Abstract
In humans, cobalamin or vitamin B12 is delivered to two target enzymes via a complex intracellular trafficking pathway comprising transporters and chaperones. CblC (or MMACHC) is a processing chaperone that catalyzes an early step in this trafficking pathway. CblC removes the upper axial ligand of cobalamin derivatives, forming an intermediate in the pathway that is subsequently converted to the active cofactor derivatives. Mutations in the cblC gene lead to methylmalonic aciduria and homocystinuria. Here, we report that nitrosylcobalamin (NOCbl), which was developed as an antiproliferative reagent, and is purported to cause cell death by virtue of releasing nitric oxide, is highly unstable in air and is rapidly oxidized to nitrocobalamin (NO2Cbl). We demonstrate that CblC catalyzes the GSH-dependent denitration of NO2Cbl forming 5-coordinate cob(II)alamin, which had one of two fates. It could be oxidized to aquo-cob(III)alamin or enter a futile thiol oxidase cycle forming GSH disulfide. Arg-161 in the active site of CblC suppressed the NO2Cbl-dependent thiol oxidase activity, whereas the disease-associated R161G variant stabilized cob(II)alamin and promoted futile cycling. We also report that CblC exhibits nitrite reductase activity, converting cob(I)alamin and nitrite to NOCbl. Finally, the denitration activity of CblC supported cell proliferation in the presence of NO2Cbl, which can serve as a cobalamin source. The newly described nitrite reductase and denitration activities of CblC extend its catalytic versatility, adding to its known decyanation and dealkylation activities. In summary, upon exposure to air, NOCbl is rapidly converted to NO2Cbl, which is a substrate for the B12 trafficking enzyme CblC.
Keywords: adenosylcobalamin (AdoCbl), enzyme kinetics, metal, vitamin, thiol, MMACHC, nitrosylcobalamin (NOCbl), processing chaperone, vitamin B12
Vitamin B12 or cobalamin is an essential cofactor needed by two mammalian enzymes: methionine synthase and methylmalonyl-CoA mutase (MCM) (1). Clinical genetics studies on patients with inborn errors of B12 metabolism had led to the identification of at least nine genes (cblA-G, J, and mut) (2), which hinted at the existence of a complex B12 trafficking pathway. Biochemical studies have since been providing insights into the roles of the seven auxiliary proteins that serve to transport, assimilate, and target B12 to its two known intracellular targets (3–5).
MMACHC (methylmalonic aciduria type C and homocystinuria), corresponding to the cblC class of cobalamin disorders, is the most common locus of mutations in the B12 trafficking pathway (6). Mutations in MMACHC (hereafter referred to as CblC), disrupt the synthesis of methylcobalamin (MeCbl) and 5'-deoxyadenosylcobalamin (AdoCbl), leading to combined homocystinuria and methymalonic aciduria (7). Functionally, CblC is a versatile enzyme that catalyzes diverse chemical reactions. It is involved in the early cytosolic portion of the B12 trafficking pathway and processes cobalamins with various upper ligands to a common cob(II)alamin intermediate, which is subsequently partitioned to the cytoplasmic (MeCbl) and mitochondrial (AdoCbl) branches of the trafficking pathway (Fig. 1a). Alkylcobalamins (RCbl) are cleaved via a nucleophilic displacement reaction in the presence of GSH, producing the corresponding thioether GSR, and cob(I)alamin (Equation 1) that is rapidly oxidized to cob(II)alamin (8).
| (Eq. 1) |
| (Eq. 2) |
| (Eq. 3) |
Figure 1.
CblC structure and activity. a, CblC catalyzes the GSH-dependent dealkylation of alkylcobalamins, the reductive decyanation of CNCbl and as described in this study (red arrow), the GSH-dependent denitration of NO2Cbl. b, the structure of human CblC showing 2,4-difluorophenylethynylcobalamin bound in a base-off state (red) and GSH (yellow) (PDB code 5UOS). Arg-161 (blue) stabilizes GSH binding via hydrogen bonds (black dashed lines).
Cyanocobalamin (CNCbl) is cleaved via a reductive elimination reaction forming cob(II)alamin and cyanide (Equation 2) (8). The electron source in the decyanation reaction can be reduced flavin that is free or bound to a protein (9) or GSH (10). CblC also exhibits GSH-dependent aquocobalamin (OH2Cbl) reductase activity (Equation 3) (10).
The substrate promiscuity of CblC combined with its catalytic versatility, potentially sets up a metabolic vulnerability via futile redox cycling reactions (11, 12). Specifically, the use of GSH as a one-electron donor in Reactions 2 and 3 above, can promote superoxide formation under aerobic conditions, leading to GSH disulfide (GSSG) formation (10–12). Similarly, oxidation of the highly reactive cob(I)alamin product to cob(II)alamin with concomitant generation of superoxide, sets up a thiol oxidation cycle (13). The pathogenic R161G and R161Q mutations in human CblC significantly enhance these oxidative side reactions (11).
CblC belongs to the flavin nitroreductase superfamily, whose members use FMN or FAD as a prosthetic group (14). CblC does not bind FMN or FAD but can accept electrons from reduced flavin that is free or protein bound. The structure of human CblC with 2,4-difluorophenylethynyl-cobalamin revealed that the thiolate group of GSH is positioned for nucleophilic attack on the alkyl group (Fig. 1b) (15). Cobalamin is bound to CblC in a “base-off” state in which the tail leading to the dimethylbenzimidazole (DMB) base is embedded in a hydrophobic side pocket (14). “Base-on” versus base-off specifically refer to whether the cobalt is or is not coordinated by the endogenous DMB base on the lower (or α) face of the corrin ring.
Several groups have explored the idea of using B12 as a scaffold to deliver therapeutics for tumor imaging and cell killing (16). Nitrosylcobalamin (NOCbl) is an example of a derivative that was developed as an NO• donor to target cancer cells overexpressing the receptor for transcobalamin II, which transports B12 in circulation and is recognized by a specific cell surface receptor (17, 18). Cell death in NOCbl-treated cells was reportedly mediated via S-nitrosylation of the death receptor 4 leading to its activation (19). However, the presence of NOCbl was later called into question because OH2Cbl and NO• used to synthesize NOCbl, do not react. Instead, it was concluded that nitrite present as an impurity in NO•, led to nitrocobalamin (NO2Cbl) formation (20). In contrast, NO• and cob(II)alamin, each with an unpaired electron, react rapidly to form NOCbl (20, 21). NO• also reacts with aquocobinamide, which has a truncated DMB tail, in a two-step process in which it is initially reduced and then traps NO• (22).
Spectroscopic and crystallization studies reveal that NOCbl consists of a short Co-NO bond (1.91 Å) that is bent, and an unusually long Co-N bond to DMB (2.35 Å), resulting from the strong trans effect exerted by the NO• ligand (21, 23, 24). NOCbl exists as a hybrid of Co(III)-NO− and Co(II)-NO• resonance structures due to a considerable π backbonding interaction between the empty π* orbital of NO• and the doubly occupied 3dyz cobalt orbital (25).
Although the chemical reactions of nitrite and NO• with cobalamin have been studied in detail (20, 21, 26, 27), the biological fates of the resulting compounds are largely unknown. Herein, we have characterized the ability of CblC to process NOCbl and NO2Cbl. The rapid air oxidation of NOCbl to NO2Cbl indicates that NOCbl could not have been responsible for inhibiting cell proliferation as claimed (18, 19). We demonstrate that in vitro, CblC binds NOCbl tightly and stabilizes it against air oxidation. Neither thiols nor reductants remove the nitrosyl group from CblC-bound NOCbl. In contrast, nitrite is eliminated from CblC-bound NO2Cbl in the presence of GSH, forming cob(II)alamin or OH2Cbl under anaerobic or aerobic conditions, respectively. NO2Cbl processing by CblC is accompanied by oxidation of GSH to GSSG, which is exacerbated by the pathogenic R161G mutation. Our study demonstrates that NO2Cbl supports cell proliferation, consistent with the ability of CblC to process it.
