESCs by electroporation (Bio-Rad Gene Pulser Xcell) or MEFs by nucleofection (Lonza Nucleofector II) as per manufacturer’s instructions. Following plasmid delivery, cells were cultured for one week to allow enough time for DNA cutting and repair. Single cells were then sorted into 96-well plates by FACS, expanded, and screened by genomic PCR, Sanger sequencing (Table S1), and two-color Xist RNA FISH.
Xist deletion ESCs were generated in the parental (“WT Xist”) M. musculus/M. castaneus F2 hybrid female ESC line carrying a mutated Tsix allele previously described as “TsixTST/+” (Ogawa et al., 2008). This mutation drives selective inactivation of Xmus. Xist deletion MEFs were generated in the parental (“WT Xist”) M. musculus/M. castaneus F1 hybrid (tetraploid) female MEF line previously described as “EY.T4” . All deletion cell lines used in this study are listed in the Key Resource Table, and characterized in detail in Table S1 and Fig. S1.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-H3K27me3 | GeneTex | Cat#GTX60892 |
| Rabbit monoclonal anti-H2AK119ub | Cell Signaling | Cat#CST8240 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant mouse LIF | Sigma | Cat#ESG1107 |
| Critical Commercial Assays | ||
| NEBNext Poly(A) mRNA Magnetic Isolation Module | New England BioLabs | Cat#E7490S |
| Agencourt AMPure XP Beads | Beckman Coulter | Cat#A63881 |
| NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina | New England BioLabs | Cat#E6240S |
| NEBNext Multiplex Oligos for Illumina (Index Primers Set 1) | New England BioLabs | Cat#E7335S |
| Quant-iT PicoGreen dsDNA Reagent | Thermo Fisher Scientific | Cat#P7581 |
| NEBNext Ultra Directional RNA Library Prep Kit for Illumina | New England BioLabs | Cat#E7420S |
| Deposited Data | ||
| RNA-seq in WT Xist female mouse differentiating ESC (days 0,5,6,7,8,10) | This study | GEO: GSE135389 |
| RNA-seq in WT Xist female mouse differentiating ESC (day 14) | Colognori et al., 2019 | GEO: GSE107217 |
| RNA-seq in ΔRepB Xist female mouse differentiating ESC (days, 0,5,6,7,8,10) | This study | GEO: GSE135389 |
| RNA-seq in ΔRepB Xist female mouse differentiating ESC (day 14) | Colognori et al., 2019 | GEO: GSE107217 |
| H3K27me3 ChIP-seq in WT Xist female mouse differentiating ESC (days 0,5,8,14) | This study | GEO: GSE135389 |
| H3K27me3 ChIP-seq in ΔRepB Xist female mouse differentiating ESC (days 0,5,8,14) | This study | GEO: GSE135389 |
| H2AK119ub ChIP-seq in WT Xist female mouse differentiating ESC (days 0,5,8,14) | This study | GEO: GSE135389 |
| H2AK119ub ChIP-seq in ΔRepB Xist female mouse differentiating ESC (days 0,5,8,14) | This study | GEO: GSE135389 |
| H3K27me3 ChIP-seq in WT Xist female MEF | Colognori et al., 2019 | GEO: GSE107217 |
| H3K27me3 ChIP-seq in ΔRepA Xist female MEF | This study | GEO: GSE135389 |
| H3K27me3 ChIP-seq in ΔRepB Xist female MEF | Colognori et al., 2019 | GEO: GSE107217 |
| H2AK119ub ChIP-seq in WT Xist female MEF | Colognori et al., 2019 | GEO: GSE107217 |
| H2AK119ub ChIP-seq in ΔRepA Xist female MEF | This study | GEO: GSE135389 |
