Abstract
In recent years, progress in the field of high-throughput sequencing technology and its application to a wide variety of biological specimens has greatly advanced the discovery and cataloging of a diverse set of non-coding RNAs (ncRNAs) that have been found to have unexpected biological functions. Y RNAs are an emerging class of highly conserved, small ncRNAs. There is a growing number of reports in the literature demonstrating that Y RNAs and their fragments are not just random degradation products but are themselves bioactive molecules. This review will outline what is currently known about Y RNA including biogenesis, structure and functional roles. In addition, we will provide an overview of studies reporting the presence and functions attributed to Y RNAs in the cardiovascular system.
Keywords: Non-coding RNA, Y RNA, Cardiovascular Diseases
1. Introduction and Historical Overview
Only about 1.5 % of the human genome is made up of protein-coding genes, but at least 80% of the genome is dynamically transcribed, creating a transcriptional landscape mainly dominated by non-coding RNAs (ncRNAs) [1, 2]. Nowadays, regulatory ncRNAs, such as the well-studied microRNAs (miRNAs) or long non-coding RNAs (lncRNAs) are recognized as essential functional molecules, and have significantly impacted our understanding of development, homeostasis and disease in various fields, including cardiovascular biology [4, 5], cancer [6, 7] and metabolic disorders [8, 9]. There are numerous examples in the literature of ncRNAs demonstrated to control key genes involved in both normal development and disease [10, 11]. There are also many instances highlighting how ncRNA dysregulation is tightly linked to the pathogenesis of many human disorders, including cardiovascular diseases (CVDs) [12–14]. From this body of research, the possibility of using ncRNAs as potential therapeutic targets and diagnostic tools has generated considerable interest among academic scientists and pharma/biotechnology commercial entities alike.
While miRNA and lncRNAs have garnered the most attention among the different types of ncRNAs, recent investigations have illuminated the increasingly diverse functions of other types of ncRNAs [15–18]. Among the ncRNAs, Y RNAs are among the least studied species, but are recently gaining attention. Y RNAs were discovered in the early 1980’s, during the characterization of serum from patients with the autoimmune disease systemic lupus erythematosis (SLE) [19]. Lerner and colleagues set out to uncover the molecular nature of the targets of the antibodies produced during the autoimmune response. Initially the group immunoprecipitated nuclear cell extracts from mouse derived tumor cells using serum from SLE patients containing antibodies against the proteins Ro and La, common autoantigens and targets of the immune system in rheumatic diseases like SLE. They discovered a class of small nuclear ncRNA, U RNA, associated with ribonucleoproteins (RNPs), as autoantigens [20]. In a following study, they used whole cell extracts and reacted with serum containing antibodies to Ro and La [19]. In this second round of experiments, they identified small cytoplasmic RNAs associated with RNPs, which they termed Y RNAs, to differentiate them from the nuclear U RNAs [19]. After these initial observations, Y RNAs have been reported to participate in other ribonucleoprotein complexes, leading to the proposition that Y-RNAs may play a role in scaffolding and assembly of RNA-protein complexes, but that their diverse functions are dependent in part, on the composition of the interacting proteins present in the complexes [21]. Y RNAs are highly conserved throughout evolution, and have been found in all vertebrates [22, 23], and related orthologs have also been reported in some bacteria [24] and nematodes [25, 26], but so far not in plants, fungi or insects. Y RNAs are expressed in all human cells; however levels of different Y RNAs vary among cell types [22]. Their expression also differs among human tissue types, with high levels reported in the heart and brain and lower amounts in the liver [27]. Furthermore, differential Y RNA expression has also been found in disease, including coronary artery disease [28] and cancer [29], pointing towards potential involvement of Y RNAs in the pathogenesis of these disorders. Finally, although the field of Y RNA has developed slowly since their discovery, functional roles of Y RNAs are beginning to be unveiled in various research fields, providing evidence that these ncRNAs may themselves be bioactive, and not simply structural RNAs involved only in scaffolding and assembly. This review will focus on the current state of knowledge about the biogenesis, structure and functional roles of Y RNAs. In addition, we will provide an updated summary of examples of Y RNAs, known or suggested to be involved in cardiovascular functions.
2. Biogenesis, Structure and Localization of Y RNAs
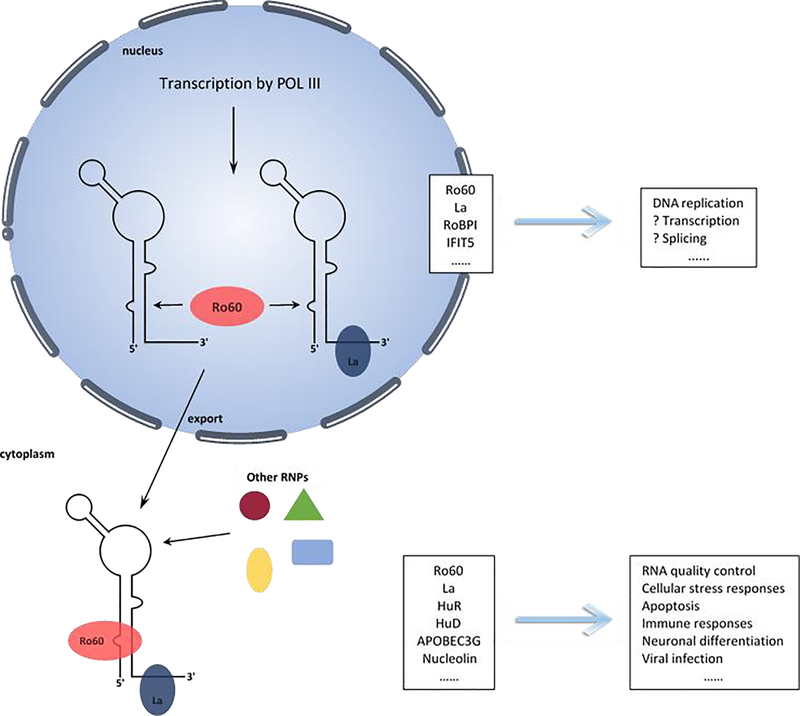
Y RNAs are encoded by individual genes and tend to reside in close proximity to each other, usually clustered on the same chromosome [30, 31]. There are four Y RNA genes denoted, Y1, Y3, Y4 and Y5 (Y2 is a truncated form of Y1) [32]. The number of individual Y RNAs differ between species. For example, in humans, all four Y RNAs are expressed, and range in size from 84 to 113 nts [33]. Rodents, on the other hand have only two Y RNAs, which are orthologs of human hY1 and hY3, but their genomes have a redundant Y5 RNA gene that is no longer expressed [34]. Both human and mouse Y RNAs have been shown to have few if any modified nucleotides [32]. Along with the annotated genes, the human genome has numerous Y RNA sequences documented as pseudogenes that are transcribed, in contrast to mice which have few [34, 35]. Y RNAs are transcribed in the nucleus by RNA polymerase III (Fig. 1) [33]. Transcription stops within a T-rich stretch, producing a 3’ oligo-uridinylated (Oligo-U) sequence, which serves as a binding site for La protein. Binding of La to the 3′ Oligo-U tail of Y-RNA protects YRNA from 3′ to 5′ exonucleolytic degradation and promotes its nuclear retention [36, 37]. In addition to La protein, the newly synthesized transcript also associates with Ro60 [22, 38]. The resulting Ro-RNP complex is then exported to the cytoplasm, which is mediated by Ran GTPase and exportin-5 [39]. Some Ro-RNPs, however, like the Y5 RNP remain in the nucleus [40], while others, such as Y3 RNA, can be exported by an alternative pathway through binding of Y3 RNP with zipcode binding protein (ZBP1), allowing export by exportin-1 [41]. It is currently unknow whether Y RNA is bound in a complex with La during the nuclear export, or it could be that La re-associates with Y-RNA after translocation (Fig. 1).
