Abstract
Oral epithelial cells (OEC) represent the first site of host interaction with viruses that infect the body through the oral route, however their innate antiviral defense mechanisms have yet to be defined. Previous studies have determined that OEC express pathogen-, damage- or danger-associated molecular patterns (PAMPs, or DAMPs), but their expression of key antiviral innate immune mediators, including type I interferons (type I IFN) and interferon-stimulated genes (ISGs) has not been studied extensively. We used the oral keratinocyte cell line, OKF6/TERT1 in the presence and absence of the viral mimics poly(I:C) and unmethylated CpG DNA, to define the expression of type I IFN and ISGs. We identified the basal expression of novel type I IFN genes IFNE and IFNK, while IFNB1 was induced by viral mimics, through the nuclear translocation of IRF3. Numerous ISGs were expressed at basal levels in OEC, with an apparent correlation between high expression and antiviral activity at the earlier stages of viral infection. Stimulation of OECs with poly(I:C) led to selective induction of ISGs, including MX1, BST2, PML, RSAD2, ISG15 and ZC3HAV1. Together, our results demonstrate that OECs exhibit a robust innate antiviral immune defense profile, which is primed to address the wide variety of pathogenic viruses that are transmitted orally.
Keywords: virus, innate immunity, oral, epithelial, interferon, interferon-stimulated gene
Introduction
Many viruses are spread via saliva and/or begin infection through the mouth, including herpesviruses (Fülöp, Larbi, & Pawelec, 2013; Looker et al., 2015; Toussirot, Roudier, Roudier, & al., 2008), coxsackievirus (Corsino & Linklater, 2018), hepatitis B virus (Scott, Snitbhan, Bancroft, Alter, & Tingpalapong, 1980), cytomegalovirus (Plosa, Esbenshade, Fuller, & Weitkamp, 2012), Epstein-Barr virus (Niedobitek, Meru, & Delecluse, 2001) and Kaposi’s sarcoma-associated herpesvirus (Cesarman, Chang, Moore, Said, & Knowles, 1995; Chang et al., 1994; Soulier et al., 1995). The site of first contact, the oral epithelial cells (OECs), act as the first line of defense against these pathogens. While specific mechanisms of how other tissues in the body respond to viruses, not much is known about the innate antiviral defenses of OECs, especially how OECs respond to actively replicating infections.
In general, host cells are able to detect the presence of certain molecular patterns, such as nucleic acid, extracellular ATP, and bacterial components, collectively termed pathogen-, damage-, or danger-associated molecular patterns (PAMPs or DAMPs) (Vénéreau, Ceriotti, & Bianchi, 2015). Originally only thought to detect evolutionarily distinct objects (Akira, Uematsu, & Takeuchi, 2006; Medzhitov & Janeway, 2000), the receptors for these molecular patterns, pattern recognition receptors (PRRs), are now hypothesized to have evolved by way of binding to objects in the environment and cell that are signals of cellular danger (Matzinger, 1994).
Regardless of the origins of the PRRs, these proteins, located either on the cell surface, endosomes, or in the cytoplasm, act as the first step toward clearing infections inside host cells. In humans, these PRRs fall under a number of categories: membrane-bound toll-like receptors (TLRs) and C-type lectin receptors (CLR) and cytoplasmic RIG-I-like receptors (RLRs), NOD-like receptors (NLRs), and dsDNA sensors (e.g. cyclic GMP-AMP synthase, cGAS) (reviewed in Brubaker, Bonham, Zanoni, & Kagan, 2015)). While these PRRs bind to a wide variety of molecular patterns, each with their own degree of promiscuity for ligands (Seong & Matzinger, 2004), their activation pathway includes a number of similar transcription factors: nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), activator protein 1 (AP-1), and interferon regulatory factors (IRFs) (Brubaker et al., 2015). These transcription factors translocate to the nucleus upon activation and lead to the expression of genes encoding for pro-inflammatory cytokines and interferons (Parkin & Cohen, 2001) as well as antimicrobial peptides, which exhibit antiviral activity (Brice & Diamond, 2019).
Type I interferons (type I IFN) are the most well studied interferons in the context of direct inhibitions of viral infections (reviewed previously (McNab, Mayer-Barber, Sher, Wack, & O’Garra, 2015)). These proteins, through either autocrine or paracrine signaling, lead to the expression of interferon-stimulated genes (ISGs). The protein products of these genes act directly to inhibit almost every aspect of the viral lifecycle and even increase the ability of NK cells to kill infected cells (Schneider, Chevillotte, & Rice, 2014; Schoggins & Rice, 2011). The goal of this study was to further define the innate antiviral defense mechanisms of OECs, specifically which type I IFN and ISGs are being expressed in these cells, and how their expression responds to DAMP stimulation that can mimic viral infection.
Materials and Methods
Cells
The oral mucosal keratinocyte cell line OKF6/Tert-1(Dickson et al., 2000) was cultured in keratinocyte serum-free media (K-SFM) (Gibco) supplemented with 2 mM L-glutamine (Corning), 1 IU/ml penicillin (Corning), 100 μg/ml streptomycin (Corning), 75 μg/ml bovine pituitary extract (Gibco), 0.3 mM CaCl2 (Sigma-Aldrich), and 0.6 ng/ml epidermal growth factor (EGF) (Gibco).