| H2AK119ub ChIP-seq in ΔRepB Xist female MEF | Colognori et al., 2019 | GEO: GSE107217 |
| Mendeley data | This study | http://dx.doi.org/10.17632/77fjk9p346.1 |
| Experimental Models: Cell Lines | ||
| WT Xist female ESC (TsixTST/+) | Ogawa et al., 2008 | N/A |
| ΔRepA Xist female ESC (clone 8) | This study | N/A |
| ΔRepB Xist female ESC (clone D2) | Colognori et al., 2019 | N/A |
| ΔRepAB Xist female ESC (clone 9) | This study | N/A |
| WT Xist female MEF (EY.T4) | Yildirim et al., 2011 | N/A |
| “Old” ΔRepA Xist female MEF (clone X9) | Colognori et al., 2019 | N/A |
| “New” ΔRepA Xist female MEF (clone ds4) | This study | N/A |
| ΔRepB Xist female MEF (clone 22) | Colognori et al., 2019 | N/A |
| Oligonucleotides | ||
| Oligo FISH probes used for Xist RNA FISH | Colognori et al., 2019 | N/A |
| Stellaris FISH probes used for Atrx and Mecp2 nascent RNA FISH | LGC Biosearch Technologies | Custom |
| XMP X Green Mouse Chromosome Paint | MetaSystems | Cat#D-1420–050-FI |
| gRNAs used to generate Xist deletions (see Table S1) | Integrated DNA Technologies | N/A |
| PCR primers (see Table S2) | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| pSpCas9(BB)-2A-GFP (PX461) | Ran et al., 2013 | Addgene Cat#48140 |
| pSpCas9(BB)-2A-Puro (PX459) v2.0 | Ran et al., 2013 | Addgene Cat#62988 |
| Software and Algorithms | ||
| HOMER v4.8 | Heinz et al., 2010 | http://homer.ucsd.edu/homer/index.html |
| NovoAlign v3.02 | Novocraft | http://www.novocraft.com/products/novoalign/ |
| TopHat2 v2.0.10 | Kim et al., 2013 | https://ccb.jhu.edu/software/tophat/index.shtml |
| SPP | Kharchenko et al., 2008 | http://compbio.med.harvard.edu/Supplements/ChIP-seq/ |
| featureCounts v1.5.0-p1 | Liao et al., 2014 | http://subread.sourceforge.net |
| SAMtools v1.4.1 | Li et al., 2009 | http://samtools.sourceforge.net/ |
Cell culture
MEFs were grown in medium containing DMEM, high glucose, GlutaMAX supplement, pyruvate (Thermo Fisher Scientific), 10% FBS (Sigma), 25 mM HEPES pH 7.2–7.5 (Thermo Fisher Scientific), 1× MEM non-essential amino acids (Thermo Fisher Scientific), 1× Pen/Strep (Thermo Fisher Scientific), and 0.1 mM βME (Thermo Fisher Scientific) at 37°C with 5% CO2. ESCs were grown on γ-irradiated MEF feeders in medium containing DMEM, high glucose, GlutaMAX supplement, pyruvate, 15% Hyclone FBS (Sigma), 25 mM HEPES pH 7.2–7.5, 1× MEM non-essential amino acids, 1x Pen/Strep, 0.1 mM βME, and 500 U/mL ESGRO recombinant mouse Leukemia Inhibitory Factor (LIF) protein (Sigma, ESG1107) at 37°C with 5% CO2. LIF was excluded from the medium during ESC differentiation procedures (see below).
ESC differentiation
Undifferentiated ESCs were grown on γ-irradiated MEF feeders for 3 days, after which ESC colonies were trypsinized and feeders removed (day 0). ESCs were then switched to medium lacking LIF and grown in suspension for 4 days, forming embryoid bodies (EBs). EBs were cytospun onto glass slides or settled down onto gelatin-coated coverslips at day 4 and allowed to further differentiate until the indicated timepoints.