Figure 1.
Y RNAs cellular functions based on the association with RNA binding proteins (RNPs).
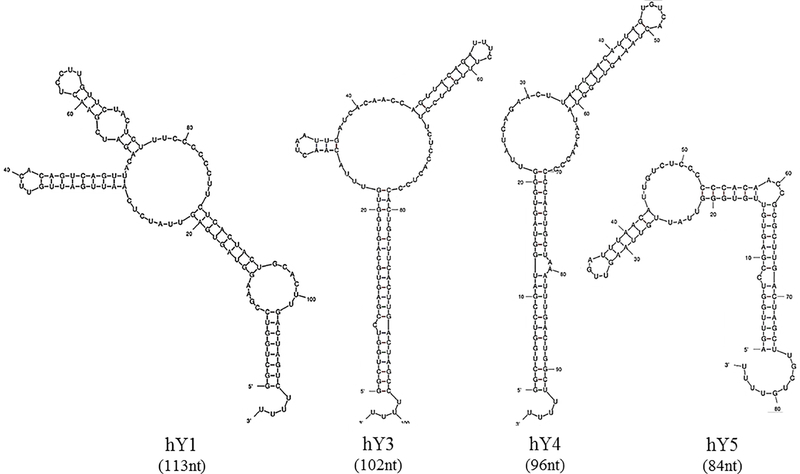
Structural experiments have revealed that Y RNAs fold into distinct hairpin-containing structures, formed by base-pairing the 3’ and 5’ ends of the mature form of Y RNA (Fig. 2) [42, 43]. A defining feature of vertebrate Y RNAs is that these RNAs have at least two main stems each, separated by a large pyrimidine-rich single stranded loop [33], but individual Y-RNAs differ slightly in their primary and secondary structures [43]. The Y RNA stems are usually not ideal double strands. Often within the stem, there is also a bulged helix region. One of these, the main stem at the 5’/3’ end, is a highly conserved cytosine bulge, which is the high affinity binding site for Ro60 [44]. It has been shown that mutation or cleavage of this cite inhibits Ro60 binding and disrupts the entire Y RNA folding [36, 44, 45]. In addition, it has been demonstrated that the lower stem structure of Y RNA is important for efficient nuclear export, as cleavage of the lower stem of hY1 RNA in Xenopus laevis oocyte blocks export of the Y RNA into the cytoplasm [39].
Figure 2.
Full-length human Y RNA (hY) secondary structure. Structures are drawn using Mfold software [3].
Ro60 is both nuclear and cytoplasmic [38, 46] and is found in most animal cells and in some bacteria [45]. Immunoprecipitation studies from human and mouse cells have shown that most Y RNAs exist as Ro60 RNP complexes [33, 38]. Ro60 binding to Y RNA was found to be important for successful Y RNA nuclear export [36] and to stabilize Y RNA in the cytoplasm, as siRNAs mediated knockdown of Ro60 in human keratinocytes was shown to significantly reduce Y RNA levels [47]. In contrast to the highly conserved lower stem region, the loops of Y RNAs differ greatly among individual Y RNAs and are quite flexible in nature [42]. Importantly, the loop domain has been reported to bind various proteins such as polypyrimidine tract-binding protein (PTB/hnRNP I) [48], nucleolin [49], and ZBP1 (Fig. 1) [50]. The effects of Y RNA interactions with these proteins are largely unknown, but it has been proposed that they could impact Y RNA localization and function [51]. Evidence for this was provided in experiments demonstrating that depletion of mouse IGF2BP1 and its chicken ortholog ZBP1, both of which directly associate with Y3, leads to nuclear accumulation of Ro60 and Y3, suggesting participation of these proteins in the nuclear export of Ro60/Y RNA complex [41, 52]. On the other hand, Y RNA itself can affect the subcellular distribution of Y RNA binding proteins. This was supported by the finding that depletion of Y RNA in mouse cells results in nuclear accumulation of Ro60, while binding of Y RNA to a Ro60 nuclear localization signal sequence promotes retaining of the RoRNPs in the cytosol [21, 45].
There are discrepancies in the literature in terms of the relative Y RNA distribution in the cytoplasm and nucleus, which may be attributed to the different experimental procedures used in the studies and/or the physiological state of the cells [53–55]. Initial cell fractionation studies in mammalian cultures and X laevis oocytes showed that Y RNAs were primarily cytoplasmic [38, 56, 57]. Later it was demonstrated that human and mouse Y5 localizes mainly to the nucleus, while Y1, Y3 and Y4 were found to be mostly cytoplasmic [40]. By using, ultrastructural and in situ hybridization experiments in human cells, others have demonstrated that Y RNAs can accumulate in both the nucleus and the cytosol at distinct compartments [58, 59]. Furthermore, in an in vitro system, where G1 phase template nuclei were incubated with fluorescently-labeled hY RNAs, it was found that all four hY RNAs bind with chromatin. Moreover, hY5 was recruited mostly to the nucleoli, while hY1, hY3 and hY4 were found to bind predominantly early replicating euchromatin [55]. It was also shown that the loop domain directs the targeting of of hY RNAs to euchromatin, since hY RNAs with mutated loop domains were shown to bind unselectively to chromatin, suggesting that this part of the hY RNA structure is important for promoting selective Y RNA chromatin association [55]. Despite inconsistent reports with regards to Y RNA distribution, growing evidence now demonstrate that Y RNAs are present in both the cytoplasm and nucleus of eukaryotic cells.
It was reported that the subcellular localization of Y RNA within the cell can be cell cycle dependent and can change during environmental stress [55, 60, 61]. Indeed, both Y RNAs and Ro60 were found to accumulate in the nucleus upon UV irradiation or oxidative stress in several organisms [21, 41, 60, 61]. This observation suggests a possible stress dependent role of Ro60/Y RNA, but on the other hand the accumulation of the complex in the nucleus can also result from suppression of the nuclear export, triggered by stress induced impairment of the RanGTP gradient, as this was shown to be the case for other proteins [62].
Y RNAs have also been detected in various retroviruses including murine leukemia virus (MLV) and human immunodeficiency virus (HIV) [63, 64]. These viruses are known to incorporate other ncRNAs as well including tRNA and miRNAs [65, 66]. The mechanisms of viral encapsulation are not very well understood, but it has been suggested that this process happens when the newly synthesized host Y RNA are still in the nucleus and seems to be independent of Ro60 binding [64]. Whether Y RNAs modulate viral function is still unknown, however several proteins involved in virus infection such as YBX1, hnRNP K and nucleolin were reported to bind to Y RNAs [67]. Consistent with this, it has been suggested that Y RNAs may promote an antiviral immune response by stimulating TLR7 [68] in the newly infected cells, or may act as scaffolds for virus packaging [63, 69].