Results and discussion
CblC binds NOCbl in the base-off state and stabilizes it against oxidation
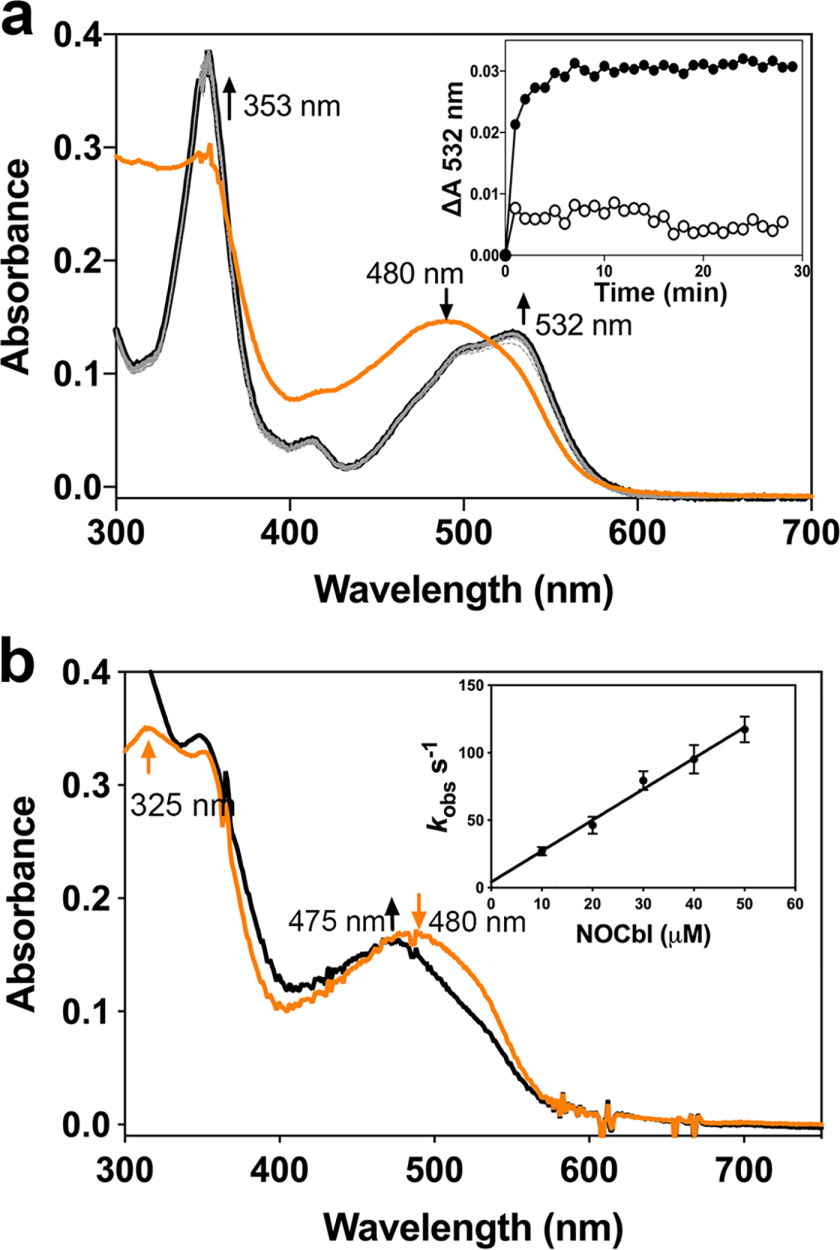
To determine the chemical species culpable for the purported antiproliferative effects of NOCbl (18), we first tested the stability of free NOCbl in aqueous solution versus bound to CblC. NOCbl (λmax = 480 nm) (Fig. 2a, orange trace), which is stable at pH 7.4 under anaerobic conditions, was rapidly oxidized to NO2Cbl (λmax = 353 nm, 532 nm; black trace) upon exposure to air.
Figure 2.
CblC protects NOCbl against oxidation. a, oxidation of NOCbl to NO2Cbl was observed when an anaerobic solution of NOCbl (20 μm in 100 mm HEPES, pH 7.4, 150 mm KCl, 10% glycerol; orange trace), was exposed to air. The final spectrum (black) represents NO2Cbl generated via oxidation of NOCbl. Inset, the kinetics of oxidation of free NOCbl (•) or bound to CblC (30 μm) (∘) was monitored at 532 nm and 25 °C. b, mixing NOCbl (20 μm; orange trace) with CblC (30 μm; black trace) under anaerobic conditions led to a slight blue shift in the absorption spectrum. From the linear dependence of the kobs for NOCbl binding to CblC, values for kon = 2.3 ± 0.1 μm−1 s−1 and koff = 3.7 ± 2.3 s−1 were obtained. The data represent the mean ± S.D. of 3 independent experiments.
Addition of CblC to an anaerobic solution of NOCbl resulted in a slight blue shift (Fig. 2b, black trace). We assign this spectral shift to the conversion of free base-on NOCbl to CblC-bound base-off NOCbl, based on the close similarity to the spectrum of NOCbl at pH 3.0 (20). The pKa for protonation of the DMB base in NOCbl is 5.1 (20). At pH 3.0, the spectrum of NOCbl therefore corresponds to that of the base-off species. The kinetics of NOCbl binding to CblC was monitored at 520 nm by stopped-flow spectroscopy. From the dependence of kobs on the NOCbl concentration (Fig. 2b, inset), the values for kon = 2.3 ± 0.1 μm−1 s−1 and koff = 3.7 ± 2.3 s−1 were obtained. From these values, the KD for NOCbl binding to CblC was estimated to be 1.6 μm.
In contrast to alkylcobalamins and CNCbl, NOCbl bound to CblC was unreactive toward GSH or other reductants (NADPH/methionine synthase reductase), suggesting that CblC is unable to process NOCbl (not shown). The lack of reactivity can be explained by the strong σ-donating NO group, which contributes an anionic character to the β-ligand (Co(III)-NO−) (25).
CblC catalyzes the GSH-dependent denitration of NO2Cbl
The instability of NOCbl in aerobic solution (Fig. 2a) suggests that the oxidized NO2Cbl product rather than the added NOCbl would have been taken up by cultured cells (19). We therefore assessed the possible cellular fate of NO2Cbl in the B12 trafficking pathway by focusing on CblC because it is proposed to bind B12 as it enters the cytoplasmic compartment (4).
Binding of NO2Cbl to CblC (Fig. 3a, red trace) did not elicit significant spectral changes compared with its spectrum in solution (λmax = 353 nm, 532 nm). Isothermal titration calorimetry yielded a binding stoichiometry of 0.928 ± 0.003 mole NO2Cbl per mole of CblC•GSMe and a KD of 0.89 ± 0.03 μm (Fig. 3b).
Figure 3.

