Abstract
Human T-cell leukemia virus type 1 (HTLV-1) causes incurable adult T-cell leukemia and HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP). Patients with HAM/TSP have increased levels of HTLV-1-infected cells compared with asymptomatic HTLV-1 carriers. However, the roles of cellular genes in HTLV-1-infected CD4+ T cells await discovery. We performed microarray analysis of CD4+ T cells from HAM/TSP patients and found that the ABL1 is an important gene in HAM/TSP. ABL1 is a known survival factor for T- and B-lymphocytes and is part of the fused gene (BCR-ABL) known to be responsible for chronic myelogenous leukemia (CML). ABL1 tyrosine kinase inhibitors (TKIs), including imatinib, nilotinib, and dasatinib, are used clinically for treating CML. To evaluate whether ABL1 is indeed important for HAM/TSP, we investigated the effect of TKIs on HTLV-1-infected cells. We developed a propidium monoazide-HTLV-1 viability quantitative PCR assay, which distinguishes DNA from live cells and dead cells. Using this method, we were able to measure the HTLV-1 proviral load (PVL) in live cells alone when peripheral blood mononuclear cells (PBMCs) from HAM/TSP cases were treated with TKIs. Treating the PBMCs with nilotinib or dasatinib induced significant reductions in PVL (21.0% and 17.5%, respectively) in live cells. Furthermore, ABL1 siRNA transfection reduced cell viability in HTLV-1-infected cell lines, but not in uninfected cell lines. A retrospective survey based on our clinical records found a rare case of HAM/TSP who also suffered from CML. The patient showed an 84.2% PVL reduction after CML treatment with imatinib. We conclude that inhibiting the ABL1 tyrosine kinase specifically reduced the PVL in PBMCs from patients with HAM/TSP, suggesting that ABL1 is an important gene for the survival of HTLV-1-infected cells and that TKIs may be potential therapeutic agents for HAM/TSP.
Author summary
Human T-cell leukemia virus type 1 (HTLV-1) is integrated as a provirus in the genomic DNA mainly of CD4+ T cell population in the infected people. HTLV-1-infected CD4+ T cells are transmitted via breast milk, semen, and blood transfusions. HTLV-1 is endemic in Japan, the Middle East, Africa, Caribbean islands, and Central and South America. A small proportion of infected people develop adult T-cell leukemia, HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP), and other diseases. HAM/TSP, a chronic neuroinflammatory disorder, is characterized by spastic paraparesis and urinary disturbance. HTLV-1-infected CD4+ T cells infiltrate the spinal cord and cause inflammation, which results in such neurological symptoms. We have identified the tyrosine kinase gene ABL1 as a gene frequently found in the signal transduction pathways in HTLV-1-infected CD4+ T cells. Therefore, ABL1 appears to be important in the pathogenesis of HAM/TSP. Inhibiting ABL1 with tyrosine kinase inhibitors (TKIs), which is used for chronic myelogenous leukemia (CML), reduced the proviral load (PVL) in vitro. We found a rare case of a patient with HAM/TSP and CML by our clinical records, who showed a decrease in PVL after TKI treatment for CML. Hence, TKIs are potential therapeutic agents for HAM/TSP.
Introduction
Human T-cell leukemia virus type 1 (HTLV-1) mainly infects CD4+ T cells in which its virus genome is integrated into the genomic DNA of the cells. There are 1.08 million infected individuals in Japan, and 5–10 million worldwide. Most of them are asymptomatic HTLV-1 carriers (ACs), but occasionally, some individuals develop adult T-cell leukemia, HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP), HTLV-1-associated uveitis, and other related diseases [1–5]. The HTLV-1 genome includes tax, a regulatory gene coded in the plus strand, and hbz, a transcription factor coded in the minus strand. Both viral genes have been investigated in relation to HTLV-1-related diseases [6,7]. HAM/TSP is a neuroinflammatory disease characterized by spastic paraparesis as well as urinary disturbance caused by the inflammation of the spinal cord induced by infiltration of HTLV-1-infected CD4+ T cells [8–10]. Initial studies of HAM/TSP have revealed no disease-specific sequences of HTLV-1 causative of the disease, but a subtype of HTLV-1 partially increases the risk of this disease [11,12]. Many of the risk factors in terms of susceptible genes and the restricting factors for HAM/TSP were identified by investigations of cellular genes, but the most important risk factor for HAM/TSP is an increased HTLV-1 proviral load (PVL) in the peripheral blood [13–16]. The PVL reflects the number of HTLV-1-infected cells in it. In the spinal cords of patients with HAM/TSP, HTLV-1-infected CD4+ T cells were found [8–10,17]. Therefore, reducing the PVL is an essential part of the treatment for HAM/TSP. A recent clinical trial of an anti-CCR4 antibody against HAM/TSP is in progress [18]. However, several clinical studies, including such antibody therapy, are far from the complete control of HTLV-1-infection in HAM/TSP.
In the present study, we investigated the cellular gene expression profiles using a whole human genome DNA microarray with CD4+ T cells (the major in vivo reservoir of HTLV-1), from patients with HAM/TSP, AC, or negative controls (NCs). By combining array data processing to refine the differentially expressed genes (DEGs) and pathway analysis, we searched the significant pathways and genes for HAM/TSP.
Herein, our data suggest that ABL1 gene may play an important role in HAM/TSP and that inhibition of ABL1 tyrosine kinase with TKIs reduces the PVL. These indicate that TKIs, which are known as agents for CML treatment, are potential therapeutic agents for HAM/TSP.
Materials and methods
Subjects
The diagnosis of NCs was made when serum anti-HTLV-1 antibody was negative (less than 16×) by particle agglutination (PA) method [19]. Diagnosis of HAM/TSP was made according to the World Health Organization criteria by neurologists belonging to the Department of Neurology and Geriatrics of Kagoshima University Hospital. Subjects who were positive for anti-HTLV-1 antibody but had no neurological symptoms were defined as ACs. We used cryopreserved peripheral blood mononuclear cell (PBMC) samples for microarray analysis from four patients with HAM/TSP, four ACs, and four NCs, from whom we obtained written informed consent. The Ethics Committee of Kagoshima University Hospital approved this study. The statistics of the subjects are summarized in Table 1. There was no significant difference in age among three groups by ANOVA (P = 0.801), and the mean PVL of the HAM/TSP group was significantly higher than that of the AC group by Mann-Whitney U test (P = 0.021). We randomly chose samples for the experiment, and there occurred a difference in sex.
Table 1. Summary of subjects for microarray analysis.
| NC | AC | HAM/TSP | |
|---|---|---|---|
| Number | 4 | 4 | 4 |
| Sex (male: female) | 2: 2 | 1: 3 | 2: 2 |
| Age (mean ± SD) (y.o.) | 52.0 ± 13.9 | 52.3 ± 4.9 | 60.0 ± 7.4 |
| Proviral load (copies/104 PBMC) | N/A* | 325.8 ± 155.3 | 2066.0 ± 403.0** |
*N/A: not applicable.
**: Significant difference in comparison between HAM/TSP and AC by Mann-Whitney U test (P = 0.021). NC: negative control. AC: asymptomatic HTLV-1 carrier. HAM/TSP: HTLV-1-associated myelopathy/tropical spastic paraparesis. SD: standard deviation. y.o.: years old.
Microarray experiment
CD4+ T cells were separated from PBMCs using the Human CD4+ T cell Isolation Kit (Miltenyi Biotech KK., Tokyo, JAPAN). Total RNA from these cells was extracted. After quality checking by an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, U.S.A.), 50 μg of total RNA was pre-amplified and labeled with cyanine 3-CTP. Complementary DNA was hybridized onto the G4112F Whole Human Genome cDNA microarray 44K × 4plex ver.1.0 (Agilent Technologies). After hybridization, the raw images were acquired and converted into fluorescence intensity data. The data were imported to GeneSpring GX Software ver. 7.3.1 (Agilent Technologies).
Array data processing and refinement of the genes
Following the Agilent 1-color manual instructions (Agilent Technologies), the raw signal intensities below 5.0 were set to 5.0. Then all the signal intensities were log2 transformed and normalized. To compare the array data from the three groups (NCs, ACs, and HAM/TSP cases), we performed one-way ANOVA at a significant level of P < 0.01. Among the significant genes, we extracted significant DEGs that were up-regulated higher than or down-regulated less than 2-fold change when compared with NCs. There are no clear criteria to decide the threshold for fold change in the analysis of array data. We used 2-fold according to previous studies [20]. Then we analyzed the DEGs by cluster analysis to create heat maps, and those DEGs were assigned to a Venn diagram to identify specific cellular genes for HAM/TSP. The raw data of the microarray experiment was deposited to the Gene Expression Omnibus repository at National Center for Biotechnology Information (GSE132666).
Pathway analysis
The specific cellular genes for HAM/TSP were mapped to the TRANSPATH database (BIOBASE GmbH, Wolfenbuettel, Germany) by ExPlain software (BIOBASE). Pathway analysis was performed using a 2 × 2 contingency table between HAM/TSP and non-HAM/TSP (AC and NC). We extracted the significant pathways for the specific cellular genes. To obtain genes that more frequently appeared than by chance among the specific pathways, we performed a chi-square test or a Fisher’s exact test using a 2 × 2 contingency table for each case by the numbers of genes included in the specific pathways. Then we performed an upstream analysis (BIOBASE). From the sequences in the promoter windows (-1000 to 100 bases from the transcriptional start sites) of the DEGs with more than 2-fold change (Yes-set) and less than 1.1-fold change (No-set), the weighted consensus sequences were calculated, and a transcriptional factor binding site search was performed using the TRANSFAC transcriptional factor binding sites database (BIOBASE) [21]. The results were extracted by the condition of the frequency ratio of the transcriptional binding sites in the Yes-site/No-site > 1.5, Fisher’s exact test (P < 0.05) for the number of transcriptional binding sites in the Yes-set, and the Fisher’s exact test (P < 0.05) of the matched promoters and binding sites.