3. Biological Functions of Y RNAs: Ongoing Work
Although Y RNAs were initially discovered more than three decades ago, a relatively small number of studies have been published regarding their biological roles, with most of the literature describing their structure and protein interactions. Reported functions of Y RNAs include involvement in DNA replication [51], and regulation of RNA stability and cellular stress responses [70, 71]. Y RNAs were identified as being essential factors for initiation of chromosomal DNA replication in cell-free reactions, in which isolated G1-phase nuclei were incubated with fractioned cellular extracts from actively proliferating human cells [72]. Using this system, Christov et al found that adding the fraction containing purified Y RNA subtypes, increased the proportion of replicating nuclei in a dose-dependent manner [72]. Depletion of hY1-RNA by RNA interference, inhibited cell proliferation and reduced significantly the percentages of human cells in S-phase, during which DNA replication occurs [72]. Further studies by the same group discovered that Y RNAs function redundantly with regards to their role in DNA replication, as any of the four hY RNAs, in the absence of the others, is sufficient to stimulate DNA replication [72]. Another interesting finding is that inhibition of replication, resulting from siRNA-mediated degradation of a hY RNA, can be rescued by adding any vertebrate Y RNA, but not non-vertebrate Y RNAs [73]. The functional redundancy observed among the hY RNAs was attributed to the presence of an evolutionary conserved upper stem region of vertebrate Y RNAs, which was shown to be necessary and sufficient for Y RNA function in DNA replication [73, 74]. In contrast, the loop domains and the lower stem of Y RNAs seem unessential with regards to Y RNA role in DNA replication, as DNA replication remained unaffected when they were mutated [73]. As mentioned earlier, Y RNA binds to euchromatin throughout the cell cycle. Evidence was provided that this association increases during S-phase and declines in G1 phase or mitosis [75]. Furthermore, the dynamics of Y RNA association with chromatin was shown to correlate with that of the origin replication complex (ORC), implying that Y RNAs and ORC likely participate in a common functional pathway [75]. Constituently, all four hY RNAs were shown to interact with members of the ORC, as well as with proteins participating in the initiation of DNA replication, but not with proteins involved in DNA replication elongation, suggesting that Y RNAs act specifically in the initiation stage of replication [55, 76]. Consistent with a function in DNA replication and cell proliferation, Y RNA levels were found to correlate with the proliferative condition of the cells [75] and to increase in solid human tumors compared to healthy controls [77]. In another study, however, it was reported that mouse Ro60 knockout cells, which had about 30-fold lower Y-RNA levels, did not demonstrate decreased growth rates, indicating that chromosomal replication can still occur in the absence of Ro60 [61, 78]. And therefore, while there are increasing examples in the literature supporting a role of Y RNA in DNA replication, the exact molecular mechanisms by which Y RNA regulates DNA replication are still unknown.
Growing amount of experimental data support a role for YRNAs in RNA stability and quality control via its interaction with Ro60. It has been shown that Ro60 binds and process [79, 80] defective non-coding RNAs such as misfolded 5S rRNA, pre-tRNAs and U2 snRNA in various organisms, including C. elegans, X. laevis, and mouse [61, 79, 81, 82]. It was found that under normal conditions Y RNAs compete with misfolded RNAs for association with Ro60 [44]. Structural and biochemical studies have shown that Y RNAs bind to the outer surface of Ro60, while misfolded RNAs pass through the Ro60 cavity and also bind to the Ro60 outer surface at portions that overlap with the Y RNA-binding region [70, 79]. Because, Y RNAs bind Ro60 with higher affinity and sequence complementarity compared to misfolded RNAs, the ‘steric occlusion’ model was proposed suggesting that bound Y RNAs sterically hinders the binding of misfolded RNAs to Ro60. [70, 79]. And thus, it appears that Y RNAs serve as guards for Ro60 function in regulating RNA quality, permitting access only when needed [80, 83]. Consistently, under conditions of stress, such as exposure to UV-irradiation, Ro60/Y RNA complex can function as cellular stress sensors. This was supported by the finding that Ro60 can dissociate from Y RNAs and rescue misfolded RNAs, thereby helping cellular recovery [45]. The mechanism that regulates dissociation of Y RNAs from Ro, however, is currently unknown. Furthermore, it has been showed that Y RNAs can not only regulate Ro60 access to RNA substrates, but also can contribute to recognition of misfolded ncRNAs and recruit other proteins involved in RNA metabolism [83, 84]. In the bacterium D. radiodurans, Y RNA function as a scaffold, tethering the exonuclease PNPase to the bacterial ortholog of Ro (ro-sixty related, Rsr), thus forming an RNA degradation complex which mediates RNA decay [47, 83]. In contrast to prokaryotes, in mammals PNPases reside inside mitochondria and since Ro60 RNPs are predominantly cytosolic, it is possible that PNPases RNA degradation machine in mammalian cells does not form. It was suggested, however, that Y RNA can scaffold Ro60 to other proteins playing a role in RNA metabolism, including helicases, exoribonucleases or RNA chaperones [47]. With regards to eukaryotes, however, association of exoribonucleases with RoRNPs and their Y RNAs to this date have not been reported. In fact, one study, using tandem affinity purifications of mouse Ro60, reported that it was unable to detect association with any ribonuclease [41].
To this date, by mostly pull down assays, more than 20 proteins with roles in the regulation of various cellular processes, have been reported to interact with Y RNAs [85]. For example, Argonaute (Ago) and MOV10 have key functions in miRNA-mediated gene silencing, others like HuR influence cytokine production, and still others regulate mRNA transcripts splicing or processing, virus infection and innate immunity. The effects from these interactions are mostly unknown, however, it has been proposed that they could impose specific cellular functions through binding with Y RNAs (Fig. 1). In addition, not all of the identified proteins were shown to associate with all four hY RNAs, implying that depending on their bound proteins, different Y-RNAs could have different functions. [29, 80]. Support for this notion was provided in several studies. For example, HuR, a protein specifically expressed in neuronal cells, was demonstrated to influence cytokine production by a mechanism involving association of HuR with Y3-RNA and AU-rich elements in mRNA transcripts [86, 87]. Similarly, CPSF, a protein with a role in mRNA splicing, was shown to regulate human histone-H3 mRNA synthesis and processing in cooperation with a fragment derived from Y3 RNA [88]. Accordingly, within the group of proteins reported to directly interact with Y RNAs, there are some, such as calcineurin and nulceolin, with documented roles in the heart [89–91]. It is interesting, whether Y RNA interaction with these proteins is of functional significance, e.g., influencing specific processes in the heart.
Interestingly, most of the reported roles of Y RNAs involve nuclear functions. But at steady state Y RNAs are predominantly cytoplasmic. Therefore, it is conceivable that Y RNAs could play functions in modulating not only nuclear but cytoplasmic mRNAs as well. In this line of thought, one can speculate that in addition to Ro60, other RBPs could also be sequestered by Y RNAs in a similar way. Thus they may serve an analogous function to long ncRNAs, for instance, which have been reported to regulate gene expression by acting as scaffolds or sponges [92, 93]. In support to this notion, recently it was found that Y3 RNA can function as a molecular sponge for the HuD enhancer [94]. HuD plays a role in regulating neuronal cell fate by promoting gene expression through interacting with many mRNAs involved in motor neuron neurogenesis [95]. It was found that binding of Y3 RNA to HuD, altered HuD localization, limiting HuD access to the polysomal compartment, which subsequently reduced expression of the involved mRNAs (PMID: [94]). Future investigations on the Y RNA compartment specific function would be interesting and useful, as it could shed more light on the Y RNA roles in regulating cytoplasmic mRNA functions. In addition to understanding the cellular role of Y RNA, future work is also needed to inform on the physiological significance of Y RNAs during normal and pathological conditions, for example by using genetic models.