RNA Library Construction for Sequencing
RNA of cells from three biological replicates per condition was isolated using the RNEasy Plus Mini kit (Qiagen, 74136). TruSeq RNA library construction was performed at the Interdisciplinary Center for Biotechnology Research (ICBR) Gene Expression Core, University of Florida (UF). RNA quantitation was done by the QUBIT (ThermoFisher), purity was evaluated on a NanoDrop Spectrophotometer (NanoDrop Technologies, Inc.), and sample quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc). Protein-free, intact total RNA (RIN>7) was used for library construction using the reagents provided in the Illumina TruSeq RNA sample preparation kit and following the manufacturer’s low-sample number protocol (Part # 15026495 Rev. F). Briefly, 0.1–1.0 μg of RNA was fragmented using a divalent cations solution and incubation at 94°C. This step was followed by first strand cDNA synthesis using reverse transcriptase and random primers. Synthesis of ds cDNA was done using the 2nd strand master mix provided in the kit, followed by end-repair and dA-tailing. At this point, Illumina adaptors were ligated to the sample. Finally, library was enriched by 10–12 cycles of amplification using barcodes-containing primers (6 bp). The final library product was purified by Agencourt AMPure beads (Beckman Coulter, catalog # A63881). The library size and mass were assessed by analysis in the Bioanalyzer. Typically, a 200–1000 broad library peak was observed with the highest peak at ~500 bp. Quantitative PCR was used to validate the library’s functionality, using the KAPA library quantification kit (Kapa Biosystems, catalog number: KK4824) and monitoring on the ABI7900HT real-time PCR system.
RNA Sequencing (RNASeq)
Sequencing was performed using the reagents provided in the Illumina 150-cycles (2×75 paired-end format), high output NextSeq500 sequencing kit (Cat# FC-404–1005). Ten μl of library (4 nM) is mixed with 10 μl 0.1 N NaOH for 5 minutes, then the library is diluted to 20 pM in HT1 buffer. A final dilution to 1.3–1.5 pM is made with HT1 buffer for a final volume of 1 ml. A volume of 600 μl is loaded onto the reagent cartridge provided in the kit for sequencing. Runs are set by choosing the ‘Generate FASTQ only’ workflow in Illumina Experiment Manager (Illumina, USA). Under these run conditions, cluster density typically falls in the 200–220 K/mm2 range. Each NextSeq500 generated approximately 400 million single-end reads with ~80 % of reads having a greater than Q30 quality score. Up to 12 barcoded samples were pooled for sequencing on a single run, with an expected output of ~33 million pair-reads per sample. The reads that passed Illumina quality control filtering were used as raw data for further bioinformatics analysis. Transcripts designated as interferon-stimulated genes and type I interferon were selected for further examination.
RT-qPCR
OKF6/Tert-1 cells were treated with either 1 μg/ml double-stranded RNA mimetic poly(I:C) (Invivogen, tlrl-picw) in cell culture media for up to six hours. RNA of cells was isolated using the RNEasy Plus Mini kit (Qiagen, 74136) and subsequently converted to cDNA via reverse transcription using the iScript Supermix (Bio-Rad, 1708840). qPCR was performed on the cDNA using SsoAdvanced SYBR Green (Bio-Rad, 172–5274) in the CFX96-C1000 Touch thermocycler (Bio-Rad). Relative expression of type I interferon or interferon-stimulated genes were measured using the 2−ΔΔCq calculations (Winer, Kwang, Jung, Shackel, & Williams, 1999) in the CFX Manager software (Bio-Rad) with ACTB serving as the reference gene. Primers sequences were generated using the PrimerQuest tool (IDT). The list of primers used in these experiments can be found in Table 1.
Table 1.
Primers used for RT-qPCR
| Gene | Primer Sequence 5’−3’ |
|---|---|
| ACTB | F: GGATCAGCAAGCAGGAGTATG |
| R: AGAAAGGGTGTAACGCAACTAA | |
| BST2 | F: CTGGATGGCATCTACTTCGTATG |
| R: CAGGAGCACCAGAATTCCTATC | |
| IFNA4 | F: GAGGCCGAAGTTCAAGGTTAT |
| R: AGCACGGCCATCAGTAAAG | |
| IFNB1 | F: GCCGCATTGACCATCTATGA |
| R: GCCAGGAGGTTCTCAACAATAG | |
| IFNK | F: CTGCAATACACCCAACCTATGA |
| R: GAAGGTGTGTTGGCTGAAGA | |
| ISG15 | F: GAGGCAGCGAACTCATCTTT |
| R: CCAGCATCTTCACCGTCAG | |
| MX1 | F: CTGGTGCTGAAACTGAAGAAAC |
| R: TACCTCTGAAGCATCCGAAATC | |
| PML | F: GCTAAGGCATGGCTGAGATT |
| R: AGGGACTCAGAATACAGGAGAG | |
| RSAD2 | F: TGGTGAGGTTCTGCAAAGTAG |
| R: GTCACAGGAGATAGCGAGAATG | |
| ZAP | F: GCAGTCCGAGCGGAATTTA |
| R: CAGTCCAGAGAGTTCGTGATTT |
Immunofluorescence Microscopy
OKF6/Tert-1 cells were plated at 30,000 cells/well of an 8-well chamber slide. Cells were treated with either or 5 μM unmethylated CpG DNA (Invivogen, tlrl-2216) or dH2O vehicle control for two hours. After treatment, cells were washed with PBS and fixed with ice-cold acetone for ten minutes at −20°C. After blocking cells overnight with 2 % v/v BSA/PBS, rabbit anti-human IRF3 IgG antibody (Abcam, 25950) at 1 μg/ml in 2 % v/v BSA/PBS was added to cells for two hours at room temperature. Secondary antibody goat anti-rabbit IgG-Alexa Fluor 488 (Thermo Fisher, A-11008) at 4 μg/ml in 2 % v/v BSA/PBS was then added to cells for two hours at room temperature. Cells were then mounted with Fluoroshield mounting medium with DAPI (Abcam, 104139) for ten minutes before the cover glass was sealed. The Zeiss Axiovert 200 M microscope with a Zeiss AxioCam MRm camera using a 20x objective was used to capture fluorescent images. Cells were washed three times with PBS between each step. Nuclear localization of IRF3 for fifty cells per condition was analyzed using ImageJ (Eliceiri, Schneider, Rasband, & Eliceiri, 2012).