Quantitative reverse transcriptase PCR for ABL1 mRNA
To evaluate the expression level of the ABL1 mRNA in enriched HTLV-1-infected cells, we performed real-time quantitative PCR (qPCR) on ABL1 mRNA in triplicate using the comparative Ct method [22]. Total RNA was extracted from 5×106 CD4+ T cells from 32 individuals, including NCs (N = 7), ACs (N = 14), and HAM/TSP patients (N = 11), and 1 μl of total RNA was transcribed into cDNA. Primer sequences were as follows: ABL1vβ mRNA forward, 5’-GCCTGCCCTGCATTTTATC-3’; and reverse, 5’-ATGCTACTGGCCGCTGAA-3’; GAPDH mRNA transcript variant 2 forward, 5’-GACTAACCCTGCGCTCCTG-3’; and reverse, 5’-GCCCAATACGACCAAATCAG-3’. GAPDH was used as an internal control. cDNA (100 ng) was amplified with 25 μl of Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA, U.S.A.) and 200 nM each of the ABL1vβ mRNA sense and antisense primers. Samples and calibrator (Jurkat cell cDNA 100 ng) were subjected to the PCR on the StepOnePlus Real-time PCR system (Applied Biosystems). The amplification cycles were as follows: 50°C, 2 min; 95°C, 10 min; 40 cycles of 95°C, 15 sec, and 60°C for 1 min. The level of ABL1 mRNA in the query sample was calculated relative to the calibrator standard by the comparative Ct method [22]. The calibrator contained 100 ng of cDNA of the Jurkat cell line at 99.0% viability. To compare the relative mean expression levels of ABL1 mRNA, we performed a one-way ANOVA and Student’s t-test (significance level, P < 0.05) for both tests.
Cell viability
C91/PL [23] and MT-2 [24] are HTLV-1-infected human T-cell lines, and Molt-4 [25] and Jurkat [26] are uninfected human T-cell lines. We also prepared PBMCs from a patient with HAM/TSP. Those cells were seeded in triplicate onto 96-well plates at a density of 5×103 cells/well. After 48 h cultivation in RPMI 1640 medium with various concentrations of imatinib, nilotinib, and dasatinib, viable cells were counted using XTT Cell Proliferation Kit II (Roche Diagnostics K.K., Tokyo, Japan). Cell viabilities were analyzed in a spectrophotometer and were estimated using the following formula:
PMA pretreatment of the PBMCs treated with TKIs
Cryopreserved PBMCs from six patients with HAM/TSP were thawed. The cell concentrations were counted by the trypan blue exclusion method. PBMCs at 95.0% viability or higher were adjusted to 1×106 cells/ml and incubated in 2 ml of RPMI 1640 with or without 500 nM nilotinib or 100 nM dasatinib in a 12-well polystyrene dish for 48 h in a 5% CO2 incubator. A control well was prepared for each PBMC sample in the same condition without TKI but with 0.006% dimethyl sulfoxide (DMSO), the concentration of which was the same as that in the TKI-treated wells. Each PBMC from the same patient with HAM/TSP were treated with the control (DMSO), nilotinib, or dasatinib. After 48 h, cells were harvested and divided into two. Each of the half samples was pretreated with 50 μM of propidium monoazide (PMA: 3-Amino-8-azido-5-{3-[diethyl(methyl)ammonio]propyl}-6-Phenylphenanthridinium) (Biotium Inc., Fremont, CA, U.S.A.) for 5 min, then photo-crosslinked by exposure to halogen lamp. Each of the other half samples was not pretreated. After that, genomic DNA from all the samples was extracted. PMA is a membrane-impermeable nucleic acid-intercalating fluorescent dye and forms covalent bond by halogen lamp exposure. In PCR with PMA pretreatment, DNA from dead or dying cells is intercalated and is not amplified. PMA pretreatment was performed to quantify tax gene numbers only in the live cells.
Conventional real-time PCR for quantitation of HTLV-1 PVLs
HTLV-1 carries the tax gene in the pX region of its genome [27]. Using 20 ng of DNA from samples not treated with PMA, we performed real-time qPCR to calculate the HTLV-1 PVL using TaqMan probes and primers for the tax gene and β-actin (ACTB, internal control) as previously described [16]. Real-time qPCR was performed in triplicate with a 25 μl volume for each measurement, using TaqMan Universal Master Mix II (Thermo Fisher Scientific KK.) and the StepOnePlus Real-time PCR system (Applied Biosystems). The cycle threshold was automatically defined by the thermal cycler. Thermal cycling involved one cycle of 50°C for 2 min and 95°C for 10 min; and 45 cycles of 95°C for 15 sec, and 60°C for 1 min. PVL was given as: PVL = (HTLV-1 tax amount)/((β-actin amount)/2)×104 (copies/104 cells)
PMA-HTLV-1 viability qPCR for PVL in live PBMCs (PVLlive)
A new technique, PMA viability PCR has been used to detect live microorganisms in the food and environmental sciences field [28–30]. The new PCR amplifies target in the DNA from live cells but not that from dead cells in the mixture condition. To evaluate the effects of the TKI drug candidates on the HTLV-1-infected cells, PBMCs from HAM/TSP patients were treated with or without the drugs, PMA pretreatment was done, and DNA was extracted from the cells. The novel qPCR assay was performed in the same way as conventional qPCR with standardization using standard curves for tax and β-actin, and we have tentatively designated it as ‘PMA-HTLV-1-viability qPCR’. Amplification was performed in triplicate using DNA (20 ng) from the PMA pretreated samples. The cycle threshold was automatically defined by the thermal cycler. The HTLV-1 PVL in live PBMCs (PVLlive) is given as:
ABL1 tyrosine kinase assay
Using ADP-Glo Kinase Assay Kit (Promega, Madison, WI., U.S.A) and ABL1 Kinase Enzyme System (Promega, Madison, WI., U.S.A) including Ablitide, a specific peptide substrate for ABL1 tyrosine kinase, ABL1 tyrosine kinase assay was performed according to the manufacturer’s protocol. For evaluating the effect of TKIs, cell lysate extracted after 48 h of incubation with TKIs (either of imatinib 5 μM, nilotinib 500 nM, or dasatinib 100 nM) was applied for this assay in a 96-well plate.
ABL1 knockdown by siRNA
We performed siRNA experiment for ABL1 mRNA in HTLV-1-infected and uninfected cell lines. ABL1 HSS100045 siRNA (Thermo Fisher Scientific K.K., Tokyo, Japan), mixture of Line 1 (5’-CCAACUACGACAAGUGGGAGAUGGA-3’) and Line 2 (5’-UCCAUCUCCCAUUUGUCGUAGUUGG-3’) target ABL1 mRNA variant α and β exon 4 sequence, very close region to the kinase domain of ABL1 tyrosine kinase. HTLV-1-infected cell lines (MT-2, HUT102, and C91/PL) and uninfected cell lines (Molt-4 and CEM) were transfected with either of siRNA, RNAi Negative Control (Thermo Fisher Scientific K.K., Tokyo, Japan), or blank control using 1 μl of Lipofectamine RNAiMAX (Thermo Fisher Scientific K.K., Tokyo, Japan) per 600 μl of Opti-MEM I Reduced Serum Medium (Thermo Fisher Scientific K.K., Tokyo, Japan). Cells treated with 50 pmol/ml of siRNA were plated at a concentration of 1.67×106 cells/ml. After incubation for 48 h at 37°C in a CO2 incubator, cells were harvested, and cell viabilities were evaluated by the trypan blue exclusion method. RNA and cell lysate were also prepared from the cells. Transfection efficiency was assayed by RT-PCR. Ten nanograms of cDNA was applied for RT-PCR using primers for ABL1 mRNA variant β (forward: 5’-AAATGGACTGCACCCGAGAG-3’ and reverse: 5’-CTCATGGTCCAGAGGATCGC-3’) and ACTB mRNA (forward: 5’-ACACTGTGCCCATCTACGAC-3’ and reverse: 5’-TGGCCATCTCTTGCTCGAA-3’). The gel image was analyzed by Image J software.
Statistics
All statistical analyses were performed using Statcel ver.4.0 (OMS Publishing, Tokorozawa, Japan) and GraphPad Prism 6J (GraphPad Software, San Diego, CA, U.S.A.).
Results
Specific cellular genes for HAM/TSP extracted by microarray analysis
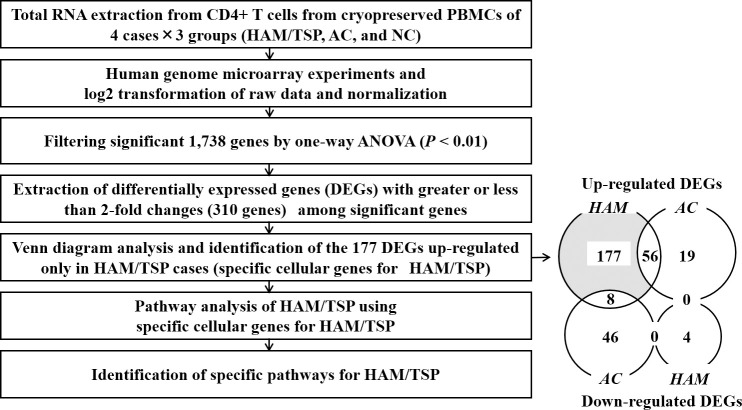
Through filtering and setting a fold change threshold of DEGs, we extracted 310 genes. By mapping to a Venn diagram, the DEGs were refined to the genes assigned to the area in the diagram where the up-regulated genes and the down-regulated genes belonging to only HAM/TSP. The refined genes were thought to be most important for HAM/TSP. We finally extracted significant 177 DEGs that were up-regulated only in HAM/TSP (Fig 1) and mapped to the shaded area and listed by their fold-change order in Table 2. Pseudogenes, discontinued or replaced genes, and unannotated genes were excluded from the list, resulting in information being available for 109 of the 177 DEGs. As shown in the down-regulated genes list of Table 2, among 4 down-regulated DEGs, 2 of them were not defined, and the remaining 2 were genes whose information was very limited. Therefore, we did not perform a subsequent investigation on these down-regulated genes.
Fig 1. Refinement of specific cellular genes for HAM/TSP.
To identify the specific cellular genes and pathways involved in the pathogenesis of HAM/TSP, we performed human genome microarray experiment and data processing according to the presented workflow. Each number in Venn’s diagram represents the number of genes that belong to each area. HAM/TSP: HTLV-1-associated myelopathy, AC: asymptomatic HTLV-1 carrier, NC: negative control.
Table 2. List of specific genes for HAM/TSP.