Y RNAs do not solely exist in their full length. A growing number of RNA sequencing studies have identified small RNA fragments of about 25–35 nt, comprising parts of Y RNAs [96, 97]. Fragmentation, performed by RNAse L [98] was shown to increase in apoptotic cells and upon activation of the innate immune system [99, 100]. Interestingly, it was found that these Y RNA fragments were still associated with Ro60 and La, suggesting that the binding of the proteins could have a protective role, preventing Y RNA degradation by exonucleases [99]. Despite the fact that Y RNA fragmentation increases significantly during apoptosis, Y RNA products have also been detected in non-apoptotic, proliferating cells, at amounts comparable to that of known miRNAs [100]. Since Y RNA fragments are produced from conserved ends of the hairpin containing Y RNAs, and their structure resembles that of the pre-miRNAs, some of these Y RNA fragments were originally annotated as a new kind of miRNA [101, 102]. Experimental validation of Y RNA encoded regulatory microRNAs, however, has not been reported to date [99]. Studies have shown that the biogenesis of Y RNA fragments is independent of the classical miRNA biogenesis pathway [100], evidenced by the findings that Y RNA fragments do not interact with Ago proteins [100, 103] and their processing is independent of Dicer [104]. Moreover, luciferase reporter assay studies have shown that reporter mRNA constructs could not be suppressed by Y RNAs, unlike miRNAs, further providing evidence that the identified fragments are not produced by the canonical miRNA pathway and do not act as miRNAs [101]. Apart from cells, Y RNA fragments have also been reported in various biofluids, either as cell-free RNPs or within extracellular vesicles and were shown to comprise a significant fraction of the RNA component in human serum and plasma [105–107]. Furthermore, differences in circulating Y RNA products were detected in disease [28, 106, 108]. For instance, increased levels of 3’ Y-RNA fragments were found in the plasma of patients with breast cancer [106], while full length and 5′ fragments of hY4 were elevated in chronic lymphocytic leukemia (CLL) patients, compared with healthy controls [108]. Elevated amounts of extracellular Y-RNA products have also been detected in patients with atherosclerosis and coronary artery disease [28]. These findings together with the observation that similar to miRNAs, Y RNA fragments are stable in human plasma [28], suggest that they may be further investigated as biomarkers for disease. Whether these Y RNA fragments are biologically significant is still unclear, but for some of them functional relevance has recently been ascribed, e.g., in the context of heart disease and cancer [108–111].
4. Y RNAs in Cardiovascular Biology and Disease
Given that Y RNAs have only recently started to gain more attention, knowledge about their roles in the heart is still limited, and the full impact of this field on the cardiovascular system is yet to be determined. Here we highlight examples of Y RNAs, known or suggested to be involved in cardiovascular functions. The few reports that exist only hint at the Y RNA roles that are yet to be revealed and which potentially may lead to the identification of novel therapeutic targets.
Myocardial infarction (MI), and subsequent ischemic heart failure remains a significant contributor to the global burden of cardiovascular disease. Patients who survive MI often will develop heart failure (HF) and will consequently be at increased risk for premature death [112]. Cardiosphere-derived cells (CDCs) are stem cells derived from cardiac tissue itself that have shown promising clinical results in reducing infarct size and improving cardiovascular function [113, 114]. They had been initially hypothesized to improve tissue repair and boost cardiac function by triggering native cardiomyocyte proliferation, recruiting endogenous progenitor cells and exerting potent anti-inflammatory, antifibrotic and angiogenic effects [115–117]. The regenerative benefits of CDCs in vivo are shown to be mediated mostly via paracrine effects, particularly through secretion of exosomes (EVs) [118–120]. These endogenous vesicles mediate intercellular communication, transferring various cargo of RNAs, proteins and lipids [120](Valadi H., 2007). Y RNAs were shown to be particularly abundant in EVs [120]. In a recent study, it was shown that after tRNAs, Y RNAs and their fragments make up the largest portion of small RNAs in CDC-EVs, accounting for about 20% of the total small exosomal RNAs [109]. One particular 5’ fragment of Y4-RNA was found to be specifically enriched, being the most abundant individual small RNA present in CDC-EVs, compared to normal human dermal fibroblast EVs [109]. It was demonstrated that Y4-fragment can be transferred to bone-marrow derived macrophages via EVs and this was shown to promote cardioprotection by altering gene expression. In an in vitro setting, Y4-fragment was shown to act indirectly on cardiomyocytes by modulating the cytokine profile secreted by macrophages. In particular, overexpression of Y4-fragment in macrophages, when cocultured with oxidatively stressed cardiomyocytes, was shown to suppress cardiomyocyte death by inducing strong and prolonged upregulation of the anti-inflammatory cytokine IL10. Furthermore, similar cardioprotective response was observed in vivo when it was demonstrated that administration of this Y4-product triggered IL10 release and reduced infarct size in a rat model of ischemia/reperfusion injury (I/R). Finally, evidence was provided that the abundance of Y4- fragments in CDC-exosomes correlated with the CDCs functional benefit of mitigating damage after myocardial infarction [109].
Later, in a follow up study from the same group, their initial findings were further expanded by demonstrating that the same highly abundant Y4-fragment promotes beneficial effects in a mouse model of cardiac hypertrophy induced by Ang II infusion [110]. Intravenous administration of Y4-fragment was shown to mitigate the progression of LV remodeling by decreasing cardiac hypertrophy, fibrosis and inflammation in the murine hypertensive model. Some of the benefits on the heart were attributed to the increased levels of IL-10 protein detected in the plasma after systemic injection of Y4-fragment. Y4-fragment was shown to replenish normal levels of IL-10 in heart after Ang II infusion. It was proposed that the expression of Y4-fragment in heart after administration promotes effects at the site of injury likely via activation of cardiac-resident macrophages. Y4-fragment was shown to suppress inflammatory response by reducing the levels of the proinflammatory cytokines IL1b and IL6 in the heart and decreasing expression of CD68 and F4/80 markers of infiltrating inflammatory macrophages. Adding to initial findings, it was demonstrated that the release of IL-10 by macrophages induced by Y4-fragment, counteracts Ang II effect in cultured cardiomyocytes and cardiac fibroblasts. Decreased atrial natriuretic peptide (ANP) expression in cultured cardiomyocytes or IL6 in cardiac fibroblasts upon Ang II treatment were only detected in the presence of macrophage-conditioned media containing increased Y4-fragment, demonstrating again evidence of the direct Y4-fragment effect on macrophage activation and underscoring the importance of cellular cross-talk during myocardial damage. Additionally, the favorable actions of Y4-fragment on the heart were detected in the absence of elevated blood pressure, suggesting that Y4-fragment prevents Ang II local actions on the heart without affecting its hemodynamic effects. Importantly, both the Y4-fragment and CDC-EVs were demonstrated to act in the same direction, producing similar beneficial effect on the heart, suggesting that CDC cardioprotective properties are at least in part mediated by this single fragment, further implying that the fragment may be useful therapeutically either by itself or at enhanced levels in EVs [110].
