Abstract
Objective:
This work focused on the function role and underlying mechanism of BLACAT1 in regulating the radiosensitivity of head and neck squamous cell carcinoma (HNSCC) cells via PSEN1.
Methods:
BLACAT1 and PSEN1 expression in HNSCC tissues and cells were measured by qRT-PCR. Kaplan–Meier method and Spearman’s correlation analysis determined the prognostic roles and association of BLCAT1 and PSEN1 in HNSCC. The impacts of BLACAT1 and PSEN1, alone and in combination, on radiosensitivity of HNSCC cells were separately assessed through CCK-8, colony formation, flow cytometry, western blot and γH2AX foci staining assays.
Results:
Our study disclosed that BLACAT1 and PSEN1 were both in association with poor prognosis and radioresistance of HNSCC cells. BLACAT1 knockdown improved the radiosensitivity of HNSCC cells by changing cellular activities containing repressed cell viability, accelerated cell apoptosis, induced cell cycle arrest, and stimulated DNA damage response. Further, we found that PSEN1 was positively correlated with BLACAT1. Rescue assays confirmed that BLACAT1 regulated the radiosensitivity of HNSCC cells by modulating PSEN1.
Conclusion:
We revealed that BLACAT1 knockdown enhanced radioresistance of HNSCC cells via regulating PSEN1, exposing the probable target role of BLACAT1 in HNSCC.
Advances in knowledge:
This was the first time that the pivotal role of BLACAT1 was investigated in HNSCC, which provided a novel therapeutic direction for HNSCC patients.
Introduction
Head and neck squamous cell carcinoma (HNSCC)is one of the most common cancers worldwide1 and is associated with high mortality and recurrence rates.2 Despite progress in treatment, the overall survival rate of HNSCC patients at 5 years remains at about 50%.3 Radiotherapy (RT) is currently the dominating non-surgical approach for advanced HNSCC patients.4 Nevertheless, the efficacy of RT is limited due to radioresistance of tumors. Therefore, identifying novel therapeutic targets to alleviate radioresistance is of great significance for gaining better outcomes of HNSCC patients.
Long non-coding RNAs (lncRNAs) are known as highly conserved transcripts with the length of over 200 nucleotides (nt) and no protein-coding potential.5 It has been indicated that pivotal roles have been played by lncRNAs in many biological courses of tumors, such as cell proliferation, cell apoptosis and cancer progression, and even the radiosensitivity of cancer cells.6,7 More and more evidence has shown that lncRNAs are commonly dysregulated in various cancers and involved in tumorigenesis and cellular processes of cancers, including HNSCC.7,8 For example, upregulation of lncRNA H19/miR-675 speeds up tumorigenesis of HNSCC.8 HuR and lncRNA HOTAIR accelerate HNSCC progression and metastasis.9 LncRNA MIR205HG facilitates cell proliferation by depleting miR-590–3p in HNSCC.10
LncRNA bladder cancer associated transcript 1 (BLACAT1) is located at chromosome 1q32.1, and BLACAT1 has already been demonstrated to represent its carcinogenic effects in many tumors, such as cervical cancer,11 small cell carcinoma,12 colorectal cancer,13 and non-small cell lung cancer.14 Recent studies have revealed that BLACAT1 expression is dramatically correlated with clinical parameters of numerous cancers, such as relapse and survival,15 which is not investigated in HNSCC. It is worth noting that the influence of BLACAT1 on RT in tumors has yet to be probed.
Presenilin 1 (PSEN1) are postulated to mediate the amyloid precursor protein (APP) processing by their impacts on γ-secretase, an enzyme which cleaves APP. Elevated expression of PSEN1 has been elucidated to be associated with inferior prognosis and radiosensitivity in cancers, involving liver cancer and oral cancer.16,17 However, the role of PSEN1 in regulating radiosensitivity of HNSCC cells is still unknown.
The purpose of this present study was to probe the expression profile and underlying mechanism of BLACAT1 in modulating radiosensitivity of HNSCC cells. Our study displayed that elevated expression of BLACAT1 was in association with poor prognosis and radiosensitivity of HNSCC patients. BLACAT1 was silenced in HNSCC cells so that we can monitor the changes in cell viability, apoptosis, cell cycle kinetics and DNA damage response. Afterwards, the regulatory mechanism of BLACAT1 was determined in HNSCC. In summary, this study investigated the interaction between BLACAT1 and PSEN1 and their role in regulating the radiosensitivity of HNSCC cells.
Methods and materials
Clinical tissue samples
73 matched HNSCC tissues and adjacent non-cancerous tissues from patients with surgical treatment were randomly collected from the Department of Otolaryngology-Head and Neck Surgery, Gansu Provincial Cancer Hospital. All tissue specimens were instantly frozen in liquid nitrogen and maintained at −80℃ for later use. Patients had received radiation therapy or chemotherapy before were completely unacceptable in this study. Standard written consent from each patient and the approval of the Hospital Ethic Review Committees were both gained.
Cell lines and cell culture
Five tumorigenic cell lines (SCC25, FaDu, HN13, SCC4 and HN30) and one human immortalized normal mucosal cell line (DOK) were bought from the Chinese Academy of Sciences Cell Bank. HNSCC cell lines were cultivated in Dulbecco’s Minimum Essential Medium (DMEM; Gibco, USA) consisting of 10% fetal bovine serum (FBS, Gibco,Grand Island, NY), 0.08 mg ml−1 streptomycin and 80 U ml−1 penicillin, while DOK cells were grown in complete medium (RPMI-1640) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA), penicillin (100 U ml−1), streptomycin (100 mg ml−1), 2 mM glutamine and 25 mM 4-(2-hydroxyethyl)−1-piperazineethanesulphonic acid (HEPES). All cell lines were grown to monolayers in a 10 cm dish and kept at 37℃ in an incubator in a moist atmosphere with 5% CO2. Cells in exponential growth phase were utilized for subsequent experiments.
Cell transfection
To achieve 30–50% confluence before transfection, SCC25 and Fadu cells were seeded into six-well plates at least 24 h in advance. And to knock down BLACAT1 expression, following the manufacturer’s protocols SCC25 and Fadu were transfected by specific siRNAs respectively using Lipofectamine RNAiMAX Reagent (Invitrogen). The siRNAs were obtained from gene pharma (Shanghai, China), involving one negative control siRNA (si-NC) and two siRNAs against BLACAT1 (si-BLACAT1#1 and si-BLACAT#2). The target sequences for siRNAs against BLACAT1 were as follow: si-BLACAT1#1,5′-AGGCUGGUUUCUGCCCUCAUCCUUU-3′; si-BLACAT1#2, 5′-GCCCAGCUUCUAGUCCUCUCCUUAU-3′. To upregulate PSEN1 expression, the PSEN1 coding sequence was subcloned into pcDNA3.1 vectors (Invitrogen; Thermo Fisher Scientific, Inc.), forming pcDNA3.1-PSEN1 vectors, compared with pcDNA3.1 vectors. Sequentially, the plasmids were infected into HNSCC cells with Lipofectamine® 2000 in consistent with the protocols.
