Abstract
Noncoding RNAs (ncRNAs) longer than 200 nucleotides are rare in bacteria, likely because bacterial genomes are under strong evolutionary pressures to maintain a small genome size. Of the long ncRNAs unique to bacteria, the OLE (ornate, large, extremophilic) RNA class is among the largest and most structurally complex. OLE RNAs form a ribonucleoprotein (RNP) complex by partnering with at least two proteins, OapA and OapB, that directly bind OLE RNA. The biochemical functions of the OLE RNP complex remain unknown, but are required for proper adaptation to certain environmental stresses, such as cold temperatures, short chain alcohols, and high magnesium concentrations. In the current study, we used electrophoretic mobility shift assays to examine the binding of OLE RNA fragments by OapB and found that OapB recognizes a small subregion of OLE RNA, including stem P13, with a dissociation constant (KD) of ∼700 pm. Analyses with mutated RNA constructs, and the application of in vitro selection, revealed that strong binding of OLE RNA by OapB requires a stem containing a precisely located single-nucleotide bulge and a GNRA tetraloop. Although the vast majority of bacteria with the ole gene also have the oapB gene, there are many whose genomes contain oapB but lack ole, suggesting that OapB has other RNA partners in some species that might exhibit similar structural features.
Keywords: ribonuclear protein (RNP), ribozyme, RNA-binding protein, RNA-protein interaction, Bacillus, Gram-positive bacteria, OapA, OLE RNA, OLE-associated protein, YbzG
Noncoding RNAs (ncRNAs) greater than 200 nucleotides in length are rare in the bacterial domain of life (1, 2), perhaps because most bacterial species are under extreme evolutionary pressure to maintain small, efficient genomes with little tolerance for producing long transcripts other than mRNAs. Intriguingly, the common classes of large and highly conserved bacterial ncRNAs reported to date mostly represent RNA-based enzymes such as ribosomal RNAs (3), group I (4), and group II (5) self-splicing ribozymes, and RNase P endonucleases (6). It has been proposed that some ribozyme classes might be evolutionary descendants of catalytic polymers that were present in RNA World organisms, before the emergence of proteins (7). Thus, reports of the existence of unusually well-conserved and long ncRNAs in bacteria (8–10) provide opportunities to study novel ribozyme classes with potentially ancient types of biochemical functions.
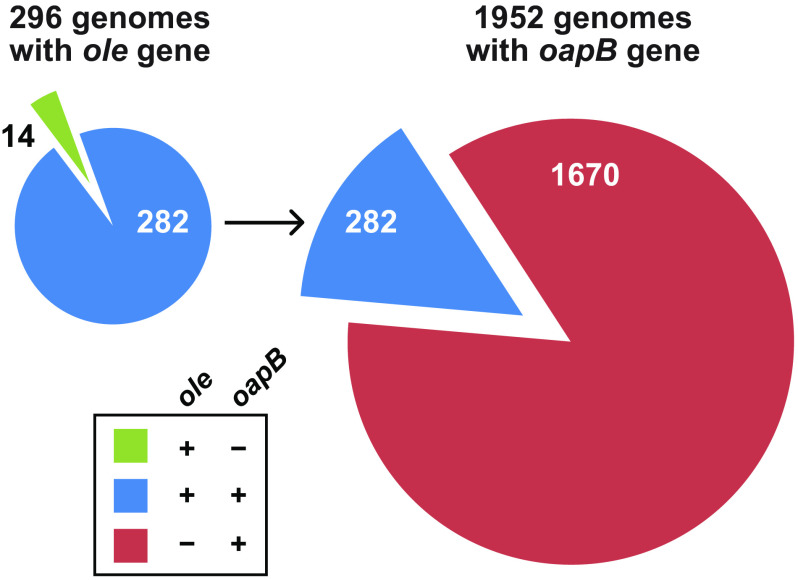
We previously reported the existence of OLE (ornate, large, extremophilic) RNAs, which represent a class of unusually long (∼600 nucleotide) bacterial ncRNA transcripts with numerous highly conserved sequence and structural features (8, 11, 12). Currently, we detect the presence of nucleotide sequences matching the OLE RNA consensus sequence and secondary structure model in the genomes of 296 Gram-positive bacterial species, and in several hundred additional metagenomic DNA sequences (12). This distribution among various Firmicutes, particularly in extremophilic and anaerobic species of Bacilli and Clostridia, makes OLE RNA one of the most widespread large ncRNAs in bacteria whose biochemical function has yet to be determined.
Although the precise biological function of OLE RNA is unknown, various lines of evidence indicate that the RNA participates in the formation of an abundant ribonucleoprotein (RNP) complex. For example, the gene for OLE RNA (ole) is almost always located upstream of a gene (oapA) coding for a 21-kDa transmembrane protein of unknown function, termed OLE-associated protein A (OapA) (8, 13). The ole and oapA genes are mutually inclusive (each only residing in genomes that include the other), suggesting that they are functionally linked. Indeed, electrophoretic mobility shift assays (EMSAs) were used to demonstrate that OapA homodimers bind to OLE RNA by recognizing two short palindromic sequences, among other contacts with the RNA (14).
Moreover, it was shown that the OLE-OapA RNP complex localizes to cell membranes (14), presumably due to the predicted transmembrane helices of OapA (13, 14). In previous work, we developed protocols to efficiently genetically alter the alkaliphile Bacillus halodurans and created knockouts of ole and oapA (15, 16). Genetic disruption of the ole gene, oapA gene, or both, yields strains that exhibit growth and survival defects when exposed to stresses such as cold (20 °C) (16), short-chain alcohols such as ethanol (5% v/v) (11), or slightly elevated Mg2+ concentrations (8 mm) (12). These observations, and other clues from its genetic context (8), suggest that OLE RNA has a key function in maintaining cell membrane integrity and osmotic homeostasis.
Intriguingly, the stress phenotypes noted above are substantially worsened through mutation of a highly conserved DXXXD amino acid sequence in OapA (where X is less conserved) to the sequence AXXXA (referred to as OapA protein mutation 1 or “PM1”) (13). These phenotypes are only seen when OLE RNA is also present. These findings are notable for three reasons. First, the fact that PM1 causes more severe versions of the same phenotypes compared with an oapA or ole knockout (KO) suggests that the OLE RNP complex has one or more other constituents whose function is disrupted by the PM1 changes. Second, the loss of the severe phenotype in PM1 when OLE RNA is absent indicates that the RNA is essential for the stress-related functions of the RNP complex. Third, the severity of the stress phenotypes offers a powerful selective pressure to pursue genetic selections for mutations elsewhere in the genome that convert the severe phenotypes of a PM1 strain to the milder stress phenotypes experienced by the ole and/or oapA KO strains.