Y RNAs have also been implicated in cardiac neonatal lupus (NL) [121]. Cardiac NL is an autoimmune disease in which tissue injury in the fetus is believed to be related to the transplacental passage of maternal autoantibodies targeting Ro/SSA (Sjögren syndrome type A antigen) and/or La/SSB (Sjögren syndrome type B antigen) ribonuclear proteins [122–124]. The most common cardiac manifestation of cardiac NL is complete congenital heart block (CHB) that may be accompanied by valvular abnormalities, endocardial fibroelastosis, and/or dilated cardiomyopathy [125, 126]. Increased physiological apoptosis has been proposed as a link between anti-Ro60 antibodies and injury, providing a mechanism by which these typically inaccessible Ro and La antigens can be translocated to the surface of the heart fetal cells and become available to maternal antibodies to initiate injury [127–129]. Evidence was provided that Y3 RNA is required for Ro60 membrane translocation and subsequent formation of immune complexes capable of inducing a toll-like receptor (TLR) dependent proinflammatory cascade [121]. In this study, two experimental approaches were used to address the dependence of Y RNA for Ro60 trafficking to the cell membrane: siRNA-mediated knockdown of mouse Y3 RNA and a single point mutation of Ro60 that blocks RNA binding. It was found that depletion of Y3 RNA in murine fibroblasts undergoing apoptosis prevented cell surface translocation of Ro60, and similarly the mutated Ro60 (unbound Y RNA) construct but not the wild type form was unable to get exposed on the cell surface. Based on these findings, it was suggested that Y3 RNA masks a nuclear localization signal on Ro60, allowing for translocation and surface accessibility of cytosolic Ro60/mY3 RNA complexes. Moreover, upon opsonization with anti-Ro60 antibodies, these apoptotic fibroblasts promoted TLR7 dependent TNFα secretion from cocultured macrophages. Interestingly, apoptosis is known to promote caspase-dependent cleavage of Y RNAs, which somewhat conflicts the findings in this study [99]. However, as mentioned earlier, the small fragments produced during apoptotic driven degradation arise from the most highly conserved part of the Y RNA and remain associated with Ro60 [99]. It is therefore possible that these Y RNA products would still be capable of blocking the Ro60 nuclear localization signal, allowing and even facilitating surface accessibility of cytosolic Ro60/mY3 RNA complexes and may be even sufficient to induce proinflammatory response [130].
Complementing the findings of this study, again in the context of autoimmune associated CHB, another paper demonstrated that the complex of human Ro60-associated Y3 RNA with anti-Ro60 IgG is required for activation of both TLR7 and FcyR dependent proinflammatory responses [131]. Macrophages transfected with both synthetic hY3 RNA that binds Ro60 and an immune complex produced by incubation of hY3/Ro60 with IgG from a CHB mother (anti-Ro60 present in serum) was shown to induce strong TNF-α release. Moreover, collagen secretion and fibrosis markers were markedly increased in cultured fetal cardiac fibroblasts exposed to supernatants of macrophages transfected with hY3 RNA. In contrast to healthy heart, immunohistological evaluation of autopsy tissue from a fetus diagnosed with CHB revealed TLR7 expression in the conduction system with TLR7 positive cells located near the atrioventricular groove. Based on the findings, Ro60-hY3 was proposed as a link between inflammation and fetal cardiac fibrosis in CHB, providing a mechanism by which hY3/Ro60 binds to anti-Ro antibodies and form immune complexes capable of inducing immune response which promotes fibrosis [131].
In addition to modulating inflammatory responses during cardiac injury, Y RNAs have also been shown to induce apoptosis in atherosclerosis [132]. In atherosclerosis, cholesterol deposition into the arterial wall triggers increased accumulation of macrophages and promotes apoptosis in these cells [133]. The induction of macrophage apoptosis speeds up the progression of atherosclerosis by promoting the development of a necrotic core, which on the other hand contributes to plaque disruption and acute thrombosis [134, 135]. Significant upregulation of fragmented Y RNA has been detected in the medium of cultured macrophages treated with lipids, as well as in the blood of mouse models for atherosclerosis and in the serum of patients with coronary artery disease (CAD) [28]. In addition, biostatistical analysis associated fragmented Y-RNA with atherosclerosis burden by demonstrating a strong positive correlation between the levels of one specific 5’ fragment of Y1-RNA (RNY1–5p) into the serum and CAD risk [28]. Importantly, RNY1–5p was shown to associate better with CAD status in comparison to some CAD-specific miRNAs (e.g., miRNA-17, miRNA-133, miRNA-155), suggesting potential diagnostic application of RNY1–5p measurement as a biomarker for CAD risk assessment [28]. In another study, it was demonstrated that macrophage activation upon treatment with atherogenic lipids triggers the processing of Y RNAs into small Y RNA products in these cells [132]. In vitro gain- and loss-of-function studies showed that Y RNA fragments, but not full-length Y RNAs from which they were derived, activated both caspase 3 and NF-κB pathways, promoting cell death and inflammation in the macrophages [132]. In addition, it was found that not only intracellular but also extracellular Y RNA fragments induce apoptosis and inflammation in macrophages [132]. Specifically, it was shown that extracellular affinity purified Y RNA fragment/Ro60 complex from apoptotic HEK293T cells could trigger cell death in macrophages. Interestingly, synthetic Y RNA fragment alone was not able to induce macrophage activation, implying that the fragments must be bound in a complex with Ro60 in order to get incorporated in the cell and subsequently produce a biological response [132]. Based on these findings, Y RNA fragment/Ro60 complex released by macrophages could contribute to the progression of CAD by participating in a negative feed-back loop in which increasing number of macrophages die by apoptosis in the lipid abundant arterial wall and by this possibly reinforce the pathogenesis of atherosclerosis [132].
5. Conclusion
In summary, Y RNAs have been shown to participate in a range of cellular processes including DNA replication, RNA quality control and cellular stress responses. More recently, accumulating evidence suggest functional involvement of Y RNAs in disease such as cancer and immune related pathologies. Roles of Y RNAs in cardiovascular biology are also starting to emerge. In this context, Y RNAs have been reported to mediate both beneficial and adverse effects on the cardiovascular system. However, investigations on Y RNAs are still at a very early stage, and there are many questions that remain to be answered. Improved functional and mechanistic understanding of Y RNAs will provide valuable insights into normal human physiology and disease pathogenesis.