CblC catalyzes the GSH-dependent denitration of NO2Cbl. a, the spectrum NO2Cbl (20 μm, red trace) bound to CblC (30 μm) in aerobic buffer (100 mm HEPES, 150 mm KCl, 10% glycerol, pH 7.4), changed upon addition of 1 mm GSH, indicating formation of OH2Cbl (black trace) with absorption maxima at 355, 495, and 525 nm. b, representative ITC titration for binding of NO2Cbl to CblC in the presence of GSMe at 20 °C. The data were fit to a one-site model and yielded the following values: KD = 0.89 ± 0.03 μm, ΔH = −8.2 ± 0.7 kcal mol−1, and TΔS = −0.5 ± 0.1 kcal mol−1. c, addition of GSH (1 mm) to a, but under anaerobic conditions (red trace), resulted in the slow accumulation of cob(II)alamin (474 nm, black trace) bound to CblC. Inset, the change in absorption at 353 nm plotted versus time, yielded kobs = 0.12 ± 0.01 min−1. d, EPR spectra of base-off 5-coordinate cob(II)alamin from top to bottom: (i) free cob(II)alamin (100 μm) in 100 mm HEPES, 150 mm KCl, 10% glycerol, pH adjusted to <3 with concentrated HCl, (ii) cob(II)alamin (100 μm) bound to WT CblC (120 μm), (iii and iv) cob(II)alamin formed after addition of GSH (1 mm) to NO2Cbl (100 μm) bound to WT CblC (iii) or R161G CblC (iv) (120 μm CblC each) under anaerobic or aerobic conditions, respectively. A.U., absorbance unit. e, scheme showing the postulated mechanism for the CblC-catalyzed denitration reaction.
Addition of GSH to CblC-bound NO2Cbl under aerobic conditions resulted in the immediate formation of OH2Cbl as evidenced by the appearance of the characteristic spectral features at 355 and 525 nm (Fig. 3a, black trace). In contrast, addition of GSH under anaerobic conditions resulted in the slow accumulation of cob(II)alamin with an absorption maximum at 474 nm and isosbestic points at 390 and 495 nm (Fig. 3c, black trace). The EPR spectrum of the product confirmed the presence of 5-coordinate, base-off cob(II)alamin (Fig. 3d).
A kobs of 0.12 ± 0.01 min−1 at pH 7.4 and 20 °C was estimated from the time dependence of the change in absorbance at 353 nm (Fig. 3c, inset). For comparison, the GSH-dependent dealkylation rates for the biologically relevant cobalamin derivatives are 0.2 ± 0.01 min−1 with MeCbl and 0.0030 ± 0.0001 min−1 with AdoCbl at pH 8.0 and 20 °C (9). The decyanation rate with CNCbl is 0.10 ± 0.004 min−1 at pH 7.0 and 20 °C in the presence of NADPH and the flavoprotein, methionine synthase reductase (8).
In solution, GSH does not react directly with NO2Cbl. Instead, the nitrite-to-water ligand substitution occurs via a dissociative interchange mechanism (Equation 4) (28). NO2Cbl, like other cobalamins with inorganic β-axial ligands, exists in equilibrium with OH2Cbl (29), which reacts with GSH (0.5 mm) forming glutathionyl-cobalamin (GSCbl). The rate constant for the overall reaction is ∼8 × 10−3 s−1 at pH 7.0 and 25 °C (28).
| (Eq. 4) |
Based on the accumulation of cob(II)alamin under anaerobic conditions, we propose that reductive elimination of nitrite from CblC-bound NO2Cbl occurs in the presence of GSH (Equation 5). The one-electron reduction of free NO2Cbl at pH 7.0 occurs at −151 mV and is irreversible (26). The other product of this reaction, GS• could react rapidly with a second molecule of GSH forming the radical anion , or with oxygen forming the peroxysulfenyl radical, GSOO• (Equations 6 and 7). The faster denitration kinetics under aerobic versus anaerobic conditions (Fig. 3, a versus c) suggests that the equilibrium in Equation 5 is shifted to the right via kinetic coupling, i.e. via the further reaction of GS• in Equations 6 and 7 or Equation 8.
| (Eq. 5) |
| (Eq. 6) |
| (Eq. 7) |
| (Eq. 8) |
GSSG is formed during the CblC-catalyzed denitration of NO2Cbl in the presence of GSH
The denitration reaction (Equation 5) is similar to that proposed for the reductive elimination of cyanide from CNCbl (Equation 2) in the presence of reductants including GSH (8). The immediate product of either reductive elimination reaction is cob(II)alamin. The lack of cob(II)alamin stabilization during denitration under aerobic conditions suggests that the initially formed cob(II)alamin is rapidly oxidized to OH2Cbl. The reaction of GS•, , and GSOO• with O2 and/or another mole of GSH have been described (Equations 7 and 9–11), and leads to the formation of GSSG as a major product in addition to GSH sulfonic acid (GSO3H) and other reactive sulfur species as minor products (30, 31).
| (Eq. 9) |
| (Eq. 10) |
| (Eq. 11) |
GSSG is therefore an expected byproduct of CblC-catalyzed and GSH-dependent denitration under aerobic conditions. GSSG formation was assessed by a coupled GSH reductase assay as described previously (Fig. 4a) (12). The kcat for GSSG formation by WT CblC was estimated to be 1.6 ± 0.1 min−1. In comparison, the kcat values for GSSG formation from MeCbl and CNCbl are 0.43 ± 0.03 and 0.16 ± 0.03 min−1, respectively (Table 1). The production of GSSG in vast stoichiometric excess over NO2Cbl in the reaction mixture, is ascribed to redox cycling, as discussed later.
Figure 4.
Stabilization of cob(II)alamin during aerobic denitration of NO2Cbl by R161G CblC and production of GSSG. a, GSSG formation during the denitration of NO2Cbl (20 μm) bound to CblC (40 μm) in anaerobic buffer (0.1M HEPES, pH 7.4, 150 mm KCl, 10% glycerol) treated with GSH (10 mm) at 20 °C. b, an aerobic solution of NO2Cbl (20 μm) bound to R161G CblC (30 μm, red trace) stabilized the cob(II)alamin product (black trace) following addition of GSH (10 mm).
Table 1.
Summary of CblC-catalyzed GSSG production rates
| Cobalamin | Wildtype CbC | R161G CblC |
|---|---|---|
| kobs min−1 | ||
| NO2Cbl | 1.6 ± 0.1 | 0.5 ± 0.1 |
| MeCbl | 0.43 ± 0.03 | 5.5 ± 0.2 |
| CNCbl | 0.16 ± 0.03 | NDa |
aND, not determined.
The pathogenic R161G CblC mutant stabilizes cob(II)alamin
Substitutions at the conserved active site Arg-161 residue to glycine or glutamine are the most common missense mutations associated with the cblC disorder (6). The R161G and R161Q mutations are associated with early and late onset of the disease, respectively (32). Both mutations weaken GSH binding and decrease the dealkylation but not the decyanation activity of CblC (11). In contrast to WT CblC where H2OCbl formation was observed immediately upon aerobic denitration (Fig. 3a) the R161G mutant stabilized cob(II)alamin under aerobic conditions (Fig. 4b). The EPR spectrum confirmed formation of the 5-coordinate, base-off cob(II)alamin as a reaction product (Fig. 3d). GSSG analysis revealed a ∼5-fold higher rate of GSSG formation by the R161G mutant (7.5 ± 0.1 min−1) compared with the WT protein (Table 1).
The stabilization of cob(II)alamin during the denitration reaction was reminiscent of the behavior of the R161G/R161Q mutants during reduction of OH2Cbl in the presence of GSH (11). Aerobic stabilization of the cob(II)alamin product was correlated with a ∼2- and 4-fold enhancement in GSH oxidation to GSSG by the R161Q and R161G mutants, respectively.
Nitrite reductase activity of CblC
A number of heme proteins including globins, cytochrome c, and cystathionine β-synthase exhibit nitrite reductase activity, catalyzing the one-electron reduction of nitrite to NO• (33–37). We tested whether cob(I)alamin, a powerful reductant with a midpoint redox potential of −500 mV for the base-off cob(II)alamin/cob(I)alamin couple (38), can reduce nitrite to NO•, when bound to CblC.