METHOD DETAILS
Oligo FISH probes
Oligo FISH probes for Xist RepA, RepB, or exon 7 were previously described (Colognori et al., 2019). Briefly, probe sequences were designed using tools available online (https://www.idtdna.com/calc/analyzer). Amino-ddUTP (Kerafast) was added to the 3’-ends of pooled oligos by Terminal Transferase (New England BioLabs) as per manufacturer’s instructions. Oligos were purified by phenol/chloroform extraction, concentrated by ethanol precipitation, resuspended in 0.1 M sodium borate, and labeled with Cy3B (GE Healthcare) or Alexa647 NHS-ester (Life Technologies). After another ethanol precipitation, labeled oligos were resuspended in water and labeling efficiency was evaluated by absorbance using NanoDrop (Thermo Fisher Scientific). Custom Stellaris FISH probes (LGC Biosearch Technologies) were designed against the first intron of Atrx or Mecp2 using the Stellaris RNA FISH probe designer available online (www.biosearchtech.com/stellarisdesigner) and labeled with Quasar570 dye.
RNA FISH
Cells were cytospun onto glass slides or settled down onto gelatin-coated coverslips and rinsed with PBS. They were permeabilized with cold CSKT buffer (100 mM NaCl, 300 mM sucrose, 10 mM PIPES, 3 mM MgCl2, 0.5% Triton X-100, pH 6.8) for 10 min and then fixed with 4% paraformaldehyde for 10 min at room temp. Cells were rinsed with PBS and dehydrated in a series of increasing ethanol concentrations. Labeled oligo probe pool (1–5 nM for Xist RNA FISH, 100 nM for Atrx or Mecp2 nascent RNA FISH) was added to hybridization buffer containing 25% formamide, 2x SSC, 10% dextran sulfate, and nonspecific competitor (0.1 mg/mL mouse Cot-1 DNA [Thermo Fisher Scientific]). Hybridization was performed in a humidified chamber at 37°C overnight. After being washed once in 25% formamide/2x SSC at 37°C for 20 min and three times in 2x SSC at 37°C for 5 min each, cells were mounted for wide-field fluorescent imaging. Nuclei were counter-stained with Hoechst 33342 (Life Technologies).
X-chromosome painting
Cells were cytospun onto glass slides or settled down onto gelatin-coated coverslips, rinsed with PBS, treated with RNase A (0.5 mg/mL in PBS) at 37°C for 40 min to remove RNA signal, and denatured for DNA FISH in 70% formamide/2x SSC at 80°C for 15 min. Slides were quenched in ice cold 70% ethanol and dehydrated in a series of increasing ethanol concentrations. 1:10 (v/v) XMP X Green mouse chromosome paint (MetaSystems, D-1420–050-FI) was added to hybridization buffer containing 50% formamide, 2× SSC, 10% dextran sulfate, and 0.2 mg/mL mouse Cot-1 DNA (Thermo Fisher Scientific) and denatured at 95°C for 10 min. Hybridization was performed in a humidified chamber at 37°C overnight. After being washed once in 0.2× SSC at 65°C for 10 min and three times in 2x SSC at room temp for 5 min each, cells were mounted for wide-field fluorescent imaging. Nuclei were counter-stained with Hoechst 33342 (Life Technologies).
IF/RNA FISH
Cells were cytospun onto glass slides or settled down onto gelatin-coated coverslips and rinsed with PBS. They were permeabilized with cold CSKT buffer (100 mM NaCl, 300 mM sucrose, 10 mM PIPES, 3 mM MgCl2, 0.5% Triton X-100, pH 6.8) for 10 min and then fixed with 4% paraformaldehyde for 10 min at room temp. After being blocked for 30 min in PBS/1% BSA supplemented with 10 mM ribonucleoside vanadyl complex (New England BioLabs), primary antibodies were added and allowed to incubate at room temp for 1 hr. Cells were washed three times with PBS/0.05% Tween-20 at room temp for 5 min each. After incubating with dye conjugated secondary antibody for 30 min at room temp, cells were washed again with PBS/0.05% Tween-20 at room temp for 5 min each. Cells were post-fixed in 4% paraformaldehyde and dehydrated in a series of increasing ethanol concentrations. Xist RNA FISH was then performed as described above.