Results
Basal Expression of Oral Epithelial Cell Interferon-stimulated Genes and Type I Interferon
To determine which, if any, ISGs were expressed in oral epithelial cells, RNA sequencing (RNASeq) was performed using RNA from untreated oral keratinocyte cell line OKF6/Tert-1. Many of the canonical ISGs were found to be expressed at baseline conditions (Table 2). Interestingly, the degree of expression of ISGs seemed to be highest with those having functions in earlier aspects of viral infection (entry, transcription, and translation) and lowest with those who act at later points in the defense against viruses (e.g. pro-NK cell functions). Surprisingly, this high level of basal expression of ISGs is observed in the absence of prior stimulation by IFN.
Table 2.
Basal expression of Oral Epithelial Cell Interferon-Stimulated Genes
| ISG | High Base Mean | Rank of all transcripts (out of 23,557) | Inhibition | Expression Level | Viral Inhibitions |
|---|---|---|---|---|---|
| ZC3HAV1 (ZAP) | 5326.347284 | 1279 | Transcription | High | Entry, Transcription, Replication |
| OAS3 | 3785.979053 | 1765 | Replication/Transcription | ||
| SP100 | 2147.193058 | 2681 | Replication/Transcription | ||
| IFITM2 | 1694.801492 | 3095 | Entry | ||
| PML | 1622.004495 | 3185 | Replication/Transcription | ||
| IFIT2 | 1009.976154 | 3972 | Transcription | ||
| IFITM3 | 650.3736959 | 4725 | Entry | Medium | Viral Protein Function, Exit |
| ISG15 | 548.096075 | 4998 | Viral Protein Function | ||
| BST2 (Tetherin) | 465.888201 | 5239 | Exit | ||
| ISG20 | 397.6777008 | 5514 | Transcription | ||
| OAS2 | 305.0472818 | 5964 | Replication/Transcription | ||
| APOBEC3G | 248.470772 | 6332 | Replication/Transcription | ||
| IFITM1 | 245.6929232 | 6352 | Entry | ||
| OAS1 | 74.7362237 | 8733 | Replication/Transcription | Low | Post-entry, Pro-NK cell |
| TRIM5 | 54.19553982 | 9471 | Post-entry | ||
| MX2 | 52.96851709 | 9524 | Post-entry | ||
| ADAR | 43.92095899 | 9970 | Replication/Transcription | ||
| IFIT1 | 41.49804198 | 10135 | Transcription | ||
| OASL | 39.05579773 | 10295 | Replication/Transcription | ||
| RSAD2 (Viperin) | 27.16107836 | 11268 | Entry/Exit | ||
| MICB | 25.12520191 | 11495 | Pro-NK cell | ||
| MICA | 20.39141631 | 12106 | Pro-NK cell | ||
| EIF2AK2 (PKR) | 19.8998665 | 12169 | Translation | ||
| IFIT3 | 14.84120697 | 13109 | Transcription | ||
| MX1 | 13.41645478 | 13433 | Post-entry | ||
| CH25H | 6.15382301 | 16147 | Entry |
Basal expression levels of ISGs from OKF6/Tert-1 cells were measured by RNASeq analysis, ranked by high base mean (base mean of most highly expressed transcript variant of particular gene from three biological replicates), and grouped based on overall expression level.
The same RNASeq data set was then analyzed to determine the expression of type I IFN at baseline conditions. As expected, neither IFNB nor most subtypes of IFNA, the major type I IFN, appear to be expressed at baseline conditions. However, other type I IFN, including IFNE and IFNK are present without stimulation (Table 3). Suggesting these type I IFN are responsible for the presence of downstream ISG expression.
Table 3.
Basal expression of Oral Epithelial Cell Type I Interferon
| Base Mean | Gene |
|---|---|
| 113.6056 | Homo sapiens interferon, epsilon (IFNE), mRNA |
| 7.783875 | Homo sapiens interferon, kappa (IFNK), mRNA |
| 0.793833 | Homo sapiens interferon, alpha 1 (IFNA1), mRNA |
| 0.282971 | Homo sapiens interferon, omega 1 (IFNW1), mRNA |
Basal expression levels of type I IFN from OKF6/Tert-1 cells were measured by RNASeq analysis and ranked based base mean from three biological replicates.
Type I Interferon Expression in Oral Epithelial Cells Upon Pattern Recognition Receptor Stimulation
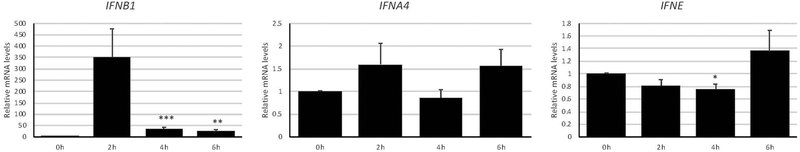
We next determined whether other type I IFN beyond those found to be expressed at baseline levels in OECs can be upregulated upon PRR stimulation. OKF6/Tert-1 treated with 1 μg/ml dsRNA mimetic poly(I:C), which signals through TLR3, for up to six hours. IFNB1 expression, but not IFNA4, was upregulated (Fig. 1). A similar spike of IFNB1 expression at two hours post-poly(I:C) treatment was observed in human corneal epithelial cells (Kumar, Zhang, & Yu, 2006). This temporal regulation of IFNB1 expression is connected to the inherent negative feedback by downstream ISGs (Imaizumi et al., 2018). The treatment, however, did not increase the expression of the type I IFN, IFNE (Fig. 1), that was already expressed at high levels at baseline (Table 3), suggesting a different mechanism leads to a sustained expression of this type I IFN that could potentially act as a way to keep certain ISGs constitutively expressed.