Mixing an anaerobic solution of CblC-bound cob(I)alamin with a characteristic peak at 390 nm (Fig. 5a, black trace), with an excess of nitrite, resulted in the conversion to a species with a broad absorption band centered at 480 nm and additional peaks at 345 and 316 nm (red trace). Isosbestic points were observed at 360, 425, and 545 nm. We assign this spectrum to base-off CblC-bound NOCbl, formed by the nitrite reduction by cob(I)alamin (Fig. 5b). From the change in absorbance at 390 or 480 nm, a kobs of 29 ± 1 and 34 ± 1 min−1 was estimated (Fig. 5a, inset). We note that an ∼3-fold excess of titanium(III) citrate was added to the reaction mixture to minimize unwanted oxidation of cob(I)alamin. Because titanium(III) citrate also reacts with sodium nitrite in solution, the dependence of the kobs on nitrite concentration could not be determined.
Figure 5.
Nitrite reductase activity of CblC. a, an anaerobic solution containing CblC (80 μm) in Buffer A loaded with cob(I)alamin (40 μm, λmax= 390 nm; black trace) was rapidly mixed 1:1 (v/v) with 1 mm sodium nitrite. Time-dependent changes consistent with the formation of NOCbl (λmax= 480 nm; red trace) were observed. Inset, the change in absorbance and 480 nm plotted versus time yielded a value of kobs = 29 ± 1 min−1 (at 390 nm) and 34 ± 1 min−1 (at 480 nm). b, scheme showing the postulated mechanism for the CblC-catalyzed nitrite reductase reaction.
In analogy to the mechanism of nitrite reduction by hemoglobin (39, 40), we postulate a two-step mechanism for the nitrite reductase activity of CblC. In the first step, a one-electron oxidation of cob(I)alamin to cob(II)alamin results in the conversion of nitrite to NO•. This is followed by the rapid recombination of NO• and cob(II)alamin, forming NOCbl (Fig. 5b).
Compared with the CblC-catalyzed reaction, the solution reaction between nitrite (HNO2) and cob(I)alamin results in the formation of cob(II)alamin and hydroxylamine (NH2OH) with a bimolecular rate constant of 1.7 × 103 m−1 s−1 at pH 7 and 25 °C (27). The reaction is postulated to involve a rate-determining two-electron reduction of HNO2 to HNO with concomitant oxidation of cob(I)alamin to OH2Cbl. HNO then reacts further with cob(I)alamin, leading to the formation of NH2OH. The overall stoichiometry of the reaction is described by Equation 12.
| (Eq. 12) |
Mechanism of NO2Cbl-induced redox cycling by CblC
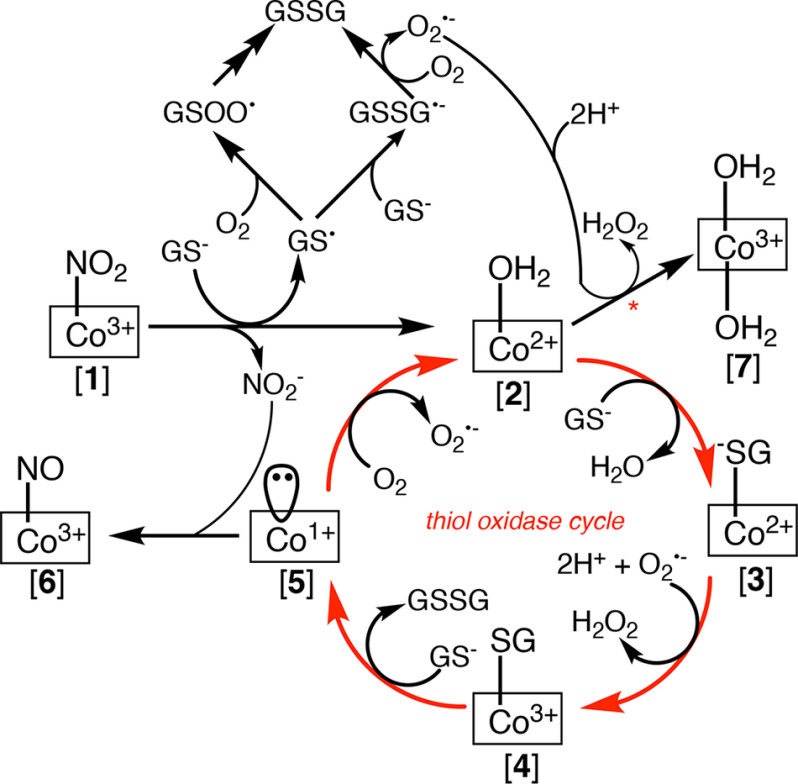
This study expands the catalytic repertoire of CblC by adding nitrite reductase and denitration of NO2Cbl to the previously characterized dealkylation, decyanation, and reduction reactions. With the exception of the nitrite reductase activity, the rest serve to remove the upper axial ligand of various cobalamin derivatives, generating either cob(I)alamin (via dealkylation) or cob(II)alamin (in the other reactions) neither of which is stabilized by human CblC (Fig. 1a). Instead, both cob(I)alamin and cob(II)alamin are oxidized to OH2Cbl in the presence of air. We demonstrate, using the denitration reaction, that the combined use of GSH as a one-electron donor and O2 as a one-electron acceptor in the cobalamin-dependent reactions catalyzed by CblC, has the potential to generate GSSG via a futile redox cycle (Fig. 6).
Figure 6.
Proposed mechanism for the denitration-fueled thiol oxidase activity of human CblC. The use of GSH as a one-electron donor sets up a futile thiol oxidase cycle under aerobic conditions leading to gratuitous formation of GSSG.
The denitration of NO2Cbl (Fig. 6, (1)) leads to cob(II)alamin. Based on EPR studies, the cob(II)alamin product is 5-coordinate, indicating the presence of an axial water ligand (2) Under aerobic conditions, CblC-bound cob(II)alamin undergoes a one-electron oxidation to cob(III)alamin. The redox potential of base-off OH2Cbl/cob(II)alamin is +514 mV (38). In comparison, the redox potentials of the and the /H2O2 couples are −330 and +890 mV at pH 7.0, respectively (41). Thus, based on redox potential considerations, we propose that oxidizes cob(II)alamin, forming OH2Cbl (7). In solution, the oxidation of cob(II)alamin by occurs with a rate constant of ∼7 × 108 m−1 s−1 at pH 7.4 and 25 °C, approaching the rate constant for the superoxide dismutase reaction (42). In the CblC reaction, the could be formed in situ during oxidation of GS• to GSSG as described in Equations 6 and 7 and in Fig. 6.
Cob(III)alamin derivatives including OH2Cbl, prefer a 6-coordinate geometry. In the crystal structure of human CblC, the α-face is hydrophilic and an ordered water is seen bridging between Ser-146 and one of the propionamide side chains of the corrin ring (14). Although the structure provides support for the accessibility of a water molecule to the α-face of the corrin, it is not known whether the water can coordinate to OH2Cbl. Instead, OH2Cbl formation from cob(II)alamin could be kinetically driven by , and the product could be 6-coordinate (as shown in Fig. 6) or 5-coordinate.
An alternate fate of 5-coordinate cob(II)alamin (2) is to undergo ligand exchange with GS− replacing water and leading to GS−-cob(II)alamin (3). The latter can undergo oxidation forming GS−-cob(III)alamin (4). Finally, a second mole of GSH can displace the thiolato ligand of (4), forming GSSG and cob(I)alamin (5), which is very rapidly oxidized to (2). Cob(II)alamin can partition between (7) and (3), with the latter perpetuating the redox cycle and promoting further thiol oxidation as observed.