DEGs up-regulated only in HAM/TSP.
| Gene Symbol | Product | Accession number | Gene ID |
Relative expression (fold change) |
Function and localization |
| MUC3A | mucin 3A, cell-surface associated | NM_005960 | 4584 | 40.6 | Unknown; O-glycosylation in S/T/P repeat |
| NAF1 | nuclear assembly factor 1 homolog (S. cerevisiae) | NM_138386 | 92345 | 13.7 | RNA binding, protein binding |
| ZC3H12D | Zinc finger CCCH-type containing 12D | NM_207360 | 340152 | 11.34 | Endonuclease activity; regulates macrophage |
| POFUT2 | Protein O-fucosyltransferase 2 isoform A | NM_015227 | 23275 | 10.71 | O-glycosylation in S/T residues in EGF or thrombospondin motifs |
| C14orf72 | Chromosome 14 open reading frame | XM_096733 | 145200 | 9.73 | hypothetical |
| CKS2 | CDC28 protein kinase regulatory subunit 2 | NM_001827 | 1164 | 9.13 | Cell cycle, cell proliferation, spindle organization |
| SMC4 | Structural maintenance of chromosome 4 | NM_005496 | 10051 | 7.38 | ATP, nucleotide, protein binding, cell cycle, division |
| SOX21 | SRY (sex determining region Y)-box 21 | NM_007084 | 11166 | 6.83 | transcription factor activity |
| KLHL21 | kelch-like protein 21 (Drosophila) | NM_014851 | 9903 | 6.81 | Protein binding |
| RABGAP1L | RAB GTPase activating protein 1-like isoform A | NM_014857 | 9910 | 6.71 | GTPase activator activity; intracellular |
| ATP1B1 | Na+/K+ -ATPase beta 1 subunit isoform a | NM_001677 | 481 | 6.68 | K ion binding; plasma membrane protein |
| WDR41 | WD repeat domain 41 | NM_018268 | 55255 | 6.49 | unknown |
| TDP1 | tyrosyl-DNA phosphodiesterase 1 | NM_018319 | 55775 | 6.39 | DNA repair, binding |
| BCL2L13 | BCL2-like protein 13 (apoptosis facilitator) | NM_015367 | 23786 | 6.30 | Caspase activator activity |
| RPS6KA5 | ribosomal protein S6 kinase, 90kDa, polypeptide 5 | NM_004755 | 9252 | 6.27 | ATP binding; cytoplasm, nucleus |
| MAML1 | Mastermind-like [domain containing] 1 | NM_005491 | 9794 | 5.95 | Regulation of transcription; nucleus |
| RABEP1 | rabaptin, RAB GTPase binding effector protein 1 isoform 1 | NM_004703 | 9135 | 5.54 | membrane fusion, GTPase activator activity; cytoplasm, endosome |
| IL-6ST | IL-6 signal transducer isoform 1 precursor (gp130, oncostatin M receptor) | NM_002184 | 3572 | 5.32 | IL-6, -11, -27 receptor activity; positive regulation of T cell proliferation; plasma membrane |
| TMEM39A | Transmembrane protein 9A | NM_018266 | 55254 | 5.2 | plasma membrane; transmembrane protein |
| S100P | S100 calcium binding protein P | NM_005980 | 6286 | 5.06 | Calcium and magnesium ion binding; cytoplasm, nucleus |
| AFF4 | AFF/FMR2 family, member 4 | NM_014423 | 27125 | 4.86 | Regulation of transcription; nucleus |
| SLFN5 | schlafen family, member 5 | NM_144975 | 162394 | 4.83 | ATP, nucleotide binding; cell differentiation; nucleus |
| SOD2 | Superoxide dismutase 2, mitochondrial | NM_000636 | 6648 | 4.83 | Magnesium ion binding; nucleus |
| RILPL2 | Rab interacting lysosomal protein-like 2 | NM_145058 | 196383 | 4.76 | Identical protein binding; cytoplasm, cytosol |
| PMFBP1 | Polyamine modulated factor 1 (PMF-1) binding protein 1 | NM_031293 | 83449 | 4.63 | Unknown; cytoplasm |
| RAN | Ras related nuclear protein | NM_006325 | 5901 | 4.50 | GTP binding, GTPase activity, mitotic spindle organization; cytosol, nucleus |
| PWP1 | PWP1 homolog (S. cerevisiae), endonuclein | NM_007062 | 11137 | 4.32 | Transcription; nucleus |
| MED28 | Mediator complex subunit 28 | NM_025205 | 80306 | 4.29 | actin binding, protein binding, regulation of transcription; cytoplasm, membrane, nucleus |
| HES2 | HES2 (hairy and enhancer of split 2 (Drosophila)) protein, hes family bHLH transcription factor 2 | NM_019089 | 54626 | 4.17 | DNA binding, transcription regulator activity; nucleus |
| FAM133B | family with sequence similarity 133, member B(hypothetical protein LOC257415) | NM_152789 | 257415 | 4.14 | Unknown |
| RRN3 | RNA polymerase I-specific transcription initiation factor RRN3 (TIF-IA) | NM_018427 | 54700 | 4.06 | Regulation of transcription; nucleoplasm, nucleus |
| HERPUD1 | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 protein | NM_014685 | 9709 | 4.04 | protein binding, response to unfolded protein; ER |
| PRPS1L1 | phosphoribosyl pyrophosphate synthetase 1-like 1 | NM_175886 | 221823 | 3.86 | ATP, magnesium ion binding, kinase activity; cellular component |
| FBXO3 | F-box protein 3 | NM_012175 | 26273 | 3.81 | ubiquitin-protein ligase activity, proteolysis |
| PDCD2 | programmed cell death 2 isoform 1 | NM_002598 | 5134 | 3.75 | DNA, protein, zinc ion binding, apoptosis; cytoplasm, nucleus |
| SNAPC5 | small nuclear RNA activating complex, polypeptide 5, 19kDa subunit | NM_006049 | 10302 | 3.75 | Regulation of transcription; nucleus |
| BIRC3 | baculoviral IAP repeat-containing 3 (cIAP-2) | NM_001165 | 330 | 3.74 | ubiquitin-protein ligase activity, zinc ion binding, anti-apoptosis, signal transduction; cytoplasm, nucleus |
| ELN | elastin | NM_000501 | 2006 | 3.71 | extracellular matrix constituent; extracellular matrix |
| KRT1 | keratin 1 | NM_006121 | 3848 | 3.7 | Protein, sugar binding, receptor activity, cytoskeleton; plasma membrane |
| GALNT11 | UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) | NM_022087 | 63917 | 3.69 | polypeptide N-acetylgalactosaminyltransferase activity; Golgi |
| IKZF5 | IKAROS family zinc finger protein 5 | NM_022466 | 64376 | 3.65 | DNA, zinc ion binding; intracellular, nucleus or NOT nucleus |
| TBC1D23 | TBC1 domain family, member 23 | NM_018309 | 55773 | 3.57 | Rab GTPase activator activity; intracellular |
| MRPL41 | mitochondrial ribosomal protein L41 | NM_032477 | 64975 | 3.52 | structural constituent of ribosome, apoptosis, cell cycle, translation; mitochondria, ribosome |
| NEUROG 3 | neurogenin 3 (class A basic helix-loop-helix protein 7) | NM_020999 | 50674 | 3.49 | Nervous system development, positive regulation of |
| CIRH1A | cirrhosis, autosomal recessive 1A (cirhin) | NM_032830 | 84916 | 3.49 | Molecular function; nucleolar, nucleus |
| NDUFB3 | NADH dehydrogenase (ubiquinone) 1 beta | NM_002491 | 4709 | 3.39 | NADH dehydrogenase (ubiquinone) activity, mitochondrial electron transport; mitochondria |
| PLA2G4D | phospholipase A2, group IVD (cytosolic) | NM_178034 | 283748 | 3.37 | calcium ion binding, hydrolase activity, phospholipase A2 activity; Cytosol, cytoplasm |
| PTBP1 | polypyrimidine tract binding protein 1 | NM_002819 | 5725 | 3.36 | RNA splicing; nucleolous, nucleoplasm, nucleus |
| HAND1 | heart and neural crest derivatives expressed 1, bHLHa27 | NM_004821 | 9421 | 3.34 | protein heterodimerization activity, transcription; nucleus |
| GMCL1 | germ cell-less homolog 1 (Drosophila) | NM_178439 | 64395 | 3.34 | protein binding, cell differentiation; nucleus |
| FAT2 | FAT tumor suppressor homolog 2 (Drosophila) | NM_001447 | 2196 | 3.29 | calcium ion, protein binding, cell adhesion; plasma membrane |
| WARS2 | tryptophanyl tRNA synthetase 2, mitochondrial | NM_201263 | 10352 | 3.29 | ATP, nucleotide binding, tryptophan-tRNA ligase activity; cytoplasm, mitochondria |
| CDX4 | caudal type homeobox transcription factor 4 | NM_005193 | 1046 | 3.29 | Sequence-specific DNA binding, transcription factor; nucleus |
| TDG | thymine-DNA glycosylase | NM_003211 | 6996 | 3.27 | DNA N-glycosylase activity, damaged DNA, mismatched DNA binding; nucleus |
| CCDC85C | coiled-coil domain containing [protein] 85C | NM_001144995 | 317762 | 3.27 | Unknown |
| ABL1 | c-abl oncogene 1, receptor tyrosine kinase | NM_007313 | 25 | 3.26 | Apoptosis, cell cycle, cell adhesion, actin cytoskeleton organization, peptidyl-tyrosine phosphorylation, transcription, signal transduction; intracellular |
| PRLH | Prolactin releasing hormone | NM_015893 | 51052 | 3.25 | Hormone activity; cytoplasm, extracellular |
| OASL | 2'-5'-oligoadenylate synthetase-like isoform a | NM_003733 | 8638 | 3.24 | ATP, DNA, double-stranded RNA binding, thyroid hormone receptor binding, transferase activity; cytoplasm, nucleus |
| HCP5 | HLA complex P5 | NM_006674 | 10866 | 3.23 | Defense response; plasma membrane |
| CCT6A | chaperonin containing TCP1, subunit 6A isoform a | NM_001762 | 908 | 3.22 | ATP, nucleotide, unfolded protein binding; cytoplasm |
| SMAD4 | SMAD family member 4 | NM_005359 | 4089 | 3.16 | R-SMAD, collagen binding, BMP signaling; cytosol, cytoplasm, nucleoplasm, nucleus |
| SMN2 | survival of motor neuron 2, centromeric | NM_022877 | 6607 | 3.15 | RNA, protein binding, RNA, mRNA splicing, cell death; cytosol, nucleus |
| CCL13 | chemokine (C-C motif) ligand 13 | NM_005408 | 6357 | 3.14 | chemokine activity, signal transducer activity, cell-cell signaling, inflammatory response; plasma membrane |
| TRIM23 | tripartite motif-containing 23 (ADP-ribosylation factor domain protein 1 isoform alpha) | NM_001656 | 373 | 3.14 | GDP, GTP, nucleotide binding; Golgi, cytoplasm |