Quantitative real-time PCR (qRT-PCR)
Total RNA was harvested from tissues and cells with TRIZOL reagent (Invitrogen, Carlsbad, CA) and then reversely transcribed into cDNA using PrimeScript Reverse Transcriptase (Takara, Dalian, China) based on the manufacturer’s instructions. The qRT-PCR was performed with the SYBR Green chemistry and Bulge-LoopTM primers (RiboBio, Guangzhou, China) in the ABI 7900HT Sequence Detection machine (ABI Applied Biosystems, Foster City, CA). The primers used were as the following: BLACAT1, 5’-GTCTCTGCCCTTTTGAGCCT-3’ (sense), and 5’-GTGGCTGCAGTGTCATACCT-3’ (antisense); PSEN1, 5’-TATGGCAGAAGGAGACCCG-3’ (sense), and 5’-TATGGCAGAAGGAGACCCG-3’ (antisense); GAPDH, 5’-GGGAAACTGTGGCGTGAT-3’ (sense), and 5’-GAGTGGGTGTCGCTGTTGA-3’ (antisense). Expression of lncRNA and mRNA were normalized using glyceraldehydes-3-phosphate dehydrogenase (GAPDH) and counted by the comparative threshold cycle (2-ΔΔCT) method.
Cell counting kit (CCK-8) assay
The cell proliferation was evaluated using CCK-8 (Dojindo Laboratories, Kumamoto, Japan). The principle of CCK-8 assay is that WST-8 can be reverted to hydrosoluble formazan through dehydrogenases from mitochondria of cells. Higher cell viability produces deeper colors and OD values. For this assay, cells with the density of 2 × 103 cells/well were cultivated in complete medium (100 µl) in 96-well flat-bottomed plates. Furthermore, cells were incubated for whole night for cell attachment and recovery, followed by transfection with si-NC or si-BLACAT1#1 for 1, 2, 3 and 4 days. The cells in each well were cultured with CCK-8 solution (10 µl) and exposed to 0 or 4 Gy radiation. Absorbance was measured at 450 nm using a microplate reader (Bio-Rad, USA, Hercules, CA) and the proliferation curve was constructed based on absorbance and time. Cell viability was computed as follows: cell viability = [A (experimental group) − A (blank)] /[A (control group) − A (blank)]. In A (experimental group), the absorbance of wells with experimented cells and CCK-8 solution was examined. In A (blank) group, the absorbance of wells with only medium and CCK-8 solution was measured. In A (control group), the absorbance of wells with control group cells and CCK-8 solution was estimated.
Colony formation assay
Transfected cells (400 cells per well) were plated in six-well plates at 37℃ containning 10% FBS-supplemented RPMI 1640 and exposed to the indicated doses of irradiation (0, 2, 4, 6 or 8 Gy). Post 2 weeks, cells were washed twice through phosphate-buffered saline (PBS), followed by 30 min of fixation with 4% paraformaldehyde for and then stained with Giemsa (ECM550; Chemicon, CA) for 20 min. Manually, individual clones above 50 cells were numbered. The survival fraction (SF) was counted as the percentage of plating efficiency for irradiated and non-irradiated cells. Clone-forming efficiency of each cell was calculated in accordance with the formula: clone forming efficiency = the number of colonies/the number of inoculated cells × 100%.
Flow cytometry
For cell cycle analysis, cells in exponential growth were gathered and treated with the Cell Cycle Staining Kit (Multi Sciences, Hangzhou, China). Post-washing by PBS, cells were fastened using 70% ice-cold ethanol and cultivated with Cell Cycle Staining Kit in the dark place for half an hour. Lastly, the proportions of cells in different phases were dissected by flow cytometry. Regarding cell apoptosis analysis, cells transiently transfected with si-BLACAT1#1 were extracted through trypsinization at 24 h post-transfection, collected 24 h post 4 Gy irradiation and then washed twice using cold PBS (10 mM) and resuspended in 1 × binding buffer (BD Biosciences, San José, CA). Double-staining was carried out using a FITC-Annexin V Apoptosis Detection Kit (BD Biosciences) under the provider's recommendations. Cells which were viable, dead, early apoptotic and advanced apoptotic were analyzed and quantified utilizing a flow cytometer (FACScan; BD Biosciences) and Cell Quest software (BD Sciences: ref.19). The apoptosis rate of transfected HNSCC cells was calculated with formula: cell apoptosis rate = (early apoptotic cells + advanced apoptotic cells)/total cell number ×100%.
Immunofluorescent staining of γH2AX foci
Cells fixation was performed at specified times (1, 6 and 24 h) 1 d post-irradiation. After washing once by PBS in chamber slides and fixation in 4% formaldehyde at ambient temperature for 10 min, cells were then permeabilized and blocked for 1 h utilizing blocking buffer (0.2% Triton X-100 in PBS and 5% normal goat serum), followed by incubation with anti-γH2AX (1:100; Upstate Biotechnology) all night at 48℃. Then cells were rinsed twice by PBS, grown with fluorescein isothiocyanate–labeled rabbit antibody antimouse (1:100) for about 1 h, and washed with PBS twice. Following the counterstain of the nuclei by 4’,6-diamidino-2-phenylindole (DAPI; 1 µg ml−1) for nearly 30 min, cells were overlapped by antifade solution (Fisher) and mounted. To measure the slides, Leica Digital Module R XA fluorescent microscope was applied. The images from a Photometrics Sensys charge-coupled device camera (Roper Scientific) were imported into and dissected via IP Labs image analysis software (Scanalytics). γH2AX foci staining in each group was evaluated in at least 50 cells.
Western blot
Cells were lyzed by RIPA protein extraction reagent from Beyotime (Beijing, China) with phenylmethylsulfonyl fluoride (Roche, Basel, Switzerland) and a protease inhibitor cocktail. The protein was quantified using the Bio-Rad protein assay kit. About 50 µg of protein extract was separated on 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), then transferred onto nitrocellulose membrane (Sigma, San Francisco, CA). The blots were cultivated at 4℃ with 0.2 µg ml−1 of primary antibodies overnight. Following three washes, membranes were cultured with secondary antibody anti IgG (Promega, USA,Madison,WI) at room temperature for 1 h. ECL chromogenic substrate was utilized to visualize the bands and the densitometry (Quantity one software; Bio-Rad, Hercules, CA) was for quantifying intensity of the bands. The primary antibodies were anti-PSEN1 (Cell Signaling Technology, USA, Boston, MA) and anti GAPDH (Santa Cruz, CA).
Statistical analysis
Applying SPSS software v. 19.0 (SPSS, Chicago, IL) and Graph-pad prism software, statistical analysis was carried out and presented. Differences between two independent groups were analyzed by Student's t-test and for multiple group comparison, one-way (comparison between groups with one variable) or two-way ANOVA (comparison between groups with two variables) was utilized followed by Student–Newman–Keuls Q test. The Kaplan–Meier analysis and log-rank test were applied for overall survival (OS) and Spearman’s correlation analysis was adopted for the relationship between BLACAT1 and PSEN1. All experiments were repeated independently at least three times. All data were represented as means ± standard deviation (SD). p-value less than 0.05 was statistically significant.