Cob(I)alamin (5) can potentially react with nitrite forming NOCbl (6). The reaction of cob(I)alamin with nitrite occurs with a rate constant of 29 ± 1 min−1 and is unlikely to be significant under physiological conditions, because cob(I)alamin is oxidized very rapidly. Furthermore, given the stability of CblC-bound NOCbl, the nitrite reductase activity of CblC is unlikely to be NO• source.
The intermediates proposed in the redox cycle triggered by the denitration activity of human CblC are analogous to those formed during the dealkylation-triggered redox cycling catalyzed by the Caenorhabditis elegans CblC (12). Evidence of the GS−-cob(III)alamin intermediate (4) and for its rate-limiting dethiolation by a second mole of GSH leading to cob(I)alamin (5) was provided by kinetic, spectroscopic, and computational analysis (12). Furthermore, the rapid oxidation of cob(II)alamin (2) bound to the C. elegans CblC by , forming OH2Cbl, supported the feasibility of the proposed reactive oxygen species-dependent oxidation mechanism. The C. elegans CblC exhibits robust thiol oxidase activity, which leads to O2 scrubbing and remarkably, to the stabilization of cob(I)alamin in a reaction mixture that was originally aerobic (13).
The enhanced oxidation of GSH by the R161G CblC mutant reveals a role for this arginine residue in promoting partitioning of the cob(II)alamin (2) intermediate to OH2Cbl (7). We propose that Arg-161 in WT CblC inhibits the approach of GSH to the cobalt ion, which is needed for the β-ligand exchange step, i.e. the conversion of (2) to (3). Mutation of Arg-161 to glycine or glutamine weakens this gating function and promotes futile cycling, leading to GSSG formation.
NO2Cbl supports B12-dependent cell proliferation
The ability of CblC to process NO2Cbl, predicted that it could support B12-dependent cell proliferation. On the other hand, the millimolar concentrations of GSSG generated during CblC-catalyzed denitration of NO2Cbl suggested that the resulting thiol oxidase activity could set up a metabolic vulnerability and potentially be anti-proliferative. We tested these contrasting predictions from the in vitro experiments by monitoring the effect of NO2Cbl in cultured cells.
During rapid proliferation as seen with malignant cell lines in culture, folate utilization is prioritized for DNA synthesis and formation of 5-CH3-tetrahydrofolate, a substrate for B12-dependent methionine synthase is limited. Instead, cells rely on the ready availability of methionine in the culture medium to support cell growth (43). Methionine synthase catalyzes the methyltransfer from 5-CH3-tetrahydrofolate to homocysteine, forming tetrahydrofolate and methionine with MeCbl as an intermediate (44). We therefore monitored proliferation of human colorectal adenocarcinoma HT-29 cells in medium lacking methionine but supplemented with homocystine (Met− Hcy2+), as described previously (45), to enforce B12 dependence.
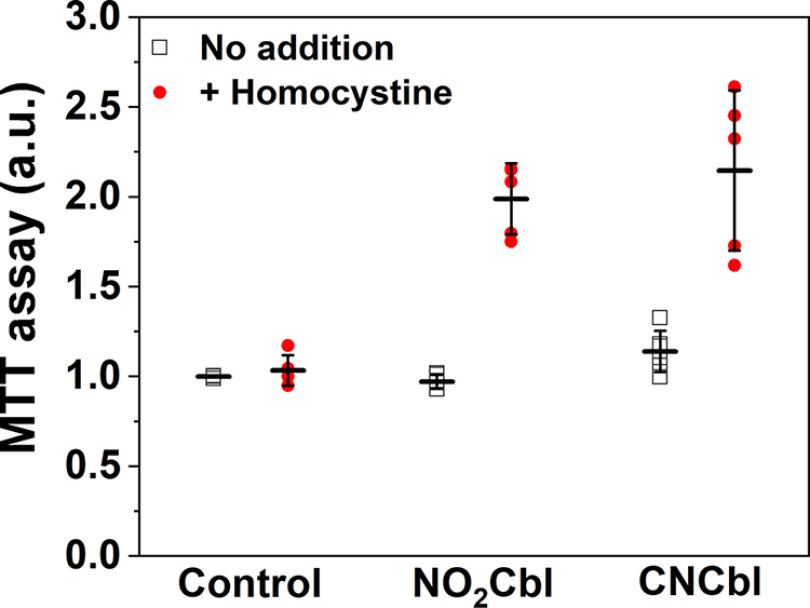
The viability of cells maintained for 2 days in Met− Hcy2+ medium supplemented with NO2Cbl or CNCbl was assessed using the MTT assay. Supplementation of Met− Hcy2+ medium with either NO2Cbl or CNCbl promoted cell proliferation (Fig. 7). This result supports our in vitro data that NO2Cbl can be processed by CblC for subsequent synthesis of MeCbl needed by methionine synthase and demonstrates that the potential antiproliferative effect of NO2Cbl is not expressed in these cells. We reason that the flux of cobalamin through the trafficking pathway is low, and that the level of NO2Cbl-dependent GSH oxidation is insufficient to impair cell growth. Our results contrast with the antiproliferative effects purportedly ascribed to NOCbl, which as this and previous studies have shown, would have been rapidly oxidized to NO2Cbl (17–19).
Figure 7.
NO2Cbl promotes cell proliferation. Human colorectal adenocarcinoma HT29 cells were seeded overnight in Met−, DMEM before addition of NO2Cbl or CNCbl (0.2 mm) in the absence (black squares) or presence (red dots) of homocystine (0.1 mm). The cells were grown for 2 days before viability was assessed by the MTT assay. The results are normalized to cells grown in the Met– medium without any supplements. The data represent the mean ± S.D. of at least 5 independent experiments.
In summary we have demonstrated that the sensitivity of NOCbl to oxidation in air leads to its rapid conversion to NO2Cbl, which is a substrate for the B12 processing enzyme, CblC. Curiously, whereas NOCbl is stabilized upon binding to CblC, NO2Cbl undergoes denitration in the presence of the co-substrate GSH, forming cob(II)alamin. The latter is partitioned into a futile thiol oxidative cycle, generating GSSG or converted to OH2Cbl via oxidation. Processing of NO2Cbl by CblC makes it available for B12-dependent cellular functions, supporting proliferation. Arg-161 in the active site suppresses the thiol oxidase activity in WT CblC limiting the potential metabolic liability associated with this activity. The R161G mutation on the other hand, enhances the thiol oxidase activity, which might be a contributing factor to the enhanced oxidative stress reported in fibroblasts from CblC patients (46).
Experimental procedures
Materials
All chemicals were purchased from Sigma-Aldrich or Fisher unless otherwise specified. DEA-NONOate was purchased from Cayman Chemical (MI).
Synthesis of nitrosylcobalamin
NOCbl was synthesized under strictly anaerobic conditions as described previously (24). DEA-NONOate (10 mg) was dissolved in 100 μl of 10 mm NaOH and mixed with OH2Cbl•HCl (40 mg) dissolved in 0.4 ml of 0.1 m TES buffer, pH 7.4. The reaction was incubated for 3 h at room temperature and the absorption spectrum was recorded to monitor completion of the reaction (NOCbl ε478nm = 6.9 mm−1 cm−1). NOCbl was precipitated by dripping cold acetone into the reaction mixture. The precipitate was washed with 1 ml of acetone and the purified sample was lyophilized and stored at −80 °C.