| PFKFB2 | 6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase 2 isoform a | NM_006212 | 5208 | 3.13 | ATP, nucleotide binding, 6-phophofrukto-2-kinase hydrolase, kinase, transferase activity; cytosol |
| ERN1 | ER to nucleus signaling 1 isoform 2 | NM_152461 | 2081 | 3.08 | Endonuclease activity, cell cycle arrest, mRNA processing, regulation of transcription; ER, nuclear inner membrane |
| GABPB2 | GA binding protein transcription factor, beta subunit 2 isoform beta 1 | NM_005254 | 126626 | 3.07 | protein binding, transcription factor activity; nucleus |
| RAPGEF1 | Rap guanine nucleotide exchange factor (GEF) 1 (guanine nucleotide-releasing factor 2 isoform b | NM_198679 | 2889 | 3.06 | SH3 domain binding, guanyl-nucleotide exchange factor activity, protein binding; cytosol, endosome, intracellular |
| YTHDC1 | YTH domain containing protein 1 | NM_133370 | 91746 | 3.04 | RNA splicing, mRNA processing; nucleus |
| MORF4L1 | mortality factor 4 like 1 | NM_206839 | 10933 | 3.03 | DNA repair, chromatin modification, histone H2A acetylation, histone H4 acetylation, histone deacetylation; nucleus |
| SOX3 | SRY (sex determining region Y)-box 3 | NM_005634 | 6658 | 3.02 | DNA binding; nucleus |
| SNRPG | small nuclear ribonucleoprotein polypeptide G | NM_003096 | 6637 | 2.99 | RNA binding, RNA splicing, protein binding; cytosol, nucleus |
| POLR2K | DNA directed RNA polymerase II polypeptide | NM_005034 | 5440 | 2.98 | DNA binding, DNA-directed RNA polymerase activity, zinc ion binding; nucleoplasm, nucleus |
| ZMAT2 | zinc finger, matrin type 2 | NM_144723 | 153527 | 2.97 | DNA binding, zinc ion binding; intracellular, nucleus |
| AATK | apoptosis-associated tyrosine kinase | AK131529 | 9625 | 2.95 | ATP, nucleotide binding, transferase; cytoplasm, mitochondria |
| TSPAN13 | Tetraspanin 13 (TM4SF13, NET-6) | NM_014399 | 27075 | 2.90 | Plasma membrane |
| TAAR5 | trace amine associated receptor 5 (putative neurotransmitter receptor) | NM_003967 | 9038 | 2.88 | G-protein coupled receptor activity; plasma membrane |
| TMEM126A | transmembrane protein 126A | NM_032273 | 84233 | 2.87 | Molecular function; plasma membrane, mitochondria |
| COMP | cartilage oligomeric matrix protein precursor | NM_000095 | 1311 | 2.85 | calcium ion binding, ECM structure, cell adhesion; extracellular |
| ULBP2 | UL16 binding protein 2 | NM_025217 | 80328 | 2.80 | MHC class I receptor, NK cell activation; plasma membrane |
| ADAM7 | a disintegrin and metalloproteinase domain | NM_003817 | 8756 | 2.76 | metalloendopeptidase activity, zinc ion binding, proteolysis; plasma membrane |
| HUS1 | HUS1 checkpoint protein | NM_004507 | 3364 | 2.75 | protein binding, protein serine/threonine kinase activity, DNA repair, cell cycle; cytoplasm, nucleus |
| SH3KBP1 | SH3-domain kinase binding protein 1 (Homo sapiens migration-inducing gene 18 protein) | NM_031892 | 30011 | 2.74 | SH3 domain binding, protein binding, apoptosis, cell-cell signaling, endocytosis, focal adhesion; plasma membrane |
| PPM1B | protein phosphatase, Mg2+/Mn2+ dependent, 1B | NM_002706 | 5495 | 2.7 | hydrolase activity, magnesium ion binding, protein binding, protein serine/threonine phosphatase activity |
| SMURF1 | SMAD specific E3 ubiquitin protein ligase 1 | NM_020429 | 57154 | 2.68 | ubiquitin-protein ligase activity; cytoplasm, plasma membrane |
| SMPX | small muscle protein, X-linked | NM_014332 | 23676 | 2.63 | striated muscle contraction; cytoplasm, nucleus |
| ADAM21 | ADAM metallopeptidase domain 21 preproprotein | NM_003813 | 8747 | 2.63 | zinc ion binding, metalloendopeptidase activity, proteolysis, single fertilization; membrane |
| IGFALS | insulin-like growth factor binding protein, acid labile subunit | NM_004970 | 3483 | 2.62 | IGF binding, protein C-terminus binding, cell adhesion; extracellular region, microsome, soluble fraction |
| NPM1 | nucleophosmin (nucleolar phosphoprotein B23, numatrin) 1 isoform 2 | NM_199185 | 4869 | 2.62 | NF-kappaB, Tat protein, RNA, histone binding, protein hetero- or homodimerization, anti-apoptosis, cell-aging |
| ZNF407 | zinc finger protein 407 (KIAA1703 protein) | NM_017757 | 55628 | 2.61 | DNA, zinc ion binding, transcription; intracellular, nucleus |
| HLA-E | MHC, class I, E precursor | NM_005516 | 3133 | 2.61 | MHC class I receptor activity, antigen processing and presentation; plasma membrane |
| MOBKL3 | Mps One Binder kinase activator-like 3 (yeast) (preimplantation protein 3 isoform 1) | NM_015387 | 25843 | 2.59 | zinc ion, protein binding; Golgi, cytoplasma, membrane |
| B9D2 | B9 protein domain 2 (hypothetical LOC80776) | NM_030578 | 80776 | 2.57 | cell projection organization; cilium, microtubule basal body, nucleus |
| PEX6 | peroxisomal biogenesis factor 6 | NM_000287 | 5190 | 2.54 | ATPase activity, ATP, nucleotide, protein C-terminus binding; cytoplasm, cytosol, peroxisome |
| YWHAZ | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | NM_145690 | 7534 | 2.52 | protein domain specific binding, transcription factor binding, anti- apoptosis; cytoplasm, mitochondria, nucleus |
| SUPV3L1 | suppressor of var1, 3-like 1 (S. cerevisiae) | NM_003171 | 6832 | 2.49 | ATP, DNA, RNA, nucleotide binding, DNA helicase activity, hydrolase activity, DNA duplex unwinding; nucleus, mitochondria |
| EIF2S1 | eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa | NM_004094 | 1965 | 2.48 | RNA, protein binding, translation initiation factor activity; cytoplasm, cytosol, nucleus |
| TUBA1B | tubulin, alpha 1b | NM_006082 | 10376 | 2.48 | GTP, protein binding, GTPase activity, microtubule cytoskeleton organization; microtubule |
| HNRPCL1 | heterogeneous nuclear ribonucleoprotein C-like 1 | NM_0010136 | 343069 | 2.48 | RNA binding, nucleotide binding; nucleus |
| POLB | polymerase (DNA directed), beta | NM_002690 | 5423 | 2.45 | DNA-directed DNA polymerase activity, damaged DNA binding, enzyme binding, lyase activity; cytoplasm, nucleoplasm, nucleus, spindle microtubule |
| SRPK3 | SFRS protein kinase 3 | NM_014370 | 26576 | 2.45 | ATP, nucleotide, protein binding, protein serine/threonine kinase activity, transferase activity; cellular component |
| TSPYL1 | TSPY-like 1 | NM_00309 | 7259 | 2.44 | Molecular function; intracellular, nucleolus, nucleus |
| RAI2 | retinoic acid induced 2 | NM_021785 | 10742 | 2.44 | Molecular function, embryonic development; cellular component |
| UQCR11 | ubiquinol-cytochrome c reductase, complex III subunit XI | NM_006830 | 10975 | 2.44 | electron carrier activity, ubiquinol-cytochrome-c reduction activity; mitochondria |
| POLDIP3 | polymerase (DNA-directed), delta interacting protein 3 | NM_032311 | 84271 | 2.41 | RNA binding, nucleotide binding, protein binding; nucleus |
| RPS14 | ribosomal protein S14 | NM_005617 | 6208 | 2.38 | RNA, mRNA 5'-UTR, protein binding, constituent of ribosome, translation regulator activity; cytosol, nucleolus, ribosome |
| SMCHD1 | structural maintenance of chromosomes flexible hinge domain containing 1 | NM_015295 | 23347 | 2.36 | ATP, protein binding; chromosome |
| HBS1L | HBS1(S. cerevisiae) -like translational GTPase | NM_006620 | 10767 | 2.32 | GTP, nucleotide binding, GTPase activity, translation elongation factor activity |
| SLC4A3 | solute carrier family 4, anion exchanger, member 3 | NM_001326559 | 6508 | 2.22 | anion transmembrane transporter activity; inorganic anion exchanger activity; transporter activity; plasma membrane |
| DEGs down-regulated only in HAM/TSP | |||||
| Gene Symbol | Product | Accession number | Gene ID |
Relative expression (fold change) |
Function and localization |
| N/A | ih08h09.x1 Human insulinoma Homo sapiens cDNA 3', mRNA sequence | BM310182.1 | N/A | 0.30 | Unknown. Transcribed locus from EST. |
| N/A | Homo sapiens cDNA FLJ37957 fis, clone CTONG2009 529 | AK095276.1 | N/A | 0.32 | Unknown. hypothetical protein LOC283588. |
| MAP7D3 | MAP7 domain containing 3 [Homo sapiens] | AL832120 | 79649 | 0.36 | Member of MAP7 (microtubule-associated protein 7) family. microtubule polymerization and stabilization; cancer growth and metastasis. |
| ARFGEF2 | ADP-ribosylation factor guanine nucleotide-exchange factor 2 [Homo sapiens] | NM_006420 | 10564 | 0.49 | Intracellular vesicular trafficking. activation of ARFs by accelerating replacement of bound GDP with GTP and is involved in Golgi transport; guanine-nucleotide exchange activity and also brefeldin A inhibition. |
The accession numbers are numbers of the longest variant as representative when there are multiple variants. N/A: not applicable.
ABL1 tyrosine kinase gene was suggested as an important gene for HAM/TSP
We extracted 12 significant pathways as specific pathways for HAM/TSP (Table 3). We obtained the frequently-appearing genes in the significant pathways. The ABL1 tyrosine kinase gene clearly stood out as being commonly included in 11 of the 12 pathways. However, its mRNA expression level was situated in the medium ranking of the up-regulated genes list (Table 2). Upstream analysis of the 177 DEGs did not converge to a few transcription factors (data not shown).