In one such genetic selection (13), 6 of 10 isolated B. halodurans strains acquired genomic mutations in the gene ybzG to overcome the severe PM1 phenotypes. The YbzG protein had not been characterized previously but was known to carry a KOW motif (17), which is a putative RNA-binding domain. Indeed, it has recently been demonstrated that the YbzG protein from B. halodurans selectively binds full-length OLE RNA, and it was thus renamed OLE-associated protein B (OapB) (13). Additionally, the H57Y mutant version of OapB, encountered in two independent PM1-resistant strains from the genetic selection, does not bind OLE RNA. These data demonstrate that OapB plays an essential role in forming the functional OLE RNP complex in B. halodurans.
Furthermore, OapB was found to bind a 160-nucleotide fragment of OLE RNA containing base-paired regions P12 through P15, suggesting that the RNA-binding target of OapB is limited to this OLE substructure (13). In the current study, we further examined the interaction of OLE RNA and OapB to establish the precise nucleotides and structural features required for forming the OLE-OapB RNP complex. Specifically, we employed EMSAs with truncated and mutated RNA constructs to localize the binding sites for OapB. In addition, we used an in vitro selection strategy to more precisely define the sequence and structural features of OLE RNA important for high-affinity binding by OapB. Our findings provide a more detailed understanding of the structures involved in the function of this large bacterial ncRNA.
Results and discussion
The oapB gene is found in most organisms that contain OLE RNA and in many Gram-positive bacteria that lack OLE RNA
Recently reported data (13) suggests that the partnership between OLE RNA and OapB (protein formerly called YbzG) is essential for the biological function of the OLE RNP complex in B. halodurans. Specifically, mutations to OapB were found to overcome the strong growth and survival deficiencies of the OapA PM1 strain as noted above. Likewise, deletion of the gene for OLE RNA similarly overcomes the PM1 phenotype. These results suggest that an intact OLE RNP complex is needed for cells to exhibit the strong PM1 phenotype, and that both OLE RNA and OapB are necessary to form a functional complex.
Unfortunately, we have not yet been able to produce a viable genetic deletion of the oapB gene in B. halodurans to determine if the loss of OapB alone is phenotypically equivalent to strains lacking OapA or OLE RNA. The oapB gene is located between two essential genes: map (type 1 methionyl aminopeptidase) and infA (translation initiation factor IF-1) (18, 19). Replacement of the entire oapB gene with a selectable marker gene might affect expression of the two flanking genes, causing propagation of the intended KO strain to fail due to polar effects, or other reasons unrelated to the function of OapB, as we suspect that oapB is a nonessential gene.
Regardless, there remain several questions about the natural role(s) of OapB. For example, do OLE RNAs from all species form a complex with OapB? Does OapB have a functional role beyond serving as a partner in the formation of OLE RNP complexes? To date, there are no additional validated functions for OapB proteins in any other species. Therefore, these uncertainties prompted us to conduct a quantitative examination of the co-occurrences of the ole and oapB genes, which can yield clues relevant to these questions.
To evaluate the distributions of ole and oapB genes, we used a bioinformatic analysis approach both to expand the number of representatives and identify instances of co-occurrences. Specifically, by using BLAST, we compiled a list of oapB-containing organisms and compared it to a previously generated list of ole-containing organisms (13). Representatives of the oapB gene are found exclusively in Firmicutes, with 1,952 examples in annotated genomes (Fig. 1, right). Importantly, over 85% of oapB-containing genomes lack ole. This indicates that, in species lacking OLE RNA, OapB has a separate biological role. Presumably, this role involves RNA binding because these additional OapB representatives are also generally very small (∼100 amino acids), and only carry well-conserved KOW RNA-binding domains. However, oapB is known to be nonessential in Bacillus subtilis (18, 19). Additionally, a mutation to the B. halodurans oapB start codon resulted in a viable strain with no growth defects under normal laboratory culture conditions (13). Thus, OapB function might be important in B. subtilis and other organisms lacking OLE RNA only under certain stress conditions.
Figure 1.
The ole and oapB genes frequently co-occur in bacterial genomes. Left, the gene for OLE RNA is present in the genomes of 296 bacterial species whose DNA has been completely sequenced. Of these, 282 species also carry a gene for OapB. Right, the gene for OapB is more widespread among bacteria than is the gene for OLE RNA.
Of the 296 fully sequenced bacterial genomes that were found to carry a representative ole gene, all but 14 species also harbor an oapB gene, corresponding to a greater than 95% co-occurrence (Fig. 1, left). Among the 14 species that lack oapB, no clear differences are observed in the structure or nucleotide conservation of OLE RNA in the P12 to P15 region (Fig. S1). Of the species that contain both ole and oapB it is unknown how many utilize OapB as a necessary factor in assembling a functional OLE RNP complex. Given the importance of OapB to the function of the B. halodurans OLE RNP complex and its broader importance to species lacking OLE RNA, we sought to further define the RNA-binding site of this protein.
The primary OapB-binding site includes the P13 region of the B. halodurans OLE RNA
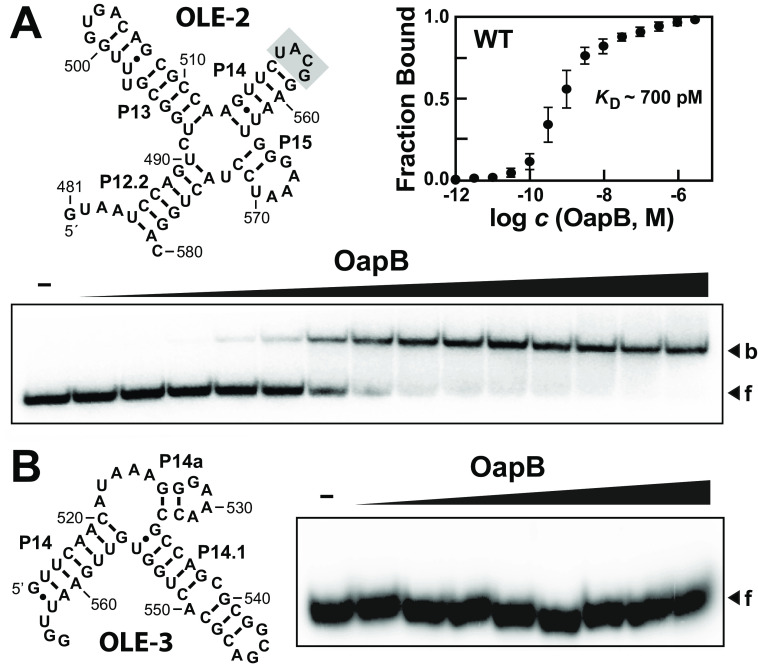
Previous work demonstrated that OapB interacts with a 162-nucleotide construct spanning positions 449-608 of the full-length WT B. halodurans OLE RNA (13). A dissociation constant (KD) of ∼60 nm reported for the 449-608 OLE RNA construct provides a basis of comparison for the assessment of the function of various RNA constructs examined in the current study. Initially, we created a 100-nucleotide construct, called OLE-1 (Fig. 2A), which encompasses positions 481-580 of the WT OLE RNA. By generating and evaluating EMSA data using trace amounts of 32P-labeled RNA, it was determined that OLE-1 appears to be bound by two OapB proteins in a manner indicative of two separate binding events (Fig. 2B).