Microscopy
For wide-field fluorescent imaging, cells were observed on a Nikon 90i microscope equipped with 60x/1.4 N.A. VC objective lens, Orca ER CCD camera (Hamamatsu), and Volocity software (Perkin Elmer).
Antibodies
The following primary antibodies were used for ChIP-seq and IF: H3K27me3 (GeneTex, GTX60892) and H2AK119ub (Cell Signaling, CST8240). Dye-conjugated secondary antibodies were purchased from Life Technologies.
(Allele-specific) qPCR and RT-qPCR
For RT-qPCR, RNA was isolated from cells using TRIzol Reagent (Thermo Fisher Scientific) as per manufacturer’s instructions. Genomic DNA was removed using TURBO DNase from the TURBO DNA-free Kit (Thermo Fisher Scientific). After inactivating TURBO DNase with DNase Inactivation Reagent (also enclosed in TURBO DNA-free Kit), RNA was reverse transcribed into cDNA using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific) with random primers (Promega, C118A) at 25°C for 5 min, 50°C for 1 hr, and enzyme inactivated at 85°C for 15 min. Depending on the experiment, qPCR was performed on cDNA or genomic DNA using iTaq Universal SYBR Green Supermix (Bio-Rad) in a CFX96 Real-Time PCR Detection System (Bio-Rad). For allele-specific detection, primers were designed to target genetic variants within each gene, as previously described (Glaab and Skopek, 1999; Li et al., 2004). The relative abundance of alleles was calculated using the formula: cas/mus fold difference = 2^(Ctmus − Ctcas), and corrected for primer bias/efficiency by comparing to standard curves using pure cas, mus, or hybrid cas/mus genomic DNA as previously described (Pinter et al., 2015). Primer sequences are listed in Table S2.
ChIP-seq
Cells were cross-linked in PBS with 1% formaldehyde at room temp for 10 min with rotation at 1 million cells/mL, and quenched with 0.125 M glycine for 5 min. Cross-linked cells were washed twice with cold PBS, pelleted, and snap-frozen in liquid nitrogen. 10 million cross-linked cells per ChIP were thawed on ice and resuspended in 1 mL buffer 1 (50 nM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA, 0.5% NP-40, 0.25% Triton X-100, 10% glycerol, 1× cOmplete EDTA-free protease inhibitor cocktail [Roche]), and rotated for 10 min at 4°C. Nuclei were pelleted by centrifugation at 1,000 g for 5 min at 4°C, resuspended in 1 mL buffer 2 (10 mM HEPES pH 7.5, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1× cOmplete EDTA-free protease inhibitor cocktail) supplemented with 0.2 mg/mL RNase A (Thermo Fisher Scientific), and rotated for 10 min at 4°C. Nuclei were pelleted by centrifugation at 1,000 g for 5 min at 4°C and resuspended in 1.3 mL buffer 3 (10 mM HEPES pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1× cOmplete EDTA-free protease inhibitor cocktail, 0.1% sodium deoxycholate, 0.5% sarkosyl). Nuclei were sonicated (Qsonica Q800 Sonicator) in polystyrene tubes at 45% power reading, 30 sec on/30 sec off for a total sonication time of 4 min at 4°C. Triton X-100 was added to the lysate to 1%, which was then centrifuged for 10 min at 16,000 g to remove debris. The lysate was pre-cleared for 2 hr at 4°C with rotation using 20 μL Dynabeads Protein G (Thermo Fisher Scientific) pre-washed with PBS/0.5% BSA. After saving 10% as “input” sample, the pre-cleared lysate was combined with 20 μL Dynabeads Protein G pre-bound to 2 μg antibody (H3K27me3, GeneTex GTX60892; H2AK119ub, Cell Signaling CST8240), and rotated overnight at 4°C. Afterwards, beads were washed five times with wash buffer (50 mM HEPES pH 7.5, 0.5 M LiCl, 1 mM EDTA, 1% NP-40, 0.7% sodium deoxycholate), once with TEN buffer (10 mM Tris pH 8.0, 1 mM EDTA, 50 mM NaCl), and once with TE buffer (10 mM Tris pH 8.0, 1 mM EDTA). Input sample and beads containing ChIP material were resuspended in 400 μL TES buffer (50 mM Tris pH 8.0, 10 mM EDTA, 1% SDS) supplemented with 0.5 mg/mL Proteinase K (Sigma) and incubated for 1 hr at 55°C and then for >3 hr at 65°C to reverse cross-links. DNA was purified by phenol-chloroform extraction and quantified with Quant-iT PicoGreen dsDNA Reagent (Thermo Fisher Scientific). Input and ChIP-seq libraries were prepared using NEBNext ChIP-seq Library Prep Master Mix Set for Illumina (New England BioLabs) as per manufacturer’s instructions. Libraries were sequenced on Illumina HiSeq2000 (high-throughput run) or HiSeq2500 (rapid run), generating ~50 million 50-nt paired-end reads per sample.