Results
Upregulated BLACAT1 was associated with poor prognosis and radioresistance in head and neck squamous cell carcinoma
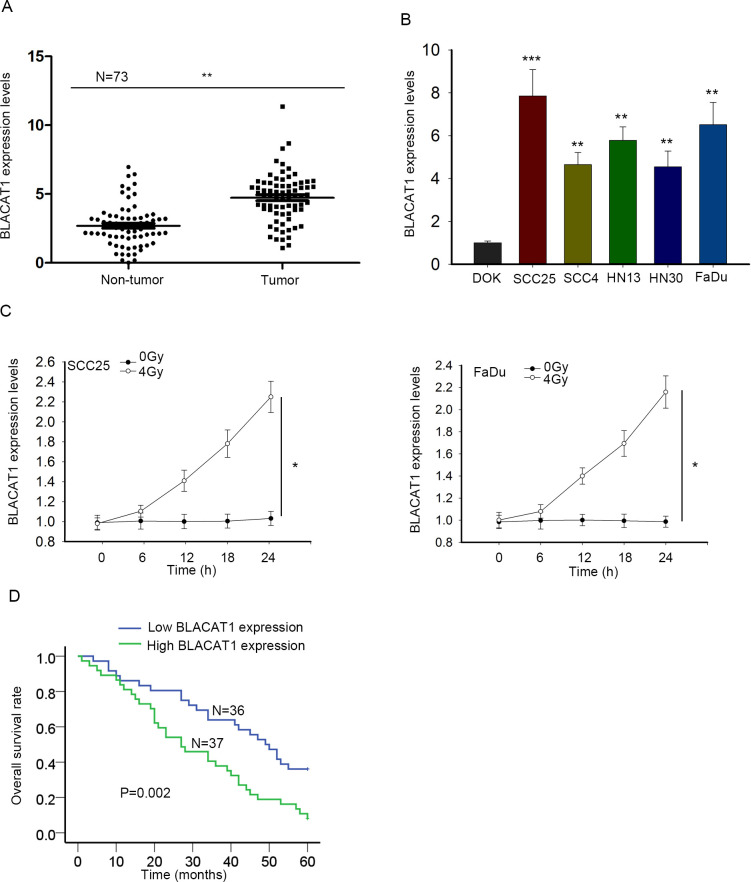
To explore whether BLACAT1 was implicated in the tumorigenesis of HNSCC, we gathered clinical HNSCC tissue samples and firstly examined BLACAT1 expression in 73 pairs of HNSCC tissues and corresponding non-cancerous tissues utilizing qRT-PCR. As exhibited in Figure 1A, BLACAT1 expression level was obviously higher in HNSCC tissues relative to normal tissues. We continued to detect BLACAT1 expression in five HNSCC cell lines (SCC25, SCC4, HN13, HN30 and FaDu,) and one human normal DOK cells using qRT-PCR, among which SCC25 and FaDu cells showed the top two highest expression of BLACAT1 and then were chosen for the conduction of further assays (Figure 1B). To detect whether BLACAT1 expression was in correlation with radiosensitivity, qRT-PCR was applied to test BLACAT1 expression level in SCC25 and FaDu cells. BLACAT1 levels in SCC25 and FaDu cells were overtly increased when cells were treated with 4 Gy irradiation (Figure 1C). Patients were divided into high/low BLACAT1 groups based on the median of BLACAT1 expression in HNSCC patients. The Kaplan–Meier analysis revealed that HNSCC patients with higher BLACAT1 expression had a lower overall survival when compared to those with lower BLACAT1 expression (Figure 1D). Then, the association of BLACAT1 with the patient clinical features was dissected. Clinical characteristics of HNSCC patients demonstrated that high expression of BLACAT1 in HNSCC was dramatically correlated with clinical stage (p = 0.015), tumor stage (p = 0.001) and nodal stage (p = 0.004), but not with other factors as presented in Table 1. Besides, as exhibited in Table 2, BLACAT1 levels and tumor stages were both positive and independent prognostic markers in HNSCC. All of these data revealed that BLACAT1 might take part in the development of radiation resistance in HNSCC progression and be an independent prognosis marker in patients with HNSCC.
Figure 1.
Upregulated BLACAT1 was associated with poor prognosis and radioresistance in head and neck squamous cell carcinoma. (A) qRT-PCR analysis of BLACAT1 expression in 73 paired HNSCC tumor tissues and corresponding normal tissues. (B) BLACAT1 expression was tested by qRT-PCR in five HNSCC cell lines (SCC25, SCC4, HN13, HN30 and FaDu) and one normal DOK cell line. (C) SCC25 and FaDu cells were exposed to 4 Gy irradiation. BLACAT1 expression was estimated by qRT-PCR. (D) Kaplan–Meier analysis of overall survival rate in BLACAT1 high (N = 37) or low (N = 36) group. * p < 0.05, ** p < 0.01 and *** p < 0.001 vs. control group. HNSCC, head and neck squamous cell carcinoma; PCR, polymerized chain reaction.
Table 1.
Correlation between BLACAT1 expression and clinical features. (n = 73)
| Variable | BLACAT1 Expression | p-value | |
|---|---|---|---|
| Low | Hhigh | ||
| Gender | |||
| Male | 23 | 26 | 0.624 |
| Female | 13 | 11 | |
| Primary site | |||
| Oropharynx | 19 | 18 | 0.816 |
| Larynx/hypopharynx | 17 | 19 | |
| Performance status | |||
| 0/1 | 10 | 12 | 0.800 |
| 2 | 26 | 25 | |
| Clinical stage | |||
| III | 18 | 8 | 0.015** |
| IV | 18 | 29 | |
| Tumor stage | |||
| T1/T2 | 25 | 11 | 0.001** |
| T3/T4 | 11 | 26 | |
| Nodal stage | |||
| N0/N1 | 21 | 9 | 0.004** |
| N2/N3 | 15 | 28 | |
| Resectability | |||
| Resectable | 12 | 15 | 0.629 |
| Unresectable | 24 | 22 | |
Low/high by the sample mean. Pearson χ2 test.
**p < 0.01 was considered statistically significant.
Table 2.
Multivariate analysis of prognostic parameters in patients with HNSCC by Cox regression analysis
| Variable | Group | HR | CI (95%) | p-value |
|---|---|---|---|---|
| Gender | ||||
| Male | 1.132 | 0.559–2.29 | 0.731 | |
| Female | ||||
| Primary site | ||||
| Oropharynx | 1.184 | 0.623–2.248 | 0.606 | |
| Larynx/hypopharynx | ||||
| Performance status | ||||
| 0/1 | 1.36 | 0.704–2.629 | 0.36 | |
| 2 | ||||
| Clinical stage | ||||
| III | 0.993 | 0.553–1.784 | 0.982 | |
| IV | ||||
| Tumor stage | ||||
| T1/T2 | 2.099 | 1.131–3.896 | 0.019a | |
| T3/T4 | ||||
| Nodal stage | ||||
| N0/N1 | 0.869 | 0.454–1.661 | 0.671 | |
| N2/N3 | ||||
| Resectability | ||||
| Resectable | 0.642 | 0.354–1.164 | 0.144 | |
| BLACAT1 level | Unresectable | |||
| Low | 2.156 | 1.181–3.935 | 0.012a | |
| High |
HNSCC, head and neck squamous cell carcinoma; HR, hazard ratio.