Figure 1. Type I interferon expression in oral epithelial cells upon pattern recognition receptor stimulation.
OKF6/Tert-1 cells were treated with 1 μg/ml poly(I:C) for up to six hours and measured for changes in type I interferon expression by RT-qPCR. Bar graphs and error bars represent the means and standard errors of measurement (SEM) of three independent experiments, each with three biological replicates per condition. *p < 0.05, **p < 0.005, and ***p < 0.0005 compared to 0h, using a two-tailed, unpaired Student’s t-test. All data analysis was performed using GraphPad Prism version 8.2.0 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com.
Oral Epithelial Cell IRF3 Localization Upon Pattern Recognition Receptor Stimulation
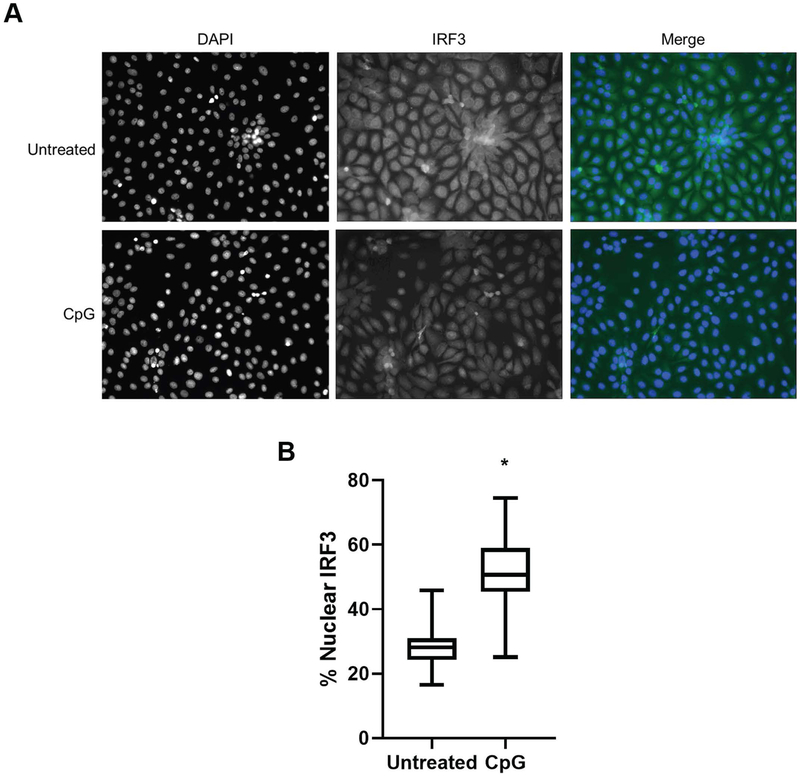
Since viral mimetics and TLR agonists poly(I:C) (Fig. 1) and unmethylated CpG DNA (data not shown) increased the expression of IFNB1, we next assessed the effect of the TLR9 ligand on the movement to the nucleus of IRF3, a transcription factor associated with both IFNB1 and IFNA regulation. After two hours, IRF3 appeared to localize to the nucleus upon unmethylated CpG DNA treatment compared to dH2O vehicle control, as measured by immunofluorescence microscopy (Fig. 2). This movement of the IRF3 transcription factor is most likely connected to the increase in type I IFN expression we observed when treating cells with TLR agonists.
Figure 2. Oral epithelial cell IRF3 localization upon pattern recognition receptor stimulation.
OKF6/Tert-1 cells were treated with with 5 μM unmethylated CpG DNA for two hours. Localization of IRF3, either within or outside the nucleus as determined by co-localization with DAPI staining, was determined by immunofluoresence microscopy (A). Percentage of IRF3 localized to the nuclei of cells was measured for fifty cells per condition (B). *p < 0.0001 compared to the untreated condition, a two-tailed, unpaired Student’s t-test after determining data from both conditions were normally distributed using the Shapiro-Wilk test.
Interferon-stimulated Gene Expression in Oral Epithelial Cells Upon Pattern Recognition Receptor Stimulation
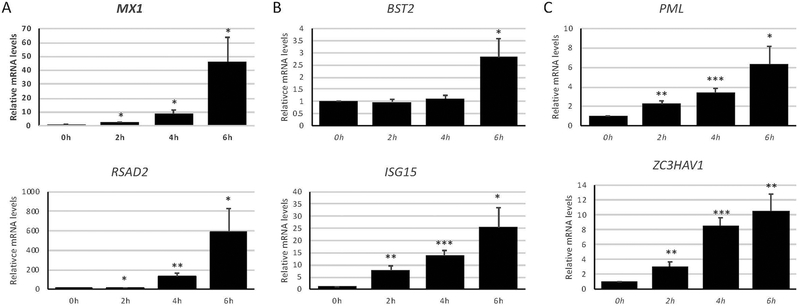
To measure downstream effects of IRF3 movement to the nucleus and type I IFN expression, we next determined the effect of the TLR3 agonist poly(I:C) on ISG expression. A number of ISGs were found to be upregulated upon poly(I:C) treatment of OKF6/Tert-1 cells (Fig. 3). Interestingly, each of the tested ISGs originally categorized in either the “Low” or “Medium” expression level groups at basal conditions were dramatically increased over time when treated with poly(I:C) compared to those ISGs in the “High” basal expression category (Fig. 3A and B compared to Fig. 3C). This difference suggests that OECs have evolved to express at high levels certain ISGs that defend against early aspects of viral infection and, upon stimulation by DAMPs associated with active viral replication, can upregulate other ISGs that dampen later aspects of the infection.