Table 3. Specific pathways for HAM/TSP.
| Pathway ID of TRANSPATHa database | Pathway nameb | Group sizec | Number of hits in groupd | Number of hits expectede | P-valuef |
|---|---|---|---|---|---|
| CH000003804 | ABL → TOPBP1 | 3 | 2 | 1 | 0.00021 |
| CH000000870 | ABL → Rad52 | 4 | 2 | 1 | 0.00043 |
| CH000000972 | ABL—/ Bcl-xL | 4 | 2 | 1 | 0.00043 |
| CH000003546 | ABL → Caspase-9 | 4 | 2 | 1 | 0.00043 |
| CH000000867 | ABL → p73α | 5 | 2 | 1 | 0.00071 |
| CH000000908 | Caspase-8 → ABL | 6 | 2 | 1 | 0.0011 |
| CH000000997 | Ubc9 —/ p73α | 7 | 2 | 1 | 0.0015 |
| CH000000895 | Fas → ABL | 10 | 2 | 1 | 0.0031 |
| CH000000977 | ABL → p53 | 11 | 2 | 1 | 0.0038 |
| CH000000869 | p73 pathway | 24 | 2 | 1 | 0.018 |
| CH000000711 | TGFβ pathway | 76 | 3 | 1 | 0.026 |
| CH000000879 | Caspase network | 93 | 3 | 1 | 0.044 |
a: TRANSPATH is a manually curated database of signal transduction pathways.
b: Pathway name and genes are described as in TRANSPATH. ABL: c-abl oncogene 1, non-receptor tyrosine kinase; TOPBP1: DNA topoisomerase II binding protein 1; RAD52: RAD52 (S. cerevisiae) homolog, DNA repair protein; TP73: tumor protein p73 isoform ɑ; Ubc9: UBE21 (ubiquitin-conjugating enzyme E2I). Symbols in pathway name represent type of reaction (e.g. →: facilitation; —/: inhibition).
c: Group size: the total number of genes, proteins, and metabolites which exist within the pathway of interest.
d: Number of hits in group: number of the cellular genes of interest (e.g. up-regulated DEGs only in HAM/TSP), which exist within the pathway chosen by a statistical test.
e: Number of hits expected: number of the cellular genes of interest that are included in the pathway chosen by statistical test by chance.
f. P-value of two-tailed Fisher’s exact test.
Verification of up-regulation of ABL1 mRNA by quantitative RT-PCR
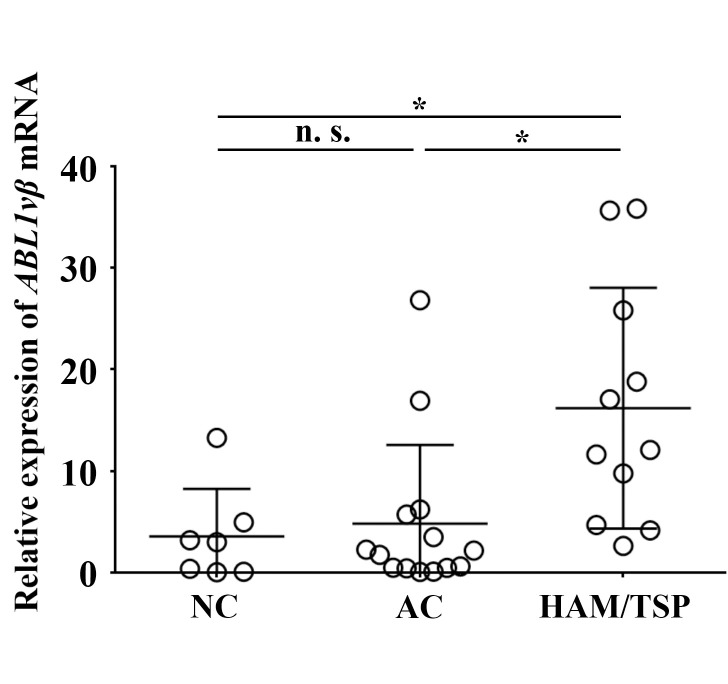
To verify the microarray results where the ABL1 mRNA expression level in CD4+ T cells in the HAM/TSP cases was up-regulated in comparison with ACs or NCs, we performed qRT-PCR. The relative expression level of ABL1 mRNA was significantly higher in HAM/TSP patients than in NCs (P = 0.013) and ACs (P = 0.029) by Student’s t-test (Fig 2).
Fig 2. ABL1 gene expression in CD4+ T cells by quantitative RT-PCR.
The total RNA extracted from CD4+ T cells from the subjects including NC (N = 7), AC (N = 14), and HAM/TSP (N = 11) was reverse-transcribed. The relative expression levels of ABL1vβ mRNA were calculated by the comparative Ct method, and 100 ng cDNA of Jurkat cell line was used for the calibrator gene. GAPDH mRNA was used as an internal control. The relative mean expression level of ABL1 mRNA was 3.56 for NC, 4.83 for AC, and 16.19 for HAM/TSP group. Student’s t-test was used. Bars represent mean ± standard deviation (SD). *: significant at P < 0.05. n.s.: not significant.
The effect of TKI treatment on cell viability of HTLV-1-infected cell lines and PBMCs from HAM/TSP patients
To investigate whether inhibition of ABL1 tyrosine kinase has an effect on HTLV-1-infected or uninfected cell lines and PBMCs from HAM/TSP cases, we performed an XTT assay (Fig 3). Both HTLV-1 uninfected cell lines (Jurkat and Molt-4) and HTLV-1-infected cell lines (C91/PL and MT-2) were similarly viable when the cells were treated with various concentrations of imatinib (Fig 3A). However, the HTLV-1-infected cell lines had relatively low viability over a wide range of nilotinib or dasatinib concentrations (Fig 3B and 3C), suggesting that both inhibitors more selectively induced cell death for HTLV-1-infected cells compared with imatinib. Therefore, we chose nilotinib and dasatinib for the next XTT assay to explore the appropriate concentration of each TKI for PBMCs from a patient with HAM/TSP. We performed this assay with concentrations up to 5 μM (Fig 3D), which was the maximum concentration where 100% of the uninfected cell lines survived (Fig 3B and 3C). We estimated 100 nM and 500 nM to be the optimal concentrations at which the PBMCs remained viable at more than 80% for dasatinib and nilotinib, respectively (Fig 3D). Using these data, the IC50 of TKIs for each cell line in the XTT assay was calculated (S1 Table).
Fig 3. Viability of cell lines and HAM/TSP PBMC treated with ABL1 tyrosine kinase inhibitors (TKIs).
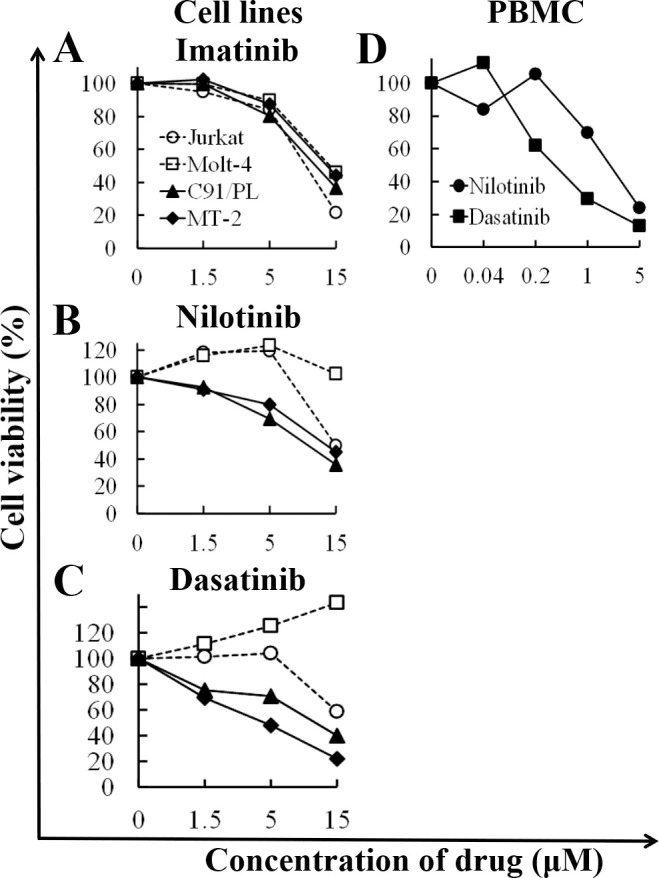
HTLV-1-uninfected cell lines (Jurkat and Molt-4), HTLV-1-infected cell lines (C91/PL and MT-2), and PBMC from a HAM/TSP patient were cultured with various concentrations of TKIs for 48 h. Cell viability was measured by an XTT assay in triplicate. A: viability of cell lines treated with imatinib. B: viability of cell lines treated with nilotinib. C: viability of cell lines treated with dasatinib. D: viability of HAM/TSP PBMC treated with TKIs (nilotinib or dasatinib). X- and y-axis show the concentration of TKIs (μM) and cell viability (%), respectively. Cell viability (%) represents the live cell number treated with TKIs relative to that without treatment. Open circle: Jurkat; open square: Molt-4; solid triangle: C91/PL; solid diamond: MT-2; solid circle: nilotinib; and solid square: dasatinib.
TKIs reduced HTLV-1 PVL in live PBMCs from patients with HAM/TSP
To test whether ABL1 is an important gene involved survival of HTLV-1-infected cells, we investigated PVL after the treatment of TKIs. By the conventional qPCR, there was no significant difference in the PVL with or without TKI treatments (Fig 4A). Conventional qPCR has a possibility to amplify DNA from dead cells [31]. Therefore, we developed PMA–HTLV-1 viability qPCR to distinguish live and dead cells and evaluated the effect of drugs on HTLV-1-infected cells. To validate PMA-HTLV-1 viability qPCR, we performed this method and conventional qPCR for PVL using mixtures of HUT102 live and dead cells in various ratios (S1 Fig). The Ct values of the two methods were completely coincident for 100% live cells. However, the difference of the Ct values expanded significantly with the increase of dead cell percentage. The result suggested that PMA-HTLV-1 viability PCR was more sensitive for HTLV-1-infected cell death than conventional qPCR. Using this assay, the relative percentage of PVLlive (%) showed a significant reduction of 17.5% and 21.0% after incubation with either dasatinib or nilotinib, respectively (Fig 4B).