Proportional hazards method analysis showed a positive, independent prognostic importance of BLACAT1 expression (p = 0.012).
p < 0.05 was considered statistically significant.
Knockdown of BLACAT1 improved the radiosensitivity in HNSCC cells
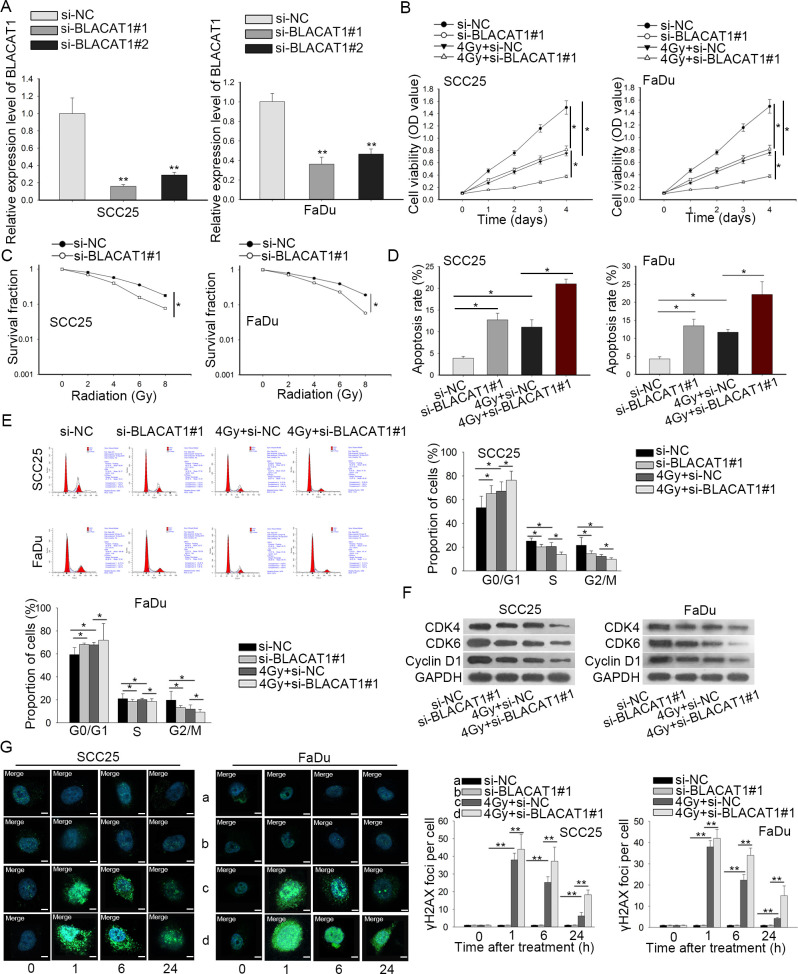
Then we investigated the function role of BLACAT1 in HNSCC through lost-of-function assays. For the upregulation of BLACAT1 in HNSCC cells, si-BLACAT1#1, si-BLACAT1#2 and negative control si-NC were stably transfected into SCC25 and FaDu cells. As displayed in Figure 2A, BLACAT1 was significantly silenced in SCC25 and FaDu cells, as estimated by qRT-PCR. Thereafter, the influence of BLACAT1 on cell proliferation was analyzed by CCK-8 and colony formation assays, respectively. The proliferation capacity was abundantly inhibited when SCC25 and FaDu cells were transfected with si-BLACAT1#1. Besides, 4 Gy irradiation significantly suppressed cell proliferation of SCC25 and FaDu cells and this effect could be strengthened by BLACAT1 knockdown (Figure 2B). Similarly, colony formation assay showed that increasing the radiation dose caused a significant decrease of colony survival fraction in both SCC25 and FaDu cells, which could be further reinforced by BLACAT1 downregulation (Figure 2C). Furthermore, flow cytometry analysis suggested that the silencing of BLACAT1 induced more apoptotic SCC25 and FaDu cells and after 4 Gy irradiation, cell apoptosis rate could be increased by downregulation of BLACAT1 (Figure 2D). The radiosensitivity of cells increases upon the lack of DNA repair and the induction of DNA damage. The cell cycle of eukaryocytes comprising cell growth, DNA replication and cell division was examined. In flow cytometry analysis, cell cycle was arrested at G0/G1 phases when BLACAT1 was downregulated or cells were exposed to 4 Gy irradiation, and this phenomenon was exacerbated when cells were given the two treatments together. These data elucidated that the inhibition of cell cycle arrest phenotype attributed to BLACAT1 knockdown enhanced radiosensitivity of HNSCC cells (Figure 2E). Additionally, western blot measured the protein levels of G0/G1 phase-related proteins, which revealed that protein levels of CDK4, CDK6 and Cyclin D1 were reduced through 4 Gy treatment or knockdown of BLACAT1. These phenomena were more obvious when cells were given two treatments (Figure 2F). Subsequently, DNA damage was assessed through immunofluorescent staining of γH2AX foci. Interestingly, silencing of BLACAT1 alone had no apparent influence on DNA damage (Figure 2G, row 1–2). However, cells with BLACAT1 inhibition were more likely to be damaged under irradiation treatment, evidenced by much more γH2AX foci observed in such SCC25 and FaDu cells (Figure 2G, row 3–4). All the results illustrated that BLACAT1 silencing improved the sensitivity of HNSCC cells to radiation.
Figure 2.
Knockdown of BLACAT1 improved the radiosensitivity of HNSCC cells. (A) The inference efficiency of si-BLACAT1#1 or si-BLACAT1#2 in SCC25 and FaDu cells was detected by qRT-PCR. (B) CCK-8 assay evaluated the viability of SCC25 and FaDu cells with BLACAT1 silence under 0 or 4 Gy irradiation, compared to NC groups. (C) Colony survival fraction of SCC25 and FaDu cells was assessed by colony formation assay after different doses of irradiation, individually. (D–E) Under 0 or 4 Gy irradiation, flow cytometry analyses of cell apoptosis and cell cycle in BLACAT1-silenced SCC25 and FaDu cells. (F) Western blotting analysis quantified cell cycle-related proteins. (G) Representative micrograph images from cells suffering four treatments (left panel). The quantified data were presented (right panel). Magnification at 1000 ×. * p < 0.05 and ** p < 0.01 vs. control group. HNSCC, head and neck squamous cell carcinoma; PCR, polymerized chain reaction.