Figure 3. Interferon-stimulated gene expression in oral epithelial cells upon pattern recognition receptor stimulation.
OKF6/Tert-1 cells were treated with 1 μg/ml poly(I:C) for up to six hours and measured for changes in interferon-stimulated gene (ISG) expression by RT-qPCR. Measured genes were divided into each ISG basal expression group, according to Table 2: Low (A), Medium (B), and High (C). Bar graphs and error bars represent the means and standard errors of measurement (SEM) of three independent experiments, each with three biological replicates per condition. *p < 0.05, **p < 0.005, and ***p < 0.0005 compared to 0h, using a two-tailed, unpaired Student’s t-test.
Discussion
The cells in the oral cavity are one of the first to be infected by pathogens that are spread orally, including a number of viruses (Corsino & Linklater, 2018; Fülöp et al., 2013; Looker et al., 2015; Scott et al., 1980; Toussirot et al., 2008). In addition to the constitutive and induced production of host defense peptides (HDPs) with antiviral activities (Brice, Toth, & Diamond, 2018), cells can also express ISGs upon recognition of signals associated with dangerous levels of viral replication and subsequent production of type I IFN (Brubaker et al., 2015; McNab et al., 2015; Schneider et al., 2014; Schoggins & Rice, 2011). This intracellular innate pathway is so crucial for the early antiviral immune response that viruses have evolved ways to counteract this specific process (Schulz & Mossman, 2016), including KSHV, which dedicates about one quarter of its genome to subverting this pathway and its downstream effects (Lee, Lee, Chaudhary, Gill, & Jung, 2010). Similarly, deficiencies in type I IFN have also been utilized to study viral infections including the common use of Vero cells, which do not successfully secrete IFN-α or IFN-β (Desmyter, Melnick, & Rawls, 1968), and IFNAR knockout mice (Müller et al., 1994). Despite this pathway garnering lots of attention in other tissues of the body, role of the type I IFN/ISG pathway in OECs is not as well studied. The results described here represent the first analysis of overall OEC type I IFN and ISG expression.
Much of the previous work of oral epithelial cell innate immunity has revolved around antibacterial aspects (McClure & Massari, 2014; Shnawa, 2016; Sugawara, Uehara, Tamai, & Takada, 2002), with the majority of IFN research involving the ability of OEC-produced IFN-γ, a type II IFN, to prime other OECs to secrete cytokines necessary for T cell and macrophage activation in the clearance of pathogenic, dysbiotic bacterial infections in the oral cavity (Mutlu, Scully, & Prime, 1991; Suga et al., 2013; Sugawara, Uehara, & Takada, 2002; Yumoto et al., 2001). While IFN-α and IFN-β have been shown to be secreted by OECs previously (Cervantes et al., 2016; Teixeira, Zhao, Kinane, & Benakanakere, 2019), our results demonstrate the ability of OECs to express not only IFNB and subtypes of IFNA, but also (for the first time) other lesser-studied type I IFN genes: IFNK and IFNE (Table 3). IFNA and IFNB, but not IFNK, induction by virus-related DAMPs, suggests a different role for IFNK and IFNE as potential constitutive activators of antiviral immunity in preparation for viral infections before the cell recognizes dangerous levels of viral replication. This early antiviral response that precedes, and independent of, canonical IFN signaling has been observed previously (Paladino, Cummings, Noyce, & Mossman, 2006) and may involve IFNK and IFNE signaling.
Similar to type I IFN research in OECs, not much is known about the expression of ISGs. This study represents the first transcriptome-wide analysis of OEC ISGs. The protein products of these ISGs have direct antiviral effects, with an overarching trend toward cell apoptosis to prevent productive viral infection (Schneider et al., 2014; Schoggins & Rice, 2011). A surprisingly high amount of ISGs are expressed in OECs without stimulation (Table 2). Interestingly, the expression of these ISGs is arranged in such a way that earlier aspects of viral replication are primed to be inhibited, while ISGs that address later steps of infection are not as highly expressed. This skewing towards preventing the first steps of viral infection make sense evolutionarily in that it is more energetically favorable to only produce ISGs and downstream proteins that affect the first steps of infection as opposed to all ISGs at the same time. Selective ISG expression at basal conditions also does not put unnecessary apoptotic stress on uninfected cells. As for how these ISGs are expressed without PAMP stimulation, the IFNK and IFNE expression observed at basal conditions may be responsible for the presence of a number of ISGs found without DAMP stimulation (Table 2). However, more testing is needed to determine not only if IFN-κ and IFN-ε are produced and lead to ISG expression, but also if these proteins have functional roles in the context of an actual viral infection.