ChIP-seq analysis
To account for the hybrid character of our cell lines, adaptor-trimmed reads were separately aligned to custom mus/129 and cas genomes using NovoAlign (Novocraft), then mapped back to reference mm9 genome using SNPs (Pinter et al., 2012). This generated three tracks: composite (comp) of all reads, and two allele-specific tracks using only allele-specifically mappable reads. After allele-specific alignment, input-subtracted allele-specific ChIP-seq tracks were generated using SPP (Kharchenko et al., 2008), with smoothing using 1-kb windows recorded every 500 bp, as previously described (Wang et al., 2018). To account for different sequencing depths for ChIP-seq, samples differing by >10% were compensated by random downsampling with SAMtools (Li et al., 2009). The densities of H2AK119ub and H3K27me3 over gene bodies versus intergenic regions, and genes subject to XCI versus non-expressed on the X chromosome were computed by Homer (Heinz et al., 2010). For the timecourse ChIP-seq analysis in differentiating ESCs, genes subject to XCI were defined as having non-zero FPKM in RNA-seq for both undifferentiated WT and ΔRepB ESCs, and non-expressed genes as having zero FPKM in any of the two datasets. For MEF ChIP-seq analysis, genes subject to XCI were defined as having non-zero FPKM in RNA-seq for WT MEFs (expressed on Xa but not Xi), and non-expressed genes as having zero FPKM (not expressed on Xa or Xi). Xist and Tsix, escapee genes, and regions too short (<200 bp) or unmappable were excluded from the analysis. H2AK119ub and H3K27me3 densities were displayed as boxplots produced using R and ggplot2. p values determined by Wilcoxon ranked sum test (two-sided).
RNA-seq
Total cell RNA was extracted using TRIzol Reagent (Thermo Fisher Scientific), from which mRNA was isolated using NEBNext Poly(A) mRNA magnetic isolation module (New England BioLabs) as per manufacturer’s instructions. RNA-seq libraries were prepared using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England BioLabs) as per manufacturer’s instructions. Libraries were sequenced on Illumina HiSeq2000 (high-throughput run) or HiSeq2500 (rapid run), generating ~50 million 50-nt paired-end reads per sample.
RNA-seq analysis
PCR duplicates were removed and reads separately aligned to custom mus/129 and cas genomes using TopHat2 (Kim et al., 2013). Final allele-specific mapping to reference mm9 genome was generated based on SNPs (Pinter et al., 2012). Only uniquely aligned concordantly mapped sequences were used in downstream analysis. Counts per gene were calculated using featureCounts (Liao et al., 2014). Using MatLab (MathWorks), library sizes were normalized and genes with insufficient allelic information (<13 allele-specific reads) were removed. We also removed potentially miscalled genes from our alignment pipeline, defined as genes incorrectly assigned to mus from a pure cas RNA-seq library. Allele-specific RPKM was calculated using allelic ratio (allele-specific counts) applied to comp RPKM (total counts). Genes with comp RPKM<1 or overlapping unmappable regions were excluded from the analysis, along with Xist and Tsix.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters including the statistical tests used and the values of n, p, and R are reported in the figures, figure legends, or associated main texts. Statistical significance is determined by the value of p < 0.05 by the indicated tests. For microscope images, n generally refers to the total number of counted cells or Xist clouds.