Synthesis of nitrocobalamin
NO2Cbl was prepared using a slight modification of a previously published protocol (47). The reaction was carried out in the dark at 0 °C. In a 1.5-ml sample tube, OH2Cbl•HCl (11.2 mg) dissolved in 150 µl of 100 mm MES buffer, pH 6.0, was mixed with 8.8 µl of a 1.5 m NaNO2 solution in 100 mm MES, pH 6.0. The reaction was incubated for 2.5 h at 0 °C and the reaction mixture was dripped into acetone to precipitate NO2Cbl. The precipitate was washed with 1 ml of acetone (three times) and the product was lyophilized. NO2Cbl (10.2 mg, 92% yield) was obtained as a purple powder.
Expression and purification of CblC
Recombinant ΔC244 human CblC was expressed and purified as previously described (9). The ΔC244 CblC mutants (R161G) was expressed and purified as previously described (11). The protein was dialyzed into a Buffer A containing 100 mm HEPES, pH 7.4, 150 mm KCl, and 10% glycerol and further purified by size exclusion chromatography (Superdex 200, GE Healthcare). The purified protein was flash frozen in liquid nitrogen and stored at −80 °C. All assays were performed in Buffer A unless otherwise specified.
Oxidation of NOCbl to NO2Cbl
A 150-μl solution of NOCbl (40 μm) or NOCbl (40 μm) mixed with CblC (60 μm) was prepared in Buffer A under anaerobic conditions in a sealed cuvette. An initial UV-visible spectrum was recorded, and the reaction was initiated by the addition of 150 μl of aerobic Buffer A (300 μl final volume). Spectra were recorded every minute for 30 min at 20 °C. The t1/2 of NOCbl alone or in the presence of CblC was estimated by plotting the change in absorbance at 532 nm versus time.
Isothermal titration calorimetry
ITC experiments were performed using a Microcal VP-ITC (GE Healthcare). CblC (35 μm) was titrated with 37 × 8-μl injections of NO2Cbl (300 μm) in Buffer A containing 1 mm GSMe at 20 °C. To determine the equilibrium dissociation constant (KD), and binding enthalpy (ΔH°), the calorimetric signals were integrated, and data were analyzed with the Microcal ORIGIN software using a single site binding model.
Reactions of CblC-bound NO2Cbl with GSH
The reaction of CblC-bound NO2Cbl with GSH was monitored under aerobic and anaerobic conditions at 20 °C on a spectrophotometer connected to a temperature-controlled water bath. The 150-μl reaction mixture contained CblC (30 μm) and NO2Cbl (20 μm) in Buffer A and the reaction was initiated by the addition of GSH (1 mm). The change in absorbance at 353 nm was plotted as a function of time and the kinetic trace was fit to a single exponential decay to obtain the rate constant for denitration.
Quantification of GSSG by a coupled GSH reductase assay
Dealkylation of CblC (40 μm)-bound B12 (20 μm) in the presence of GSH (10 mm) was carried out at 20 °C under aerobic conditions. The reactions were stopped at the desired time points (0-60 min) by precipitating the protein with an equal volume of metaphosphoric acid solution (16.8 mg/ml of metaphosphoric acid, 2 mg/ml of EDTA, and 9 mg/ml of NaCl). The samples were then treated and analyzed by a coupled GSH reductase assay as previously described (12). When the R161G mutant was used, the protein and B12 concentrations were decreased to 10 and 5 μm, respectively.
EPR spectroscopy
EPR spectra were recorded on a Bruker EMX 300 spectrometer equipped with a Bruker 4201 cavity and a ColdEdge cryostat. The temperature was controlled by an Oxford Instruments MercuryiTC temperature controller. EPR spectra were recorded at 80 K with the following parameters: 9.38 GHz microwave frequency, 2 milliwatt power, 10 G modulation amplitude, 100 kHz modulation frequency, 3000 G sweep width centered at 3500 G, 164 ms conversion time, and 82 ms time constant. Five scans were collected per measurement.
NO2Cbl (100 μm) was added to CblC (120 μm) in Buffer A under anaerobic conditions. GSH (1 mm) was added to the reaction mixture and incubated for 30-45 min. The sample was transferred to an EPR tube sealed and flash frozen in liquid nitrogen. NO2Cbl (100 μm) was added to R161G CblC (120 μm) in Buffer A under aerobic conditions. GSH (10 mm) was added to the reaction mixture and incubated for ∼1 h. The sample was transferred to an EPR tube, sealed, and flash frozen in liquid nitrogen.
Stopped-flow spectroscopy
Rapid-mixing spectroscopic experiments were carried out at 20 °C, in an anaerobic chamber (<0.5 ppm O2), using an Applied Photophysics SX.MV18 spectrometer equipped with a diode array detector.
Binding of NOCbl to CblC was monitored by rapidly mixing CblC (7 μm after mixing) with varying concentrations of NOCbl (10-50 μm after mixing). The change in absorbance at 520 nm was monitored for 1 s. The kinetic traces were fit using the Applied Photophysics Pro-Data Viewer application to determine the kobs values. The kobs was plotted versus substrate concentration to determine kon (slope), koff (y intercept), and KD (koff/kon) values.
The nitrite reductase activity was monitored in an anaerobic mixture containing CblC (80 μm) and cob(II)alamin (40 μm) in Buffer A to which titanium(III) citrate (150 μm) was added. The concentration of the resulting CblC-bound cob(I)alamin was determined at 390 nm (ε390nm = 28 mm−1 cm−1). Then, NaNO2 (1 mm) in anaerobic Buffer A was rapidly mixed with the CblC-bound cob(I)alamin. Kinetic traces at select wavelengths were fitted to single exponential change using the Applied Photophysics Pro-Data Viewer software.
Cell proliferation assay with B12 treatment
HT29 cells line were maintained in DMEM with high glucose and pyruvate (Gibco, 11995), supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. Passage 15-30 HT29 cells (ATCC, Manassas, VA) were used in the experiments, which were performed in a 5% CO2 incubator at 37 °C. Cells (125 × 103 cells/well) were seeded in 12-well plates containing 1 ml of Met−, DMEM (Gibco, 21013) per well, supplemented with 4 mm glutamine, 0.2 mm l-cystine, 10% fetal bovine serum, and 1% penicillin and streptomycin. Following overnight incubation, 0.2 mm CNCbl or NO2Cbl ± 0.1 mm l-homocystine, or an equal volume of PBS was added to the medium. The cells were then grown for 2 days, and cell viability was assessed using the MTT assay as described previously (48). The readings were normalized to the value obtained with cells grown in the absence of B12 or homocystine supplementation.
Data availability
All data are contained within the manuscript.
Author contributions—R. M., Z. L., and R. B. conceptualization; R. M. and R. B. resources; R. M. and Z. L. data curation; R. M., Z. L., C. G., M. R., and R. B. formal analysis; R. M., Z. L., and R. B. funding acquisition; R. M. and Z. L. writing-original draft; R. M., Z. L., C. G., M. R., and R. B. writing-review and editing; Z. L., C. G., M. R., and R. B. investigation.
Funding and additional information–This work was supported in part by a grant from the National Institutes of Health Grant DK45776 (to R. B.) and the American Heart Association Grant 19POST34370113 (to R. M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no competing interests.
- MCM
- methylmalonyl-CoA mutase
- MeCbl
- methylcobalamin
- AdoCbl
- 5′-deoxyadenosyl cobalamin
- GSMe
- glutathione methylester
- NOCbl
- nitrosylcobalamin
- NO2Cbl
- nitrocobalamin
- GSCbl
- glutathionyl-cobalamin
- DMB
- dimethylbenzimidazole
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- TES
- 2-{[lsqb]2-hydroxy-1,1-bis(hydroxymethyl)ethyl[rsqb]amino}ethanesulfonic acid
- ITC
- isothermal titration calorimetry
- DMEM
- Dulbecco's modified Eagle's medium.