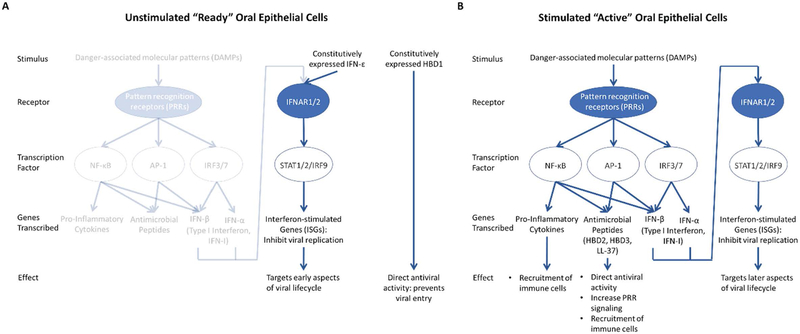
The upregulation, in addition to expression, of these ISGs has similarly not been well described in OECs. Representative genes for each group of basally expressed ISGs, according to Table 2, also exhibited similar upregulation patterns after stimulation with poly(I:C). MX1 and RSAD2, members of the “Low” basal expression group of ISGs, lead to inhibition of post-entry and exit stages of viral infection (Haller, Staeheli, & Kochs, 2007; Seo, Yaneva, & Cresswell, 2011). These genes were the most robustly induced group after DAMP stimulation (Fig. 3A). Members of the “Medium” basal expression group, BST2 and ISG15, are part of earlier aspects of viral infection, including prevention of viral protein functioning (le Tortorec, Willey, & Neil, 2011; Morales & Lenschow, 2013). These genes were the next highly expressed after poly(I:C) stimulation (Fig. 3B). The “High” basal expression group ISGs tested, PML and ZC3HAV1, are part of the antiviral pathway targeting viral replication and transcription (Everett & Chelbi-Alix, 2007.; Gao, Guo, & Goff, 2002), upstream of the other ISGs tested. In line with the trend established with the other two groups, these genes were least upregulated after poly(I:C) stimulation (Fig. 3C). The overall upregulation upon viral DAMP stimulation of the originally lower expressed ISGs, and a less robust increase of those ISGs of higher expression at basal conditions, underscores the hypothesis that OECs can exist as either “ready” to be infected or “active” in their antiviral activities, each with a different group of type I IFN and ISGs—in addition to pro-inflammatory cytokines and host defense peptides—orchestrating those actions (Fig. 4). More transcriptomic studies are necessary to determine exactly if, when, and how these states are defined and fluctuate.
Figure 4. Proposed model of oral epithelial cell innate antiviral defenses.
Oral epithelial cells (OECs) either exist as unstimulated and “ready” for a potential viral infection (A) or stimulated and “active” in the defense against a replicating virus (B). (A) “Ready” OECs are not stimulated by danger-associated molecular patterns (DAMPs) and, therefore, do not express pro-inflammatory cytokines. However, despite not expressing the full repertoire of either type I interferon (type I IFN) or antimicrobial peptides (also known as host defense peptides, HDPs), OECs do express some of these proteins constitutively, leading to direct antiviral activity mediated by human beta defensin 1 (HBD1) (Brice & Diamond, 2019; Zhao, Wang, & Lehrer, 1996) and certain interferon-stimulated genes (ISGs) that target early aspects of the viral lifecycle. (B) OECs become “active” when stimulated with DAMPs, leading to transcription of a larger number of pro-inflammatory cytokines, HDPs, and type I IFN, each with their own antiviral effects.
Taken together, our results suggest that OECs express a certain subset of type I IFN and ISGs, while others can be induced upon different stresses associated with productive viral infections. The expressions of these antiviral genes point toward a natural defense mechanism evolved to protect against viral infections that begin their pathogenesis in the mouth. While these genes, and their protein products, have been previously characterized in other tissues (Schneider et al., 2014; Schoggins & Rice, 2011) and even used as therapies for viral infections in general (Galvani, Griffiths, & Cawley, 1988; Hsu, Chao, Lin, Chen, & Kao, 2015; Rivero-Juárez, Frias, & Rivero, 2016; Smith et al., 1999), more remains to be studied in OECs, the first cells viruses encounter when orally infecting individuals. Understanding the intricacies of our endogenous type I IFN and ISGs in OECs will eventually lead to targets for either preventative measures or therapies for diseases associated with oral viral infections.
Acknowledgements
This work was supported by grants from the US Public Health Service: NIH 1R01DE22723 (GD) and 5T90DE021990-07 (DCB).
Footnotes
The authors have no conflicts of interest to declare.