Figure 2.
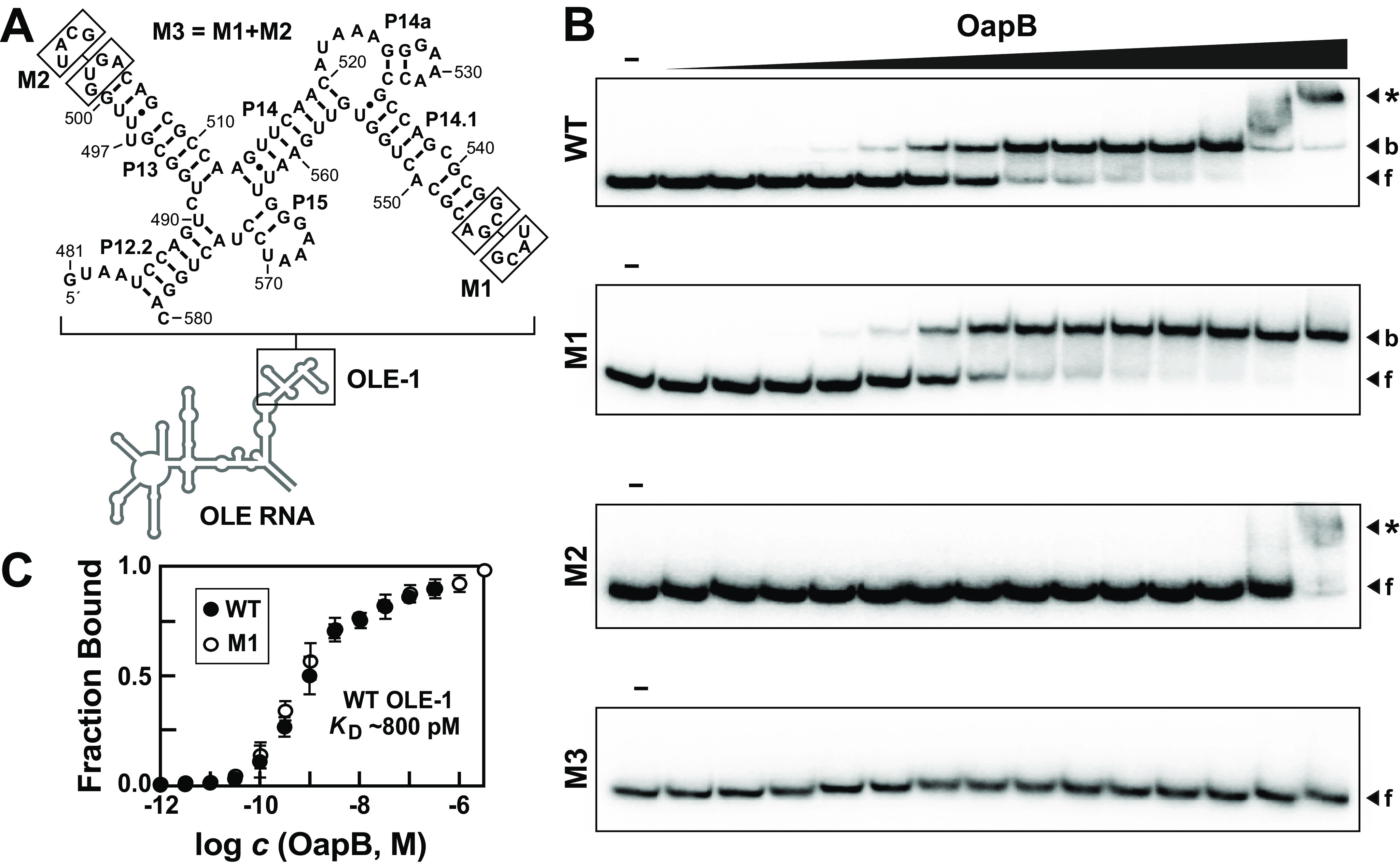
EMSA data for OapB binding to OLE-1, a 100-nucleotide fragment of B. halodurans OLE RNA. A, Sequence and secondary structure model for WT OLE-1 and various mutants. Boxed nucleotides are altered as depicted for mutant constructs M1 and M2. Construct M3 combines the mutations from M1 and M2. B, representative EMSA autoradiograms derived by incubating trace amounts of 5ˊ-32P-labeled RNA constructs as indicated either in the absence (–) of OapB, or in the presence of various concentrations of OapB ranging from 1 pm to 3.2 μm (in half-log increments). Bands corresponding to unbound or “free” RNA (f), bound RNA (b), and RNA undergoing a supershift (*) are annotated accordingly. C, binding curves for WT and M1 OLE-1 RNA constructs with OapB derived from EMSA data as depicted in B. The plot for WT OLE-1 RNA excludes the highest two OapB concentrations examined to avoid the effects of the supershift. Error bars represent the standard error of the data based on at least three replicate experiments. When absent, error bars are smaller than the symbols used to represent the value.
The initial shift in RNA electrophoretic mobility exhibits an OapB concentration-response curve that is consistent with a stoichiometry of 1:1 and a KD of ∼800 pm (Fig. 2C). The second event causes a further reduction in mobility of the RNA, or a “supershift,” suggesting that a second OapB protein is binding to the existing RNA-OapB complex, albeit with a KD that appears to be in the micromolar range. Under our assay conditions, we do not see evidence of cooperative binding between OLE-1 and two OapB proteins.
A prominent characteristic of OLE RNAs encompassed by the OLE-1 construct is the presence of a semi-repetitive architecture, primarily represented by the P13 and P14.1 stems, and the regions immediately flanking these stems (Fig. 2A). For example, both stems are formed by 8 base pairs that are interrupted by one or two unpaired nucleotides. In addition, both hairpins are closed by GNRA tetraloops. Loop sequences conforming to this consensus are commonly present in naturally occurring structured RNAs, where they can both stabilize base-pairing interactions of adjoining stems (20), and present unique tertiary folding opportunities by docking with tetraloop receptor structures (21, 22). Given the architectural similarity of P13 and P14.1, we hypothesized that an OapB molecule might individually bind to each of these repeated substructures.