High expression of PSEN1 was also related to poor prognosis and radioresistance in HNSCC
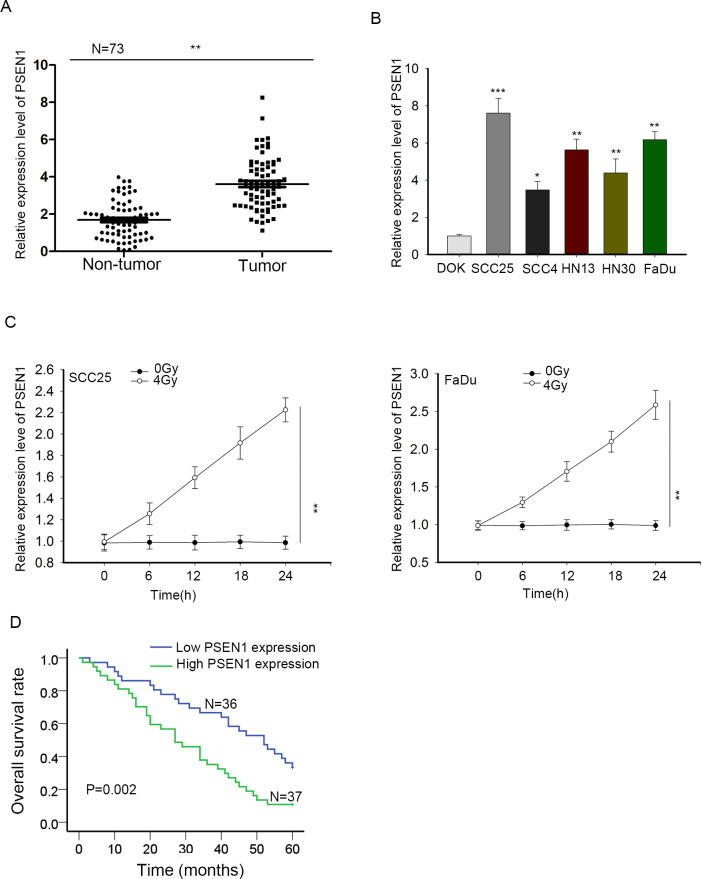
LncRNAs have been recognized to regulate gene expression and further affect cellular activities of tumors.18 Based on the radioresistance function of BLACAT1 in HNSCC cells, we further explored PSEN1, a protein that has already been demonstrated to have the property of resisting RT but never investigated in HNSCC. We speculated boldly that BLACAT1 could regulate PSEN1 to resist radiation. Firstly, we dissected the role of PSEN1 in HNSCC. qRT-PCR analyses were used for detecting PSEN1 expression levels in HNSCC tissues and adjacent normal tissues as well as in HNSCC cells and normal DOK cells, among which PSEN1 overexpression merely occurred in HNSCC tissues and cells (Figure 3A–B). Then we investigated the response of PSEN1 to irradiation through evaluating PSEN1 expression under 4 Gy radiation by qRT-PCR. The result presented that PSEN1 expression in SCC25 and FaDu cells was going higher in 4 Gy radiation than that in 0 Gy radiation (Figure 3C). Finally, Kaplan–Meier analysis was applied again for the correlation between PSEN1 expression and overall survival rate of patients with HNSCC. It uncovered a positive correlation between high PSEN1 expression and low overall survival (Figure 3D). Briefly, PSEN1 overexpression predicted poor prognosis and was associated with radioresistance of HNSCC.
Figure 3.
High expression of PSEN1 was also related to poor prognosis and radioresistance in HNSCC. (A) PSEN1 expression in HNSCC tissues and noncancerous tissues was examined through qRT-PCR. (B) qRT-PCR detection of PSEN1 expression in HNSCC cell lines and normal immortalized DOK cells. (C) Under 4 Gy irradiation, the expression of PSEN1 in SCC25 and FaDu cells was dissected by qRT-PCR. (D) The correlation between PSEN1 expression and HNSCC patient overall survival by Kaplan-Meier analysis. * p < 0.05, ** p < 0.01 and *** p < 0.001 vs. control group. HNSCC, head and neck squamous cell carcinoma; PCR, polymerized chain reaction.
BLACAT1 regulated PSEN1 in mRNA and protein levels
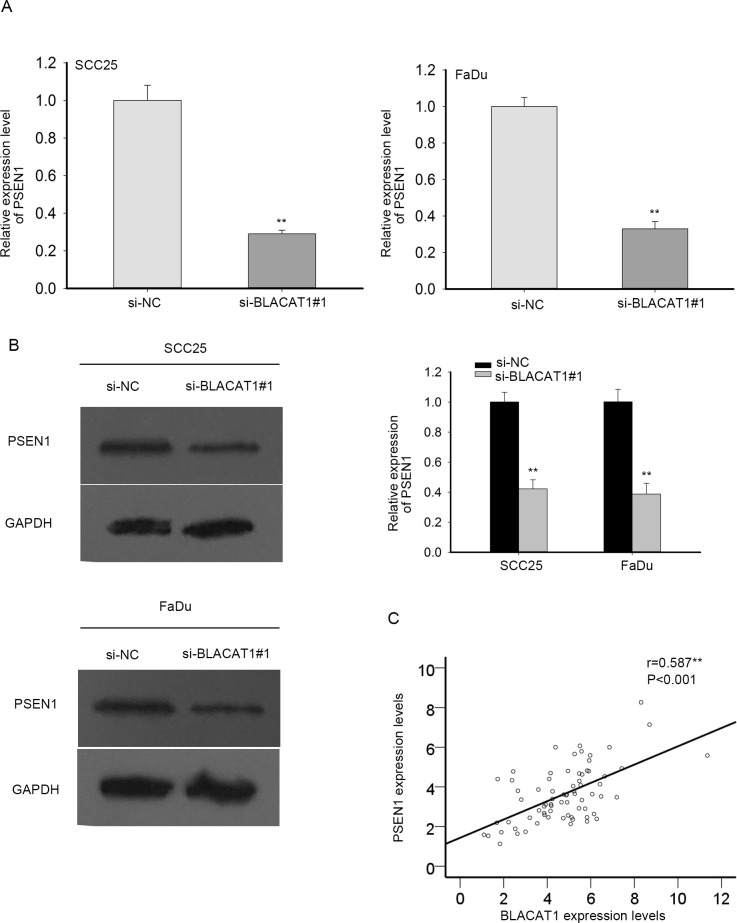
Further, we probed the regulation of BLACAT1 on PSEN1. In SCC25 and FaDu cells, PSEN1 mRNA and protein were separately assessed via qRT-PCR and western blot experiments. PSEN1 mRNA and protein expression was relatively downregulated when BLACAT1 was knocked down in SCC25 and FaDu cells in comparison with negative control (Figure 4A–B). Finally, we resorted to the Spearman’s correlation analysis and found that PSEN1 expression was positively related to BLACAT1 expression (Figure 4C). Thus, BLACAT1 mediated the mRNA and protein levels of PSEN1 positively.
Figure 4.
BLACAT1 regulated PSEN1 in mRNA and protein levels. (A-B) In SCC25 and FaDu cells, PSEN1 mRNA and protein after BLACAT1 was knocked down were estimated by qRT-PCR and western blot, individually. (C) Spearman’s correlation analysis of the association of PSEN1 with BLACAT1. **P < 0.01 vs. control group.