References Cited
- Akira S, Uematsu S, & Takeuchi O (2006). Pathogen recognition and innate immunity. Cell, 124(4), 783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Brice DC, & Diamond G (2019). Antiviral Activities of Human Host Defense Peptides. Current Medicinal Chemistry, 26 10.2174/0929867326666190805151654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice DC, Toth Z, & Diamond G (2018). LL-37 disrupts the Kaposi’s sarcoma-associated herpesvirus envelope and inhibits infection in oral epithelial cells. Antiviral Research, 158, 25–33. 10.1016/J.ANTIVIRAL.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker SW, Bonham KS, Zanoni I, & Kagan JC (2015). Innate immune pattern recognition: a cell biological perspective. Annual Review of Immunology, 33, 257–290. 10.1146/annurev-immunol-032414-112240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes CAC, Oliveira LMS, Manfrere KCG, Lima JF, Pereira NZ, Duarte AJS, & Sato MN (2016). Antiviral factors and type I/III interferon expression associated with regulatory factors in the oral epithelial cells from HIV-1-serodiscordant couples. Scientific Reports, 6(1), 25875 10.1038/srep25875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, & Knowles DM (1995). Kaposi’s Sarcoma–Associated Herpesvirus-Like DNA Sequences in AIDS-Related Body-Cavity–Based Lymphomas. New England Journal of Medicine, 332(18), 1186–1191. 10.1056/NEJM199505043321802 [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, & Moore PS (1994). Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma Science (New York, N.Y.), Vol. 266, pp. 1865–1869. 10.1126/science.7997879 [DOI] [PubMed] [Google Scholar]
- Corsino CB, & Linklater DR (2018). Herpangina. In StatPearls. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/29939569 [PubMed]
- Desmyter J, Melnick JL, & Rawls WE (1968). Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). Journal of Virology, 2(10), 955–961. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/4302013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson M. a, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg R. a, … Rheinwald JG (2000). Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Molecular and Cellular Biology, 20(4), 1436–1447. 10.1128/MCB.20.4.1436-1447.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliceiri K, Schneider CA, Rasband WS, & Eliceiri KW (2012). NIH Image to ImageJ : 25 years of image analysis HISTORICAL commentary NIH Image to ImageJ : 25 years of image analysis. Nature Methods, 9(7), 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, & Chelbi-Alix MK (n.d.). PML and PML nuclear bodies: implications in antiviral defence. Biochimie, 89(6–7), 819–830. 10.1016/j.biochi.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Fülöp T, Larbi A, & Pawelec G (2013). Human T Cell Aging and the Impact of Persistent Viral Infections. Frontiers in Immunology, 4, 271 10.3389/fimmu.2013.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani D, Griffiths SD, & Cawley JC (1988). Interferon for treatment: the dust settles. British Medical Journal (Clinical Research Ed.), 296(6636), 1554–1556. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/2456124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Guo X, & Goff SP (2002). Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science (New York, N.Y.), 297(5587), 1703–1706. 10.1126/science.1074276 [DOI] [PubMed] [Google Scholar]
- Haller O, Staeheli P, & Kochs G (2007). Interferon-induced Mx proteins in antiviral host defense. Biochimie. 10.1016/j.biochi.2007.04.015 [DOI] [PubMed] [Google Scholar]
- Hsu C-S, Chao Y-C, Lin HH, Chen D-S, & Kao J-H (2015). Systematic Review: Impact of Interferon-based Therapy on HCV-related Hepatocellular Carcinoma. Scientific Reports, 5, 9954 10.1038/srep09954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Sassa N, Kawaguchi S, Matsumiya T, Yoshida H, Seya K, … Tanaka H (2018). Interferon-stimulated gene 60 (ISG60) constitutes a negative feedback loop in the downstream of TLR3 signaling in hCMEC/D3 cells. Journal of Neuroimmunology. 10.1016/j.jneuroim.2018.08.016 [DOI] [PubMed] [Google Scholar]
- Kumar A, Zhang J, & Yu FSX (2006). Toll-like receptor 3 agonist poly(I:C)-induced antiviral response in human corneal epithelial cells. Immunology. 10.1111/j.1365-2567.2005.02258.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Tortorec A, Willey S, & Neil SJD (2011). Antiviral inhibition of enveloped virus release by Tetherin/BST-2: Action and counteraction. Viruses. 10.3390/v3050520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-R, Lee S, Chaudhary PM, Gill P, & Jung JU (2010). Immune evasion by Kaposi’s sarcoma-associated herpesvirus. Future Microbiology, 5(9), 1349–1365. 10.2217/fmb.10.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looker KJ, Magaret AS, May MT, Turner KME, Vickerman P, Gottlieb SL, & Newman LM (2015). Global and Regional Estimates of Prevalent and Incident Herpes Simplex Virus Type 1 Infections in 2012. PLOS ONE, 10(10), e0140765 10.1371/journal.pone.0140765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P (1994). Tolerance, Danger, and the Extended Family. Annual Review of Immunology, 12(1), 991–1045. 10.1146/annurev.iy.12.040194.005015 [DOI] [PubMed] [Google Scholar]
- McClure R, & Massari P (2014). TLR-Dependent Human Mucosal Epithelial Cell Responses to Microbial Pathogens. Frontiers in Immunology, 5, 386 10.3389/fimmu.2014.00386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F, Mayer-Barber K, Sher A, Wack A, & O’Garra A (2015). Type I interferons in infectious disease. Nature Reviews Immunology, 15(2), 87–103. 10.1038/nri3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, & Janeway C (2000). Innate Immunity. New England Journal of Medicine, 343(5), 338–344. 10.1056/NEJM200008033430506 [DOI] [PubMed] [Google Scholar]
- Morales DJ, & Lenschow DJ (2013). The antiviral activities of ISG15. Journal of Molecular Biology, 425(24), 4995–5008. 10.1016/j.jmb.2013.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, & Aguet M (1994). Functional role of type I and type II interferons in antiviral defense. Science (New York, N.Y.), 264(5167), 1918–1921. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8009221 [DOI] [PubMed] [Google Scholar]