We observed that replacement of the highly conserved GNRA tetraloop linking the left and right shoulders of base-paired region P14.1 with an alternative stable UNCG tetraloop (23) (mutant construct M1, Fig. 2A) causes a loss of the second (weaker) OapB binding event, but has no effect on the first (stronger) binding interaction (Fig. 2B). The OapB-binding curves and the KD values for the WT and M1 constructs are essentially identical (Fig. 2C). In contrast, replacement of the GNRA tetraloop linking the shoulders of P13 with a UNCG sequence (mutant construct M2) causes a loss of the first, but not the second OapB-binding interaction. A construct wherein both GNRA loops were replaced (M3) exhibits no OapB binding, even at concentrations as high as 3.2 μm (Fig. 2B), again demonstrating the importance of the tetraloop sequences for formation of the OLE-OapB RNP complex.
To further establish the specific portions of OLE RNA required for the OapB-binding events, we created RNA constructs OLE-2 and OLE-3 to examine each binding site in isolation. Construct OLE-2, encompassing P13 and flanking regions, exhibits robust single-event binding by OapB, again with a KD value (∼700 pm) that is essentially identical to that observed for the OLE-1 construct (Fig. 3A). In contrast, we did not observe binding of OapB to construct OLE-3, which encompasses P14.1 and its flanking regions (Fig. 3B). Given the small size of OLE-3 and its strong predicted secondary structure features, it is unlikely that the absence OapB binding is due to RNA misfolding.
Figure 3.
An OLE RNA fragment containing P13 and flanking regions is sufficient to form a complex with OapB. A, characteristics of OapB binding to the OLE-2 RNA construct, which lacks the P14.1 region. Shaded nucleotides identify a UNCG tetraloop that was used in place of a portion of P14 and all of P14.1. OapB concentrations range from 1 pm to 1 μm (half-log increments). Additional annotations are as described in the legend to Fig. 2. B, OapB fails to bind the OLE-3 RNA construct, which lacks the P13 region. OapB concentrations range from 320 pm to 1 μm (half-log increments). Additional annotations are as described in the legend to Fig. 2.
A mutated version of the OLE-2 construct also was examined, wherein the P13 GNRA tetraloop was replaced with a UNCG tetraloop (OLE-4) (Fig. S2A). This alteration results in the complete loss of OapB binding, which matches that observed for the OLE-1 M2 mutant and further demonstrates the requirement of the P13 GNRA tetraloop. However, a construct called OLE-5 encompassing only the P13 stem plus a partial P14 stem does not serve as a binding site for OapB (Fig. S2B). These findings demonstrate that the P13 stem and GNRA tetraloop alone are insufficient for high-affinity OapB binding.
To observe if these effects hold true for the binding of full-length OLE RNA to OapB, a construct (FL-OLE M2) was made wherein the P13 GNRA tetraloop was replaced with a UNCG tetraloop. Comparison of the WT (FL-OLE) and mutated full-length OLE RNA by EMSA (Fig. S3) reveals that disruption of the P13 GNRA tetraloop leads to a reduction in affinity to OapB, although the effects are less substantial than in truncated constructs due to the overall poorer affinity of the full-length RNA.
Taken together, these results localize the high-affinity OapB-binding site to P13, but only when embedded in its larger natural structural context. Although the P14.1 region is similar in appearance to P13, the poor affinity binding of OapB to construct OLE-1 (∼10−3 weaker) and the lack of binding to OLE-3 casts doubt on the biological relevance of this second, weaker interaction with P14.1. In its native cellular context, the OLE RNP might involve the binding of two OapB proteins, but our current in vitro data supports only one strong RNA-binding site with no evidence for cooperative interactions between two OapB molecules.
A single bulged nucleotide is essential for high-affinity binding of P13 by OapB
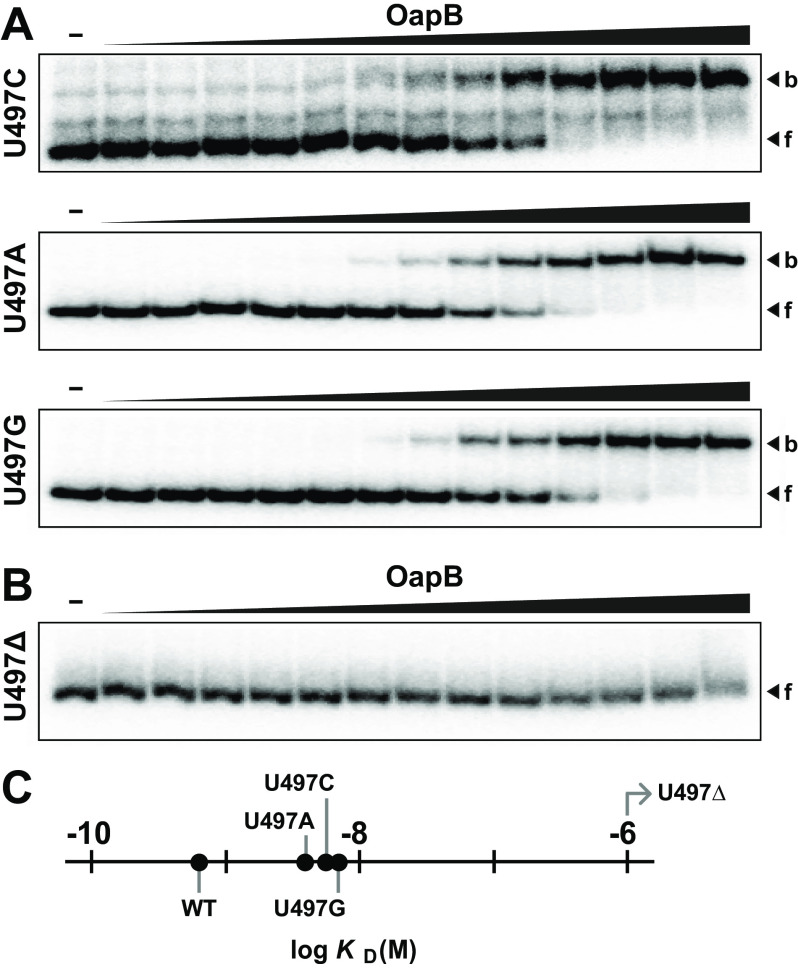
Upon confirming that the P13 region forms a high-affinity OapB-binding site compared with the P14.1 region, we further examined these two regions to determine what differences might explain our results. The preceding experiments highlight the critical importance of the GNRA tetraloops for both binding sites, although additional binding site discrimination must rely on other features of these two regions. The most obvious difference is that P13 contains a single bulged nucleotide in the stem (U497), whereas P14.1 contains a mismatched pair of nucleotides (C539 and A550) (Fig. 2A). To assess the importance of U497, constructs were made in which this nucleotide was either mutated or deleted. The mutation of U497 to any of the other three common nucleotides only weakens binding (Fig. 4A), whereas deletion of this bulged nucleotide completely eliminates binding (Fig. 4B). Specifically, the KD values range from 4 to 7 nm for constructs wherein the nucleotide identity at position 497 has been changed (Fig. 4C), compared with 700 pm for the WT OLE-2 construct (Fig. 2C).