BLACAT1 regulated the radiosensitivity of HNSCC cells through modulating PSEN1
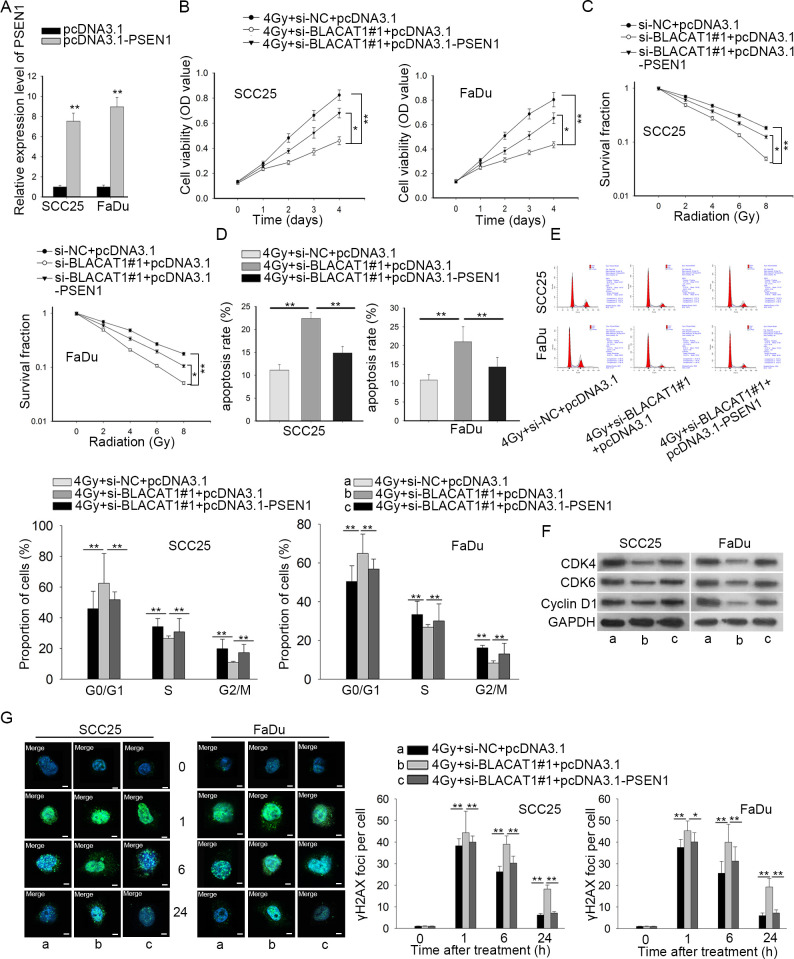
Based on the regulation of BLACAT1 on PSEN1 expression, we hypothesized that BLACAT1 affected the radiosensitivity of HNSCC cells by regulating PSEN1. To certify our hypothesis, we conducted rescue assays. SCC25 and FaDu cells co-treated with si-NC +pcDNA3.1, si-BLACAT1#1 + pcDNA3.1 or si-BLACAT1#1 + pcDNA3.1-PSEN1 were exposed to 4 Gy radiation. The transfection efficacy of pcDNA3.1-PSEN1 was ensured through qRT-PCR (Figure 5A). CCK-8 assay results discovered that BLACAT1 silence remarkably decreased cell proliferation while overexpression of PSEN1 reversed this effect in SCC25 and FaDu cells (Figure 5B). Besides, PSEN1 overexpression recuperated the si-BLACAT1#1-inhibited survival fraction of HNSCC cells, as estimated by colony formation experiment (Figure 5C). Moreover, flow cytometry exhibited that BLACAT1 downregulation strikingly induced cell apoptosis and cell cycle G0/G1 arrest, which could be rescued after PSEN1 was upregulated (Figure 5D–E). Western blot assays validated the same result as BLACAT1 inhibition-decreased CDK4, CDK6 and Cyclin D1 protein were regained after PSEN1 up-regulation (Figure 5F). The cell cycle kinetics that took place by overexpressing PSEN1 rescued the stimulated radiosensitivity attributed to the cell cycle arrest phenotype by BLACAT1 knockdown. Immunofluorescence staining showed that under irradiation, the formation of γH2AX foci in both cells was facilitated by knockdown of BLACAT1, but was later retarded by promotion of PSEN1 (Figure 5G). Taken together, all of these data suggested that BLACAT1 downregulation improved radiosensitivity of HNSCC cells by modulating PSEN1.
Figure 5.
BLACAT1 regulated the radiosensitivity of HNSCC cells through modulating PSEN1. SCC25 and FaDu cells were co-transfected with si-NC +pcDNA3.1, si-BLACAT1#1 + pcDNA3.1 or si-BLACAT1#1 + pcDNA3.1-PSEN1. Post 24 h, different irradiation treatment was applied. (A) The overexpression efficiency was validated through qRT-PCR. (B) Cell viability of stably transfected SCC25 and FaDu cells under 4 Gy radiation was measured by CCK-8 assay. (C) Colony formation assay was adopted to examine the colonization ability of SCC25 and FaDu cells after transfection. (D–E) Apoptosis and cell cycle in SCC25 and FaDu cells were evaluated through flow cytometry. (F) Cell cycle-associated proteins in three groups were assessed through western blot. (G) Representative images and quantification of immunofluorescence of γH2AX foci in three groups, respectively. Magnification at 1000 ×. * p < 0.05 and ** p < 0.01 vs. control group. PCR, polymerized chain reaction.
Discussion
HNSCC with devastating morbidity exhibits high recurrence rate, mortality rate and poor survival rate,19,20 hence the treatment planning for HNSCC remains a challenge.20,21 Although the application of RT is an accessible treatment for cancers, including cervical cancer,22 gastric cancer, oesophageal carcinoma23 and HNSCC,4 the therapeutic outcomes of RT on HNSCC patients are still unsatisfactory as a result of tumor radioresistance.24–26 Therefore, it is necessary for us to explore more molecular biomarkers in HNSCC. Our study was to find more feasible therapies for patients with HNSCC.
An increasing number of studies have claimed that lncRNAs with aberrant expressions get involved in modulating the sensitivity of cancer cells to RT,27–29 including HNSCC cells.30 LncRNA Bladder cancer associated transcript 1 (BLACAT1), located in chromosome 1q32.1, has been identified as an oncogene in small cell carcinoma,12 and it has been revealed as an independent prognostic and a diagnostic biomarker of some common cancers.13,15 The role of BLACAT1 in HNSCC and its impact on RT have never been elucidated before. The present study revealed that BLACAT1 was upregulated in HNSCC and in close association with radioresistance and poor prognosis of HNSCC patients.
Previous studies have demonstrated that BLACAT1 could regulate several cellular activities of tumors cell proliferation, migration and invasion.11,13,14 Then we investigated the impact of BLACAT1 on biological activities of HNSCC cells by lost-of-function assays. Consistent with researches above, we also discovered the promotion of BLACAT1 in HNSCC cell proliferation. More importantly, we further unveiled that BLACAT1 was a positive mediator of radioresistance in HNSCC as well. Previously, though researchers uncovered the role of BLACAT1 in enhancing chemoresistance in several cancers,31,32 its impact on the response of cancer cells to irradiation was scarcely studied before. It was the first time that the influence of BLACAT1 on radiosensitivity of HNSCC cells was dissected.
Accumulating evidence has suggested that lncRNAs can function in carcinomas by regulating proteins.33,34 Here, PSEN1 protein, which has never been explored in HNSCC, is acknowledged for its function against RT or chemotherapy in liver cancer16 and oesophageal cancer.17 We purposed to interrogate the possible relationship between BLACAT1 and PSEN1. Intriguingly, our study uncovered that PSEN1 expression was similarly high in HNSCC tissues and cells, and was closely correlated with both radioresistance and poor prognosis of HNSCC patients. This disclosed the prognostic and diagnostic significance of PSEN1 in the clinical treatment of HNSCC. Moreover, BLACAT1 could mediate the mRNA and protein level of PSEN1 and was in positive correlation with PSEN1. The correlation between BLACAT1 and PSEN1 in HNSCC was, for the first time, exposed. Finally, rescue assays were conducted to validate that BLACAT1 boosted radioresistance of HNSCC cells by controlling the expression of PSEN1.
In brief, this study uncovered BLACAT1 mediated the radiosensitivity of HNSCC cells via regulating PSEN1. Moreover, as one of the core components of γ-secretase complex, PSEN1 protein exerts an indispensable function in Notch signaling whose activation is essential for the differentiation and maintenance of cancer stem-like cells in multiple cancer types including HNSCC.35–37 On this basis, we speculated that BLACAT1/PSEN1 axis might affect the stem cell-like properties of HNSCC cells since stemness plays an important part in tumorigenesis and resistance formation.38–40
In conclusion, the results of our research exposed new insights into the radiation resistance mechanism underlying BLACAT1 in HNSCC. There are some limitations in this study as we did not totally expound the deep modulation mechanism of BLACAT1 on PSEN1, and we merely performed in vitro assays but not in vivo assays, which may make our paper under suspicion. Significantly, it was a great pity of this work that the effect of BLACAT1/PSEN1 axis on the stemness of HNSCC cells was unexplored, which might be the topic of our next research. Thus, deeper investigations are indispensable in the future.
Footnotes
Acknowledgment: We thank all members of our laboratories for sincere advice and support.
Funding: This study was supported by Subject of Gansu administration of traditional Chinese medicine: GZK-2017–40.
Contributor Information
Caixia Gou, Email: Caixia_Gou4523@163.com.
Pengbing Han, Email: Pengbing_Han1h66@163.com.
Jin Li, Email: JinLi_Li239grg@163.com.
Liying Gao, Email: liying_Gao_68ff@163.com.
Xuejuan Ji, Email: ji71714700477@163.com.
Fang Dong, Email: Fang_Dong_387DF@163.com.
Qun Su, Email: Qun_Su214suqun@163.com.
Yanping Zhang, Email: biping0408@163.com.
Xiaofeng Liu, Email: fengni4144@163.com.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015; 65: 5–29. doi: 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 2. Feldman R, Gatalica Z, Knezetic J, Reddy S, Nathan C-A, Javadi N, et al. Molecular profiling of head and neck squamous cell carcinoma. Head Neck 2016; 38 Suppl 1(Suppl 1): E1625–38. doi: 10.1002/hed.24290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Datta J, Islam M, Dutta S, Roy S, Pan Q, Teknos TN. Suberoylanilide hydroxamic acid inhibits growth of head and neck cancer cell lines by reactivation of tumor suppressor microRNAs. Oral Oncol 2016; 56: 32–9. doi: 10.1016/j.oraloncology.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grégoire V, Langendijk JA, Nuyts S. Advances in radiotherapy for head and neck cancer. JCO 2015; 33: 3277–84. doi: 10.1200/JCO.2015.61.2994 [DOI] [PubMed] [Google Scholar]
- 5. Wang Y, Li Y, Yang Z, Liu K, Wang D. Genome-Wide microarray analysis of long non-coding RNAs in eutopic secretory endometrium with endometriosis. Cell Physiol Biochem 2015; 37: 2231–45. doi: 10.1159/000438579 [DOI] [PubMed] [Google Scholar]
- 6. Luo G, Wang M, Wu X, Tao D, Xiao X, Wang L, et al. Long non-coding RNA MEG3 inhibits cell proliferation and induces apoptosis in prostate cancer. Cell Physiol Biochem 2015; 37: 2209–20. doi: 10.1159/000438577 [DOI] [PubMed] [Google Scholar]
- 7. Zhou X-L, Wang W-W, Zhu W-G, Yu C-H, Tao G-Z, Wu Q-Q, et al. High expression of long non-coding RNA AFAP1-AS1 predicts chemoradioresistance and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Mol Carcinog 2016; 55: 2095–105. doi: 10.1002/mc.22454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guan G-F, Zhang D-J, Wen L-J, Xin D, Liu Y, Yu D-J, et al. Overexpression of lncRNA H19/miR-675 promotes tumorigenesis in head and neck squamous cell carcinoma. Int J Med Sci 2016; 13: 914–22. doi: 10.7150/ijms.16571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu C-Z, Jiang C, Wu Q, Liu L, Yan X, Shi R. A feed-forward regulatory loop between HuR and the long noncoding RNA HOTAIR promotes head and neck squamous cell carcinoma progression and metastasis. Cell Physiol Biochem 2016; 40: 1039–51. doi: 10.1159/000453160 [DOI] [PubMed] [Google Scholar]
- 10. Di Agostino S, Valenti F, Sacconi A, Fontemaggi G, Pallocca M, Pulito C, et al. Long non-coding MIR205HG depletes Hsa-miR-590-3p leading to unrestrained proliferation in head and neck squamous cell carcinoma. Theranostics 2018; 8: 1850–68. doi: 10.7150/thno.22167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang C-H, Li Y-H, Tian H-L, Bao X-X, Wang Z-M. Long non-coding RNA BLACAT1 promotes cell proliferation, migration and invasion in cervical cancer through activation of Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci 2018; 22: 3002–9. doi: 10.26355/eurrev_201805_15057 [DOI] [PubMed] [Google Scholar]
- 12. Chen W, Hang Y, Xu W, Wu J, Chen L, Chen J, et al. BLACAT1 predicts poor prognosis and serves as oncogenic lncRNA in small-cell lung cancer. Journal of cellular biochemistry 2018;. [DOI] [PubMed] [Google Scholar]
- 13. Su J, Zhang E, Han L, Yin D, Liu Z, He X, et al. Long noncoding RNA BLACAT1 indicates a poor prognosis of colorectal cancer and affects cell proliferation by epigenetically silencing of p15. Cell Death Dis 2017; 8: e2665. doi: 10.1038/cddis.2017.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye J-R, Liu L, Zheng F. Long noncoding RNA bladder cancer associated transcript 1 promotes the proliferation, migration, and invasion of nonsmall cell lung cancer through sponging miR-144. DNA Cell Biol 2017; 36: 845–52. doi: 10.1089/dna.2017.3854 [DOI] [PubMed] [Google Scholar]
- 15. Chen X, Dai M, Zhu H, Li J, Huang Z, Liu X, et al. Evaluation on the diagnostic and prognostic values of long non-coding RNA BLACAT1 in common types of human cancer. Mol Cancer 2017; 16: 160. doi: 10.1186/s12943-017-0728-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ma H, Yuan L, Li W, Xu K, Yang L. The lncRNA H19/miR-193a-3p axis modifies the radio-resistance and chemotherapeutic tolerance of hepatocellular carcinoma cells by targeting PSEN1. J Cell Biochem 2018; 119: 8325–35. doi: 10.1002/jcb.26883 [DOI] [PubMed] [Google Scholar]
- 17. Meng F, Qian L, Lv L, Ding B, Zhou G, Cheng X, et al. miR-193A-3P regulation of chemoradiation resistance in oesophageal cancer cells via the PSEN1 gene. Gene 2016; 579: 139–45. doi: 10.1016/j.gene.2015.12.060 [DOI] [PubMed] [Google Scholar]
- 18. Shen Y, Liu S, Fan J, Jin Y, Tian B, Zheng X, et al. Nuclear retention of the lncRNA SNHG1 by doxorubicin attenuates hnRNPC-p53 protein interactions. EMBO Rep 2017; 18: 536–48. doi: 10.15252/embr.201643139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shkeir O, Athanassiou-Papaefthymiou M, Lapadatescu M, Papagerakis P, Czerwinski MJ, Bradford CR, et al. In vitro cytokine release profile: predictive value for metastatic potential in head and neck squamous cell carcinomas. Head Neck 2013; 35: 1542–50. doi: 10.1002/hed.23191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sweeny L, Hartman YE, Zinn KR, Prudent JR, Marshall DJ, Shekhani MS, et al. A novel extracellular drug conjugate significantly inhibits head and neck squamous cell carcinoma. Oral Oncol 2013; 49: 991–7. doi: 10.1016/j.oraloncology.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beck TN, Smith CH, Flieder DB, Galloway TJ, Ridge JA, Golemis EA, et al. Head and neck squamous cell carcinoma: ambiguous human papillomavirus status, elevated p16, and deleted retinoblastoma 1. Head Neck 2017; 39: E34–9. doi: 10.1002/hed.24604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vordermark D. Radiotherapy of cervical cancer. Oncol Res Treat 2016; 39: 516–20. doi: 10.1159/000448902 [DOI] [PubMed] [Google Scholar]
- 23. Créhange G, Huguet F, Quero L, N'Guyen TV, Mirabel X, Lacornerie T. Radiotherapy in cancers of the oesophagus, the gastric cardia and the stomach. Cancer Radiother 2016; 20 Suppl(20 Suppl): S161–8. doi: 10.1016/j.canrad.2016.07.039 [DOI] [PubMed] [Google Scholar]
- 24. Perri F, Pacelli R, Della Vittoria Scarpati G, Cella L, Giuliano M, Caponigro F, et al. Radioresistance in head and neck squamous cell carcinoma: biological bases and therapeutic implications. Head Neck 2015; 37: 763–70. doi: 10.1002/hed.23837 [DOI] [PubMed] [Google Scholar]
- 25. Amoils M, Lee CS, Sunwoo J, Aasi SZ, Hara W, Kim J, et al. Node-Positive cutaneous squamous cell carcinoma of the head and neck: survival, high-risk features, and adjuvant chemoradiotherapy outcomes. Head Neck 2017; 39: 881–5. doi: 10.1002/hed.24692 [DOI] [PubMed] [Google Scholar]
- 26. Bur AM, Lin A, Weinstein GS. Adjuvant radiotherapy for early head and neck squamous cell carcinoma with perineural invasion: a systematic review. Head Neck 2016; 38 Suppl 1(Suppl 1): E2350–7. doi: 10.1002/hed.24295 [DOI] [PubMed] [Google Scholar]
- 27. Chi H-C, Tsai C-Y, Tsai M-M, Yeh C-T, Lin K-H. Roles of long noncoding RNAs in recurrence and metastasis of Radiotherapy-Resistant cancer stem cells. Int J Mol Sci 2017; 18: 1903. doi: 10.3390/ijms18091903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang H, Hu X, Zhang H, Li W. Down-Regulation of lncRNA TUG1 enhances radiosensitivity in bladder cancer via suppressing HMGB1 expression. Radiat Oncol 2017; 12: 65. doi: 10.1186/s13014-017-0802-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li G, Liu Y, Liu C, Su Z, Ren S, Wang Y, et al. Genome-Wide analyses of long noncoding RNA expression profiles correlated with radioresistance in nasopharyngeal carcinoma via next-generation deep sequencing. BMC Cancer 2016; 16: 719. doi: 10.1186/s12885-016-2755-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou J, Cao S, Li W, Wei D, Wang Z, Li G, et al. Time-Course differential lncRNA and mRNA expressions in radioresistant hypopharyngeal cancer cells. Oncotarget 2017; 8: 40994–1010. doi: 10.18632/oncotarget.17343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shu D, Xu Y, Chen W. Knockdown of lncRNA BLACAT1 reverses the resistance of afatinib to non-small cell lung cancer via modulating STAT3 signalling. J Drug Target 2019; 109: 1–7. doi: 10.1080/1061186X.2019.1650368 [DOI] [PubMed] [Google Scholar]
- 32. Wu X, Zheng Y, Han B, Dong X. Long noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin resistance of gastric cancer via sponging miR-361. Biomed Pharmacother 2018; 99: 832–8. doi: 10.1016/j.biopha.2018.01.130 [DOI] [PubMed] [Google Scholar]
- 33. She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep 2016; 36: 2673–80. doi: 10.3892/or.2016.5105 [DOI] [PubMed] [Google Scholar]
- 34. Li J, Chen C, Ma X, Geng G, Liu B, Zhang Y, et al. Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing Tat protein degradation. Nat Commun 2016; 7: 11730. doi: 10.1038/ncomms11730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J, Sullenger BA, Rich JN. Notch Signaling in Cancer Stem Cells : Reichrath J, Reichrath S. New York, NY: Springer US: Notch Signaling in Embryology and Cancer; 2012. 174–85. [DOI] [PubMed] [Google Scholar]
- 36. Shrivastava S, Steele R, Sowadski M, Crawford SE, Varvares M, Ray RB. Identification of molecular signature of head and neck cancer stem-like cells. Sci Rep 2015; 5: 7819. doi: 10.1038/srep07819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee SH, Do SI, Lee HJ, Kang HJ, Koo BS, Lim YC. Notch1 signaling contributes to stemness in head and neck squamous cell carcinoma. Lab Invest 2016; 96: 508–16. doi: 10.1038/labinvest.2015.163 [DOI] [PubMed] [Google Scholar]
- 38. Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature 2004; 432: 324–31. doi: 10.1038/nature03100 [DOI] [PubMed] [Google Scholar]
- 39. Maugeri-Sacca M, Vigneri P, De Maria R, Research CC. Cancer stem cells and chemosensitivity. Clinical Cancer Research 2011; 17: 4942–7. doi: 10.1158/1078-0432.CCR-10-2538 [DOI] [PubMed] [Google Scholar]
- 40. Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer 2008; 8: 545–54. doi: 10.1038/nrc2419 [DOI] [PubMed] [Google Scholar]