- Mutlu S, Scully C, & Prime SS (1991). Effect of IFN-gamma on the expression of MHC class I and class II antigens in a human malignant oral epithelial cell line. Journal of Oral Pathology & Medicine : Official Publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology, 20(5), 218–221. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/1906105 [DOI] [PubMed] [Google Scholar]
- Niedobitek G, Meru N, & Delecluse HJ (2001). Epstein-Barr virus infection and human malignancies. International Journal of Experimental Pathology, 82(3), 149–170. 10.1046/J.1365-2613.2001.IEP0082-0149-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladino P, Cummings DT, Noyce RS, & Mossman KL (2006). The IFN-independent response to virus particle entry provides a first line of antiviral defense that is independent of TLRs and retinoic acid-inducible gene I. Journal of Immunology (Baltimore, Md. : 1950), 177(11), 8008–8016. 10.4049/JIMMUNOL.177.11.8008 [DOI] [PubMed] [Google Scholar]
- Parkin J, & Cohen B (2001). An overview of the immune system. Lancet (London, England), 357(9270), 1777–1789. 10.1016/S0140-6736(00)04904-7 [DOI] [PubMed] [Google Scholar]
- Plosa EJ, Esbenshade JC, Fuller MP, & Weitkamp J-H (2012). Cytomegalovirus Infection. Pediatrics in Review, 33(4), 156–163. 10.1542/pir.33-4-156 [DOI] [PubMed] [Google Scholar]
- Rivero-Juárez A, Frias M, & Rivero A (2016). Current views on interferon therapy for HIV. Expert Opinion on Biological Therapy, 16(9), 1135–1142. 10.1080/14712598.2016.1196180 [DOI] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, & Rice CM (2014). Interferon-stimulated genes: a complex web of host defenses. Annual Review of Immunology, 32, 513–545. 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, & Rice CM (2011). Interferon-stimulated genes and their antiviral effector functions. Current Opinion in Virology, 1(6), 519–525. 10.1016/j.coviro.2011.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz KS, & Mossman KL (2016). Viral Evasion Strategies in Type I IFN Signaling – A Summary of Recent Developments. Frontiers in Immunology, 7, 498 10.3389/fimmu.2016.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RM, Snitbhan R, Bancroft WH, Alter HJ, & Tingpalapong M (1980). Experimental transmission of hepatitis B virus by semen and saliva. The Journal of Infectious Diseases, 142(1), 67–71. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7400629 [DOI] [PubMed] [Google Scholar]
- Seo JY, Yaneva R, & Cresswell P (2011). Viperin: A multifunctional, interferon-inducible protein that regulates virus replication. Cell Host and Microbe. 10.1016/j.chom.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong S-Y, & Matzinger P (2004). Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nature Reviews Immunology, 4(6), 469–478. 10.1038/nri1372 [DOI] [PubMed] [Google Scholar]
- Shnawa IM (2016). Oral Epithelial Cytokines. International Journal of Vaccines & Vaccination, 2(2), 1–0. 10.15406/IJVV.2016.2.00026 [DOI] [Google Scholar]
- Smith JK, Siddiqui AA, Krishnaswamy GA, Dykes R, Berk SL, Magee M, … Cummins J (1999). Oral Use of Interferon-a Stimulates ISG-15 Transcription and Production by Human Buccal Epithelial Cells. In JOURNAL OF INTERFERON AND CYTOKINE RESEARCH (Vol. 19). Retrieved from Mary Ann Liebert, Inc website: www.liebertpub.com [DOI] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, … Degos L (1995). Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood, 86(4), 1276–1280. Retrieved from http://www.bloodjournal.org/content/86/4/1276.abstract [PubMed] [Google Scholar]
- Suga T, Mitani A, Mogi M, Kikuchi T, Fujimura T, Takeda H, … Noguchi T (2013). Aggregatibacter actinomycetemcomitans lipopolysaccharide stimulated epithelial cells produce interleukin-15 that regulates T cell activation. Archives of Oral Biology, 58(10), 1541–1548. 10.1016/j.archoralbio.2013.06.020 [DOI] [PubMed] [Google Scholar]
- Sugawara S, Uehara A, & Takada H (2002). Priming of human oral epithelial cells by interferon-γ to secrete cytokines in response to lipopolysaccharides, lipoteichoic acids and peptidoglycans. Journal of Medical Microbiology, 51(8), 626–634. 10.1099/0022-1317-51-8-626 [DOI] [PubMed] [Google Scholar]
- Sugawara S, Uehara A, Tamai R, & Takada H (2002). Innate immune responses in oral mucosa. Journal of Endotoxin Research, 8(6), 465–468. 10.1179/096805102125001082 [DOI] [PubMed] [Google Scholar]
- Teixeira H, Zhao J, Kinane DF, & Benakanakere MR (2019). IFN-β secretion is through TLR3 but not TLR4 in human gingival epithelial cells. Molecular Immunology. 10.1016/j.molimm.2019.03.006 [DOI] [PubMed] [Google Scholar]
- Toussirot É, Roudier J, Roudier C, & al., et. (2008). Epstein–Barr virus in autoimmune diseases. Best Practice & Research Clinical Rheumatology, 22(5), 883–896. 10.1016/j.berh.2008.09.007 [DOI] [PubMed] [Google Scholar]
- Vénéreau E, Ceriotti C, & Bianchi ME (2015). DAMPs from Cell Death to New Life. Frontiers in Immunology, 6, 422 10.3389/fimmu.2015.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer J, Kwang C, Jung S, Shackel I, & Williams PM (1999). Development and Validation of Real-Time Quantitative Reverse Transcriptase-Polymerase Chain Reaction for Monitoring Gene Expression in Cardiac Myocytes in Vitro. In Analytical Biochemistry (Vol. 270). Retrieved from http://www.idealibrary.comon [DOI] [PubMed] [Google Scholar]
- Yumoto H, Nakae H, Yamada M, Fujinaka K, Shinohara C, Ebisu S, & Matsuo T (2001). Soluble products from Eikenella corrodens stimulate oral epithelial cells to induce inflammatory mediators. Oral Microbiology and Immunology, 16(5), 296–305. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11555307 [DOI] [PubMed] [Google Scholar]
- Zhao C, Wang I, & Lehrer RI (1996). Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Letters. 10.1016/0014-5793(96)01123-4 [DOI] [PubMed] [Google Scholar]