Figure 4.
A bulged nucleotide in P13 is required for OapB binding. A, EMSA autoradiograms of OapB binding to OLE-2 constructs wherein U497 has been mutated to C, A, or G, respectively. OapB concentrations range from 1 pm to 1 μm (half-log increments). Additional annotations are as described in the legend to Fig. 2B. B, EMSA autoradiogram of OapB binding to an OLE-2 construct wherein nucleotide 497 has been deleted (U497Δ). C, plot of the KD values for OapB binding to the WT OLE-2 construct, and for the various constructs with alterations at position 497. Note that the U497Δ construct has a KD value that is poorer than 10−6 m. Data were derived from the EMSA assay data depicted in A and B.
These findings suggest that U497 might form one or more additional contacts with OapB, thereby resulting in a tighter RNA-protein complex. However, a far greater contributor to binding affinity is the presence of a bulge in the P13 helix. Perhaps the protein makes direct contact with the ribose and/or phosphate moieties of this bulged nucleotide, or perhaps the binding site of the protein prefers a distorted or bent RNA helix that results from the bulged nucleotide at this location.
In vitro selection reveals nucleotides critical for high-affinity OapB binding to OLE RNA
To generate a more detailed consensus sequence and structural model for the OapB-binding site, we created a mutagenized RNA population that presents a single, high-affinity binding site based on the OLE-2 RNA construct (Fig. 3A). The mutagenized version of this construct, called OLE-6 (Fig. 5A), was formed by synthesizing a DNA population (Table S1) for in vitro transcription that carried a central 56-nucleotide region with 6% sequence degeneracy. This region was flanked by invariant 15-nucleotide primer-binding sites to enable reverse transcription and subsequent PCR to amplify RNAs that were selected for their ability to be bound by OapB. The initial population (generation zero or “G0”) included ∼6 × 1011 molecules (1 pmol), and is expected to fully sample all possible sequences with 6 or fewer mutations relative to the WT sequence.
Figure 5.
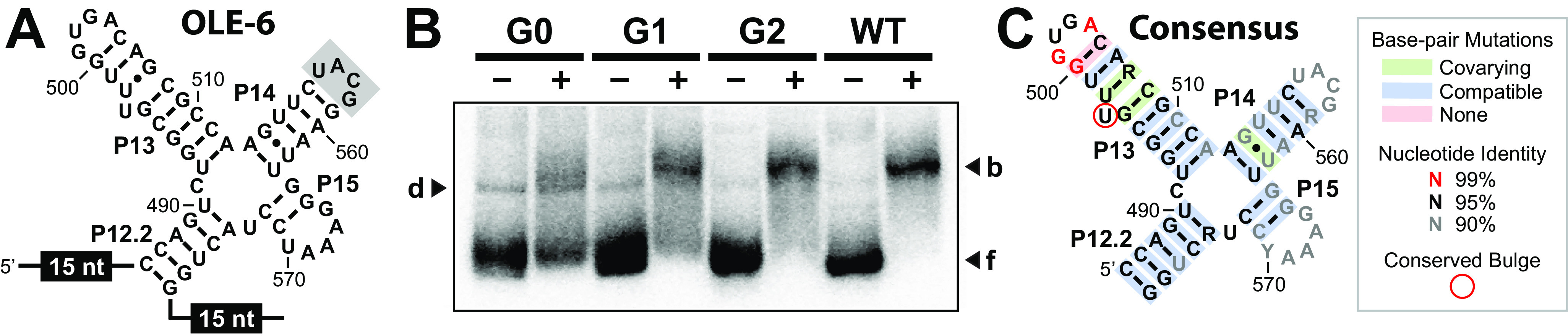
In vitro selection of mutant OLE-derived RNAs that are bound by OapB. A, sequence and secondary structure model for the in vitro selection construct OLE-6, wherein the nucleotides depicted were mutagenized to a level of 6% degeneracy. Circled numbers identify the primer-binding sites and the shaded nucleotides identify the UNCG tetraloop replacing the natural intervening sequences of OLE RNA. B, EMSA results for G0 through G2 RNA populations, and for the unmodified parental (P) OLE-6 construct. A band representing a putative protein-independent RNA dimer (d) is annotated. Additional annotations are as described in the legend to Fig. 2. C, consensus model for the high-affinity OapB-binding site based on the B. halodurans OLE RNP system. The consensus model was prepared by R2R using a covariation threshold of 10% (26). Red nucleotides are >99% conserved among the RNAs in the G2 population with 500 or more representatives.
Starting from the G0 population, two rounds of in vitro selection were performed, which were sufficient to produce a G2 population that is bound by OapB with characteristics that are comparable to that observed for WT OLE-6 RNA (Fig. 5B). A minor band (d, Fig. 5B) appears to be dimers of OLE-6 that have mobility similar to that of the RNA-protein complex. However, the mobility of the dimer was sufficiently different from that of RNAs bound by OapB, to exclude RNA-only dimers from amplification in subsequent generations.
The G2 population was then subjected to RNA-Seq, and the results were used to generate a consensus model for the OapB-binding site (Fig. 5C). The overall secondary structure appears to be retained by the vast majority of the members of the G2 population, wherein the most highly conserved nucleotides are present in the P13 stem-loop substructure. In particular, the GNRA tetraloop and the terminal base pair of the P13 stem are conserved to a greater extent than any other region, with G501 of this tetraloop appearing in 100% of the sequence reads represented 500 times or more. Finally, although the U nucleotide identity at position 497 was not strictly conserved, the presence of the bulge at this location was retained. These results are consistent with our earlier data revealing that the bulge is essential for high-affinity binding by OapB, whereas the nucleotide identity is not (Fig. 4).
Bioinformatics search for RNA sequences that match the OapB-binding site consensus
After obtaining the consensus model for the RNA-binding site of B. halodurans OapB, we searched for matches to the model by employing comparative sequence analyses (24) of B. halodurans (Table S2) and B. subtilis (Table S3) genomes. All candidate OapB-binding sites with an E value of less than or equal to 1 reside within protein-coding genes. These candidates appear to be false positives, as coding regions are an uncommon location for protein-RNA interactions to occur. Furthermore, there is no apparent trend regarding gene functionality, and there is no overlap in the specific genes identified from the B. halodurans and B. subtilis genome analyses.