Fig 4. The effect of TKIs to PVL in total PBMC or in live PBMC from HAM /TSP patients.
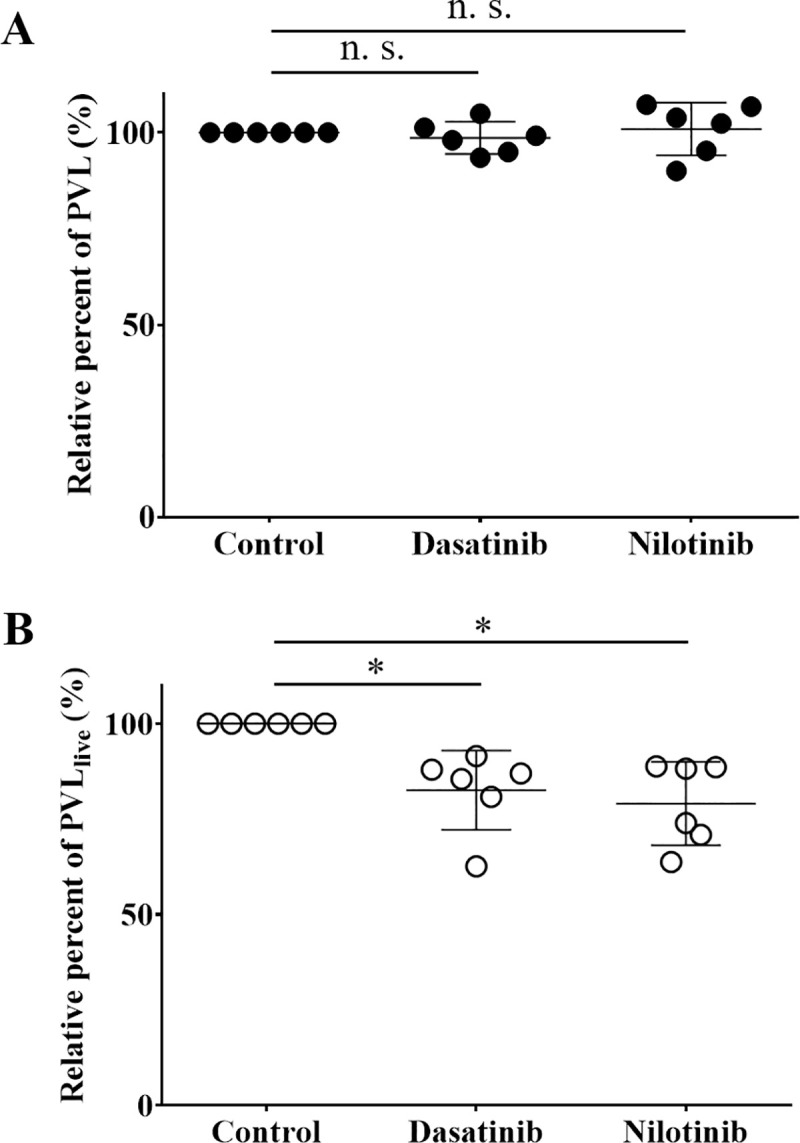
PBMC from patients with HAM/TSP were cultured with 0.006% dimethyl sulfoxide (control), 100 nM dasatinib, or 500 nM nilotinib for 48 h and were harvested. Genomic DNA was extracted from the cells and then subjected to both conventional qPCR for PVL (A) and PMA-HTLV-1 viability qPCR (B). The former counts PVL in total PBMC (a mixture of live and dead cells), while the latter enables the quantification of the PVL only in live PBMC (PVLlive). A: conventional qPCR for PVL. Solid circles represent PVL relative to that of the control, and bars represent means and SD. There was no significant difference in the relative PVL in the cells treated with or without TKI. B: PMA-HTLV-1 viability qPCR to quantify PVLs only in live cells (PVLlive). Open circles represent PVLlive relative to that of the control, and bars represent means and SD. The mean PVLlive of samples treated with dasatinib or nilotinib were significantly reduced at 17.5% and 21.0% relative to the control, respectively. Student’s t-test was used (N = 6). *: significant at P < 0.05; n.s.: not significant.
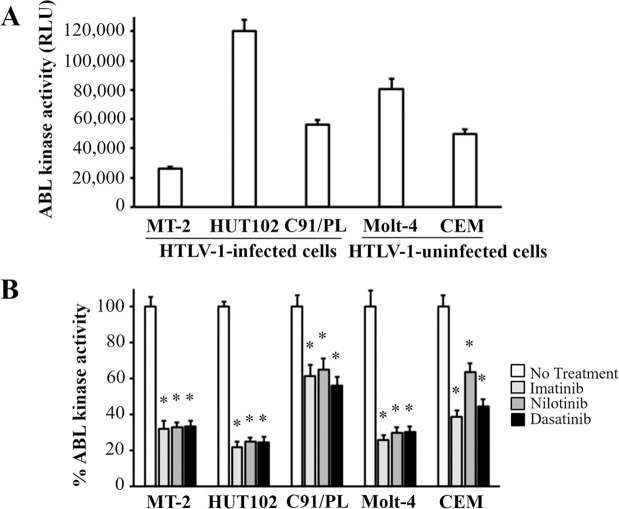
TKIs suppressed ABL1 tyrosine kinase activity in both HTLV-1-infected or uninfected cell lines
At first, we measured the kinase activities in HTLV-1-infected and uninfected cell lines and found that the kinase activities varied among cell lines (Fig 5A). Next, we investigated the effect of TKIs on kinase activities. We found that ABL1 tyrosine kinase activities in all cell lines treated with TKIs were reduced compared with those of cell lines without treatment (Fig 5B).
Fig 5. Effect of TKIs on ABL1 tyrosine kinase activity of HTLV-1-infected or uninfected cell lines.
A: Basal level of ABL1 tyrosine kinase activity of HTLV-1-infected or uninfected cell lines. B: Relative ABL1 tyrosine kinase activity (%) of cells treated with or without TKIs for 48 h. Error bars represent SD. Asterisks in panel B indicate statistically significant (P < 0.05, paired t-test) when compared with that of the cells without treatment (no treatment).
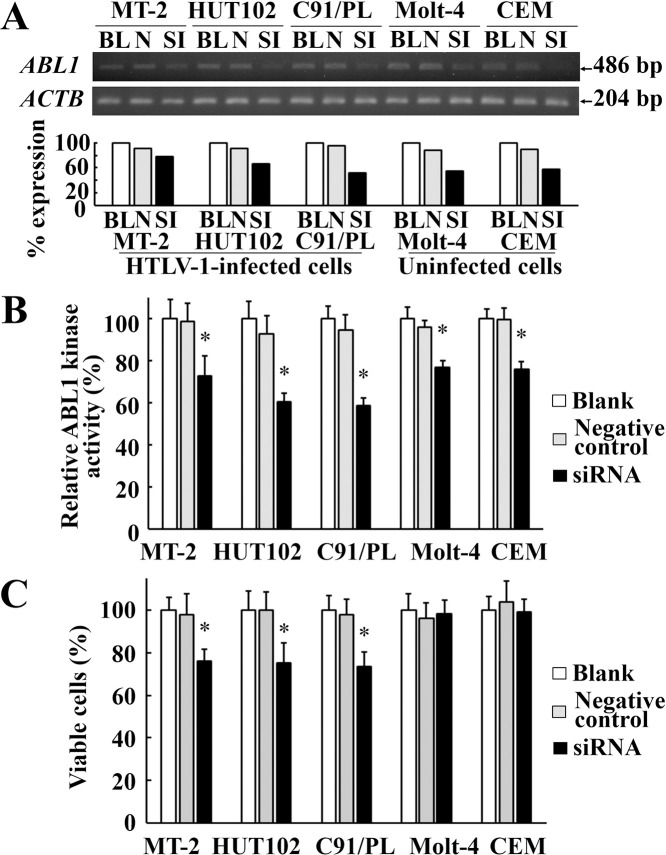
Knockdown of ABL1 mRNA by siRNA reduced cell viability in HTLV-1-infected cell lines
To investigate whether TKIs really target ABL1 tyrosine kinase and cause a reduction in HTLV-1 PVL, we performed a siRNA experiment targeting the kinase region of ABL1. After ABL1 siRNA transfection, ABL1 expression was reduced in HTLV-1-infected cell lines (MT-2, HUT102, and C91/PL) as well as in uninfected cell lines (Molt-4 and CEM) (Fig 6A), and ABL1 tyrosine kinase activities were also reduced in all cell lines (Fig 6B). In a cell viability assay, viable cells were significantly reduced in HTLV-1-infected cell lines, but not in the uninfected cell lines (Fig 6C).
Fig 6. ABL1 mRNA knockdown by siRNA in HTLV-1-infected or uninfected cell lines.
A: RT-PCR of HTLV-1-infected or uninfected cell lines after ABL1 siRNA transfection. Upper and lower gel images represent amplicon of ABL1 mRNA and ACTB mRNA, respectively. The bar graph under the gel images indicates relative expression of ABL1 mRNA standardized by ACTB mRNA. BL, N, and SI represent blank control, negative control siRNA, and ABL1 siRNA, respectively. B: Relative ABL1 tyrosine kinase activities of HTLV-1-infected or uninfected cell lines after ABL1 siRNA transfection. Grey and solid bars indicate the relative ABL1 kinase activities in each cell line treated with negative control siRNA or ABL1 siRNA to those without siRNA treatment (white bars), respectively. Error bars represent SD. Asterisks indicate statistically significant (P < 0.05, paired t-test) when compared with those treated with negative control siRNA (gray bars). C: Cell viabilities of HTLV-1-infected or uninfected cell lines after ABL1 siRNA transfection. Grey and solid bars represent the relative percent of viable cells in each cell line treated with negative control siRNA or ABL1 siRNA to those without siRNA treatment (white bars), respectively. Error bars represent SD. Asterisks indicate statistically significant (P < 0.05, paired t-test) when compared with those treated with negative control siRNA (gray bars).
Presentation of a HAM/TSP case complicated by CML that was treated with imatinib
As a retrospective survey based on our clinical records, we searched for HTLV-1-positive CML cases where the PVL was estimated before and after the treatment of the CML with a TKI. We found only one case who had both CML and HAM/TSP. The patient was a 52-year-old male diagnosed as having HAM/TSP. Osame’s Motor Disability Score (OMDS) was 6/13 for this patient (i.e., able to walk more than 10 m with a cane and holding on to something) [32]. After prednisolone administration (5 mg/day), the OMDS became 4/13 (i.e., walking without a cane but a handrail is necessary when climbing steps). Eight years later, he was diagnosed with CML because of leukocytosis, a Philadelphia-positive chromosome, and a low neutrophil alkaline phosphatase score. On admission, the OMDS was 6/13, the PVL was 2,844 copies/104 in PBMCs, and neopterin in the cerebral spinal fluid was 86.2 nmol/ml (the normal value is ≦30). He was administered imatinib (400 mg/day) for CML treatment for the first month, followed by 300 mg/day for 30 months. After imatinib treatment, he showed molecular genetics-based remission of the CML. Additionally, the OMDS improved from 6/13 to 5/13 (i.e., he was able to walk while holding on to something). Likewise, the PVL decreased from 2,844 to 448 copies/104 PBMCs (an 84.2% reduction). Severe adverse effects by the imatinib treatment were not seen during the whole treatment period.
Discussion
We identified the ABL1 tyrosine kinase gene as being important in HAM/TSP. The pathway analysis revealed that ABL1 gene was involved in many of the relevant pathways for HAM/TSP specific genes extracted by the microarray data. The ABL1 expression level was relatively high in the CD4+ T cells from HAM/TSP patients and ranged in the list for the specific genes for HAM/TSP (Table 2). The finding of the up-regulation of ABL1 mRNA in HTLV-1-infected cells is supported by a study of other researchers [33]. In addition, we showed that ABL1 tyrosine kinase inhibitors, dasatinib and nilotinib, reduced HTLV-1 PVL in cultured PBMC from the HAM/TSP patients (Fig 4B). In cell viability assay, the TKIs preferentially reduced the viability of HTLV-1-infected cell lines rather than uninfected cell lines (Fig 3A–3C). The addition of TKIs induced the significant reduction in ABL1 kinase activity in both of HTLV-1-infected or uninfected cell lines (Fig 5B). At the same time, siRNA experiment revealed that the knockdown of ABL1 mRNA induced cell death in HTLV-1-infected cell lines, but not in uninfected cell lines (Fig 6C). These data suggest that ABL1 tyrosine kinase inhibitors reduced PVL and preferentially induced cell death in HTLV-1-infected cells. These data are consistent with the clinical case, who was diagnosed CML in the clinical course of HAM/TSP and showed an 84.2% reduction in PVL after imatinib treatment for CML. These in vitro and clinical data suggest that ABL1 tyrosine kinase inhibitors may be therapeutic agents for HAM/TSP.
HAM/TSP patients show higher PVLs compared with ACs [16]. PVL is known to be associated with disease exacerbation [34–36], rapid disease progression, and many aspects of the central nerve system (CNS) inflammation in HAM/TSP [8–10]. These findings strongly indicate that PVL reduction is crucial for improving the neurological dysfunctions in patients with HAM/TSP. For example, treatments with interferon-α or anti-CCR4 antibody are reported to reduce PVLs somewhat [18, 37]. Therefore, another treatment to reduce the PVL, such as TKIs shown in this study, is needed for HAM/TSP.
The three TKIs used in the present study reduced the ABL1 kinase activity in all tested cell lines (Fig 5B). Imatinib reduced cell viability in both HTLV-1-infected (C91/PL and MT-2) and uninfected cell lines (Jurkat and Molt-4), while nilotinib and dasatinib showed a relatively specific reduction of cell viability for HTLV-1-infected cells (Fig 3). These results were similar to previous findings on in vitro activity of TKIs, in which nilotinib and dasatinib are more sensitive for ABL1 tyrosine kinase activity compared to imatinib [38]. Nilotinib and dasatinib are blood-brain barrier-permeable, but imatinib is impermeable [39–41]. As a treatment for HAM/TSP, nilotinib and dasatinib may be more appropriate therapeutic agents than imatinib for inducing death of HTLV-1-infected cells infiltrating the spinal cord, which may result in reducing inflammation in the CNS.
TKIs reduced cell viability of HTLV-1infected cell lines (Fig 3) and PVL in PBMC from HAM/TSP patients (Fig 4B). However, it was unclear whether these effects occurred via specific inhibition of ABL1 tyrosine kinase. In our study, the TKIs reduced the ABL1 kinase activities in HTLV-1-infected or uninfected T cell lines (Fig 5B). In addition, in the ABL1 siRNA experiment targeting ABL1 tyrosine kinase, cell viabilities were significantly reduced in the HTLV-1-infected cell lines, but not in the uninfected cell lines (Fig 6C). Although we cannot completely deny the possibility that death in HTLV-1-infected cells occurred via the off-target effect of the TKIs, these results suggest that TKIs targeted ABL1 tyrosine kinase and induced cell death in HTLV-1-infected cells which showed the increased expression of ABL1 gene.
QPCR for PVL quantitation is a standard method for estimating HTLV-1 provirus in genomic DNA from PBMC. However, when cells are dead during a short incubation with a drug, it was known that DNA extracted from the dead cells could be amplified by the qPCR [31]. In our experiment, as shown in Fig 4A, conventional PCR did not show any inhibitory effects of TKIs on PVL in cultured PBMC from HAM/TSP patients. Therefore, we developed a novel technique, PMA-HTLV-1 viability qPCR, with PMA pretreatment before amplification. Our preliminary experiment for this method showed apparent differences in Ct values, depending on the ratio of dead cells in samples (S1 Fig). When the ratio of dead cells was increased, the Ct value in the case with PMA treatment became high compared to that without PMA treatment, suggesting that the treatment made it possible to evaluate PVL in live cells more sensitive. By applying the method to clinical samples with TKI treatment, we could more accurately evaluate PVL in live cells than the conventional qPCR and found that the TKIs reduced PVL (Fig 4B). Of course, there is a limitation that the PMA-HTLV-1 viability qPCR amplified some of DNA even from 100% dead cells, to a lesser extent compared to the conventional qPCR (S1 Fig). Our data suggest that PMA-HTLV-1 viability qPCR can be expanded to an application that quantifies another target in the mixture of live and dead cells.
Conclusions
We identified the ABL1 tyrosine kinase gene as being important in HAM/TSP by combining microarray and pathway analysis. The addition of the ABL1 tyrosine kinase inhibitors, nilotinib, and dasatinib, into the PBMC culture significantly reduced the PVL in live PBMCs by 21.0% and 17.5%, respectively. The knockdown of ABL1 mRNA induced cell death in HTLV-1-infected cell lines. We also found a HAM/TSP case who was complicated by CML. He showed an 84.2% reduction in PVL after imatinib treatment. These data suggest that inhibiting ABL1 tyrosine kinase by TKI can reduce HTLV-1 PVL in patients with HAM/TSP.
Ethics statement
The Ethics Committee of Kagoshima University Hospital, Japan, approved this study. All human subjects were adults except one. We obtained written informed consent from all subjects. We explained to the child about blood sampling and obtained written informed consent from him with his parents. This report does not contain any individual data.
Supporting information
(DOCX)
To evaluate whether the mixture of live and dead cells and PMA pretreatment affect HTLV-1 PVL measurement, we induced dead cells by digitonin treatment at 300 ng/ml for 2 h and made five different cell mixture (percent ratios of live /dead cells were 100/0, 75/25, 50/50, 25/75, and 0/100) using HTLV-1-infected cell line HUT102. We divided these cell mixtures into two groups: one was pretreated with PMA (which enters only dead cells and fix the DNA), and the other was not. Genomic DNA was extracted and we performed quantitative PCR targeting HTLV-1 pX gene in triplicate and repeated twice. Error bars represent standard error of the mean.
(TIF)
Acknowledgments
We wish to thank Joint Research Laboratory, Kagoshima University Graduate School of Medical and Dental Sciences, for the use of their facilities. We also thank Sandra Cheesman, Ph.D., from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Data Availability
The raw data of microarray experiments were deposited and available from the Gene Expression Ominibus (GEO) repository at National Center for Biotechnology Information (NCBI) (GEO DataSets Series accession number: GSE132666) located at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132666.
Funding Statement
This work was supported by Grant-in-Aid for Scientific Research of Ministry of Health, Labour, and Welfare, 201231013B was awarded to Shuji Izumo. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, et al. (1980) Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci U S A 77: 7415–7419. 10.1073/pnas.77.12.7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakao K, Ohba N (1993) Clinical features of HTLV-I associated uveitis. Br J Ophthalmol 77: 274–279. 10.1136/bjo.77.5.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H (1977) Adult T-cell leukemia: clinical and hematologic features of 16 cases. Blood 50: 481–492. 10.1182/blood-2016-10-746677 [DOI] [PubMed] [Google Scholar]
- 4.Gessain A, Barin F, Vernant JC, Gout O, Maurs L, et al. (1985) Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet 2: 407–410. 10.1016/s0140-6736(85)92734-5 [DOI] [PubMed] [Google Scholar]
- 5.Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, et al. (1986) HTLV-I associated myelopathy, a new clinical entity. Lancet 1: 1031–1032. 10.1016/S0140-6736(86)91298-5 [DOI] [PubMed] [Google Scholar]
- 6.Giam CZ, Jeang KT (2007) HTLV-1 Tax and adult T-cell leukemia. Front Biosci 12: 1496–1507. 10.2741/2163 [DOI] [PubMed] [Google Scholar]
- 7.Saito M, Matsuzaki T, Satou Y, Yasunaga J, Saito K, et al. (2009) In vivo expression of the HBZ gene of HTLV-1 correlates with proviral load, inflammatory markers and disease severity in HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP). Retrovirology 6: 19 10.1186/1742-4690-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moritoyo T, Izumo S, Moritoyo H, Tanaka Y, Kiyomatsu Y, et al. (1999) Detection of human T-lymphotropic virus type I p40tax protein in cerebrospinal fluid cells from patients with human T-lymphotropic virus type I-associated myelopathy/tropical spastic paraparesis. J Neurovirol 5: 241–248. 10.3109/13550289909015810 [DOI] [PubMed] [Google Scholar]
- 9.Matsuoka E, Takenouchi N, Hashimoto K, Kashio N, Moritoyo T, et al. (1998) Perivascular T cells are infected with HTLV-I in the spinal cord lesions with HTLV-I-associated myelopathy/tropical spastic paraparesis: double staining of immunohistochemistry and polymerase chain reaction in situ hybridization. Acta Neuropathol 96: 340–346. 10.1007/s004010050903 [DOI] [PubMed] [Google Scholar]
- 10.Matsuura E, Kubota R, Tanaka Y, Takashima H, Izumo S (2015) Visualization of HTLV-1-specific cytotoxic T lymphocytes in the spinal cords of patients with HTLV-1-associated myelopathy/tropical spastic paraparesis. J Neuropathol Exp Neurol 74: 2–14. 10.1097/NEN.0000000000000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kira J, Koyanagi Y, Yamada T, Itoyama Y, Tateishi J, et al. (1994) Sequence heterogeneity of HTLV-I proviral DNA in the central nervous system of patients with HTLV-I-associated myelopathy. Ann Neurol 36: 149–156. 10.1002/ana.410360206 [DOI] [PubMed] [Google Scholar]
- 12.Furukawa Y, Yamashita M, Usuku K, Izumo S, Nakagawa M, et al. (2000) Phylogenetic subgroups of human T cell lymphotropic virus (HTLV) type I in the tax gene and their association with different risks for HTLV-I-associated myelopathy/tropical spastic paraparesis. J Infect Dis 182: 1343–1349. 10.1086/315897 [DOI] [PubMed] [Google Scholar]
- 13.Jeffery KJ, Usuku K, Hall SE, Matsumoto W, Taylor GP, et al. (1999) HLA alleles determine human T-lymphotropic virus-I (HTLV-I) proviral load and the risk of HTLV-I-associated myelopathy. Proc Natl Acad Sci U S A 96: 3848–3853. 10.1073/pnas.96.7.3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kodama D, Saito M, Matsumoto W, Sabouri AH, Izumo S, et al. (2004) Longer dinucleotide repeat polymorphism in matrix metalloproteinase-9 (MMP-9) gene promoter which correlates with higher HTLV-I Tax mediated transcriptional activity influences the risk of HTLV-I associated myelopathy/tropical spastic paraparesis (HAM/TSP). J Neuroimmunol 156: 188–194. 10.1016/j.jneuroim.2004.07.007 [DOI] [PubMed] [Google Scholar]
- 15.Sabouri AH, Saito M, Lloyd AL, Vine AM, Witkover AW, et al. (2004) Polymorphism in the interleukin-10 promoter affects both provirus load and the risk of human T lymphotropic virus type I-associated myelopathy/tropical spastic paraparesis. J Infect Dis 190: 1279–1285. 10.1086/423942 [DOI] [PubMed] [Google Scholar]
- 16.Nagai M, Usuku K, Matsumoto W, Kodama D, Takenouchi N, et al. (1998) Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J Neurovirol 4: 586–593. 10.3109/13550289809114225 [DOI] [PubMed] [Google Scholar]
- 17.Kamihira S, Dateki N, Sugahara K, Hayashi T, Harasawa H, et al. (2003) Significance of HTLV-1 proviral load quantification by real-time PCR as a surrogate marker for HTLV-1-infected cell count. Clin Lab Haematol 25: 111–117. 10.1046/j.1365-2257.2003.00503.x [DOI] [PubMed] [Google Scholar]
- 18.Sato T, Coler-Reilly ALG, Yagishita N, Araya N, Inoue E, et al. (2018) Mogamulizumab (Anti-CCR4) in HTLV-1-Associated Myelopathy. N Engl J Med 378: 529–538. 10.1056/NEJMoa1704827 [DOI] [PubMed] [Google Scholar]
- 19.Ikeda M, Fujino R, Matsui T, Yoshida T, Komoda H, et al. (1984) A new agglutination test for serum antibodies to adult T-cell leukemia virus. Gan 75: 845–848. [PubMed] [Google Scholar]
- 20.Petersen D, Chandramouli GV, Geoghegan J, Hilburn J, Paarlberg J, et al. (2005) Three microarray platforms: an analysis of their concordance in profiling gene expression. BMC Genomics 6: 63 10.1186/1471-2164-6-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wingender E, Dietze P, Karas H, Knuppel R (1996) TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res 24: 238–241. 10.1093/nar/24.1.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H (2006) Quantitative real-time RT-PCR data analysis: current concepts and the novel "gene expression's CT difference" formula. J Mol Med (Berl) 84: 901–910. 10.1007/s00109-006-0097-6 [DOI] [PubMed] [Google Scholar]
- 23.Popovic M, Sarin PS, Robert-Gurroff M, Kalyanaraman VS, Mann D, et al. (1983) Isolation and transmission of human retrovirus (human t-cell leukemia virus). Science 219: 856–859. 10.1126/science.6600519 [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi I, Kubonishi I, Yoshimoto S, Akagi T, Ohtsuki Y, et al. (1981) Type C virus particles in a cord T-cell line derived by co-cultivating normal human cord leukocytes and human leukaemic T cells. Nature 294: 770–771. 10.1038/294770a0 [DOI] [PubMed] [Google Scholar]
- 25.Drexler HG, Gaedicke G, Minowada J (1987) T-leukemia cell lines CCRF-CEM, HPB-ALL, JM and MOLT-4: changes in isoenzyme profiles during induction of differentiation. Blut 54: 79–87. 10.1007/BF00321034 [DOI] [PubMed] [Google Scholar]
- 26.Schneider U, Schwenk HU, Bornkamm G (1977) Characterization of EBV-genome negative "null" and "T" cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int J Cancer 19: 621–626. 10.1002/ijc.2910190505 [DOI] [PubMed] [Google Scholar]
- 27.Seiki M, Hattori S, Hirayama Y, Yoshida M (1983) Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci U S A 80: 3618–3622. 10.1073/pnas.80.12.3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nocker A, Sossa KE, Camper AK (2007) Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J Microbiol Methods 70: 252–260. 10.1016/j.mimet.2007.04.014 [DOI] [PubMed] [Google Scholar]
- 29.Nocker A, Sossa-Fernandez P, Burr MD, Camper AK (2007) Use of propidium monoazide for live/dead distinction in microbial ecology. Appl Environ Microbiol 73: 5111–5117. 10.1128/AEM.02987-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Contreras PJ, Urrutia H, Sossa K, Nocker A (2011) Effect of PCR amplicon length on suppressing signals from membrane-compromised cells by propidium monoazide treatment. J Microbiol Methods 87: 89–95. 10.1016/j.mimet.2011.07.016 [DOI] [PubMed] [Google Scholar]
- 31.Choi JJ, Reich CF 3rd, Pisetsky DS (2004) Release of DNA from dead and dying lymphocyte and monocyte cell lines in vitro. Scand J Immunol 60: 159–166. 10.1111/j.0300-9475.2004.01470.x [DOI] [PubMed] [Google Scholar]
- 32.Osame M (1990) Review of WHO Kagoshima meeting and diagnostic guidelines for HAM/TSP In: Human retrovirology: HTLV (ed. Blattner W), pp. 191–197, Raven Press, New York. [Google Scholar]
- 33.de La Fuente C, Deng L, Santiago F, Arce L, Wang L, et al. (2000) Gene expression array of HTLV type 1-infected T cells: Up-regulation of transcription factors and cell cycle genes. AIDS Res Hum Retroviruses 16: 1695–1700. 10.1089/08892220050193164 [DOI] [PubMed] [Google Scholar]
- 34.Kubota R, Nagai M, Kawanishi T, Osame M, Jacobson S (2000) Increased HTLV type 1 tax specific CD8+ cells in HTLV type 1-asociated myelopathy/tropical spastic paraparesis: correlation with HTLV type 1 proviral load. AIDS Res Hum Retroviruses 16: 1705–1709. 10.1089/08892220050193182 [DOI] [PubMed] [Google Scholar]
- 35.Takenouchi N, Yamano Y, Usuku K, Osame M, Izumo S (2003) Usefulness of proviral load measurement for monitoring of disease activity in individual patients with human T-lymphotropic virus type I-associated myelopathy/tropical spastic paraparesis. J Neurovirol 9: 29–35. 10.1080/13550280390173418 [DOI] [PubMed] [Google Scholar]
- 36.Hayashi D, Kubota R, Takenouchi N, Nakamura T, Umehara F, et al. (2008) Accumulation of human T-lymphotropic virus type I (HTLV-I)-infected cells in the cerebrospinal fluid during the exacerbation of HTLV-I-associated myelopathy. J Neurovirol 14: 459–463. 10.1080/13550280802178538 [DOI] [PubMed] [Google Scholar]
- 37.Saito M, Nakagawa M, Kaseda S, Matsuzaki T, Jonosono M, et al. (2004) Decreased human T lymphotropic virus type I (HTLV-I) provirus load and alteration in T cell phenotype after interferon-alpha therapy for HTLV-I-associated myelopathy/tropical spastic paraparesis. J Infect Dis 189: 29–40. 10.1086/380101 [DOI] [PubMed] [Google Scholar]
- 38.O'Hare T, Walters DK, Stoffregen EP, Jia T, Manley PW, et al. (2005) In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res 65: 4500–4505. 10.1158/0008-5472.CAN-05-0259 [DOI] [PubMed] [Google Scholar]
- 39.Senior K (2003) Gleevec does not cross blood-brain barrier. Lancet Oncol 4: 198 10.1016/s1470-2045(03)01050-7 [DOI] [PubMed] [Google Scholar]
- 40.Karuppagounder SS, Brahmachari S, Lee Y, Dawson VL, Dawson TM, et al. (2014) The c-Abl inhibitor, nilotinib, protects dopaminergic neurons in a preclinical animal model of Parkinson's disease. Sci Rep 4: 4874 10.1038/srep04874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porkka K, Koskenvesa P, Lundan T, Rimpilainen J, Mustjoki S, et al. (2008) Dasatinib crosses the blood-brain barrier and is an efficient therapy for central nervous system Philadelphia chromosome-positive leukemia. Blood 112: 1005–1012. 10.1182/blood-2008-02-140665 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
To evaluate whether the mixture of live and dead cells and PMA pretreatment affect HTLV-1 PVL measurement, we induced dead cells by digitonin treatment at 300 ng/ml for 2 h and made five different cell mixture (percent ratios of live /dead cells were 100/0, 75/25, 50/50, 25/75, and 0/100) using HTLV-1-infected cell line HUT102. We divided these cell mixtures into two groups: one was pretreated with PMA (which enters only dead cells and fix the DNA), and the other was not. Genomic DNA was extracted and we performed quantitative PCR targeting HTLV-1 pX gene in triplicate and repeated twice. Error bars represent standard error of the mean.
(TIF)
Data Availability Statement
The raw data of microarray experiments were deposited and available from the Gene Expression Ominibus (GEO) repository at National Center for Biotechnology Information (NCBI) (GEO DataSets Series accession number: GSE132666) located at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132666.