Acknowledgements
SD is funded by RO1 HL122547 and AHA SFRN grant.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Birney E, Evolutionary genomics: come fly with us. Nature, 2007. 450(7167): p. 184–5. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, et al. , The transcriptional landscape of the mammalian genome. Science, 2005. 309(5740): p. 1559–63. [DOI] [PubMed] [Google Scholar]
- 3.Zuker M, Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res, 2003. 31(13): p. 3406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thum T, MicroRNA therapeutics in cardiovascular medicine. EMBO Mol Med, 2012. 4(1): p. 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greco S, et al. , Long Noncoding RNAs and Cardiac Disease. Antioxid Redox Signal, 2018. 29(9): p. 880–901. [DOI] [PubMed] [Google Scholar]
- 6.Huarte M, The emerging role of lncRNAs in cancer. Nat Med, 2015. 21(11): p. 1253–61. [DOI] [PubMed] [Google Scholar]
- 7.Hayes J, Peruzzi PP, and Lawler S, MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med, 2014. 20(8): p. 460–9. [DOI] [PubMed] [Google Scholar]
- 8.Fan B, et al. , MicroRNA and Diabetic Complications: A Clinical Perspective. Antioxid Redox Signal, 2018. 29(11): p. 1041–1063. [DOI] [PubMed] [Google Scholar]
- 9.Giroud M and Scheideler M, Long Non-Coding RNAs in Metabolic Organs and Energy Homeostasis. Int J Mol Sci, 2017. 18(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sayed D and Abdellatif M, MicroRNAs in development and disease. Physiol Rev, 2011. 91(3): p. 827–87. [DOI] [PubMed] [Google Scholar]
- 11.Ng SY, et al. , Long noncoding RNAs in development and disease of the central nervous system. Trends Genet, 2013. 29(8): p. 461–8. [DOI] [PubMed] [Google Scholar]
- 12.Duan L, et al. , miRNA-1: functional roles and dysregulation in heart disease. Mol Biosyst, 2014. 10(11): p. 2775–82. [DOI] [PubMed] [Google Scholar]
- 13.Iorio MV and Croce CM, MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med, 2012. 4(3): p. 143–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iorio MV and Croce CM, Causes and consequences of microRNA dysregulation. Cancer J, 2012. 18(3): p. 215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbott JA, Francklyn CS, and Robey-Bond SM, Transfer RNA and human disease. Front Genet, 2014. 5: p. 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, et al. , Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther, 2013. 21(2): p. 368–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozata DM, et al. , PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet, 2019. 20(2): p. 89–108. [DOI] [PubMed] [Google Scholar]
- 18.Assumpcao CB, et al. , The role of piRNA and its potential clinical implications in cancer. Epigenomics, 2015. 7(6): p. 975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lerner MR, et al. , Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science, 1981. 211(4480): p. 400–2. [DOI] [PubMed] [Google Scholar]
- 20.Lerner MR and Steitz JA, Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A, 1979. 76(11): p. 5495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sim S, et al. , The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell, 2009. 20(5): p. 1555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruijn GJ, et al. , Ro RNP associated Y RNAs are highly conserved among mammals. Biochim Biophys Acta, 1993. 1216(3): p. 395–401. [DOI] [PubMed] [Google Scholar]
- 23.Mosig A, et al. , Evolution of the vertebrate Y RNA cluster. Theory Biosci, 2007. 126(1): p. 9–14. [DOI] [PubMed] [Google Scholar]
- 24.Sim S and Wolin SL, Bacterial Y RNAs: Gates, Tethers, and tRNA Mimics. Microbiol Spectr, 2018. 6(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Horn DJ, et al. , Caenorhabditis elegans embryos contain only one major species of Ro RNP. RNA, 1995. 1(3): p. 293–303. [PMC free article] [PubMed] [Google Scholar]
- 26.Boria I, et al. , Nematode sbRNAs: homologs of vertebrate Y RNAs. J Mol Evol, 2010. 70(4): p. 346–58. [DOI] [PubMed] [Google Scholar]
- 27.Wolin SL and Steitz JA, The Ro small cytoplasmic ribonucleoproteins: identification of the antigenic protein and its binding site on the Ro RNAs. Proc Natl Acad Sci U S A, 1984. 81(7): p. 1996–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Repetto E, et al. , RNY-derived small RNAs as a signature of coronary artery disease. BMC Med, 2015. 13: p. 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langley AR, et al. , Ribonucleoprotein particles containing non-coding Y RNAs, Ro60, La and nucleolin are not required for Y RNA function in DNA replication. PLoS One, 2010. 5(10): p. e13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maraia RJ, et al. , The human Y4 small cytoplasmic RNA gene is controlled by upstream elements and resides on chromosome 7 with all other hY scRNA genes. Nucleic Acids Res, 1994. 22(15): p. 3045–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maraia R, et al. , Gene encoding human Ro-associated autoantigen Y5 RNA. Nucleic Acids Res, 1996. 24(18): p. 3552–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendrick JP, et al. , Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol, 1981. 1(12): p. 1138–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolin SL and Steitz JA, Genes for two small cytoplasmic Ro RNAs are adjacent and appear to be single-copy in the human genome. Cell, 1983. 32(3): p. 735–44. [DOI] [PubMed] [Google Scholar]
- 34.Perreault J, et al. , Retropseudogenes derived from the human Ro/SS-A autoantigen-associated hY RNAs. Nucleic Acids Res, 2005. 33(6): p. 2032–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perreault J, Perreault JP, and Boire G, Ro-associated Y RNAs in metazoans: evolution and diversification. Mol Biol Evol, 2007. 24(8): p. 1678–89. [DOI] [PubMed] [Google Scholar]
- 36.Simons FH, et al. , The interactions with Ro60 and La differentially affect nuclear export of hY1 RNA. RNA, 1996. 2(3): p. 264–73. [PMC free article] [PubMed] [Google Scholar]
- 37.Wolin SL and Cedervall T, The La protein. Annu Rev Biochem, 2002. 71: p. 375–403. [DOI] [PubMed] [Google Scholar]
- 38.Peek R, et al. , Subcellular distribution of Ro ribonucleoprotein complexes and their constituents. J Cell Sci, 1993. 106 (Pt 3): p. 929–35. [DOI] [PubMed] [Google Scholar]
- 39.Rutjes SA, et al. , Identification of a novel cis-acting RNA element involved in nuclear export of hY RNAs. RNA, 2001. 7(5): p. 741–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gendron M, Roberge D, and Boire G, Heterogeneity of human Ro ribonucleoproteins (RNPS): nuclear retention of Ro RNPS containing the human hY5 RNA in human and mouse cells. Clin Exp Immunol, 2001. 125(1): p. 162–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sim S, et al. , The zipcode-binding protein ZBP1 influences the subcellular location of the Ro 60-kDa autoantigen and the noncoding Y3 RNA. RNA, 2012. 18(1): p. 100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teunissen SW, et al. , Conserved features of Y RNAs: a comparison of experimentally derived secondary structures. Nucleic Acids Res, 2000. 28(2): p. 610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Gelder CW, et al. , Common structural features of the Ro RNP associated hY1 and hY5 RNAs. Nucleic Acids Res, 1994. 22(13): p. 2498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green CD, et al. , Binding of the 60-kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA, 1998. 4(7): p. 750–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sim S and Wolin SL, Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley Interdiscip Rev RNA, 2011. 2(5): p. 686–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia PZ, et al. , The particulate (speckled-like thread) nuclear staining pattern: species and cellular distribution of Ro/SSA antigen. J Clin Lab Immunol, 1987. 22(3): p. 101–5. [PubMed] [Google Scholar]
- 47.Wolin SL, et al. , Non-coding Y RNAs as tethers and gates: Insights from bacteria. RNA Biol, 2013. 10(10): p. 1602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fabini G, et al. , The heterogeneous nuclear ribonucleoproteins I and K interact with a subset of the ro ribonucleoprotein-associated Y RNAs in vitro and in vivo. J Biol Chem, 2001. 276(23): p. 20711–8. [DOI] [PubMed] [Google Scholar]
- 49.Fouraux MA, et al. , Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J Mol Biol, 2002. 320(3): p. 475–88. [DOI] [PubMed] [Google Scholar]
- 50.Kohn M, et al. , Near-infrared (NIR) dye-labeled RNAs identify binding of ZBP1 to the noncoding Y3-RNA. RNA, 2010. 16(7): p. 1420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowalski MP, Baylis HA, and Krude T, Non-coding stem-bulge RNAs are required for cell proliferation and embryonic development in C. elegans. J Cell Sci, 2015. 128(11): p. 2118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolin SL, Sim S, and Chen X, Nuclear noncoding RNA surveillance: is the end in sight? Trends Genet, 2012. 28(7): p. 306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hall AE and Dalmay T, Discovery of novel small RNAs in the quest to unravel genome complexity. Biochem Soc Trans, 2013. 41(4): p. 866–70. [DOI] [PubMed] [Google Scholar]
- 54.Pruijn GJ, Simons FH, and van Venrooij WJ, Intracellular localization and nucleocytoplasmic transport of Ro RNP components. Eur J Cell Biol, 1997. 74(2): p. 123–32. [PubMed] [Google Scholar]
- 55.Zhang AT, et al. , Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J Cell Sci, 2011. 124(Pt 12): p. 2058–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Brien CA, Margelot K, and Wolin SL, Xenopus Ro ribonucleoproteins: members of an evolutionarily conserved class of cytoplasmic ribonucleoproteins. Proc Natl Acad Sci U S A, 1993. 90(15): p. 7250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simons FH, Pruijn GJ, and van Venrooij WJ, Analysis of the intracellular localization and assembly of Ro ribonucleoprotein particles by microinjection into Xenopus laevis oocytes. J Cell Biol, 1994. 125(5): p. 981–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farris AD, et al. , The ultrastructural localization of 60-kDa Ro protein and human cytoplasmic RNAs: association with novel electron-dense bodies. Proc Natl Acad Sci U S A, 1997. 94(7): p. 3040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matera AG, et al. , A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol, 1995. 129(5): p. 1181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen X, Quinn AM, and Wolin SL, Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev, 2000. 14(7): p. 777–82. [PMC free article] [PubMed] [Google Scholar]
- 61.Chen X, et al. , The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr Biol, 2003. 13(24): p. 2206–11. [DOI] [PubMed] [Google Scholar]
- 62.Kohn M, Pazaitis N, and Huttelmaier S, Why YRNAs? About Versatile RNAs and Their Functions. Biomolecules, 2013. 3(1): p. 143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia EL, et al. , Packaging of host mY RNAs by murine leukemia virus may occur early in Y RNA biogenesis. J Virol, 2009. 83(23): p. 12526–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang T, et al. , 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol, 2007. 81(23): p. 13112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang Y, et al. , Incorporation of excess wild-type and mutant tRNA(3Lys) into human immunodeficiency virus type 1. J Virol, 1994. 68(12): p. 7676–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balasubramaniam M, Pandhare J, and Dash C, Are microRNAs Important Players in HIV-1 Infection? An Update. Viruses, 2018. 10(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stake M, et al. , HIV-1 and two avian retroviral 5’ untranslated regions bind orthologous human and chicken RNA binding proteins. Virology, 2015. 486: p. 307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Telesnitsky A and Wolin SL, The Host RNAs in Retroviral Particles. Viruses, 2016. 8(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eckwahl MJ, et al. , A retrovirus packages nascent host noncoding RNAs from a novel surveillance pathway. Genes Dev, 2015. 29(6): p. 646–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stein AJ, et al. , Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell, 2005. 121(4): p. 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X, et al. , Bacterial noncoding Y RNAs are widespread and mimic tRNAs. RNA, 2014. 20(11): p. 1715–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Christov CP, et al. , Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol Cell Biol, 2006. 26(18): p. 6993–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gardiner TJ, et al. , A conserved motif of vertebrate Y RNAs essential for chromosomal DNA replication. RNA, 2009. 15(7): p. 1375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang I, et al. , Nucleotide contributions to the structural integrity and DNA replication initiation activity of noncoding y RNA. Biochemistry, 2014. 53(37): p. 5848–63. [DOI] [PubMed] [Google Scholar]
- 75.Kheir E and Krude T, Non-coding Y RNAs associate with early replicating euchromatin in concordance with the origin recognition complex. J Cell Sci, 2017. 130(7): p. 1239–1250. [DOI] [PubMed] [Google Scholar]
- 76.Krude T, et al. , Y RNA functions at the initiation step of mammalian chromosomal DNA replication. J Cell Sci, 2009. 122(Pt 16): p. 2836–45. [DOI] [PubMed] [Google Scholar]
- 77.Christov CP, Trivier E, and Krude T, Noncoding human Y RNAs are overexpressed in tumours and required for cell proliferation. Br J Cancer, 2008. 98(5): p. 981–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xue D, et al. , A lupus-like syndrome develops in mice lacking the Ro 60-kDa protein, a major lupus autoantigen. Proc Natl Acad Sci U S A, 2003. 100(13): p. 7503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fuchs G, et al. , Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen. Nat Struct Mol Biol, 2006. 13(11): p. 1002–9. [DOI] [PubMed] [Google Scholar]
- 80.Hogg JR and Collins K, Human Y5 RNA specializes a Ro ribonucleoprotein for 5S ribosomal RNA quality control. Genes Dev, 2007. 21(23): p. 3067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Labbe JC, Hekimi S, and Rokeach LA, Assessing the function of the Ro ribonucleoprotein complex using Caenorhabditis elegans as a biological tool. Biochem Cell Biol, 1999. 77(4): p. 349–54. [PubMed] [Google Scholar]
- 82.O’Brien CA and Wolin SL, A possible role for the 60-kD Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes Dev, 1994. 8(23): p. 2891–903. [DOI] [PubMed] [Google Scholar]
- 83.Chen X, et al. , An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell, 2013. 153(1): p. 166–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen X, et al. , An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev, 2007. 21(11): p. 1328–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Driedonks TAP and Nolte-’t Hoen ENM, Circulating Y-RNAs in Extracellular Vesicles and Ribonucleoprotein Complexes; Implications for the Immune System. Front Immunol, 2018. 9: p. 3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katsanou V, et al. , HuR as a negative posttranscriptional modulator in inflammation. Mol Cell, 2005. 19(6): p. 777–89. [DOI] [PubMed] [Google Scholar]
- 87.Herdy B, et al. , The RNA-binding protein HuR/ELAVL1 regulates IFN-beta mRNA abundance and the type I IFN response. Eur J Immunol, 2015. 45(5): p. 1500–11. [DOI] [PubMed] [Google Scholar]
- 88.Kohn M, et al. , The Y3** ncRNA promotes the 3’ end processing of histone mRNAs. Genes Dev, 2015. 29(19): p. 1998–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilkins BJ and Molkentin JD, Calcineurin and cardiac hypertrophy: where have we been? Where are we going? J Physiol, 2002. 541(Pt 1): p. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parra V and Rothermel BA, Calcineurin signaling in the heart: The importance of time and place. J Mol Cell Cardiol, 2017. 103: p. 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang B, et al. , Nucleolin protects the heart from ischaemia-reperfusion injury by up-regulating heat shock protein 32. Cardiovasc Res, 2013. 99(1): p. 92–101. [DOI] [PubMed] [Google Scholar]
- 92.Gaiti F, et al. , Sponge Long Non-Coding RNAs Are Expressed in Specific Cell Types and Conserved Networks. Noncoding RNA, 2018. 4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang KC and Chang HY, Molecular mechanisms of long noncoding RNAs. Mol Cell, 2011. 43(6): p. 904–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tebaldi T, et al. , HuD Is a Neural Translation Enhancer Acting on mTORC1-Responsive Genes and Counteracted by the Y3 Small Non-coding RNA. Mol Cell, 2018. 71(2): p. 256–270 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bolognani F, Contente-Cuomo T, and Perrone-Bizzozero NI, Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res, 2010. 38(1): p. 117–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rother S and Meister G, Small RNAs derived from longer non-coding RNAs. Biochimie, 2011. 93(11): p. 1905–15. [DOI] [PubMed] [Google Scholar]
- 97.Tuck AC and Tollervey D, RNA in pieces. Trends Genet, 2011. 27(10): p. 422–32. [DOI] [PubMed] [Google Scholar]
- 98.Donovan J, et al. , Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA, 2017. 23(11): p. 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rutjes SA, et al. , Rapid nucleolytic degradation of the small cytoplasmic Y RNAs during apoptosis. J Biol Chem, 1999. 274(35): p. 24799–807. [DOI] [PubMed] [Google Scholar]
- 100.Nicolas FE, et al. , Biogenesis of Y RNA-derived small RNAs is independent of the microRNA pathway. FEBS Lett, 2012. 586(8): p. 1226–30. [DOI] [PubMed] [Google Scholar]
- 101.Meiri E, et al. , Discovery of microRNAs and other small RNAs in solid tumors. Nucleic Acids Res, 2010. 38(18): p. 6234–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Verhagen AP and Pruijn GJ, Are the Ro RNP-associated Y RNAs concealing micro-RNAs? Y RNA-derived miRNAs may be involved in autoimmunity. Bioessays, 2011. 33(9): p. 674–82. [DOI] [PubMed] [Google Scholar]
- 103.Chen CJ and Heard E, Small RNAs derived from structural non-coding RNAs. Methods, 2013. 63(1): p. 76–84. [DOI] [PubMed] [Google Scholar]
- 104.Langenberger D, et al. , Dicer-processed small RNAs: rules and exceptions. J Exp Zool B Mol Dev Evol, 2013. 320(1): p. 35–46. [DOI] [PubMed] [Google Scholar]
- 105.Dhahbi JM, et al. , 5’-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol Genomics, 2013. 45(21): p. 990–8. [DOI] [PubMed] [Google Scholar]
- 106.Dhahbi JM, Circulating small noncoding RNAs as biomarkers of aging. Ageing Res Rev, 2014. 17: p. 86–98. [DOI] [PubMed] [Google Scholar]
- 107.Vojtech L, et al. , Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res, 2014. 42(11): p. 7290–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Haderk F, et al. , Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci Immunol, 2017. 2(13). [DOI] [PubMed] [Google Scholar]
- 109.Cambier L, et al. , Y RNA fragment in extracellular vesicles confers cardioprotection via modulation of IL-10 expression and secretion. EMBO Mol Med, 2017. 9(3): p. 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cambier L, et al. , Angiotensin II-Induced End-Organ Damage in Mice Is Attenuated by Human Exosomes and by an Exosomal Y RNA Fragment. Hypertension, 2018. 72(2): p. 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chakrabortty SK, et al. , Extracellular vesicle-mediated transfer of processed and functional RNY5 RNA. RNA, 2015. 21(11): p. 1966–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jhund PS and McMurray JJ, Heart failure after acute myocardial infarction: a lost battle in the war on heart failure? Circulation, 2008. 118(20): p. 2019–21. [DOI] [PubMed] [Google Scholar]
- 113.Marban E and Cingolani E, Direct Reprogramming: Bypassing Stem Cells for Therapeutics. JAMA, 2015. 314(1): p. 19–20. [DOI] [PubMed] [Google Scholar]
- 114.Marban E, Breakthroughs in cell therapy for heart disease: focus on cardiosphere-derived cells. Mayo Clin Proc, 2014. 89(6): p. 850–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barile L, Milano G, and Vassalli G, Beneficial effects of exosomes secreted by cardiac-derived progenitor cells and other cell types in myocardial ischemia. Stem Cell Investig, 2017. 4: p. 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tseliou E, et al. , Fibroblasts Rendered Antifibrotic, Antiapoptotic, and Angiogenic by Priming With Cardiosphere-Derived Extracellular Membrane Vesicles. J Am Coll Cardiol, 2015. 66(6): p. 599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.de Couto G, et al. , Macrophages mediate cardioprotective cellular postconditioning in acute myocardial infarction. J Clin Invest, 2015. 125(8): p. 3147–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ibrahim A and Marban E, Exosomes: Fundamental Biology and Roles in Cardiovascular Physiology. Annu Rev Physiol, 2016. 78: p. 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vandergriff AC, et al. , Intravenous Cardiac Stem Cell-Derived Exosomes Ameliorate Cardiac Dysfunction in Doxorubicin Induced Dilated Cardiomyopathy. Stem Cells Int, 2015. 2015: p. 960926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Valadi H, et al. , Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol, 2007. 9(6): p. 654–9 [DOI] [PubMed] [Google Scholar]
- 121.Reed JH, et al. , Ro60 requires Y3 RNA for cell surface exposure and inflammation associated with cardiac manifestations of neonatal lupus. J Immunol, 2013. 191(1): p. 110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brucato A, et al. , Pregnancy outcomes in patients with autoimmune diseases and anti-Ro/SSA antibodies. Clin Rev Allergy Immunol, 2011. 40(1): p. 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brucato A, et al. , Risk of congenital complete heart block in newborns of mothers with anti-Ro/SSA antibodies detected by counterimmunoelectrophoresis: a prospective study of 100 women. Arthritis Rheum, 2001. 44(8): p. 1832–5. [DOI] [PubMed] [Google Scholar]
- 124.Buyon JP and Winchester R, Congenital complete heart block. A human model of passively acquired autoimmune injury. Arthritis Rheum, 1990. 33(5): p. 609–14. [DOI] [PubMed] [Google Scholar]
- 125.Izmirly PM, et al. , Assessment of fluorinated steroids to avert progression and mortality in anti-SSA/Ro-associated cardiac injury limited to the fetal conduction system. Ann Rheum Dis, 2016. 75(6): p. 1161–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Izmirly PM, et al. , Maternal and fetal factors associated with mortality and morbidity in a multiracial/ethnic registry of anti-SSA/Ro-associated cardiac neonatal lupus. Circulation, 2011. 124(18): p. 1927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Casciola-Rosen LA, Anhalt G, and Rosen A, Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med, 1994. 179(4): p. 1317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Miranda-Carus ME, et al. , Induction of antibodies reactive with SSA/Ro-SSB/La and development of congenital heart block in a murine model. J Immunol, 1998. 161(11): p. 5886–92. [PubMed] [Google Scholar]
- 129.Clancy RM, et al. , Impaired clearance of apoptotic cardiocytes is linked to anti-SSA/Ro and - SSB/La antibodies in the pathogenesis of congenital heart block. J Clin Invest, 2006. 116(9): p. 2413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vollmer J, et al. , Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J Exp Med, 2005. 202(11): p. 1575–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clancy RM, et al. , Ro60-associated single-stranded RNA links inflammation with fetal cardiac fibrosis via ligation of TLRs: a novel pathway to autoimmune-associated heart block. J Immunol, 2010. 184(4): p. 2148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hizir Z, et al. , RNY (YRNA)-derived small RNAs regulate cell death and inflammation in monocytes/macrophages. Cell Death Dis, 2017. 8(1): p. e2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moore KJ and Tabas I, Macrophages in the pathogenesis of atherosclerosis. Cell, 2011. 145(3): p. 341–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tabas I, Williams KJ, and Boren J, Subendothelial lipoprotein retention as the initiating process in atherosclerosis: update and therapeutic implications. Circulation, 2007. 116(16): p. 1832–44. [DOI] [PubMed] [Google Scholar]
- 135.Naghavi M, et al. , From vulnerable plaque to vulnerable patient: a call for new definitions and risk assessment strategies: Part I. Circulation, 2003. 108(14): p. 1664–72. [DOI] [PubMed] [Google Scholar]