These findings highlight the mystery regarding the predominant biological function of OapB for each species in which this protein is carried. Perhaps the B. halodurans OapB has a single function: to serve as an essential RNA-binding partner in the OLE RNP complex. The features of its RNA-binding site might be distinct for B. halodurans. If true, then the consensus model might not be useful for identifying biologically relevant binding sites in other species. Regardless, the majority of bacterial species that carry OapB do not have OLE RNAs (Fig. 1), and therefore the primary biological role for OapB in these species remains undiscovered.
Whereas the vast majority of species that carry OLE RNA also code for OapB, we have identified 14 species that appear to lack OapB (Fig. 1). Furthermore, the consensus model for the high-affinity RNA target site for B. halodurans is not strictly conserved across all OLE RNA representatives (13). These observations suggest that OapB is not an essential partner in OLE RNP complexes from all species, and that the determinants of OapB binding might have co-evolved with the RNA if OapB is more widely essential for OLE RNP function. Overall, these observations suggest that OapB might function as a remarkably small RNA-binding protein that recognizes simple sequence and structural features to assist in the folding and functioning of certain RNA transcripts. In B. halodurans, its primary function might be to assist in the formation of the OLE RNP complex, but additional functional roles cannot be ruled out.
The protein partners of OLE RNA interact with different regions of the polynucleotide
In the current study, we have specifically mapped two binding sites for OapB within OLE RNA. Both sites are located considerable distances 3ˊ relative to the specific binding sites of the previously established protein partner, OapA (14). OapA presumably binds as a homodimer and relies on two YAGNCUR consensus sequence motifs located in the P2 and P4a regions of OLE RNA. This separation suggests that both OapA and OapB might bind full-length OLE RNA molecules simultaneously (Fig. 6). It is already known that shortened OLE RNA constructs that exclude P13 are tightly bound by OapA proteins (14), whereas shortened constructs such as OLE-1 (Fig. 2) and OLE-2 (Fig. 3) that lack the two YAGNCUR consensus sequences are tightly bound by OapB. These observations are consistent with the hypothesis that both proteins can bind OLE RNA at the same time. Unfortunately, given the large shift in gel mobility upon the binding of OapA to OLE RNA (14), and given the small size of OapB, we have not been able to demonstrate simultaneous binding using the EMSA methods described herein.
Figure 6.
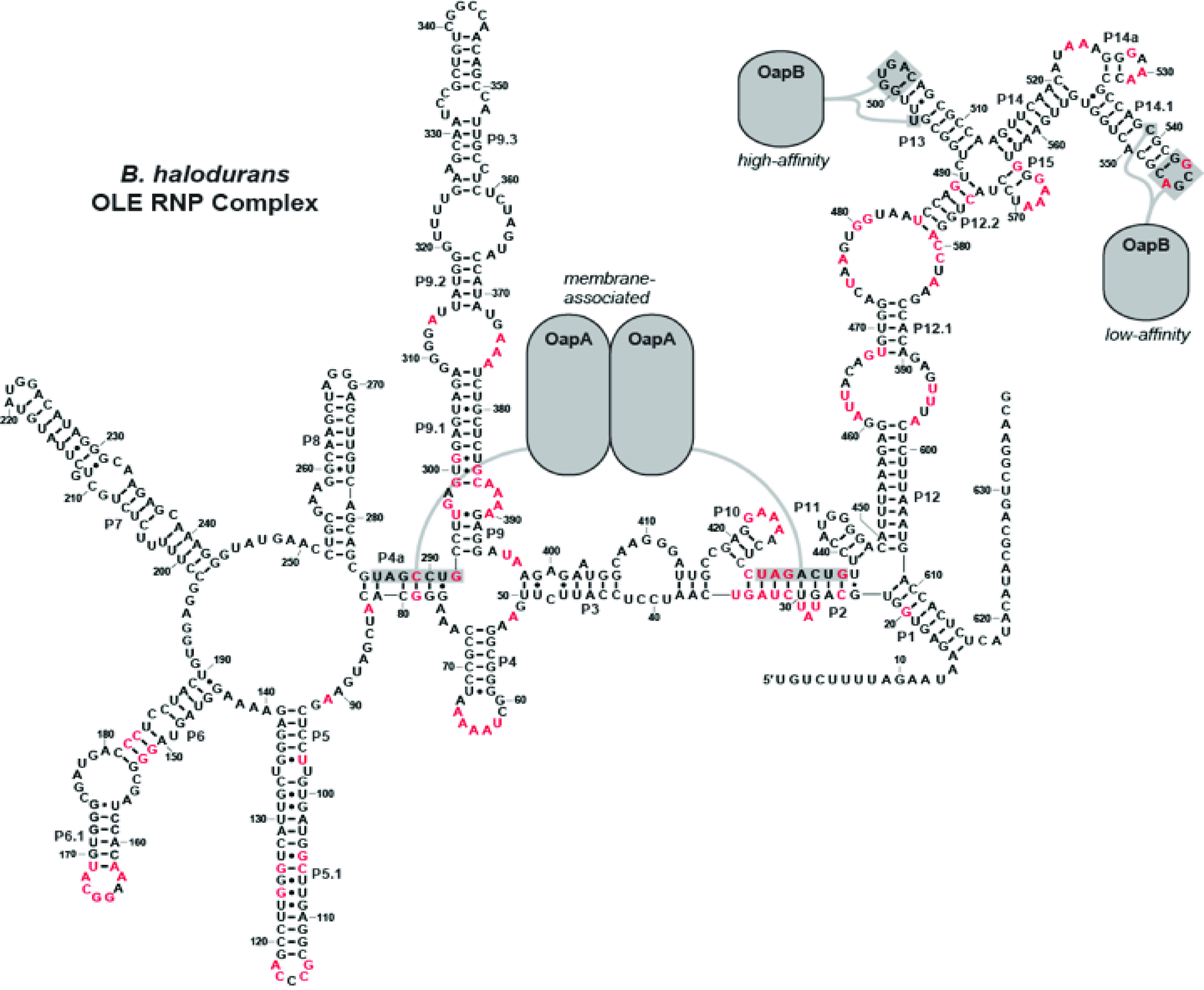
Model for an OLE RNP complex. A model for the B. halodurans OLE RNP complex showing where OapA and OapB interact. Gray shaded nucleotides are required for protein binding. Red residues are greater than 97% conserved across all OLE RNA examples (13).
Importantly, the protein-binding regions noted above encompass only small portions of OLE RNA. There are ∼80 highly conserved nucleotides that reside outside of the key molecular recognition motifs important for binding of OapA and OapB (Fig. 6). Therefore, we expect that these other well-conserved sequence and structural features are recognized by other protein factors or, perhaps more likely, are critical for OLE RNA to perform a complex biochemical function as exemplified by all other large, highly structured, and widespread ncRNAs in bacteria.
Conclusions
The OapB protein from B. halodurans binds the P13 region of OLE RNA with an affinity in the mid-picomolar range. This strong interaction is required for cells to exhibit the most severe sensitivity phenotypes when exposed to cold, short-chain alcohols, or slightly elevated Mg2+ concentrations. Despite its small size (102 amino acids), the protein is remarkably selective for its RNA-binding site. The consensus sequence for OapB binding includes a GNRA tetraloop on a stem closed with a G-C base-pair, and a bulged nucleotide (preferably uridine) located 3 base pairs away and on the 5ˊ side of the tetraloop (Fig. 5C).
Although the RNA consensus model for OapB binding is relatively simple, and there are close matches for this architecture elsewhere in the RNAs of B. halodurans (Table S2), we do not have evidence that any of these additional binding site candidates are biologically relevant. It is apparent that additional regions of the high-affinity OapB-binding site flanking the OLE RNA P13 stem are also required for complex formation (Fig. S1B). The nucleotide sequences for these additional molecular recognition determinants are not highly conserved. However, they might be sufficiently important for binding such that their absence precludes OapB from recognizing other cellular RNAs that closely mimic the P13 stem of B. halodurans OLE RNA. Regardless, OapB from most species must have other RNA targets, because more than 85% of sequenced bacterial species with the oapB gene do not contain the gene for OLE RNA. Additional experiments with species lacking OLE RNA will need to be conducted to determine the broader functions of OapB.
Because OapA and OapB only interact with small portions of OLE RNA it can be assumed that either OLE RNA is interacting with additional protein partners or that the other conserved substructures of this large ncRNA perform another function, possibly even acting as a ribozyme. Although increased understanding of the OLE-OapB complex does not readily provide additional insight into the function of the larger OLE RNP, it does lay the foundation for the eventual reconstitution of an active OLE RNP complex in vitro, allowing for the testing of specific hypotheses for biochemical function, and the further structural analysis of this unusual ncRNA.
Experimental procedures
Synthetic DNA oligonucleotides
All synthetic DNA oligonucleotides (Table S1) were purchased from Sigma-Aldrich except for DLW137, which was purchased from the Keck Oligonucleotide Synthesis facility at Yale University.
Bioinformatics
OapB representatives were identified using NCBI BLAST with B. halodurans OapB (WP_010896340.1) as the query sequence. A list of organisms that contain ole was obtained from the OLE RNA alignment file created previously (13). The oapB gene was detected in each of these genomes using NCBI BLAST.
RNA constructs
DsDNA templates for in vitro RNA transcription were prepared by overlap extension reactions. Specifically, annealing incubations were conducted with 200 pmol of each synthetic DNA strand in a mixture prepared by the addition of 10 μl of 5× First Strand Buffer (Thermo Fisher Scientific), 1 μl of a 10 mm (each) dNTP mixture, and deionized H2O in a volume of 44 μl, which was heated to 90 °C for 1 min, then cooled to 23 °C. To this mixture was added 5 μl of 0.1 m DTT and 1 μl (200 units) of Superscript II Enzyme (Thermo Fisher Scientific). The reaction was incubated at 42 °C for 1 h followed by 70 °C for 15 min. DNA templates that did not require overlap extension reactions were diluted to 4 μm and heated to 90 °C for 1 min. All DNA constructs served as templates for in vitro transcription using T7 RNA polymerase. The resulting RNAs were purified by denaturing (7 m urea) 10% PAGE, dephosphorylated, and 5ˊ-32P-radiolabeled with [γ-32P]ATP using methods described previously (14).
Binding assays
The OapB stock used in this study was purified and stored as described previously (13). RNA-binding assays with OapB were performed as described previously (13), but with two exceptions. First, OapB stock solutions were prepared in concentrations that were 10-fold higher than the final concentrations used in the binding assays, in half-log intervals. Second, bound complexes were separated from free 5ˊ-32P-labeled OLE RNA constructs by using nondenaturing 10% PAGE at 20 watts for 1–2 h.
Selection for mutant OLE RNAs that bind OapB
A single-stranded synthetic DNA construct (Table S1, DLW137) containing a T7 RNA polymerase (T7 RNAP) promoter, 56 nucleotides prepared with a mutation rate of 6%, and two 15-nucleotide segments flanking the degenerate region to allow for amplification of the selected RNA representatives was ordered from the Keck Oligonucleotide Synthesis facility at Yale University. DsDNA templates for in vitro transcription (generated by primer-extension from DLW137 and DLW140) and the corresponding RNAs were produced as described above. Binding assays were prepared as previously described (13), with 100 μm OapB and 50 μm OLE-6 RNA, and were separated by a nondenaturing 10% PAGE run for 3 h at 20 W. The band containing the OLE-OapB RNP complex was excised and subjected to crush-soak elution (200 mm NaCl, 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, pH 8.0, 1 m urea) for 2 h. The eluted RNA was concentrated by precipitation with ethanol. Primers (Table S1, DLW145 and DLW167) were used to reverse transcribe the RNA. The resulting cDNAs were then amplified by PCR and used to generate a new pool of RNAs for the next round of selection. A total of two rounds of selection were performed and the resulting G2 population was sequenced.
Construction of consensus model
The double-stranded G2 DNA population was sequenced by the Yale Center for Genome Analysis, which yielded 10 million reads. Individual sequences were reconstructed from the pairwise reads, and identical reads were grouped and sorted by prevalence. Sequences with at least 500 reads (366 unique sequences) were compiled and used to generate the consensus RNA motif for OapB binding. Clustal Omega was used to align the sequences, followed by manual editing to refine the alignment (25). The Stockholm file of aligned sequences was used to generate a consensus model for OapB binding with R2R (26). To best display the data, R2R parameters were modified to report conservation of >99% (red), >95% (black), and >90% (gray), with all other parameters set at the default values.
Search for additional OapB interacting RNAs
The Stockholm file described above was modified to include only OLE RNA nucleotides 494-510. This trimmed file was run through cmbuild and Infernal algorithms to generate a calibrated covariance model (24). The calibrated covariance model was run against the B. halodurans and B. subtilis genomes using cmsearch to generate a list of potential OapB-interacting sequences.
Data availability
All data are available in the manuscript except for RNA-Seq results and alignment files used to generate Fig. 5C and alignment files used to generate Fig. S1. These data are available upon request from Ronald Breaker (ronald.breaker@yale.edu).
Supplementary Material
Acknowledgments
We thank Michelle Peters for purifying the OapB used in this study and other members of the Breaker laboratory for helpful discussion.
This article contains supporting information.
Author contributions—D. L. W., K. A. H., and R. R. B. conceptualization; D. L. W. and K. A. H. data curation; D. L. W. formal analysis; D. L. W., K. A. H., and L. C. investigation; D. L. W., and K. A. H. methodology; D. L. W. and R. R. B. writing-original draft; D. L. W., K. A. H., L. C., and R. R. B. writing-review and editing; R. R. B. funding acquisition; R. R. B. project administration.
Funding and additional information—This work was supported by National Institutes of Health Grant F32GM116426 (to K. A. H.), National Institutes of Health Grant GM022778 (to D. L. W., K. A. H., L. C., and R. R. B.), Foundation for the National Institutes of Health Grant T32GM007223 (to D. L. W.), and the Howard Hughes Medical Institute (to D. L. W., K. A. H., L. C., and R. R. B). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of interest—None of the authors declare any related to the data presented in this article.
- ncRNA
- noncoding RNA
- OLE
- ornate, large, extremophilic
- RNP
- ribonucleoprotein
- OapA
- OLE-associated protein A
- EMSA
- electrophoretic mobility shift assay.
References
- 1. Harris K. A., and Breaker R. R. (2018) Large noncoding RNAs in bacteria. Microbiol. Spectr. 6, RWR-0005-2017 10.1128/microbiolspec.RWR-0005-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cech T. R., and Steitz J. A. (2014) The noncoding RNA revolution: trashing old rules to forge new ones. Cell 157, 77–94 10.1016/j.cell.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 3. Nissen P., Hansen J., Ban N., Moore P. B., and Steitz T. A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 10.1126/science.289.5481.920 [DOI] [PubMed] [Google Scholar]
- 4. Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., and Cech T. R. (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31, 147–157 10.1016/0092-8674(82)90414-7 [DOI] [PubMed] [Google Scholar]
- 5. Peebles C. L., Perlman P. S., Mecklenburg K. L., Petrillo M. L., Tabor J. H., Jarrell K. A., and Cheng H. L. (1986) A self-splicing RNA excises an intron lariat. Cell 44, 213–223 10.1016/0092-8674(86)90755-5 [DOI] [PubMed] [Google Scholar]
- 6. Guerrier-Takada C., Gardiner K., Marsh T., Pace N., and Altman S. (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849–857 10.1016/0092-8674(83)90117-4 [DOI] [PubMed] [Google Scholar]
- 7. Benner S. A., Ellington A. D., and Tauer A. (1989) Modern metabolism as a palimpsest of the RNA world. Proc. Natl. Acad. Sci. U.S.A. 86, 7054–7058 10.1073/pnas.86.18.7054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puerta-Fernandez E., Barrick J. E., Roth A., and Breaker R. R. (2006) Identification of a large noncoding RNA in extremophilic eubacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 19490–19495 10.1073/pnas.0607493103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weinberg Z., Perreault J., Meyer M. M., and Breaker R. R. (2009) Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature 462, 656–659 10.1038/nature08586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weinberg Z., Lünse C. E., Corbino K. A., Ames T. D., Nelson J. W., Roth A., Perkins K. R., Sherlock M. E., and Breaker R. R. (2017) Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res. 45, 10811–10823 10.1093/nar/gkx699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wallace J. G., Zhou Z., and Breaker R. R. (2012) OLE RNA protects extremophilic bacteria from alcohol toxicity. Nucleic Acids Res. 40, 6898–6907 10.1093/nar/gks352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris K. A., Odzer N. B., and Breaker R. R. (2019) Disruption of the OLE ribonucleoprotein complex causes magnesium toxicity in Bacillus halodurans. Mol. Microbiol. 112, 1552–1563 10.1111/mmi.14379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harris K. A., Zhou Z., Peters M. L., Wilkins S. G., and Breaker R. R. (2018) A second RNA-binding protein is essential for ethanol tolerance provided by the bacterial OLE ribonucleoprotein complex. Proc. Natl. Acad. Sci. U.S.A. 115, E6319–E6328 10.1073/pnas.1803191115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Block K. F., Puerta-Fernandez E., Wallace J. G., and Breaker R. R. (2011) Association of OLE RNA with bacterial membranes via an RNA-protein interaction. Mol. Microbiol. 79, 21–34 10.1111/j.1365-2958.2010.07439.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takami H., and Horikoshi K. (1999) Reidentification of facultatively alkaliphilic Bacillus sp. C-125 to Bacillus halodurans. Biosci. Biotechnol. Biochem. 63, 943–945 10.1271/bbb.63.943 [DOI] [PubMed] [Google Scholar]
- 16. Wallace J. G., and Breaker R. R. (2011) Improved genetic transformation methods for the model alkaliphile Bacillus halodurans C-125. Lett. Appl. Microbiol. 52, 430–432 10.1111/j.1472-765X.2011.03017.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kyrpides N. C., Woese C. R., and Ouzounis C. A. (1996) KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 21, 425–426 10.1016/S0968-0004(96)30036-4 [DOI] [PubMed] [Google Scholar]
- 18. Kobayashi K., Ehrlich S. D., Albertini A., Amati G., Andersen K. K., Arnaud M., Asai K., Ashikaga S., Aymerich S., Bessieres P., Boland F., Brignell S. C., Bron S., Bunai K., Chapuis J., et al. (2003) Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 100, 4678–4683 10.1073/pnas.0730515100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koo B.-M., Kritikos G., Farelli J. D., Todor H., Tong K., Kimsey H., Wapinski I., Galardini M., Cabal A., Peters J. M., Hachmann A.-B., Rudner D. Z., Allen K. N., Typas A., and Gross C. A. (2017) Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 4, 291–305e7 10.1016/j.cels.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Correll C. C., and Swinger K. (2003) Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 Å resolution. RNA 9, 355–363 10.1261/rna.2147803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Costa M., and Michel F. (1997) Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J. 16, 3289–3302 10.1093/emboj/16.11.3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fiore J. L., and Nesbitt D. J. (2013) An RNA folding motif: GNRA tetraloop-receptor interactions. Q. Rev. Biophys. 46, 223–264 10.1017/S0033583513000048 [DOI] [PubMed] [Google Scholar]
- 23. Molinaro M., and Tinoco I. Jr. (1995) Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: thermodynamic and spectroscopic applications. Nucleic Acids Res 23, 3056–3063. 10.1093/nar/23.15.3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nawrocki E. P., and Eddy S. R. (2013) Infernal 1.1 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weinberg Z., and Breaker R. R. (2011) R2R - software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 12, 3 10.1186/1471-2105-12-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript except for RNA-Seq results and alignment files used to generate Fig. 5C and alignment files used to generate Fig. S1. These data are available upon request from Ronald Breaker (ronald.breaker@yale.edu).