Supplementary Material
Figure 3. Repeat A is required for gene silencing and contributes to early Polycomb recruitment on Xi.

See also Figures S1 and S3.
(A) Repeat A/AB deletion leads to loss of an X chromosome (either Xcas or Xmus) in ~90% of cells over the course of differentiation.
(B) Allele-specific RT-qPCR showing relative expression from each allele for several X-linked genes in ΔRepA and ΔRepAB differentiating female ESCs. Error bars show standard deviation between 3 biological replicates. Two-tailed student t-test, asterisks indicate p < 0.05.
(C) Atrx and Mecp2 nascent RNA FISH combined with Xist RNA FISH in ΔRepA and ΔRepAB differentiating female ESCs. Only Xist-positive cells (XmusXcas and XmusO) were examined. Arrowheads mark positions of Xist cloud.
(D) Quantification of (C). Note that no Xist clouds are present at day 0 to indicate Xmus, but expressionis inferred by the presence of two pinpoint signals. n > 100 per time point.
(E) H2AK119ub and H3K27me3 IF combined with Xist RNA FISH in ΔRepA and ΔRepAB differentiating female ESCs. Note the diffuse Xist cloud morphology caused by disruption of Repeat B/Polycomb (Fig. 1D) (Colognori et al., 2019) appears exacerbated by additional loss of Repeat A/Polycomb (but not by Repeat A loss alone). Only Xist-positive cells (XmusXcas and XmusO) were examined.
(F) Quantification of (E). Note that no Xist clouds are present at day 0 to indicate Xmus, but cells accordingly show no focal enrichment of H2AK119ub or H3K27me3. n > 100 per time point.
HIGHLIGHTS.
X-inactivation establishment is a biphasic process requiring Xist Repeats A and B.
Polycomb complexes can initially be recruited without Repeat B.
Repeat A initiates Polycomb recruitment and X-silencing while B stabilizes them.
Frequency of X-chromosome loss (XO state) is heightened by deleting Repeat A.
ACKNOWLEDGEMENTS
We thank all members of the Lee lab for critical comments and stimulating discussions. H.S. was supported by NIH 5T32HD007396–24 and J.T.L. by NIH R01-HD097665 and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
J.T.L. is a co-founder of Translate Bio and Fulcrum Therapeutics and serves as Advisor to Skyhawk Therapeutics.
REFERENCES
- Adrianse RL, Smith K, Gatbonton-Schwager T, Sripathy SP, Lao U, Foss EJ, Boers RG, Boers JB, Gribnau J, and Bedalov A (2018). Perturbed maintenance of transcriptional repression on the inactive X-chromosome in the mouse brain after Xist deletion. Epigenetics Chromatin 11, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida M, Pintacuda G, Masui O, Koseki Y, Gdula M, Cerase A, Brown D, Mould A, Innocent C, Nakayama M, et al. (2017). PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 356, 1081–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar S, Zhu X, Ou J, Lin L, Chamberlain L, Zhu LJ, Wajapeyee N, and Green MR (2014). Genetic and pharmacological reactivation of the mammalian inactive X chromosome. Proceedings of the National Academy of Sciences of the United States of America, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousard A, Raposo AC, Zylicz JJ, Picard C, Pires VB, Qi Y, Gil C, Syx L, Chang HY, Heard E, et al. (2019). The role of Xist-mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep, e48019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, and Willard HF (1994). The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature 368, 154–156. [DOI] [PubMed] [Google Scholar]
- Carrette LLG, Blum R, Ma W, Kelleher RJ 3rd, and Lee JT (2018). Tsix-Mecp2 female mouse model for Rett syndrome reveals that low-level MECP2 expression extends life and improves neuromotor function. Proc Natl Acad Sci U S A 115, 8185–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrette LLG, Wang CY, Wei C, Press W, Ma W, Kelleher RJ 3rd, and Lee JT (2017). A mixed modality approach towards Xi reactivation for Rett syndrome and other X-linked disorders. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Hernandez AJ, Sarma K, and Lee JT (2014). Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell 55, 171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colognori D, Sunwoo H, Kriz AJ, Wang CY, and Lee JT (2019). Xist Deletional Analysis Reveals an Interdependency between Xist RNA and Polycomb Complexes for Spreading along the Inactive X. Mol Cell 74, 101–117 e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, and Jaenisch R (2001). Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol 153, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, et al. (2014). Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Mol Cell 53, 301–316. [DOI] [PubMed] [Google Scholar]
- Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, and Cech TR (2015). Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell 57, 552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, and Cech TR (2013). Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol 20, 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. (2004). Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7, 663–676. [DOI] [PubMed] [Google Scholar]
- Disteche CM (2016). Dosage compensation of the sex chromosomes and autosomes. Semin Cell Dev Biol 56, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, et al. (2016). Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaab WE, and Skopek TR (1999). A novel assay for allelic discrimination that combines the fluorogenic 5’ nuclease polymerase chain reaction (TaqMan) and mismatch amplification mutation assay. Mutat Res 430, 1–12. [DOI] [PubMed] [Google Scholar]
- Ha N, Lai LT, Chelliah R, Zhen Y, Yi Vanessa SP, Lai SK, Li HY, Ludwig A, Sandin S, Chen L, et al. (2018). Live-Cell Imaging and Functional Dissection of Xist RNA Reveal Mechanisms of X Chromosome Inactivation and Reactivation. iScience 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojfeldt JW, Laugesen A, Willumsen BM, Damhofer H, Hedehus L, Tvardovskiy A, Mohammad F, Jensen ON, and Helin K (2018). Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat Struct Mol Biol 25, 225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoki Y, Kimura N, Kanbayashi M, Amakawa Y, Ohhata T, Sasaki H, and Sado T (2009). A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development 136, 139–146. [DOI] [PubMed] [Google Scholar]
- Jegu T, Aeby E, and Lee JT (2017). The X chromosome in space. Nat Rev Genet 18, 377–389. [DOI] [PubMed] [Google Scholar]
- Jegu T, Blum R, Cochrane JC, Yang L, Wang CY, Gilles ME, Colognori D, Szanto A, Marr SK, Kingston RE, et al. (2019). Xist RNA antagonizes the SWI/SNF chromatin remodeler BRG1 on the inactive X chromosome. Nat Struct Mol Biol 26, 96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn TG, Dorafshan E, Schultheis D, Zare A, Stenberg P, Reim I, Pirrotta V, and Schwartz YB (2016). Interdependence of PRC1 and PRC2 for recruitment to Polycomb Response Elements. Nucleic Acids Res 44, 10132–10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, and Magnuson T (2006). The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet 2, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, and Bonasio R (2013). PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20, 1258–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, and Park PJ (2008). Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, and Wutz A (2004). A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2, E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DM, Trotman JB, Cherney RE, Inoue K, Schertzer MD, Bischoff SR, Cowley DO, and Calabrese JM (2019). A 5’ fragment of Xist can sequester RNA produced from adjacent genes on chromatin. Nucleic Acids Res 47, 7049–7062. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Leeb M, and Wutz A (2007). Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol 178, 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Kadura I, Fu DJ, and Watson DE (2004). Genotyping with TaqMAMA. Genomics 83, 311–320. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Minajigi A, Froberg J, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W, et al. (2015). Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minks J, Baldry SE, Yang C, Cotton AM, and Brown CJ (2013). XIST-induced silencing of flanking genes is achieved by additive action of repeat a monomers in human somatic cells. Epigenetics Chromatin 6, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova TB, Wei G, Coker H, Pintacuda G, Bowness JS, Zhang T, Almeida M, Bloechl B, Moindrot B, Carter EJ, et al. (2019). Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat Commun 10, 3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa RS, Nagao K, Igami KT, Shibata S, Shirai N, Nozaki N, Sado T, Kimura H, and Obuse C (2013). Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat Struct Mol Biol 20, 566–573. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, and Lee JT (2008). Intersection of the RNA interference and X-inactivation pathways. Science 320, 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksuz O, Narendra V, Lee CH, Descostes N, LeRoy G, Raviram R, Blumenberg L, Karch K, Rocha PP, Garcia BA, et al. (2018). Capturing the Onset of PRC2-Mediated Repressive Domain Formation. Mol Cell 70, 1149–1162 e1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda G, Wei G, Roustan C, Kirmizitas BA, Solcan N, Cerase A, Castello A, Mohammed S, Moindrot B, Nesterova TB, et al. (2017). hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Mol Cell 68, 955–969 e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter SF, Colognori D, Beliveau BJ, Sadreyev RI, Payer B, Yildirim E, Wu CT, and Lee JT (2015). Allelic Imbalance Is a Prevalent and Tissue-Specific Feature of the Mouse Transcriptome. Genetics 200, 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter SF, Sadreyev RI, Yildirim E, Jeon Y, Ohsumi TK, Borowsky M, and Lee JT (2012). Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res 22, 1864–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, and Zhang Y (2003). Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135. [DOI] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce-Tolland ME, Andersen AA, Koyfman HR, Talbot DJ, Wutz A, Tonks ID, Kay GF, and Panning B (2010). The A-repeat links ASF/SF2-dependent Xist RNA processing with random choice during X inactivation. Nat Struct Mol Biol 17, 948–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta A, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, and Wutz A (2006). Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. The EMBO journal 25, 3110–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova T, Webster Z, Peters A, Jenuwein T, Otte A, and Brockdorff N (2003). Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Developmental cell 4, 481–495. [DOI] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, and Lee JT (2013). High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy S, Leko V, Adrianse RL, Loe T, Foss EJ, Dalrymple E, Lao U, Gatbonton-Schwager T, Carter KT, Payer B, et al. (2017). Screen for reactivation of MeCP2 on the inactive X chromosome identifies the BMP/TGF-beta superfamily as a regulator of XIST expression. Proc Natl Acad Sci U S A 114, 1619–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starmer J, and Magnuson T (2009). A new model for random X chromosome inactivation. Development 136, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Colognori D, Sunwoo H, Wang D, and Lee JT (2019). PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nat Commun 10, 2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Jegu T, Chu HP, Oh HJ, and Lee JT (2018). SMCHD1 Merges Chromosome Compartments and Assists Formation of Super-Structures on the Inactive X. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mager J, Chen Y, Schneider E, Cross J, Nagy A, and Magnuson T (2001). Imprinted X inactivation maintained by a mouse Polycomb group gene. Nature genetics 28, 371–375. [DOI] [PubMed] [Google Scholar]
- Wutz A, and Jaenisch R (2000). A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5, 695–705. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, and Jaenisch R (2002). Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30, 167–174. [DOI] [PubMed] [Google Scholar]
- Yildirim E, Kirby J, Brown D, Mercier F, Sadreyev R, Scadden D, and Lee J (2013). Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152, 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L-F, Huynh K, and Lee J (2007). Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129, 693–1399. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, and Lee JT (2008). Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylicz JJ, Bousard A, Zumer K, Dossin F, Mohammad E, da Rocha ST, Schwalb B, Syx L, Dingli F, Loew D, et al. (2019). The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell 176, 182–197 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original unprocessed microscope images in this manuscript have been deposited at Mendeley Data and are available at: https://doi.org/10.17632/77fjk9p346.1
Raw high-throughput sequencing data and processed files for RNA-seq and ChIP-seq reported in this paper have been deposited at GEO under accession number: GSE135389