References
- 1. Banerjee R., and Ragsdale S. W. (2003) The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem. 72, 209–247 10.1146/annurev.biochem.72.121801.161828 [DOI] [PubMed] [Google Scholar]
- 2. Watkins D., and Rosenblatt D. S. (2011) Inborn errors of cobalamin absorption and metabolism. Am. J. Med. Genet. C Semin. Med. Genet. 157C, 33–44 10.1002/ajmg.c.30288 [DOI] [PubMed] [Google Scholar]
- 3. Banerjee R., Gherasim C., and Padovani D. (2009) The tinker, tailor, soldier in intracellular B12 trafficking. Curr. Opin. Chem. Biol. 13, 484–491 10.1016/j.cbpa.2009.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gherasim C., Lofgren M., and Banerjee R. (2013) Navigating the B12 road: assimilation, delivery and disorders of cobalamin. J. Biol. Chem. 288, 13186–13193 10.1074/jbc.R113.458810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banerjee R. (2006) B12 trafficking in mammals: a case for coenzyme escort service. ACS Chem. Biol. 1, 149–159 10.1021/cb6001174 [DOI] [PubMed] [Google Scholar]
- 6. Lerner-Ellis J. P., Anastasio N., Liu J., Coelho D., Suormala T., Stucki M., Loewy A. D., Gurd S., Grundberg E., Morel C. F., Watkins D., Baumgartner M. R., Pastinen T., Rosenblatt D. S., and Fowler B. (2009) Spectrum of mutations in MMACHC, allelic expression, and evidence for genotype-phenotype correlations. Hum. Mutat. 30, 1072–1081 10.1002/humu.21001 [DOI] [PubMed] [Google Scholar]
- 7. Lerner-Ellis J. P., Tirone J. C., Pawelek P. D., Dore C., Atkinson J. L., Watkins D., Morel C. F., Fujiwara T. M., Moras E., Hosack A. R., Dunbar G. V., Antonicka H., Forgetta V., Dobson C. M., Leclerc D., et al. (2006) Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nat. Genet. 38, 93–100 10.1038/ng1683 [DOI] [PubMed] [Google Scholar]
- 8. Kim J., Gherasim C., and Banerjee R. (2008) Decyanation of vitamin B12 by a trafficking chaperone. Proc. Natl. Acad. Sci. U.S.A. 105, 14551–14554 10.1073/pnas.0805989105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim J., Hannibal L., Gherasim C., Jacobsen D. W., and Banerjee R. (2009) A human vitamin B12 trafficking protein uses glutathione transferase activity for processing alkylcobalamins. J. Biol. Chem. 284, 33418–33424 10.1074/jbc.M109.057877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Z., Gherasim C., Lesniak N. A., and Banerjee R. (2014) Glutathione-dependent one-electron transfer reactions catalyzed by a B12 trafficking protein. J. Biol. Chem. 289, 16487–16497 10.1074/jbc.M114.567339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gherasim C., Ruetz M., Li Z., Hudolin S., and Banerjee R. (2015) Pathogenic mutations differentially affect the catalytic activities of the human B12-processing chaperone CblC and increase futile redox cycling. J. Biol. Chem. 290, 11393–11402 10.1074/jbc.M115.637132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Z., Shanmuganathan A., Ruetz M., Yamada K., Lesniak N. A., Krautler B., Brunold T. C., Koutmos M., and Banerjee R. (2017) Coordination chemistry controls the thiol oxidase activity of the B12-trafficking protein CblC. J. Biol. Chem. 292, 9733–9744 10.1074/jbc.M117.788554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Z., Lesniak N. A., and Banerjee R. (2014) Unusual aerobic stabilization of Cob(I)alamin by a B12-trafficking protein allows chemoenzymatic synthesis of organocobalamins. J. Am. Chem. Soc. 136, 16108–16111 10.1021/ja5077316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koutmos M., Gherasim C., Smith J. L., and Banerjee R. (2011) Structural basis of multifunctionality in a vitamin B12-processing enzyme. J. Biol. Chem. 286, 29780–29787 10.1074/jbc.M111.261370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruetz M., Shanmuganathan A., Gherasim C., Karasik A., Salchner R., Kieninger C., Wurst K., Banerjee R., Koutmos M., and Krautler B. (2017) Antivitamin B12 inhibition of the human B12-processing enzyme CblC: crystal structure of an inactive ternary complex with glutathione as the cosubstrate. Angew. Chem. Int. Ed. Engl. 56, 7387–7392 10.1002/anie.201701583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clardy S. M., Allis D. G., Fairchild T. J., and Doyle R. P. (2011) Vitamin B12 in drug delivery: breaking through the barriers to a B12 bioconjugate pharmaceutical. Expert Opin. Drug. Deliv. 8, 127–140 10.1517/17425247.2011.539200 [DOI] [PubMed] [Google Scholar]
- 17. Bauer J. A. (1998) Synthesis, characterization and nitric oxide release profile of nitrosylcobalamin: a potential chemotherapeutic agent. Anticancer Drugs 9, 239–244 10.1097/00001813-199803000-00006 [DOI] [PubMed] [Google Scholar]
- 18. Bauer J. A., Lupica J. A., Schmidt H., Morrison B. H., Haney R. M., Masci R. K., Lee R. M., Didonato J. A., and Lindner D. J. (2007) Nitrosylcobalamin potentiates the anti-neoplastic effects of chemotherapeutic agents via suppression of survival signaling. PLoS ONE 2, e1313 10.1371/journal.pone.0001313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang Z., Bauer J. A., Morrison B., and Lindner D. J. (2006) Nitrosylcobalamin promotes cell death via S-nitrosylation of Apo2L/TRAIL receptor DR4. Mol. Cell Biol. 26, 5588–5594 10.1128/MCB.00199-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolak M., Zahl A., Schneppensieper T., Stochel G., and van Eldik R. (2001) Kinetics and mechanism of the reversible binding of nitric oxide to reduced cobalamin B(12r) (Cob(II)alamin). J. Am. Chem. Soc. 123, 9780–9791 10.1021/ja010530a [DOI] [PubMed] [Google Scholar]
- 21. Zheng D., and Birke R. L. (2001) Spectroscopic evidence for nitric oxide binding with cob(II)alamin. J. Am. Chem. Soc. 123, 4637–4638 10.1021/ja015682k [DOI] [PubMed] [Google Scholar]
- 22. Sharma V. S., Pilz R. B., Boss G. R., and Magde D. (2003) Reactions of nitric oxide with vitamin B12 and its precursor, cobinamide. Biochemistry 42, 8900–8908 10.1021/bi034469t [DOI] [PubMed] [Google Scholar]
- 23. Hassanin H. A., El-Shahat M. F., DeBeer S., Smith C. A., and Brasch N. E. (2010) Redetermination of the X-ray structure of nitroxylcobalamin: base-on nitroxylcobalamin exhibits a remarkably long co-N(dimethylbenzimidazole) bond distance. Dalton Trans. 39, 10626–10630 10.1039/c0dt00628a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hassanin H. A., Hannibal L., Jacobsen D. W., Brown K. L., Marques H. M., and Brasch N. E. (2009) NMR spectroscopy and molecular modelling studies of nitrosylcobalamin: further evidence that the deprotonated, base-off form is important for nitrosylcobalamin in solution. Dalton Trans. 424–433 10.1039/B810895A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pallares I. G., and Brunold T. C. (2014) Spectral and electronic properties of nitrosylcobalamin. Inorg. Chem. 53, 7676–7691 10.1021/ic500986x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng D., Yan L., and Birke R. L. (2002) Electrochemical and spectral studies of the reactions of aquocobalamin with nitric oxide and nitrite ion. Inorg. Chem. 41, 2548–2555 10.1021/ic010802a [DOI] [PubMed] [Google Scholar]
- 27. Plymale N. T., Dassanayake R. S., Hassanin H. A., and Brasch N. E. (2012) Kinetic and mechanistic studies on the reactions of the reduced vitamin B12 complex cob(I)alamin with nitrite and nitrate. Eur. J. Inorg. Chem. 2012, 913–921 10.1002/ejic.201290011 [DOI] [Google Scholar]
- 28. Walker D. T., Dassanayake R. S., Garcia K. A., Mukherjee R., and Brasch N. E. (2013) Mechanistic studies on the reaction of nitrocobalamin with glutathione: kinetic evidence for formation of an aquacobalamin intermediate. Eur. J. Inorg. Chem. 2013, 3049–3053 10.1002/ejic.201300254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meier M., and van Eldik R. (1993) Ligand-substitution reactions of aquacobalamin (Vitamin B12a) revisited: conclusive evidence for the operation of a dissociative interchange mechanism. Inorg. Chem. 32, 2635–2639 10.1021/ic00064a011 [DOI] [Google Scholar]
- 30. Buettner G. R. (1993) The pecking order of free radicals and antioxidants: Lipid peroxidation, α-tocopherol, and ascorbate. Arch. Biochem. Biophys. 300, 535–543 10.1006/abbi.1993.1074 [DOI] [PubMed] [Google Scholar]
- 31. Winterbourn C. C. (2016) Revisiting the reactions of superoxide with glutathione and other thiols. Arch. Biochem. Biophys. 595, 68–71 10.1016/j.abb.2015.11.028 [DOI] [PubMed] [Google Scholar]
- 32. Froese D. S., Zhang J., Healy S., and Gravel R. A. (2009) Mechanism of vitamin B12-responsiveness in cblC methylmalonic aciduria with homocystinuria. Mol. Genet. Metab. 98, 338–343 10.1016/j.ymgme.2009.07.014 [DOI] [PubMed] [Google Scholar]
- 33. Gherasim C., Yadav P. K., Kabil O., Niu W. N., and Banerjee R. (2014) Nitrite reductase activity and inhibition of H2S biogenesis by human cystathionine β-synthase. PLoS ONE 9, e85544 10.1371/journal.pone.0085544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cosby K., Partovi K. S., Crawford J. H., Patel R. P., Reiter C. D., Martyr S., Yang B. K., Waclawiw M. A., Zalos G., Xu X., Huang K. T., Shields H., Kim-Shapiro D. B., Schechter A. N., Cannon R. O. 3rd, et al. (2003) Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nat. Med. 9, 1498–1505 10.1038/nm954 [DOI] [PubMed] [Google Scholar]
- 35. Shiva S., Huang Z., Grubina R., Sun J., Ringwood L. A., MacArthur P. H., Xu X., Murphy E., Darley-Usmar V. M., and Gladwin M. T. (2007) Deoxymyoglobin is a nitrite reductase that generates nitric oxide and regulates mitochondrial respiration. Circ. Res. 100, 654–661 10.1161/01.RES.0000260171.52224.6b [DOI] [PubMed] [Google Scholar]
- 36. Tiso M., Tejero J., Basu S., Azarov I., Wang X., Simplaceanu V., Frizzell S., Jayaraman T., Geary L., Shapiro C., Ho C., Shiva S., Kim-Shapiro D. B., and Gladwin M. T. (2011) Human neuroglobin functions as a redox-regulated nitrite reductase. J. Biol. Chem. 286, 18277–18289 10.1074/jbc.M110.159541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Basu S., Azarova N. A., Font M. D., King S. B., Hogg N., Gladwin M. T., Shiva S., and Kim-Shapiro D. B. (2008) Nitrite reductase activity of cytochrome c. J. Biol. Chem. 283, 32590–32597 10.1074/jbc.M806934200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lexa D., and Saveant J.-M. (1983) The electrochemistry of vitamin B12. Acc. Chem. Res. 16, 235–243 10.1021/ar00091a001 [DOI] [Google Scholar]
- 39. Huang K. T., Keszler A., Patel N., Patel R. P., Gladwin M. T., Kim-Shapiro D. B., and Hogg N. (2005) The reaction between nitrite and deoxyhemoglobin: reassessment of reaction kinetics and stoichiometry. J. Biol. Chem. 280, 31126–31131 10.1074/jbc.M501496200 [DOI] [PubMed] [Google Scholar]
- 40. Grubina R., Basu S., Tiso M., Kim-Shapiro D. B., and Gladwin M. T. (2008) Nitrite reductase activity of hemoglobin S (sickle) provides insight into contributions of heme redox potential versus ligand affinity. J. Biol. Chem. 283, 3628–3638 10.1074/jbc.M705222200 [DOI] [PubMed] [Google Scholar]
- 41. Wood P. M. (1988) The potential diagram for oxygen at pH 7. Biochem. J. 253, 287–289 10.1042/bj2530287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suarez-Moreira E., Yun J., Birch C. S., Williams J. H., McCaddon A., and Brasch N. E. (2009) Vitamin B(12) and redox homeostasis: cob(II)alamin reacts with superoxide at rates approaching superoxide dismutase (SOD). J. Am. Chem. Soc. 131, 15078–15079 10.1021/ja904670x [DOI] [PubMed] [Google Scholar]
- 43. Ducker G. S., and Rabinowitz J. D. (2017) One-carbon metabolism in health and disease. Cell Metab. 25, 27–42 10.1016/j.cmet.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Banerjee R. V., and Matthews R. G. (1990) Cobalamin-dependent methionine synthase. FASEB J. 4, 1450–1459 10.1096/fasebj.4.5.2407589 [DOI] [PubMed] [Google Scholar]
- 45. Prudova A., Bauman Z., Braun A., Vitvitsky V., Lu S. C., and Banerjee R. (2006) S-Adenosylmethionine stabilizes cystathionine beta-synthase and modulates redox capacity. Proc. Natl. Acad. Sci. U.S.A. 103, 6489–6494 10.1073/pnas.0509531103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Richard E., Jorge-Finnigan A., Garcia-Villoria J., Merinero B., Desviat L. R., Gort L., Briones P., Leal F., Pérez-Cerdá C., Ribes A., Ugarte M., and Perez B, and MMACHC Working Group (2009) Genetic and cellular studies of oxidative stress in methylmalonic aciduria (MMA) cobalamin deficiency type C (cblC) with homocystinuria (MMACHC). Hum. Mutat. 30, 1558–1566 10.1002/humu.21107 [DOI] [PubMed] [Google Scholar]
- 47. Suarez-Moreira E., Hannibal L., Smith C. A., Chavez R. A., Jacobsen D. W., and Brasch N. E. (2006) A simple, convenient method to synthesize cobalamins: synthesis of homocysteinylcobalamin, N-acetylcysteinylcobalamin, 2-N-acetylamino-2-carbomethoxyethanethiolatocobalamin, sulfitocobalamin and nitrocobalamin. Dalton Trans. 28, 5269–5277 10.1039/b610158e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Libiad M., Vitvitsky V., Bostelaar T., Bak D. W., Lee H. J., Sakamoto N., Fearon E., Lyssiotis C. A., Weerapana E., and Banerjee R. (2019) Hydrogen sulfide perturbs mitochondrial bioenergetics and triggers metabolic reprogramming in colon cells. J. Biol. Chem. 294, 12077–12090 10.1074/jbc.RA119.009442 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript.